Abstract
The E2 proteins of papillomaviruses regulate both viral transcription and DNA replication. The human papillomavirus type 18 (HPV18) E2 protein has been shown to repress transcription of the oncogenic E6 and E7 genes, inducing growth arrest in HeLa cells. Using HPV18 E2 fused to the green fluorescent protein (GFP), we showed that this protein was short-lived in transfected HeLa cells. Real-time microscopy experiments indicated that the E2-dependent signal increased for roughly 24 h after transfection and then rapidly disappeared, indicating that E2 was unstable in HeLa cells and could confer instability to GFP. Similar studies done with a protein lacking the transactivation domain indicated that this truncation strongly stabilizes the E2 protein. In vitro, full-length E2 or the transactivation domain alone was efficiently ubiquitinated, whereas deletion of the transactivation domain strongly decreased the ubiquitination of the E2 protein. Proteasome inhibition in cells expressing E2 increased its half-life about sevenfold, which was comparable to the half-life of the amino-terminally truncated protein. These characteristics of E2 instability were independent of the E2-mediated G1 growth arrest in HeLa cells, as they were reproduced in MCF7 cells, where E2 does not affect the cell cycle. Altogether, these experiments showed that the HPV18 E2 protein was degraded by the ubiquitin-proteasome pathway through its amino-terminal transactivation domain. Tight regulation of the stability of the HPV 18 E2 protein may be essential to avoid accumulation of a potent transcriptional repressor and antiproliferative agent during the viral vegetative cycle.
Human papillomavirus type 18 (HPV18) is one of the viruses associated with the development of cervical cancer. During the initial viral infection of susceptible cells, the E2 early protein represses the transcription of the viral oncogenes E6 and E7 by binding to sites within the promoter, interfering with the binding of two proteins essential for transcription initiation, the TATA binding protein and SP1 (6, 7, 11, 12, 36). Carcinogenic progression of productive lesions is accompanied by integration of part of the viral genome in the cellular genome disrupting the E1 and E2 open reading frames (2, 3, 5, 33). Consequently, in cervical carcinoma, expression of the two viral oncoproteins E6 and E7 is upregulated due to loss of the E2 protein. In addition, E2 is required for viral DNA replication together with the E1 viral helicase and therefore appears as a key regulator of both viral replication and viral transcription (8). Reintroduction of HPV18 E2 to cervical carcinoma cell lines by transient transfection leads to repression of the endogenous E6/E7 transcription, inducing cell cycle arrest in G1 via a mechanism involving stabilization of p53 (9, 14, 23). In addition, E2 induces cell death by apoptosis, independently of p53 (10).
E2 proteins are composed of three distinct functional domains, an amino-terminal transactivation domain and a carboxy-terminal DNA binding domain (DBD) separated by a variable hinge domain (13, 17, 27). The carboxy-terminal domain, of about 80 amino acids, binds to specific palindromic DNA sequences as highly structured dimers (22). The amino-terminal domain, of about 200 amino acids, is required for activation of both transcription and replication. Its recently determined crystal structure shows two distinct structural regions separated by a fulcrum (1, 19). Its most N-terminal part contains α helices that can interact to form dimers (1). Whether dimerization of the amino-terminal domain is of physiological relevance remains to be demonstrated, although it seems likely. The amino-terminal domain is involved in all known functions of E2, including transcriptional repression, growth arrest, and induction of apoptosis in transfected HeLa cells (10, 18).
Although E2 is a crucial regulator of the viral vegetative cycle, its detection in productive genital lesions has been elusive, while the E6 and E7 transcripts and gene products are readily detectable in the basal layers of infected lesions. Moreover, in organotypic cultures of differentiated keratinocytes, while the complete vegetative cycle of certain types of papillomaviruses could be reproduced (16, 28), expression of the E1 and E2 regulatory proteins often remained below the level of detection. We found that the level of HPV18 E2 protein overexpressed in cultured cell lines was low, even though its biological activities were readily detectable. These observations pointed to a potential instability of E2, at least under certain conditions. Many transcriptional regulators, including Myc (31), c-Jun (39), E2F (20), ATF-2 (15), and p53 (21, 24), have been shown to be unstable and tightly regulated by degradation pathways. In addition, such control of protein stability is particularly relevant regarding proteins involved in growth inhibition and apoptosis. Since E2 plays an active role in both the viral vegetative cycle and control of cell proliferation, we inferred that a crucial pathway to regulate these activities could be mediated through stability of the protein.
The green fluorescent protein (GFP) was fused to the amino terminus of full-length HPV18 E2. This GFP-E2 fusion protein was shown to retain the biological activities of the wild-type E2 as well as a similar half-life. It was transfected in HeLa cells, and the fate of the fluorescent protein was analyzed by real-time microscopy. We show here that the GFP-E2 fusion protein accumulates in the nuclei of transfected cells for the first 24 h and then rapidly disappears, while the control GFP nuclear protein is stable. The GFP-E2 fusion protein appeared to be stabilized by use of proteasome inhibitors in both HeLa and MCF7 cells, indicating the involvement of the proteasome pathway in its degradation. We found that the E2 transactivation domain carried a degradation signal since deletion of this domain led to a strong stabilization of the protein in the cells. In addition, in vitro ubiquitination experiments showed that the full-length E2 protein and the transactivation domain alone were efficiently ubiquitinated. Altogether, these data indicate that HPV18 E2 stability is controlled by the ubiquitin-proteasome degradation pathway through its amino-terminal transactivation domain.
