Abstract
For efficient and versatile use of adenovirus (Ad) as an in vivo gene therapy vector, modulation of the viral tropism is highly desirable. In this study, a novel method to genetically alter the Ad fiber tropism is described. The knob and the last 15 shaft repeats of the fiber gene were deleted and replaced with an external trimerization motif and a new cell-binding ligand, in this case the integrin-binding motif RGD. The corresponding recombinant fiber retained the basic biological functions of the natural fiber, i.e., trimerization, nuclear import, penton formation, and ligand binding. The recombinant fiber bound to integrins but failed to react with antiknob antibody. For virus production, the recombinant fiber gene was rescued into the Ad genome at the exact position of the wild-type (WT) fiber to make use of the native regulation of fiber expression. The recombinant virus Ad5/FibR7-RGD yielded plaques on 293 cells, but the spread through the monolayer was two to three times delayed compared to WT, and the ratio of infectious to physical particles was 20 times lower. Studies on virus tropism showed that Ad5/FibR7-RGD was able to infect cells which did not express the coxsackie-adenovirus receptor (CAR), but did express integrins. Ad5/FibR7-RGD virus infectivity was unchanged in the presence of antiknob antibody, which neutralized the WT virus. Ad5/FibR7-RGD virus showed an expanded tropism, which is useful when gene transfer to cells not expressing CAR is needed. The described method should also make possible the construction of Ad genetically retargeted via ligands other than RGD.
One of the general limitations for successful gene therapy today is the difficulty of achieving in vivo gene delivery to specific cells. Among several potent vectors used for gene therapy is adenovirus (Ad), which benefits from being safe, well studied, and easy to propagate (46). However, Ad has a broad tropism and infects a wide variety of cells by binding to the coxsackie B virus and Ad receptor (CAR) (3) and the major histocompatibility complex class I alpha-2 domain (16). On the other hand, cells that do not express these receptors are often refractory to Ad transduction.
Cellular binding of Ad is mediated by the fiber protein, which is anchored to the penton bases at vertices of the viral icosahedron. The fiber is a homotrimer composed of three identical fiber polypeptides arranged in a parallel orientation (39). Trimerization is absolutely crucial for the fiber to function in attachment both to the capsid and to the cellular receptor (7) and is achieved by a trimerization signal that is situated within the knob region (13, 27), which also contains the ligand that binds to the cellular receptors (24).
Viral retargeting can be divided roughly into two conceptually different strategies: (i) nongenetic retargeting and (ii) genetic retargeting involving engineering of viral proteins. Within each group, expanded as well as narrowed tropism may be achieved. The first strategy has mainly utilized bispecific antibodies or peptides that block the native binding of the fiber and redirect the virus to a new cellular receptor (9, 15).
Efforts using the second strategy include construction of a chimeric Ad5 fiber with an Ad3 knob (22), modifications of the penton base (43) or hexon proteins (41), and insertions of new amino acid (aa) motifs in the fiber knob (8, 25). However, the last approach is limited by the fact that the trimeric nature of the fiber is sensitive to genetic alterations, so that only small insertions are tolerated. As an example, the C-terminal insertion of 24 aa (25) was tolerated, while 26 aa at the same position totally disrupted the trimeric structure (45). Most of the approaches mentioned retain the native binding structure and thus broaden the viral tropism. For these vectors to work satisfactorily in vivo, tissue-specific promoters or other regulatory elements are a necessity unless ablation of the native cellular binding is achieved. Recently, Kirby et al. (21) abolished high-affinity binding to CAR by point mutations in the DG loop of the knob. However, the native conformation of the knob will still be needed, and large insertions in flexible loops such as DG or HI might be as badly tolerated as in the rest of the knob. It is therefore unlikely that the use of nonbinding fiber-knobs as molecular scaffolds or frameworks for new cell-binding ligands will be widely useful for the construction of genetically retargeted Ad.
The aim of this study was to genetically retarget Ad and simultaneously remove the cell-binding ligand. In contrast to the earlier concept of preserving and modifying the knob, we have “deknobbed” the fiber by removing the fiber sequence C-terminal of the seventh shaft repeat. This completely removes the cell-binding ligand but also the trimerization signal. To compensate for the loss of trimer formation, we inserted the neck region peptide (NRP) of human lung surfactant protein D as an external trimerization signal. This 36-residue motif self-assembles into an extremely strong, tightly associated, parallel three-stranded α-helical bundle (17). In its original lung surfactant protein D context, NRP is flanked N-terminally by collagen regions and C-terminally by C-type lectin domains, suggesting that complex structures can be placed C-terminally of NRP without disrupting the ability to trimerize. As a proof of concept, we placed the short peptide motif arginine-glycine-aspartic acid (RGD) at the C-terminal end of the fiber for binding to cell surface integrins αvβ3 and αvβ5 (44) and showed that these recombinant fibers could be rescued into functional, infectious, retargeted virions.
(Construction of knobless fibers with a new trimerization signal and a new cell-binding ligand was described by L.L. at the Cold Spring Harbor meeting on Vector Targeting Strategies, 1997.)
MATERIALS AND METHODS
Cells.
HEK-293 cells (11) were purchased from Microbix (Toronto, Ontario, Canada) and Cos7 cells, RD cells, and HeLa cells were obtained from the American Type Culture Collection (ATCC, Rockville, M.). All cultures were maintained in Iscove's medium (Gibco-BRL) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and gentamicin (50 μg/ml) (Gibco-BRL) at 37°C and 5% CO2. Spodoptera frugiperda Sf9 cells (ATCC) were propagated in TC100 medium (Gibco-BRL) with the abovementioned supplements at 28°C.
Antibodies.
Three monoclonal fiber antibodies were obligingly supplied by Jeff Engler (University of Alabama at Birmingham). Antibody 4D2.5 recognizes the conserved linear motif FNPVYP found in most Ad fiber tails (13, 14); 2A6.36 is specific for a conformational epitope present in fiber trimers within residues 17 to 61, as suggested by deletion mapping (13) and the lack of reactivity with trimers of our deletion mutant AT61 (27); 1D6.14 is an antiknob, CAR-binding blocking antibody (32) whose epitope has been mapped within residues 471 to 491 (unpublished data). Fluorescein isothiocyanate (FITC)-labeled rabbit anti-mouse immunoglobulin G (IgG) and streptavidin-horseradish peroxidase (HRP) were purchased from Dako. Anti-αvβ3 integrin (LM609) and anti-αvβ5 integrin (P1F6) monoclonal antibodies were from Chemicon International, Inc. The monoclonal antibody RL2, which is specific for O-linked GlcNAc residues (26), was obtained from Larry Gerace via Jeff Engler. Antihexon capsomers were produced by the 2Hx2 hybridoma, purchased from ATCC.
Construction of recombinant fibers.