MATERIALS AND METHODS
Construction of plasmids and recombinant adenoviruses.
The pEGFP-C1 expression plasmid (Clontech) was used to clone the E2 protein and its separate domains. The complete E2 open reading frame was cloned in frame with the carboxy terminus of GFP. Separate domains were defined by comparison of E2 primary sequences and were prepared by PCR amplification using specific primers containing EcoRI and BamHI restriction sites for cloning in frame with the carboxy terminus of GFP. Sequences of the oligonucleotides used are as follows: for the Trans domain, 5′-CGAATTCCATGGAGACACCGAAGGAAACC-3′ and 5′-CGCGGATCCACTGCACATAGAGTCATTAC-3′; for the Hinge, 5′-CGAATTCCACCAGTGACGACACGGTATCC-3′ and 5′-CGCGGATCCGTTA CCACTACAGAGTTTCC-3′; and for the DBD, 5′-CGAATTCCACTACGCCTATAATACAT-3′ and 5′-AAAGGATCCTTACATTGTCATGTATCCC-3′ (restriction sites are indicated in boldface). The nuclear localization sequence (NLS) of the simian virus 40 T antigen (SV40 TAg) was cloned between the XhoI and EcoRI restriction sites in the GFP-Trans and GFP-Hinge plasmids in frame between GFP and E2 protein with two hybridized oligonucleotides containing the sense (5′-TCGACCTCCAAAAAAGAAGAGAAAGGTA-3′) and antisense (5′-AATTTACCTTTCTCTTCTTTTTTGGAGG-3′) sequences.
The GFP-NLS expression plasmid was generated by cloning of the E2 NLS in frame between the HindIII and BamHI restriction sites at the GFP carboxy terminus. Two oligonucleotides containing the sense (5′-AGCTTTAAAATGTTTACGGTACAGATTGCGAAAAG-3′) and antisense (5′-GATCCTTTTCGCAATCTGTACCGTAAACATTTTAA-3′) sequences of the NLS were used.
Recombinant adenoviruses were constructed by bacterial recombination as described elsewhere (4). The complete expression cassettes for full-length and amino-terminally truncated E2, ΔTrans (deletion of amino acids 1 to 215), fused to GFP were prepared from the pEGFP expression plasmids and used for recombination with the adenovirus vector (kind gift from P. Yieh, Rhone Poulenc Rhorer).
The HPV18 E2 protein cloned in pET 14B (Novagen) has already been described (7). Two separate domains of E2, containing either the complete Trans or the fused Hinge and DBD domains (Hinge/DBD), were cloned between the NdeI and BamHI sites (in boldface) in the pET 14B after PCR amplification with the following primers: for the Trans domain, 5′-CAATCTAGACATATGGAGACACCGAAGGAAAAC-3′ and 5′-CGCGGATCCACTGCACATAGAGTCATTAC-3′; and for the Hinge/DBD domain, 5′-CAATCTAGACATATGACCAGTGACGACACGGTA-3′ and 5′-AAAGGATCCTTACATTGTCATGTATCCC-3′.
Cell cultures, transient transfections, and infections.
Cells were grown in Dulbecco's modified Eagle medium supplemented with 7% fetal calf serum. Transient transfections of HeLa cells were done by the standard calcium phosphate coprecipitation technique as previously described (7) with 1 or 2 μg of the GFP expression plasmids for direct observation of GFP fluorescence in living cells. For real-time microscopy, cells were transferred in a thermostated chamber under the microscope, where they were maintained for 24 h. Images were taken every 5 min in phase contrast and every 30 min in fluorescence. For chloramphenicol acetyltransferase (CAT) assays, 4 μg of the P105 CAT expression plasmid (38) was cotransfected with 2 μg of the GFP expression plasmids. CAT assays were done 40 h posttransfection as previously described (7).
Stocks of recombinant adenoviruses were prepared in 293 cells. A control recombinant virus expressing the GFP alone (Quantum) was used to assess the infectivity of the stocks. Cells were infected at a multiplicity of infection (MOI) of 20 for metabolic labeling or 100 for flow cytometry.
Proteasome inhibition.
For inhibition experiments, cells were treated 40 h after transfection with either 10 μM lactacystin or 40 μM MG132 dissolved in dimethyl sulfoxide (DMSO) or with DMSO alone for 6 to 8 h. Inhibitors were purchased from Calbiochem.
In vitro ubiquitination.
In vitro transcription-translation of the plasmids expressing E2 and its subdomains was done with the TNT T7 coupled reticulocyte lysate system (Promega). For ubiquitination assays, 1/25 of the translated lysates was incubated at 30°C with various concentrations of fresh reticulocyte lysate (0 to 4 μl for a final volume of 12 μl) or, alternatively with a constant concentration of fresh reticulocyte lysate (2 μl for a final volume of 12 μl) plus increasing concentrations of purified bovine ubiquitin (0 to 7.5 μg). Incubations were restricted to 20 min to prevent extensive degradation of the ubiquitin-targeted complexes. Polypeptide products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel.
Immunofluorescence.
HeLa cells grown on coverslips were rinsed with phosphate-buffered saline (PBS) 24 h after transfection with the expression plasmids of GFP fusion proteins and then fixed in 4% formaldehyde. After rehydration, cells were permeabilized with 0.1% Triton and incubated with a rabbit polyclonal antibody against HPV18 E2 followed by an anti-rabbit antibody coupled to Texas red.
DNA binding assays.
A double-stranded oligonucleotide, end labeled by Klenow fill-in with [32P]dATP, was used as a probe in gel shifts. It contains the core sequence of E2 binding site 2 of the HPV18 long control region, 5′-AATTGTAGTATATAAAAAAGTTAGTGACCGAAAACGGTCGGG-3′ (the conserved palindrome recognized by E2 is underlined). Binding reactions were carried out in a final volume of 20 μl in a buffer containing 12 mM HEPES (pH 7.9), 10% glycerol, 0.5 mM EDTA, 4 mM MgCl2, 60 mM KCl, 8 mM dithiothreitol, and 0.1% NP-40. Reactions containing 8 μg of proteins of total cell extracts were preincubated for 20 min at 4°C in the presence of 1 μg of poly(dI-dC) and 1 μg of salmon sperm DNA. After addition of the specific labeled probe, they were further incubated 5 min at 4°C, then loaded on a polyacrylamide gel, and run for 2 h at room temperature.
In vivo 35S labeling and immunoprecipitation.
Cells were incubated with medium deficient in methionine and cysteine and supplemented with 5% dialyzed fetal calf serum for 30 min. Radiolabeling with Trans 35S label (0.25 mCi/ml; ICN) was done for 30 min, followed by chase with complete medium for the indicated times. Treatment with proteasome inhibitors was initiated 2 h before labeling. Cell lysates were prepared in HNB buffer (0.5 M sucrose, 15 mM Tris [pH 7.5], 0.42 M KCl, 0.25 mM EDTA, 0.125 mM EGTA, 0.1% NP-40, 0.15 mM spermidine, 0.5 mM spermine, 1 mM dithiothreitol) containing protease inhibitors (Roche). The KCl concentration was decreased to 0.26 M prior to preclearing with a nonspecific rabbit antibody. Immunoprecipitation was done with a specific anti-E2 antibody directed against the C terminus of E2. Immune complexes were collected on protein A-Sepharose and washed three times with HNB (0.26 M KCl) buffer. Proteins were eluted in SDS-PAGE sample buffer, boiled, and separated by SDS-PAGE on a 10% gel. Gels were fixed, dried, and autoradiographed. Quantification was performed using a PhosphorImager and ImageQuant software (Molecular Dynamics).
Flow cytometry.
Infected cells were detached from P60 dishes by incubation for 10 min in trypsin-EDTA. After washing, the cells were fixed in 80% ethanol and then rehydrated in PBS and incubated for 1 h at 37°C with propidium iodide (10 μg/ml). Cells were analyzed with a flow cytometer (EPICS XL; Coulter). The cell cycle was analyzed with Multicycle software (Phoenix Flow System, Inc.).
RESULTS
Characterization of GFP fusion proteins with full-length E2 and separate domains.
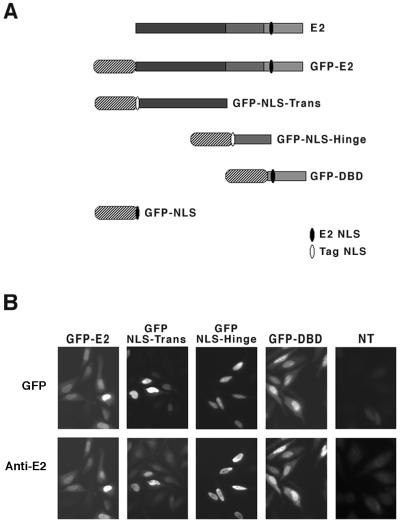
The GFP-E2 fusion protein was obtained by fusion of GFP to the amino terminus of the full-length E2 protein and expressed under the control of the cytomegalovirus promoter (Fig. 1A). The E2 protein is constituted of three structurally and functionally distinct domains: the amino-terminal domain of 206 amino acids (Trans), the carboxy-terminal domain (DBD) of 80 amino acids, and a 79-amino-acid domain (Hinge). GFP was also fused to the amino terminus of each of the three domains (Fig. 1A). We found that while the GFP-E2 and GFP-DBD fusion proteins were expressed in the nuclei of transfected cells, the GFP-Hinge fusion was cytoplasmic and the GFP-Trans fusion was expressed both in the nuclei and in the cytoplasms of transfected cells (results not shown). This is in agreement with the presence of a functional well-conserved NLS in the DBD (34). The transactivation domain appeared to contain a weak NLS, not sufficient for a strict nuclear localization, and no NLS was found in the hinge of the HPV18 E2, in contrast to a recent report indicating the presence of a strong NLS in the hinge of the HPV11 E2 protein (40). To target all of the fusion proteins to the nuclei of transfected cells, we fused a sequence containing the NLS from the SV40 TAg to the amino terminus of the Hinge and Trans domains in the corresponding GFP fusion proteins. GFP alone was also targeted to the nucleus by addition of the E2 NLS (Fig. 1A).
FIG. 1.
Schematic representation of the GFP fusion proteins and their expression in HeLa cells. (A) The three domains of E2 (gray bars) and the GFP moiety (stripped ovals) are shown. Sequence containing the E2 NLS is schematized in black, and the SV40 TAg NLS is shown in white. (B) The four constructs were transfected in HeLa cells, and their expression was checked 24 h later by either direct green fluorescence of GFP or immunofluorescence with anti-E2 antibodies revealed by Texas red as indicated. NT, not transfected.