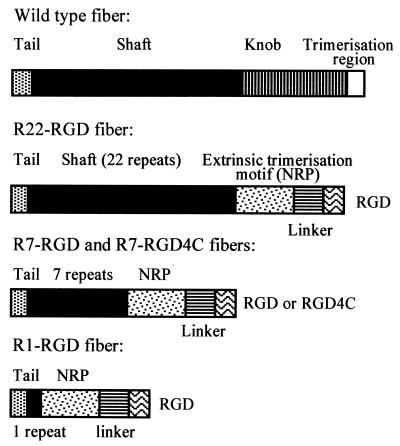
The wild-type (WT) fiber gene was amplified from pAB26 (Microbix) using the primers 5′-CTC GGA TCC GAT GAA GCG CGC AAG ACC GTC TGA A-3′ (5′ oligonucleotide) and 5′-TTC CTC GAG TTA TTC TTG GGC AAT GTA TGA-3′ (3′ oligonucleotide), introducing an upstream BamHI and a downstream XhoI site, respectively (Fig. 1). Recombinant fibers containing the tail, different numbers of shaft repeats (1, 7, or 22) followed by the external trimerization signal NRP (PDVASLRQQVAELQGQVQHLQAAFSQYKKVELFPNG) (17), a linker sequence from Staphylococcus protein A (AKKLNDAQAPKSD), and RGD were constructed by PCR amplification of the WT fiber gene, followed by splicing by overlap extension and ligation, which introduced the flanking restriction sites mentioned above (detailed information can be requested from the authors). A recombinant fiber with seven shaft repeats and the disulfide bond-containing, constrained ligand RGD4C (ACDCRGDCFCG) (30) was also constructed for comparison to the linear RGD motif. The fibers were named R1-RGD, R7-RGD, R7-RGD4C, and R22-RGD (Fig. 1), respectively, indicating the different numbers of shaft repeats preceding the cell ligand. NRP was ligated to the first repeat at the SphI site, the seventh repeat at the NheI site, and the 22nd repeat after the conserved TLWT motif at positions 400 to 403.
FIG. 1.
Schematic representation of the WT fiber and recombinant fibers R22-RGD, R7-RGD (or RGD4C), and R1-RGD. The tail fiber domain is shown by a stippled box, the shaft by a solid box, and the knob domain by a hatched box. Additional structural and functional motifs in recombinant fibers, i.e., extrinsic trimerization signal (NRP), linker peptide, and cell ligand RGD or RGD4C, are represented as indicated in the diagrams. The fiber sequence ends at aa M-61 in R1-RGD, K-156 in R7-RGD, and T-403 in R22-RGD.
Protein expression and purification.
For analysis of cellular localization in mammalian cells, recombinant fibers were cloned into pcDNA3.1 (Invitrogen, San Diego, Calif.), and Cos7 cells were transfected using Lipofectamine (Life Technologies Inc., Gaitherburg, Md.) according to the protocol of the manufacturer. Two days posttransfection, the cells were harvested, centrifuged onto cytospin slides, and dried overnight. Immunofluorescent staining was performed after fixation in 3% paraformaldehyde and permeabilization in 0.1% Triton X-100 in phosphate-buffered saline (PBS). The primary antibodies 4D2.5 and 2A6.36 were used as ascites fluids diluted to 1:1,000, while the secondary FITC-labeled anti-mouse Ig antibody was used at a dilution of 1:10. Each antibody was incubated with the specimens for 30 min at 37°C. Expression and nuclear localization were observed using a Zeiss Axioskop fluorescent microscope and photographed with a Zeiss automatic camera at ×40 magnification.
For protein production and purification, the genes coding for fibers, WT penton base, and penton base mutant R340E (19, 20) were cloned into the baculovirus Autographa californica nuclear polyhedrosis virus, using the pBacPAK9 (Novagen) or BacPAK6 (Clonetech) vector. Recombinant Ad proteins were expressed in Sf9 cells and purified according to a previously described method (5), with some modifications. The anion-exchange chromatographic step was performed using a high-performance liquid chromatography system (BioLogic DuoFlow; Bio-Rad) and a DEAE-Sepharose Fast Flow column (DFF-100; Sigma) equilibrated in 50 mM sodium phosphate buffer, pH 6.8 (PB-50). Samples (2 to 3 mg of protein) were applied to the column, and elution was obtained by applying a 0.0 to 0.6 M NaCl gradient in PB-50. Fiber protein was eluted at 200 mM salt and penton base at 250 mM salt, respectively. Protein samples were then further purified and concentrated using concentrator membranes with a 100-kDa cutoff (Vivaspin-100; Vivascience Ltd, Binbrook, Lincoln, England). Final protein concentration was estimated using the Bradford assay (Bio-Rad).
Phenotypic analysis of fiber proteins.
Recombinant fiber proteins were phenotypically analysed for their trimerization, glycosylation, and assembly in Sf9 cells with recombinant penton base to form complete pentons. Oligomerization was assayed by means of nondenaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (NDS-PAGE) and by conventional, denaturing SDS-PAGE. NDS-PAGE differed from SDS-PAGE in that the samples were not denatured by boiling in SDS sample buffer prior to electrophoresis. Glycosylation of recombinant fibers was assessed both by immunoreaction on blots using the monoclonal antibody RL2 and by chemical detection using the DIG Glycan detection kit (Roche). Assembly of fiber with penton base to form penton in vivo was assayed by coinfecting the same Sf9 cells with two recombinant AcNPV, one expressing the penton base and the other expressing the fiber protein. The presence of penton capsomer was detected in cell lysates 40 h postinfection and analyzed by PAGE in native conditions at low voltage overnight with cooling, as previously described (19). For immunological quantification of native penton, penton base, and fiber proteins, blots were reacted with the corresponding primary antibody (anti-penton base or antifiber), followed by [35S]SRL-labeled anti-mouse or anti-rabbit whole IgG secondary antibody (Amersham Pharmacia Biotech; 100 μCi/ml; 5 μCi per blot). Blots were exposed to radiographic films (Hyperfilm Beta-max; Amersham Pharmacia Biotech), and autoradiograms were scanned at 610 nm using an automatic densitometer (REP-EDC; Helena Laboratories, Beaumont, Tex.). Alternatively, protein bands were excised from blots and radioactivity was measured in a scintillation counter (Beckman LS-6500), as previously described (18).
ELISA.
Integrins αvβ3 and α5β1 (Chemicon) at 1 μg/ml in coating buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2) was used to coat 96-well plastic plates for 16 h at 4°C. After one wash in rinsing buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2), the plates were blocked with 200 μl of blocking buffer (rinsing buffer containing 4% nonfat dried milk) at room temperature for 2 h. Wells were then washed three times with rinsing buffer. Five-step dilutions (1 to 0.008 μg/ml) of recombinant fibers in blocking buffer were added to the wells in 100 μl for 1 h at room temperature. Detection of bound fibers was done with 1 μg of 4D2.5 (biotinylated and protein A purified), streptavidin-HRP diluted 1:2,000, and TMB substrate (CanAg Diagnostics, Göteborg, Sweden) per ml. Color development was stopped with 0.12 M HCl after 10 min, and plates were analyzed in a microtiter plate reader set at 450 nm.
For WT fiber detection, 10 μg of 4D2.5 (protein A purified) per ml in 50 mM sodium carbonate buffer (pH 9.6) was used to coat 96-well plates in 100-μl aliquots for 16 h at 4°C. Wells were washed once in PBS containing 0.1% Tween 20 (PBS-T) prior to addition of 200 μl of PBS containing 1% bovine serum albumin (PBS-B) for 2 h of blocking at room temperature. After three washes in PBS-T, recombinant fibers (diluted in PBS-B, 100 μl total volume) were added to the wells and further incubated for 1 h at room temperature. The CAR-binding inhibiting antiknob antibody 1D6.14 (biotinylated and protein A purified) was used at 3 μg/ml, and streptavidin-HRP was used at 1:2,000. Color development with TMB substrate and microtitration were performed as described above.
Generation of recombinant Ad genomes.
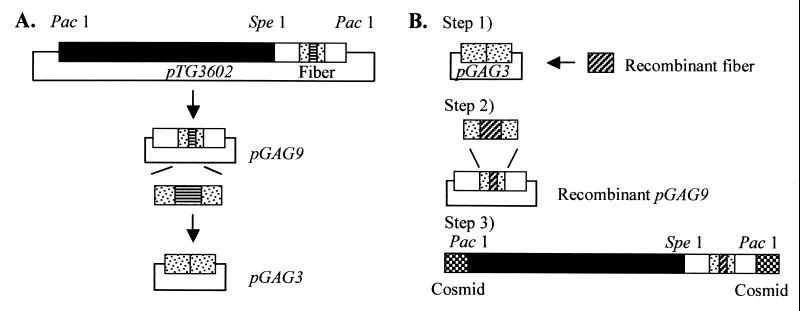
To generate recombinant Ad5 genomes, a fiber shuttle vector and a fiber rescue plasmid were constructed using pTG3602, a plasmid which contains the WT Ad5 genome with PacI restriction sites at both ends (6). This is schematically depicted in Fig. 4. To construct the shuttle vector, the SacI-KpnI fragment of 3.4 kb from 84.0 to 93.5 map units (mu), which contains the fiber gene, was cloned into pBluescript II SK(−) (Stratagene). The fiber gene was then deleted from the NdeI site, located in the fiber tail, to the MunI site, present a few nucleotides downstream of the fiber gene. The deleted region was replaced by an annealed oligonucleotide linker constituted of an XhoI site flanked by an NdeI site and a MunI site at its 5′ and 3′ ends, respectively. The resulting shuttle plasmid was referred to as pGAG3. For construction of the fiber rescue plasmid, pBluescript II SK(−) was first supplied with a PacI site. Thereafter, the SpeI-PacI fragment of 8.8 kb (75.4 to 100 mu) from pTG3602 was ligated to the pBluescript II SK(−) PacI site to generate pGAG9.
FIG. 4.
Generation of recombinant Ad5 genomes. (A) Construction of pGAG9 and pGAG3 from pTG3602. (B) Three-step rescue system. Step 1, ligation of recombinant fiber to pGAG3. Step 2, homologous recombination into pGAG9. Step 3, cosmid cloning to join the modified fragment from step 2 to the PacI-SpeI fragment of 24 kb from the recombined pAdTrackCMV/pAdEasy. Detailed procedure is described in Materials and Methods.
Fiber constructions were subcloned into pGAG3 using NdeI and XhoI. As a negative control, the empty pGAG3 was used to obtain a fiber deleted from the NheI site in the tail region to the 3′ end of the fiber gene. For further rescue into pGAG9, homologous recombination in Escherichia coli BJ5183 (recBC sbcBC) (6) was performed as described elsewhere (28). Briefly, 50 ng of pGAG9 restricted with NheI was transformed into BJ5183 by electroporation together with a 10:1 (insert-vector) molar ratio of SacI-KpnI-digested pGAG3. The recombinant pGAG9 clones were then retransformed into the Nova Blue bacterial strain (Novagen) to obtain large quantities of plasmid DNA.
To join the recombinant pGAG9 to the remaining 0 to 75.4 mu of the Ad5 genome, a cosmid cloning system (SuperCos 1 cosmid vector kit and Gigapack III Gold packaging extract; Stratagene) was used after addition of a PacI site to SuperCos1 in the BamHI site. This cosmid (SupercosA) was restricted with PacI and XbaI and treated according to the manual. Then 0.5 μg of SupercosA, 0.5 μg of PacI-SpeI-restricted recombinant pGAG9, and 0.75 μg of PacI-SpeI-restricted fragment (0 to 75.4 mu) were used for ligation. Clones were screened by PCR using primers specific for the fiber and restricted by HindIII and SpeI. Large plasmid preparations were made using a plasmid maxi kit (Qiagen).
The 0 to 75.4 mu sequence of the Ad5 genome originated from recombined pAdEasy1 and pAdTrackCMV (12), in which the deleted E1 region is replaced by a green fluorescent protein (GFP) gene under the control of the cytomegalovirus (CMV) promoter (12).
Virus generation.
Recombinant cosmid DNA was restricted with PacI, precipitated with ethanol, and resuspended in sterile H2O. Transfection into 293 cells was performed using FuGENE (Roche) according to the manufacturer's recommendations, with 2 μg of DNA and 3 μl of FuGENE in each 35-mm well. Large-scale production and CsCl gradient purification of virus were performed as previously described (10). The presence of fiber genes in virions was analyzed by PCR with primers specific for the WT and recombinant fibers. The presence of fiber proteins in virions was determined by Western blot analysis with 4D2.5 antitail antibody. Expression of GFP in infected cultures was verified by UV microscopy using a Zeiss Axioskop fluorescent microscope. Infectious titers (PFU per milliliter) were determined by plaque titration on 293 cells using an endpoint dilution method (29), and the number of physical virus particles was determined using the IDEIA Ad detection kit (Dako). The rate of virus growth was determined as follows. 293 cells (4 × 104) were infected with WT or recombinant virus at 10 PFU/cell for 1 h at 37°C. After rinsing with PBS, complete medium was added and cell samples were harvested at 24, 48, and 72 h after infection. Cell pellets were washed in PBS, resuspended in 200 μl of PBS, and freeze-thawed four times. The cell lysates were assayed for production of Ad5 proteins by the IDEIA kit, and the infectivity titer, determined by plaque titration on 293 cells as above, was plotted versus the time of infection.
Quantitative analysis of fiber content of virions.
293 cells in 24-well plates were washed once with Iscove's medium and infected with 10 PFU/cell in 100 μl of Iscove's medium. After 1 h at 37°C, the cells were washed in complete medium (Iscove's medium with 10% fetal bovine serum and 50 μg of gentamicin per ml) and incubated with complete medium at 37°C and 5% CO2. Cells were harvested at 24, 48, and 72 h postinfection, collected in 0.2 ml of Iscove's medium, and freeze-thawed four quick rounds to release virions. Virus titer was determined by plaque assay in 293 cells, and viral proteins were assayed using the IDEIA kit. Western blots of the freeze-thawed material and CsCl-purified virus were reacted with 4D2.5 and 2Hx2 to assay the fiber versus hexon content, and quantification was performed using the Scion Image program.
Gene transfer assay.
Monolayers of HeLa and RD cells in 24-well plates were infected as described above with 10 PFU of the recombinant viruses per cell, with or without different blocking agents. The antiknob antibody 1D6.14 was used at concentrations ranging from 0.001 to 10 μg/ml. Virus and antibody were mixed and diluted to 0.1 ml in Iscove's medium and incubated for 1 h at 37°C before cell infection, as above. The cells were examined by immunofluorescence (IF) microscopy at intervals during the course of infection for the appearance of GFP fluorescence. For fluorescence-activated cell sorting (FACS) analysis, the cells were harvested at 24 h postinfection and washed with ice-cold PBS, followed by fixation with 0.5% glutaraldehyde for 15 min. After three washes in PBS, the cells were analyzed for GFP expression using the FL1 emission channel in a FACScan cytometer (Becton Dickinson, San Jose, Calif.).