To test if the fusion proteins were correctly expressed, we compared the immunodetection by immunofluorescence with anti-E2 antibodies with the GFP fluorescence in the nuclei of cells transfected with plasmids expressing the full-length E2 and the separate domains fused to GFP. These experiments indicated that the GFP and immunofluorescence patterns colocalized in nuclei of transfected cells, although part of the GFP staining appeared concentrated in nucleoli that were not stained by anti-E2 antibodies (Fig. 1B). This was particularly visible for the constructs expressing GFP-Hinge and GFP-DBD, but we have no explanation for this phenomenon. Western blot analysis of total extracts of transfected cells with both the anti-E2 and anti-GFP antibodies revealed that the fusion proteins ran at their expected sizes (data not shown).
Biological activities of the fusion proteins.
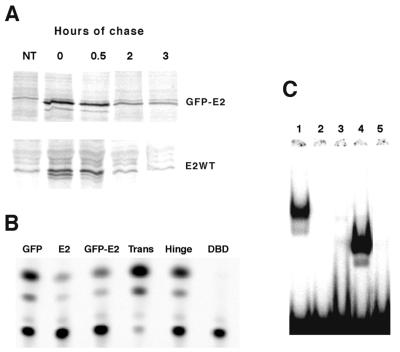
A crucial control for our experiments was to verify whether the stability of the fusion GFP-E2 protein was comparable to that of wild-type E2. We performed pulse-chase experiments using HeLa cells 24 h after transfection with expression plasmids of both proteins. These experiments showed that the two proteins are similarly stable, with relatively short half-lives of 50 min, indicating that fusion with GFP does not interfere with the stability of the E2 protein (Fig. 2A). In functional assays, the behavior of the GFP-E2 fusion protein was indistinguishable from that of wild-type E2. These assays included transcriptional activation of a synthetic thymidine kinase promoter containing six upstream E2 binding sites (37) (data not shown) and repression of the HPV18 P105 promoter (Fig. 2B), DNA binding (Fig. 2C), viral DNA replication, p53 stabilization (data not shown), and cell cycle arrest (see Fig. 5A) (9). These experiments indicated that the GFP-E2 fusion protein retained all tested functions of the E2 protein.
FIG. 2.
Biological activities of the GFP fusion proteins. (A) E2 and GFP-E2 exhibit identical half-lives in pulse-chase experiments done in transfected HeLa cells. Cells were labeled for 30 min and chased for 30 min, 2 h, or 3 h, as indicated, prior to immunoprecipitation of the E2 proteins and separation by SDS-PAGE on a 10% gel. (B) The various GFP fusion protein expression plasmids were cotransfected in HeLa cells with the HPV18 P105 CAT expression plasmid (38) as indicated at the top. Results of representative CAT assays are shown, giving conversion values of 15% for GFP alone as a control, 1 and 5% for E2 and GFP-E2, and 80% for the GFP-Trans, 15% for the GFP-Hinge, and 0.8% for GFP-DBD fusion proteins. (C) Binding assays with a DNA probe containing the E2 binding site and extracts prepared from cells transfected with GFP-E2 (lane 1), GFP-NLS-Trans (lane 2), GFP-NLS-Hinge (lane 3), GFP-DBD (lane 4), and probe alone (lane 5).
FIG. 5.
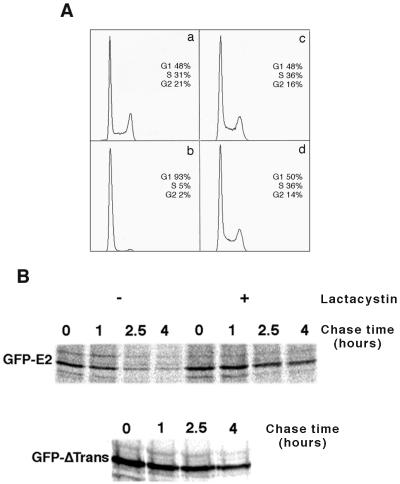
E2 degradation by the proteasome is independent of cell cycle arrest. (A) HeLa (a and b) and MCF7 (c and d) cells were infected at an MOI of 100 with adenoviruses expressing GFP alone (a and c) or the GFP-E2 fusion (b and d). The DNA content of infected cells was determined by flow cytometry. (B) Pulse-chase experiments were performed with MCF7 cells infected at an MOI of 20 with adenovirus expressing either GFP-E2 or GFP-ΔTrans, labeled for 30 min with [35S] methionine, and chased as indicated in the absence (−) or presence (+) of 50 μM lactacystin 24 h postinfection.
Functional assays performed with the separately fused domains indicated that the GFP-DBD fusion protein could bind DNA as well as the GFP-E2 protein and could repress transcription from the HPV18 P105 promoter (Fig. 2B and C). In contrast, the GFP-Trans fusion protein was inactive in DNA binding assays, whereas it activated transcription from the P105 promoter (Fig. 2B and C). This ∼5-fold activation of the P105 promoter could be attributed to its intrinsic transactivating properties and was detected as well with heterologous promoters (not shown). As expected, the hinge domain appeared inactive in these functional assays (Fig. 2B and C).
Real-time microscopy of HeLa cells transfected with GFP fusion proteins.
HeLa cells transfected with the GFP-E2 expression plasmid were analyzed by real-time microscopy for a period of 16 h, between 16 and 32 h posttransfection. Images were recorded at 5-min intervals in phase contrast and at 30-min intervals in fluorescence. We found that the fluorescence started to accumulate in the nuclei of transfected cells immediately after cell division and separation of the two sister cells. Fluorescence increased at similar rates in these two sister cells for about 7 to 8 h until around 24 h posttransfection, after which one of two events occurred: either the two sister cells expressing E2 died concomitantly by apoptosis and remained fluorescent or, in cells that stayed alive, the fluorescence sharply dropped (Fig. 3A). Therefore, these experiments confirmed that E2-mediated apoptosis is an early event occurring immediately after maximal accumulation of the protein (Fig. 3A), as previously published (10). At 28 to 30 h posttransfection, fluorescence had disappeared in all living cells, indicating that expression of the GFP-E2 protein was no longer detectable at this time due to high instability of the protein.
FIG. 3.
Real-time microscopy of HeLa cells transfected with GFP-E2 or GFP-NLS. (A) Transfected cells were subjected to real-time microscopy 16 h posttransfection. Phase-contrast and GFP fluorescent images were taken at the times after transfection indicated at the top. Appearance of the fluorescence in two sister cells arising from mitosis, which will undergo rapid apoptosis, is indicated by white arrows. Black arrows indicate living cells in which the GFP-E2 fluorescence declined. (B) Similar experiment done with the GFP-NLS, indicating that the GFP fluorescence was sustained for a longer time, at least until 36 h after transfection.
To compare the stability of proteins with similar localization, we used a nuclear GFP protein as a control for our experiments. We fused the NLS, contained within the E2 DBD and conserved among E2 proteins, to the C terminus of GFP in the GFP-C1 expression plasmid (Fig. 1A) and performed real-time microscopy with transfected HeLa cells. Cells were analyzed for a period of 23 h, between 16 to 39 h after transfection. Accumulation of fluorescence occurred for 7 to 8 h, reaching a maximum level at around 28 h posttransfection. At that time, and in contrast to the E2 fusion protein, GFP showed a steady-state level of expression for the remaining 8 h (Fig. 3B). This result was not surprising since GFP has been shown to be extremely stable, and we showed here that its nuclear localization did not alter this property.
The behavior of GFP, either alone or fused to E2, clearly indicated that the presence of the E2 moiety destabilized the fusion protein in HeLa cells. Furthermore, characteristics of the E2 expression in real-time microscopy suggested that the stability of the E2 protein might vary during the cell cycle. Indeed, although the appearance of both fluorescent proteins, either alone or fused to E2, seemed to follow cell division, accumulating immediately after mitosis, the sharp disappearance of the E2 fusion protein occurred after a constant period of time (7 to 8 h), independently of the maximum level of expression reached, while GFP alone remained stable (Fig. 3). More work is needed to establish the putative link between E2 stability and specific phases of the cell cycle.
E2 is degraded by the ubiquitin-proteasome pathway, mostly through its transactivation domain.
To check whether the instability of E2 was due to degradation by the proteasome pathway, we used proteasome inhibitors in vivo and studied the stability of GFP-E2. When used from 30 h after transfection, at a time when the GFP-E2 protein was no longer detectable in living cells, treatment with MG132 or lactacystin allowed reappearance of the GFP fluorescence signal in nuclei of transfected cells (Fig. 4A). These experiments demonstrated a significant stabilization of the fusion protein upon inhibition of the proteasome, indicating that E2 degradation occurs through this pathway.
FIG. 4.
E2 is degraded by the ubiquitin-proteasome pathway mostly through its transactivation domain (A) Proteasome inhibitors restore GFP fluorescence in nuclei of cells when added 40 h after transfection of the GFP-E2 fusion protein. Shown are phase-contrast and the corresponding fluorescent fields of living cells treated for 8 h with DMSO, 8 h with 10 μM lactacystin, and 6 h with 40 μM MG132 beginning 40 h posttransfection, as indicated. (B) Cells infected with recombinant adenovirus expressing either GFP-E2 or GFP-ΔTrans were labeled for 30 min with [35S] methionine and chased in the presence of excess of nonradioactive methionine for 1, 2.5, and 4 h, as indicated, in the absence (−) or presence (+) of 50 μM lactacystin, 24 h postinfection. (C) Ubiquitination of in vitro-translated E2, Trans domain, or Hinge/DBD domain in reticulocyte lysate with increasing concentrations of fresh lysate: 0 (lanes 1, 4, and 7), 2 (lanes 2, 5, and 8), and 4 (lanes 3, 6, and 9) μl of lysate added; 0 (lane 10) and 2 μl of lysate added (lanes 11 to 14) with the presence of 2.5 μg, 5 μg, and 7.5 μg of ubiquitin (lanes 12, 13, and 14, respectively).
Pulse-chase experiments done with the GFP-E2 fusion expressed following HeLa cells infection with recombinant adenoviruses indicated that it was strongly stabilized when the cells were treated with lactacystin (Fig. 4B). Interestingly, the stabilization of E2, due to inhibition of the proteasome, was calculated to be sevenfold, which is in close agreement with the stabilization of the amino-terminally truncated E2 (GFP-ΔTrans [Fig. 4B]).
The implication of the transactivation domain in E2 instability was confirmed by real-time microscopy. Real-time microscopy experiments performed with the GFP-NLS-Trans, GFP-NLS-Hinge, or GFP-DBD protein, indicated that the transactivation domain exhibited expression kinetics similar to that of the full-length protein, while those of the hinge and DBD domains were comparable to the kinetics of the GFP protein alone (data not shown and Fig. 3). Altogether, these results indicated that the transactivation domain of E2 is able to confer intrinsic instability to the GFP similarly to the full-length E2 protein, while the two other domains are not.