RESULTS
Phenotypic characterization of recombinant fibers with different shaft lengths.
The number of shaft repeats in fiber determines the shaft length (33) and has important implications in receptor usage (31, 38). Fiber proteins with various numbers of shaft repeats were thus compared (Fig. 1). Fiber with one single repeat (R1-RGD) was constructed to test a very short fiber; fiber with seven repeats (R7-RGD) resembled the short-shafted Ad3 fiber; and fiber with 22 repeats (R22-RGD) mimicked the Ad5 WT fiber shaft. The choice of the number of shaft repeats for the fiber of retargetable Ad5 vectors was guided by the results of phenotypic analysis of the different fiber constructs. The recombinant fibers were expressed in both mammalian and insect cells and phenotyped according to the following criteria: (i) their cellular localization, (ii) the occurrence and stability of fiber trimers, (iii) their capacity to form penton capsomers in insect cells coinfected with a recombinant baculovirus-expressing penton base, and (iv) their glycosylation status.
(i) Cellular localization and fiber trimerization.
The cellular localization and trimer status of the recombinant fibers were studied by transient expression in Cos7 cells at 48 h after transfection. The cells were fixed, reacted with monoclonal antibodies specific for fiber tail (4D2.5) and fiber trimers (2A6.36), and examined by IF microscopy. The IF patterns observed showed that all our recombinant fiber proteins were well expressed and localized in the nucleus. They were all capable of forming trimers, just like the WT fiber (data not shown).
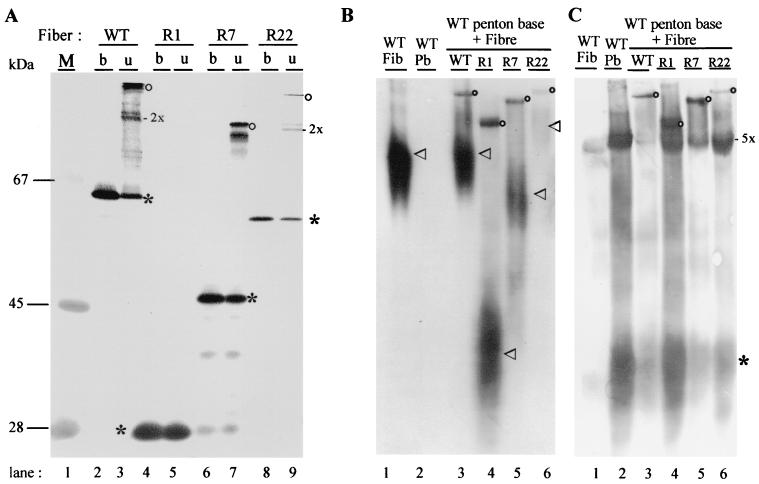
The trimerization status was also tested by electrophoresis and Western blotting of fiber samples denatured by boiling in SDS (SDS-PAGE) or unboiled (nondenatured; NDS-PAGE). The NDS-PAGE pattern for unboiled recombinant WT fiber consisted of three distinct protein species, monomers, dimers, and trimers (Fig. 2A, lane 3). For recombinant fibers R7-RGD and R22-RGD, dimers and trimers were similarly observed (Fig. 2 A, lanes 7 and 9). However, the oligomeric forms of R22-RGD were barely visible, since the R22-RGD clone was a low expressor in terms of soluble fiber protein (data not shown). In the case of R1-RGD, no trimers were visible in NDS-PAGE, and all detectable fiber occurred in monomeric form (Fig. 2A, compare lanes 4 and 5). However, in gel electrophoresis under native conditions, in the absence of SDS, fiber trimers were detected for all the recombinants, including R1-RGD, using the antitrimer antibody 2A6.36 (data not shown). Moreover, R1-RGD fiber reacted positively with 2A6.36 within the cell, as shown by IF microcopy. This strongly suggested that R1-RGD fiber did form trimers in vivo but that these trimers were unstable in vitro in the presence of SDS, even at low temperature.
FIG. 2.
Recombinant fibers, ability to trimerize (A) and to form penton capsomers (B and C). (A) Fiber trimers were analyzed by SDS-PAGE and NDS-PAGE and immunoblotting with 4D2.5 antibody as boiled (b) or unboiled (u) samples. WT fiber (lanes 2 and 3), R1-RGD (lanes 4 and 5), and R7-RGD (lanes 6 and 7) were all highly expressed in Sf9 cells as soluble recombinant proteins, whereas R22-RGD was a low expresser (lanes 8 and 9). Note that no stable fiber trimer was detectable in unboiled R1-RGD lysate (lane 5). Fiber monomers are indicated by an asterisk, dimers by 2x, and trimers by an open circle. (B and C) Assembly of fiber with penton base was assayed in lysates of Sf9 cells coexpressing recombinant fiber and penton base (16). Native proteins were electrophoresed in an 8% nondenaturing gel and electrically transferred, and blots were reacted with antitrimer fiber 2A6.36 monoclonal antibody (B) or polyclonal anti-penton base (C). Control samples of single expression of WT fiber and penton base alone are shown in lanes 1 and 2, respectively. Lanes 3 to 6, lysates from cells coexpressing WT penton base with WT fiber (lane 3), R1-RGD (lane 4), R7-RGD (lane 5), or R22-RGD (lane 6), respectively. Penton capsomer, comprised of penton base-anchored fiber, migrates as a discrete band reacting with both antifiber 2A6.36 (B) and anti-penton base antibodies (C), and is indicated by open circles. In panel B, free fibers, migrating as a diffuse band of polydisperse species, are indicated by an arrowhead. In panel C, monomers of penton base are indicated by an asterisk and pentamers (penton base capsomers) by 5x.
(ii) Assembly with penton base to form penton capsomers.
In vivo in coinfected Sf9 cells, all our recombinant fibers were able to assemble with penton base to form penton capsomers, which were detectable in gels electrophoresed under native conditions (Fig. 2B and C). Free fiber trimers migrated in the native gel as polydispersed molecules visible as a smear on immunoblots (Fig. 2B), whereas penton capsomers appeared as a sharp, slow-migrating band (Fig. 2B and C). This discrete larger protein species was revealed by both fiber and penton base antibodies, confirming that they were penton complexes (Fig. 2B and C).
The efficiency of penton assembly with our recombinant fibers was quantitatively estimated by immunoblotting of Sf9 cell lysates electrophoresed in native gels, using fiber and penton base antibodies and the corresponding radioactively labeled secondary antibody. The radioactivity was measured and compared for the penton capsomer band and the free fiber band in blots probed with antifiber antibody (Fig. 2B) or for the penton capsomer band and the free penton base band in blots probed with anti-penton base antibody (Fig. 2C). The data are shown in Table 1. With WT fiber, which is highly expressed in insect cells (27), only 5% of the fiber assembled with penton base, while the rest remained as free fiber trimers, and the penton base often appeared to be the limiting factor for penton assembly (Fig. 2C, lane 3). With R1-RGD, R7-RGD, and even R22-RGD fiber, assembly with penton base seemingly occurred with a higher efficiency and/or greater stability than with WT fiber (Table 1 and Fig. 2B, compare lanes 3, 4, 5, and 6). This would suggest that penton constituted of knobless fiber and containing the NRP trimerization signal would be more stable than WT penton capsomers constituted of penton base and full-length, knob-terminated fiber, at least in insect cells. It has already been suggested that the fiber knob domain could be responsible for a certain degree of flexibility or instability in Ad2 penton capsomer (4, 14).