We then asked whether E2 and the transactivation domain alone could be ubiquitinated in vitro in reticulocyte lysate, which is a commonly used method to identify sequences responsive to ubiquitination (29). In vitro-translated HPV18 E2 protein was efficiently ubiquitinated by addition of fresh lysate, as demonstrated by shift of the protein to high-molecular-weight species (Fig. 4C, lanes 1 to 3). In similar experiments, we found that the Trans domain was ubiquitinated as well as the full-length protein (Fig. 4C, lanes 4 to 6), while ubiquitination of the Hinge/DBD domain was much less efficient (Fig. 4C, lanes 7 to 9). To confirm that the protein shift was due to ubiquitination, the radiolabeled E2 protein was incubated with a limiting concentration of fresh reticulocyte lysate and increasing concentrations of purified bovine ubiquitin. These increasing concentrations of ubiquitin led to an increasing shift of the E2 protein (Fig. 4C, lanes 10 to 14). We conclude from these experiments that the HPV18 E2 transactivation domain contains sequences that allow efficient ubiquitination of the protein and can therefore mediate the ubiquitin-proteasome degradation of the E2 protein observed in vivo.
E2 degradation by the proteasome is independent of E2-mediated control of cell growth.
As expected from previously published data showing that expression of E2 in cervical carcinoma cell lines led to cell cycle arrest (9, 14, 23), HeLa cells infected with an adenovirus expressing the GFP-E2 fusion protein were arrested in G1 (Fig. 5A, chart b) compared to cells infected with an adenovirus expressing only GFP (Fig. 5A, chart a). To verify that the E2 degradation observed in HeLa cells was not a consequence of the E2-mediated cycle arrest, we checked E2 stability in infected MCF7, a breast carcinoma cell line not associated with HPV. At identical conditions of infection, MCF7 cells exhibited an unaltered cell cycle when expressing the GFP-E2 fusion (Fig. 5A, chart d) compared to GFP alone (Fig. 5A, chart c).
Pulse-chase experiments indicated that E2 was as unstable in MCF7 as in HeLa cells, with a half-life of about 1 h, and could be stabilized by lactacystin treatment, thus indicating that it is degraded by the proteasome (Fig. 5B). Furthermore, truncation of its amino-terminal domain led to stabilization in MCF7 cells (Fig. 5B) as well as in HeLa cells (Fig. 4B). Therefore, comparative studies of the stability of E2 in the HeLa and MCF7 cell lines showed that the proteasome-dependent degradation of the HPV E2 protein, through its amino-terminal domain, is an intrinsic property of the protein that was not a consequence of the cycle arrest observed in HeLa cells.
DISCUSSION
We have shown that the amino-terminal domain of HPV18 E2 protein is responsible for its rapid turnover in transfected or infected cells. In addition, this domain contains signals that confer instability to the GFP to which it was fused. This degradation is mediated by the ubiquitin-proteasome pathway. The lability of many regulatory proteins is crucial for their function in the cell. The instability of many transcription factors affects their regulatory properties. The E2 proteins of genital HPVs play a multifunctional role in viral infection, since they are required for regulation of both viral transcription and viral DNA replication. Furthermore, the loss of E2 function plays a determining role in carcinogenic progression of HPV-associated lesions. In this respect, it is not surprising that the HPV18 E2 protein is unstable in transfected cells. The rapid turnover of E2 could explain why the protein is not detected in benign productive lesions associated with genital HPVs, despite its crucial role in the vegetative viral cycle.
Previous studies have shown a close correlation between transcriptional activation and degradation domains in several transcription factors (31, 32). Interestingly, although sequences for transcriptional activation and for degradation overlap in E2, its transactivation function is not sufficient to mediate protein instability. We found that a point mutant of the amino-terminal transactivation domain that is impaired in transcriptional activation (10) is as unstable as the wild-type protein in HeLa cells (results not shown). This means that the link between sequences involved in transcriptional activation and degradation is not absolute, as previously discussed for the Myc protein (32). The E2 amino-terminal domain is highly structured and contains many amino acids conserved among E2 proteins. Most of these conserved amino acids appear to play a crucial structural role that makes mutational analysis of the protein very laborious. Examination of the 200 amino acids of the amino-terminal domain of HPV18 E2 revealed the presence of putative PEST sequences in the first α helix. Deletion of these sequences, in the context of the full-length protein fused to GFP, led to a protein that was not expressed properly and could therefore not be studied (results not shown). Alternative ways to dissect the sequences responsible for proteasome degradation of E2 are needed.
Our finding that the HPV18 E2 protein is degraded by a proteasome pathway is in agreement with a recent paper showing that the E2 protein from bovine papillomavirus type 1 (BPV1) is degraded by the same pathway (30). Half-lives of the two proteins are very similar, but there are some interesting differences between the two systems. One of the most intriguing differences is that the stability of BPV1 E2 is regulated by phosphorylation of a specific site in the hinge domain of the protein. BPV1 E2 phosphorylation was shown to be involved in regulating viral DNA replication or cellular transformation by BPV1 (25, 26). There are, however, no similar phosphorylation sites in the hinge of the HPV18 E2 protein, and the phosphorylation status of E2 proteins from human papillomaviruses has not been specifically addressed. In addition, the results presented here indicate unambiguously that the hinge of the HPV18 E2 protein is not involved in controlling the stability of the protein, at least in our experimental conditions. Although HPV and BPV1 E2 proteins are highly homologous and show close structural and functional similarities, they may be subjected to different regulatory pathways for their stability. A crucial difference between the two proteins is that BPV1 E2 acts mainly as a transcriptional activator of the transforming functions in the BPV1 life cycle, whereas HPV E2 is a transcriptional repressor in the HPV life cycle.