TABLE 1.
Efficiency of penton assembly in Sf9 cells coexpressing WT penton base and recombinant fibers with various shaft repeats and C-terminal RGD ligandsa
| Recombinant fiber | Avg % of total radioactivity ± SD
|
|
|---|---|---|
| Assembled fiberb | Assembled penton basec | |
| WT | 5.5 ± 0.8 | 51.4 ± 7.0 |
| R1-RGD | 9.9 ± 4.4 | 48.1 ± 15.1 |
| R7-RGD | 10.2 ± 1.5 | 27.8 ± 10.3 |
| R22-RGD | 9.9 ± 1.9 | 1.1 ± 0.4 |
Penton base and fiber proteins were coexpressed in the same Sf9 cells using two recombinant baculoviruses. Cell lysates were analyzed by gel electrophoresis run in native conditions and immunoblotting with anti-penton base and anti-fiber antibodies followed by radioactively labeled secondary antibody, as depicted in Fig. 2B and C. Results are expressed as the percentage of total radioactivity recovered from penton plus penton base and penton plus fiber bands. Data shown are the average of four determinations ± standard deviation (SD).
The amounts of fiber engaged in penton and remaining as unbound fiber were determined by counting the radioactivity associated with the bands corresponding to penton and free fiber, respectively (refer to Fig. 2B).
The amount of penton base bound to fiber versus the amount of unassembled penton base was determined by counting the radioactivity in the bands of penton and penton base, respectively (refer to Fig. 2C).
When probed with penton base antibody (Fig. 2C and Table 1), the blots of Sf9 cell lysates confirmed that none of our recombinant fibers was defective in assembly with penton base. The apparent low efficiency of assembly shown by R22-RGD fiber (1% of the total penton plus penton base signal; Fig. 2C, lane 6) likely resulted from its low expression level (Fig. 2A, lanes 8 and 9), the R22-RGD fiber protein being the limiting factor for penton assembly.
(iii) Glycosylation status of recombinant fibers.
None of our recombinant RGD fibers showed any detectable O-GlcNAc signal, as assayed by two different methods, Western blot analysis using RL2 antibody specific for peptide-linked O-GlcNAc residues and a biochemical assay using the DIG Glycan detection kit. Only WT fiber was found to be glycosylated (data not shown). This suggested that O-GlcNAc residues were dispensable for most of the known biological functions of the fiber.
Thus, considering that R22-RGD fiber was expressed at low levels and that R1-RGD self-assembled into rather unstable fiber trimers, R7-RGD was retained as the most favorable knobless fiber construct to be reintroduced into the viral genome for the generation of knobless Ad5 vectors. Further characterization therefore concerned R7-RGD fiber and R7-RGD-containing virions.
Functionality and binding specificity of fiber proteins.
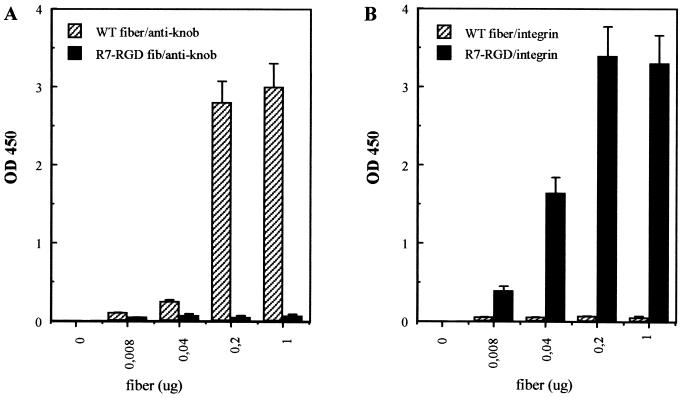
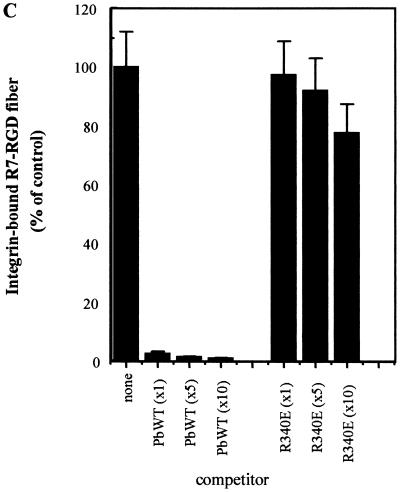
The binding function of WT and R7-RGD fibers was examined by solid-phase enzyme-linked immunosorbent assay (ELISA). Both fibers were tested for binding to antiknob monoclonal antibody 1D6.14 and integrin αvβ3. WT fiber was found to bind with a high affinity to the knob antibody compared to the absence of significant binding by the R7-RGD fiber (Fig. 3A). The reverse was observed with wells coated with αvβ3 integrin; WT fiber showed no detectable binding, while R7-RGD fiber had a high affinity for αvβ3 integrin (Fig. 3B). The specificity of binding of the R7-RGD fiber to immobilized integrin αvβ3 was also studied by ELISA in the presence of RGD peptides or penton base proteins as competitors. RGD peptides competed poorly with R7-RGD fiber for integrin binding (data not shown), probably due to a low stability of the integrin-RGD complexes in ELISA. However, WT penton base, which carries five RGD motifs, competed efficiently (Fig. 3C). In contrast, penton base mutant R340E, mutated in the RGD motifs (20), failed to compete with R7-RGD fiber, suggesting that the competition of WT penton base with R7-RGD fiber was RGD specific (Fig. 3C). Binding of WT and R7-RGD fibers was also tested on α5β1 integrins (data not shown), and the assays gave similar results as for αvβ3 integrin. Thus, our results suggested that the R7-RGD fiber had lost its affinity for the knob antibody but had gained the ability to bind to αvβ3 and α5β1 integrins. This implied that the RGD motif on the fiber was in the proper conformation and accessible for binding to cell surface-exposed αvβ3 and α5β1 integrins.
FIG. 3.
Fiber-binding properties in vitro tested by ELISA. (A) Wells were coated with anti-fiber tail monoclonal antibody 4D2.5, WT or R7-RGD fiber was added (amount of fiber protein in a total volume of 100 μl), and detection was performed using antiknob blocking antibody 1D6.14. (B) Wells were coated with αvβ3 integrin, WT or R7-RGD fiber was added, and integrin-bound fiber was detected using 4D2.5 antibody. (C) Wells were coated with αvβ3 integrin (1 μg of protein per well), followed by stepwise addition of recombinant penton base (Pb) protein (WT or R340E mutant) and R7-RGD fiber (1 μg of fiber protein per well in a total volume of 200 μl). The stoichiometric ratio of R7-RGD fiber to penton base molecules was 1:1, 1:5, and 1:10, indicated under the x axis as x1, x5, and x10, respectively. Results shown represent the mean of three separate experiments, ± SD.
Rescue of functional recombinant fiber genes into infectious virions.
To generate virions with modified fibers, a strategy was designed to insert the recombinant fiber gene exactly in the position of the fiber gene in the Ad5 chromosome so as to make use of the native regulation of fiber gene expression. This was achieved by a three-step rescue system (Fig. 4), in which the recombinant fiber was first cloned into the shuttle vector pGAG3 containing the tail and the flanking sequences of the fiber gene. As a negative control, the empty pGAG3 vector was used. The fragments were then rescued into pGAG9 (75.4 to 100 mu of the WT Ad5 genome) by homologous recombination in E. coli BJ5183. As a final step, cosmid cloning was used for ligation to the rest of the genome. For convenient detection of gene transfer, our recombinant Ad genome contained the GFP reporter gene cloned under the control of the CMV promoter in the deleted E1 region. The resulting genome was joined at both ends to the cosmid backbone by PacI and could thus be recovered as a linear DNA fragment after cleavage with PacI and transfected into cells to yield virus. On the average, approximately 20% of the cosmid colonies contained the expected recombinant Ad5 genome.
Efficiency of virus propagation.
After transfection of 293 cells, approximately 8, 16, and 16 days elapsed before plaques appeared for the viruses designated Ad5/FibWT, Ad5/FibR7-RGD, and Ad5/FibR7-RGD4C, respectively. The genome with the deleted fiber did not give any plaques. The recombinant viruses were then amplified in 293 cells, and the viruses were purified by ultracentrifugation in a self-generating CsCl gradient. After virus purification, the fiber genotype was controlled and confirmed by PCR amplification and DNA sequencing, using primers specific for WT and R7-RGD fibers. In addition, the presence of fiber proteins in virions was assayed immunologically by Western blot analysis using antifiber antibody. Although the expected fiber sequence and signal were found in our virus constructs, Ad5/FibR7-RGD and Ad5/FibR7-RGD4C virus spread through cellular monolayers occurred at a two to three times slower rate compared to Ad5/FibWT.
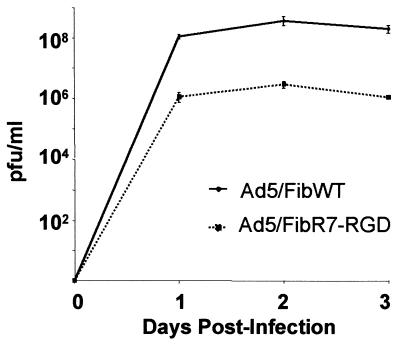
Comparison of the infectivity index of WT and recombinant virus showed that Ad5/FibR7-RGD was 20 times less infectious than Ad5/FibWT; the ratio of infectious particles (detected by fluorescent plaques and expressed as PFU) to physical particles (estimated by biochemical methods) was found to be 1:25 for Ad5/FibWT, versus 1:500 for Ad5/FibR7-RGD. Analysis of growth curves in 293 cells showed that Ad5/FibWT and Ad5/FibR7-RGD virions grew at similar rates in a complementing cell line and synthesized similar amounts of viral proteins. However, the plateau of infectious progeny yields for Ad5/FibR7-RGD was 100-fold lower than for Ad5/FibWT (Fig. 5).
FIG. 5.
Comparative growth curves of Ad5/FibWT virus (solid line) and Ad5/FibR7-RGD recombinant (dotted line). HEK-293 cells were infected with the same input multiplicity (10 PFU/cell) for 1 h at 37°C, then cells were harvested and lysed at different times postinfection, as indicated. Infectious titer of each sample was determined by fluorescent plaque assay in 293 cell monolayers. Data shown are the average of three separate experiments ± SD.
Fiber content of recombinant Ad5 virions.
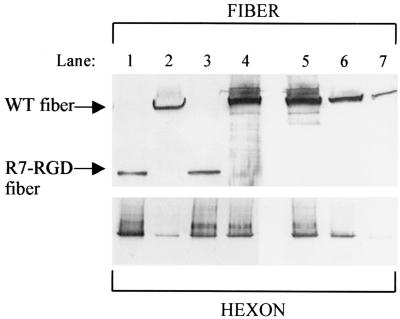
To further investigate the difference in virus propagation between Ad5/FibR7-RGD and Ad5/FibWT, 293 cells were infected with aliquots (10 PFU/cell) of each virus. The cells were harvested at different times postinfection and lysed, and the virus proteins released were assayed in the supernatants. At all time points the infected cells appeared identical, as judged visually. The IDEIA assays showed that similar amounts of viral proteins were produced by Ad5/FibR7-RGD- and Ad5/FibWT-infected cells. Protein analysis of cell lysates showed similar amounts of hexon yields, while probing for fiber revealed about 10 times less R7-RGD fiber compared to WT fiber (data not shown). CsCl-purified viruses were also probed for fiber and hexon contents in virus samples normalized for physical particle number and infectious particle number, respectively (Fig. 6). The results showed that similar amounts of fibers were found when Ad5/FibR7-RGD and Ad5/FibWT virus samples were normalized for infectious particles (as counted on 293 cells; Fig. 6, top panel, compare lanes 1 and 2). However, when virus samples were normalized for physical particles (assayed by hexon content; Fig. 6, lower panel), significantly fewer fibers were present in the Ad5/FibR7-RGD than in the Ad5/FibWT sample (Fig. 6, top panel, compare lanes 3 and 4). As shown by quantitative analysis, the difference ranged within 10- to 40-fold, depending on virus preparations. This result implied that there was a lower fiber copy number per virion in Ad5/FibR7-RGD or, alternatively, that the Ad5/FibR7-RGD virus preparation contained a mixture of fiberless particles with low infectivity (23) and fully infectious virions with normal fiber content.
FIG. 6.
Protein analysis of virus particles purified by ultracentrifugation in CsCl gradient. Virus samples from CsCl gradient fractions were analyzed by SDS-PAGE, and viral proteins were electrically transferred to nitrocellulose membranes. Blots were reacted with antifiber (4D2.5; upper panel) or antihexon (2Hx2; lower panel) antibody. Lane 1, Ad5/FibR7-RGD (5 × 105 PFU); lane 2, Ad5/FibWT (5 × 105 PFU); lane 3, Ad5/FibR7-RGD (5 × 105 PFU); lane 4, Ad5/FibWT (1 × 107 PFU); lanes 5 to 7, Ad5/FibWT at 107, 106, and 105 PFU, respectively. Note that lanes 1 and 2 had equal loads of infectious particles (PFU, as counted on 293 cells), while virus loads in lanes 3 and 4 had been normalized to equal numbers of physical particles, based on the hexon protein content (see the hexon signal in the lower panel).
Cell specificity of gene transfer mediated by recombinant Ad5.