Real-time microscopy of GFP-E2 accumulation in transfected HeLa cells suggested that the turnover of E2 might be linked to the cell cycle. We found that the protein started to accumulate in the nuclei of transfected cells following mitosis and that its concentration then increased for about 8 h. This maximal accumulation was followed by one of two events: either the cells died by apoptosis or the cells survived and the fluorescence sharply declined after a constant period of time. This observation suggests that the stability of E2 is regulated differentially between the different phases of the cell cycle. However, more work is needed to decipher the link between E2 degradation and the cell cycle. In addition, it should be noted that when expressed in HeLa cells, E2 induces a cell cycle arrest in G1, due to stabilization of the p53 protein that in turn activates p21, a negative regulator of the cell cycle (9). At 40 h after transfection of a plasmid expressing E2 in HeLa cells, accumulation of p53 and p21 is clearly detected, while the cells are arrested in G1 (reference 10 and results not shown). However, at that time, as shown here, E2 is no longer detectable. Therefore, the link between E2 stability and the cell cycle needs to be studied in a cell line in which E2 does not induce cell cycle arrest. We found that E2 is unstable and degraded by the proteasome in the MCF7 cell line, which does not express HPV genes and is not sensitive to E2-induced cell cycle arrest, thus representing a good recipient cell for further studies.
The potential link between E2 stability and cell cycle that we inferred from real-time microscopy analyses led us to propose a hypothesis regarding the control of E2 accumulation during the viral vegetative cycle in infected lesions (Fig. 6). Infected benign lesions are characterized by a hyperplasia of the epithelium due to E6/E7 expression that induces basal cell proliferation. In these cycling cells of the basal layer, E2 would be degraded in one phase of the cycle. Consequently, only low levels of E2 are expressed and viral DNA replication remains low, maintaining a low stable copy number of viral genome per cell. In addition, the transforming genes E6 and E7 are not efficiently repressed, leading to cellular proliferation. The upper layers of the lesion contain cells that left the cell cycle and are undergoing terminal differentiation and sustain late phases of the viral vegetative cycle, including viral genome replication, late gene expression, and viral maturation. In these noncycling cells, E2 would be stabilized, leading to its accumulation; this would (i) reinforce transcriptional repression of the transforming genes E6 and E7, with high amplification of the viral genomes, and (ii) induce apoptosis of recipient cells, favoring spreading of the viral progeny. In the line with the present model, a recent study showed that HPV16 E2 protein expression is highest in differentiated cells of lower-grade lesions, while it decreases with increasing grade (35). In the BPV1 model, high E2 expression was also found in a subset of differentiated cells of the upper layers of the lesions where high viral DNA replication takes place (30).
FIG. 6.
A model for the role of E2 instability in the viral vegetative cycle.
ACKNOWLEDGMENTS
This work was supported by the Association pour la Recherche contre le Cancer and the Ligue Française contre le Cancer.
We are very grateful to Sylvie Beaudenon, Jon Huibregtse, and Moshe Yaniv for critical reading of the manuscript. We thank Patrice Yieh for the gift of adenovirus vectors and helpful discussions and Alejandro Garcia-Carranca for help in the construction of recombinant adenoviruses. We are grateful to Serge Garbay for real-time microscopy and to Catherine Bonne-Andrea and Marie-Hélène Malclès for help with the in vitro ubiquitination studies.
REFERENCES
- 1.Antson A A, Burns J E, Moroz O V, Scott D J, Sanders C M, Bronstein I B, Dodson G G, Wilson K S, Maitland N J. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature. 2000;403:805–809. doi: 10.1038/35001638. [DOI] [PubMed] [Google Scholar]
- 2.Berumen J, Casas L, Segura E, Amezcua J, Garcia-Carranca A. Genome amplification of human papillomavirus types 16 and 18 in cervical carcinomas is related to the retention of E1/E2 genes. Int J Cancer. 1994;56:640–645. doi: 10.1002/ijc.2910560506. [DOI] [PubMed] [Google Scholar]
- 3.Corden S A, Sant-Cassia L J, Easton A J, Morris A G. The integration of HPV-18 DNA in cervical carcinoma. Mol Pathol. 1999;52:275–282. doi: 10.1136/mp.52.5.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crouzet J, Naudin L, Orsini C, Vigne E, Ferrero L, Le Roux A, Benoit P, Latta M, Torrent C, Branellec D, Denèfle P, Mayaux J, Perricaudet M, Yeh P. Recombinational construction in Escherichia coli of infectious adenovirus genomes. Proc Natl Acad Sci USA. 1997;94:1414–1419. doi: 10.1073/pnas.94.4.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen A P, Reid R, Campion M, Lorincz A T. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J Virol. 1991;65:606–612. doi: 10.1128/jvi.65.2.606-612.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demeret C, Desaintes C, Yaniv M, Thierry F. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J Virol. 1997;71:9343–9349. doi: 10.1128/jvi.71.12.9343-9349.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demeret C, Yaniv M, Thierry F. The E2 transcriptional repressor can compensate for Sp1 activation of the human papillomavirus type 18 early promoter. J Virol. 1994;68:7075–7082. doi: 10.1128/jvi.68.11.7075-7082.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desaintes C, Demeret C. Control of papillomavirus DNA replication and transcription. Semin Cancer Biol. 1996;7:339–347. doi: 10.1006/scbi.1996.0043. [DOI] [PubMed] [Google Scholar]
- 9.Desaintes C, Demeret C, Goyat S, Yaniv M, Thierry F. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 1997;16:504–514. doi: 10.1093/emboj/16.3.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desaintes C, Goyat S, Garbay S, Yaniv M, Thierry F. Papillomavirus E2 induces p53-independent apoptosis in HeLa cells. Oncogene. 1999;18:4538–4546. doi: 10.1038/sj.onc.1202818. [DOI] [PubMed] [Google Scholar]
- 11.Dong G, Broker T R, Chow L T. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68:1115–1127. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dostatni N, Lambert P F, Sousa R, Ham J, Howley P M, Yaniv M. The functional BPV1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 1991;5:1657–1671. doi: 10.1101/gad.5.9.1657. [DOI] [PubMed] [Google Scholar]
- 13.Dostatni N, Thierry F, Yaniv M. A dimer of BPV-1 E2 containing a protease resistant core interacts with its DNA target. EMBO J. 1988;7:3807–3816. doi: 10.1002/j.1460-2075.1988.tb03265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowhanick J J, McBride A A, Howley P M. Suppression of cellular proliferation by the papillomavirus E2 protein. J Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firestein R, Feuerstein N. Association of activating transcription factor 2 (ATF2) with the ubiquitin-conjugating enzyme hUBC9. Implication of the ubiquitin/proteasome pathway in regulation of ATF2 in T cells. J Biol Chem. 1998;273:5892–5902. doi: 10.1074/jbc.273.10.5892. [DOI] [PubMed] [Google Scholar]
- 16.Frattini M G, Lim H B, Doorbar J, Laimins L A. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J Virol. 1997;71:7068–7072. doi: 10.1128/jvi.71.9.7068-7072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giri I, Yaniv M. Structural and mutational analysis of E2 trans-activating proteins of papillomaviruses reveals three distinct functional domains. EMBO J. 1988;7:2823–2829. doi: 10.1002/j.1460-2075.1988.tb03138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodwin E C, Naeger L K, Breiding D E, Androphy E J, DiMaio D. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J Virol. 1998;72:3925–3934. doi: 10.1128/jvi.72.5.3925-3934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris S F, Botchan M R. Crystal structure of the human papillomavirus type 18 E2 activation domain. Science. 1999;284:1673–1676. doi: 10.1126/science.284.5420.1673. [DOI] [PubMed] [Google Scholar]
- 20.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 21.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 22.Hegde R S, Grossman S R, Laimins L A, Sigler P B. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 23.Hwang E S, Riese D J, Settleman J, Nilson L A, Honig J, Flynn S, DiMaio D. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J Virol. 1993;67:3720–3729. doi: 10.1128/jvi.67.7.3720-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubbutat M H, Ludwig R L, Ashcroft M, Vousden K H. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehman C W, King D S, Botchan M R. A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J Virol. 1997;71:3652–3665. doi: 10.1128/jvi.71.5.3652-3665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBride A A, Bolen J B, Howley P M. Phosphorylation sites of the E2 transcriptional regulatory proteins of bovine papillomavirus type 1. J Virol. 1989;89:5076–5085. doi: 10.1128/jvi.63.12.5076-5085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride A A, Byrne J C, Howley P M. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc Natl Acad Sci USA. 1989;86:510–514. doi: 10.1073/pnas.86.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers C, Mayer T J, Ozbun M A. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa S, Huibregtse J M. Human Scribble (vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol. 2000;20:8244–8253. [Google Scholar]
- 30.Penrose K, McBride A. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. J Virol. 2000;74:6031–6038. doi: 10.1128/jvi.74.13.6031-6038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salghetti S E, Kim S Y, Tansey W P. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salghetti S E, Muratani M, Wijnen H, Futcher B, Tansey W P. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc Natl Acad Sci USA. 2000;97:3118–3123. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 34.Skiadopoulos M H, McBride A A. The bovine papillomavirus type 1 E2 transactivator and repressor proteins use different nuclear localization signals. J Virol. 1996;70:1117–1124. doi: 10.1128/jvi.70.2.1117-1124.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevenson M, Hudson L C, Burns J E, Stewart R L, Wells M, Maitland N J. Inverse relationship between the expression of the human papillomavirus type 16 transcription factor E2 and virus DNA copy number during the progression of cervical intraneoplasia. J Gen Virol. 2000;81:1825–1832. doi: 10.1099/0022-1317-81-7-1825. [DOI] [PubMed] [Google Scholar]
- 36.Tan S H, Leong L E C, Walker P A, Bernard H-U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thierry F, Dostatni N, Arnos F, Yaniv M. Cooperative activation of transcription by bovine papillomavirus type 1 E2 can occur over a large distance. Mol Cell Biol. 1990;10:4431–4437. doi: 10.1128/mcb.10.8.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thierry F, Heard J M, Dartmann K, Yaniv M. Characterization of a transcriptional promoter of human papillomavirus 18 and modulation of its expression by simian virus 40 and adenovirus early antigens. J Virol. 1987;61:134–142. doi: 10.1128/jvi.61.1.134-142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treier M, Staszewski L, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 40.Zou N, Lin B Y, Duan F, Lee K Y, Jin G, Guan R, Yao G, Lefkowitz E J, Broker T R, Chow L T. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J Virol. 2000;74:3761–3770. doi: 10.1128/jvi.74.8.3761-3770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]