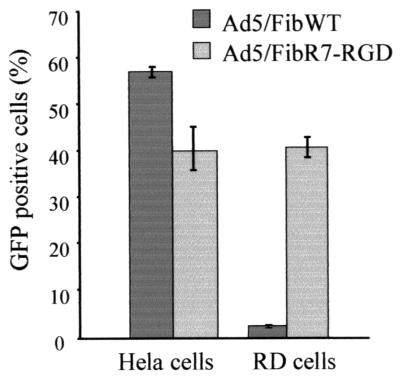
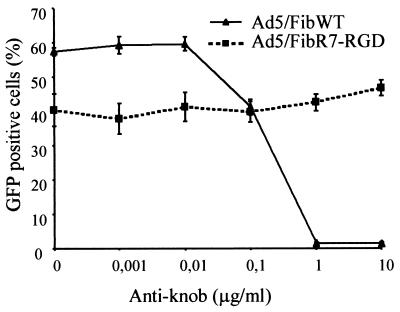
To evaluate the tropism of the recombinant viruses, the efficiency of Ad5-mediated gene transfer to HeLa cells and RD cells was assayed in the presence and absence of antiknob antibody. CAR is known to be expressed by HeLa cells but not by RD cells if they are passaged and grown to low cell density (37). The integrin αvβ5 was found to be expressed on HeLa cells, and both αvβ3 and αvβ5 integrins were found on RD cells, as detected by FACS analysis after staining with specific monoclonal antibodies (data not shown). The infectivity of Ad5/FibWT and Ad5/FibR7-RGD was determined in both HeLa and RD cells (Fig. 7). As expected, HeLa cells were fully susceptible to both Ad5/FibWT and Ad5/FibR7-RGD viruses, whereas RD cells were found to be only permissive for Ad5/FibR7-RGD and poorly permissive for Ad5/FibWT. To verify that cell binding of Ad5/FibR7-RGD virus was not dependent on the knob receptor, CAR-blocking experiments were performed using 1D6.14 antibody. As shown in Fig. 8, the Ad5/FibWT virus was readily blocked by the antiknob antibody in a dose-dependent manner, while the antibody had no effect on the Ad5/FibR7-RGD virus.
FIG. 7.
Susceptibility of HeLa (CAR positive) and RD (CAR negative) cells to Ad5/FibWT virus (dark columns) and Ad5/FibR7-RGD recombinant (light columns), as determined by the level of expression of reporter gene GFP. Cells were infected with an equal multiplicity of infection (10 PFU/cell) for 1 h at 37°C. The efficiency of Ad5-mediated gene transfer was estimated by counting the GFP-positive cells by FACS analysis at 24 h postinfection, and results are expressed as the percentage of positive, fluorescent cells (mean of three separate experiments ± SD).
FIG. 8.
Influence of neutralizing antiknob monoclonal antibody 1D6.14 on Ad5/FibWT-mediated (solid line) and Ad5/FibR7-RGD-mediated (dotted line) gene transfer to HeLa cells. Aliquots of serial dilutions of antibody were incubated with an equal multiplicity of infection (10 PFU/cell) of virus for 1 h at 37°C before infection. The level of inhibitory effect was estimated by counting the GFP-positive cells by FACS analysis at 24 h postinfection, and results are expressed as the percentage of positive, fluorescent cells (mean of three separate experiments ± SD).
DISCUSSION
This study presents a new concept for genetic retargeting of Ad and demonstrates its utility for gene transfer into cells lacking the CAR receptor. The main feature in the present reconstruction of the fiber gene is to genetically delete the fiber of its native binding specificity and to replace this with a new ligand in order to retarget the virus to a new cellular receptor. However, in order for Ad fibers to be properly assembled into functional proteins and be encapsidated into infectious virions, several requirements have to be fulfilled. For example, the fibers need to be able to form parallel homotrimers, to bind to penton base via its tail region to form penton capsomers, to localize within the correct cellular compartment, i.e., the nucleus, where capsid assembly occurs, and lastly, in the context of the virion, to cellular receptors.
The genetically modified knobless fibers that we have constructed here were designed to meet the abovementioned requirements. As evaluated from the results obtained, it seemed feasible to compensate for the lack of biological functions carried by the fiber knob at the virion level by inserting an extrinsic trimerization motif and a new cell ligand which completely changed the tropism of the virus. We introduced a 36-aa peptide from the human lung surfactant protein D (NRP) as an extrinsic trimerization motif and an RGD peptide as a cell ligand. To allow more flexibility and accessibility, a linker sequence from Staphylococcus protein A was inserted between the NRP and the cell surface ligand RGD. The recombinant fibers could be made to contain different numbers of shaft repeats. We have been able to generate infectious viruses with functional fibers containing one and seven shaft repeats (this study), as well as three and five repeats (unpublished results), but not with recombinant Ad5 fibers containing the full-length shaft (22 repeats). This was in apparent contradiction to a recent work which showed the rescue of knobless, His-Myc epitope-tagged, full-length fibers into virions (40). However, the authors had cotransfected cells with a plasmid expressing both WT fiber and a knobless, full-length fiber construct, and both fibers were thus rescued into chimaeric Ad5 virions with two distinct fiber species, WT and knobless mutant. This difference in strategy could explain the yield of infectious virus progeny using knobless full-length fibers. Among several of our shortened fiber constructs, the knobless fiber with seven shaft repeats (R7) and an RGD peptide motif at its C terminus (linear, as in R7-RGD, or constrained, as in R7-RGD4C), was found to be the one which retained the essential functions of the fiber while carrying a new cell-binding specificity. The reason for this observation is presently under investigation, but it constitutes the rationale for the use of R7 fibers in the present work.
The recombinant R7-RGD fiber was found to bind to αvβ3, αvβ5, and α5β1 integrins. As shown previously using different approaches, the RGD motif has the ability to target Ad virions to otherwise refractory cell lines after being incorporated into the fiber or the hexon protein (8, 41). The failure to block the virus with the antiknob antibody further demonstrated that Ad5/FibR7-RGD was not longer dependent on the CAR receptor for cell attachment and infection. The recombinant Ad5/FibR7-RGD and Ad5/FibR7-RGD4C viruses may therefore be used as gene transfer vectors for cells expressing integrins but not CAR (Fig. 7), thereby broadening the tropism of Ad vectors. Since the integrin ligand motif RGD is also present in the penton base (1, 2, 44), it was legitimate to ask to what extent penton base RGD could contribute to the cellular integrin binding of Ad5/FibR7-RGD, considering that the fibers are shortened and the RGD in penton base could theoretically protrude approximately to the top of the shortened fiber (36, 42). Although the participation of penton base RGD could not be excluded, some arguments suggested that the major cell-binding determinants for Ad5/FibR7-RGD still resided in its recombinant fiber capsomer. (i) Fiberless virions have been reported to be far less infectious than WT, with 10,000-fold difference in infectivity (23). In our case, Ad5/FibR7-RGD and Ad5/FibWT showed only a 20-fold difference in infectivity. (ii) When the RGD motif was replaced by the sequence of epidermal growth factor (EGF) in R7 fiber, no viable Ad5/FibR7-EGF virus could be isolated, although R7-EGF recombinant fiber self-assembled into trimers which were detected in penton complexes (Magnusson et al., unpublished data). This suggested that RGD in penton base could not compensate for the lack of initial cell binding of the virus via the fiber.
As mentioned in the Results, the amount of viral capsid proteins synthesized after infection with the same multiplicity of infection (equal number of PFU/cell) was equivalent for viruses containing the WT fiber and the R7-RGD fiber, although their respective infectivities were significantly different. Several hypotheses could be proposed to explain the 20-fold difference in infectivity between Ad5/FibWT and Ad5/FibR7-RGD viruses. (i) A lower ability for recombinant fibers to assemble with penton base compared to WT fiber and/or a lower stability of penton capsomers containing the knobless R7 fibers would be unlikely on the basis of their efficient assembly with coexpressed recombinant penton base in insect cells (Fig. 2B and C). (ii) Likewise, the recombinant fibers used in our study were C-terminally truncated, and it has been suggested that deletion of portions of the fibers could remove some function(s) necessary for virus maturation (23, 42). However, we have also constructed R7 fibers with ligands other than RGD which yield viruses that are as infectious as WT virus (Strand et al., unpublished data). This suggested that the reduction in shaft length to seven repeats does not per se seem to significantly impair the capacity of the virus to infect cells or that the influence of shaft length can vary with the ligand used.
(iii) At late times postinfection, Ad5/FibR7-RGD-infected cells yielded significantly lower levels of R7-RGD fiber than WT Ad5-infected cells (about 10-fold less), and the Ad5/FibR7-RGD virus samples contained 10 to 40 times fewer fibers per virion than WT Ad5 (Fig. 6). The low R7-RGD fiber content of infected cells was unexpected, since the tail and all the sequences upstream of the fiber gene were left intact in order not to disturb the normal regulation of fiber expression. However, if R7-RGD fiber protein was present in limiting amounts, this could negatively affect both the equilibrium of the penton assembly reaction (5) and the overall fiber content of the virus. The resulting lower number of cell-binding sites in the recombinant virus would in turn contribute to the lower infectivity of Ad5/FibR7-RGD. (iv) Even though there was a direct relationship between the low fiber content of recombinant viruses and the low fiber yields of Ad5/FibR7-RGD-infected cells, the molecular mechanisms behind the latter remain unclear. It has been suggested that O-GlcNAc might be important for Ad2 and Ad5 fiber assembly and stabilization (26), and a major O-GlcNAc site has been mapped within residues 61 and 410 of the Ad2 fiber shaft (27). Further deletion mapping analyses have narrowed down the site(s) to within residues 61 to 260 (13), then to 61 to 216 (35). Our biochemical and immunological analyses showed that recombinant knobless R7-RGD fibers expressed in Sf9 cells and the ones encapsidated into virions were not glycosylated, suggesting that both lacked a functional O-glycosylation site(s) from residues M-1 to K-156. Although most of the known functions assumed by the fiber were still apparently effectuated by our R7-RGD fiber, it could not be totally excluded that its lack of O-glycosylation would have altered one of its subtle but critical biological properties.
(v) Results from a recent comparative study of infectivity of Ad containing fiber shafts of various lengths suggested that the observed weak cell attachment of short-shafted fiber-containing Ad vectors could be due to repulsive acidic charges carried by the Ad capsid and present on the cell surface (38). (vi) Another factor that may affect the ability of RGD-targeted viruses to infect cells is the efficiency of the interaction between fiber RGD and cellular integrins in promoting virus entry (31). It has been reported (44) that the interaction between the fiber and CAR has a 30-fold-higher affinity than the penton-integrin interaction. Furthermore, the particular context in which the RGD is presented to integrins might largely influence the binding (34). It is therefore possible that other RGD-containing peptides incorporated into the fiber would yield more efficient or competent viruses than the present ones. A comparison between RGD and RGD4C fibers, which may differ in this regard, is under way. (vii) In the same line of reasoning, it is very possible that at the multiplicity of infection used with the Ad5/FibR7-RGD recombinant, surface integrins were approaching saturation with the RGD peptide in the fiber. This would lead to reduced ability of the penton RGD to interact with another integrin molecule, precluding viral penetration. Thus, at 10 PFU/cell, equivalent to 5,000 physical particles of Ad5/FibR7-RGD per cell, and based on a theoretical full copy number of fiber and penton base subunits in intact Ad5/FibR7-RGD capsids (i.e., with 12 complete apex structures), a total of 180,000 RGD/cell (5,000 × 12 × 3) would be presented by the fiber projections and 300,000 RGD/cell (5,000 × 12 × 5) would be presented by the penton base capsomers. (viii) These hypotheses are not mutually exclusive, and the factors described in iii, iv, v, vi, and vii could combine and account for the lower infectivity of Ad5/FibR7-RGD compared to WT Ad5.
It is our hope that the radical fiber-engineering approach described in this study will enable insertions of larger ligands than those that are tolerated by the native fiber knob, for example, single-chain antibodies and other complex, folded binding structures. This could be reasonably envisaged, given that conditions for correct domain folding are maintained in our recombinant fiber constructs harboring the trimerization motif NRP. Indeed, in its natural environment within the human lung surfactant protein D, NRP is bordered by N-terminal collagen regions and C-terminal C-type lectin domains (17), two domains which have structural and functional similarities with the shaft and knob domains, respectively. Our novel approach to genetic retargeting of Ad could then open up new possibilities for gene therapy.
ACKNOWLEDGMENTS
The work in Gothenburg (M.K.M. and L.L.) was supported by the Swedish Medical Research Council (grant no. K98-06X-12624-01), the Swedish Cancer Society (grant no. 0512-B99-12XCC), and the IngaBritt and Arne Lundberg Foundation (grant no. 205/96) and by grants from Got-A-Gene AB, Gothenburg, Sweden. The work in Lyon (S.S.H. and P.B.) was financially supported by the French Ministère de la Recherche et de la Technologie (PRFMMIP AO-98) and the French Foundation for Cystic Fibrosis (Vaincre la Mucoviscidose).
The expert technical contribution of Elisabeth Pettersson and Petra Strand is gratefully acknowledged. Bert Vogelstein is thanked for the generous supply of the vectors pAdEasy and pAdTrack.
ADDENDUM IN PROOF
After submission of the manuscript, an article taking a similar approach was published by Krasnykh et al. (V. Krasnykh, N. Belousova, N. Korokhov, G. Mikheeva, and D. T. Curiel, J. Virol. 75:4176–4183).
REFERENCES
- 1.Bai M, Harfe B, Freimuth P. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J Virol. 1993;67:5198–5205. doi: 10.1128/jvi.67.9.5198-5205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belin M T, Boulanger P. Involvement of cellular adhesion sequences in the attachment of adenovirus to the HeLa cell surface. J Gen Virol. 1993;74:1485–1497. doi: 10.1099/0022-1317-74-8-1485. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 4.Boudin M-L, Boulanger P. Antibody-triggered dissociation of adenovirus penton capsomer. Virology. 1981;113:781–786. doi: 10.1016/0042-6822(81)90208-7. [DOI] [PubMed] [Google Scholar]
- 5.Boudin M-L, Boulanger P. Assembly of adenovirus penton base and fiber. Virology. 1982;116:589–604. doi: 10.1016/0042-6822(82)90151-9. [DOI] [PubMed] [Google Scholar]
- 6.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chroboczek J, Ruigrok R W, Cusack S. Adenovirus fiber. Curr Top Microbiol Immunol. 1995;199:163–200. doi: 10.1007/978-3-642-79496-4_10. [DOI] [PubMed] [Google Scholar]
- 8.Dmitriev I, Krasnykh V, Miller C R, Wang M, Kashentseva E, Mikheeva G, Belousova N, Curiel D T. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas J T, Rogers B E, Rosenfeld M E, Michael S I, Feng M, Curiel D T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 10.Ebbinghaus S W, Vigneswaran N, Miller C R, Chee-Awai R A, Mayfield C A, Curiel D T, Miller D M. Efficient delivery of triplex forming oligonucleotides to tumor cells by adenovirus-polylysine complexes. Gene Ther. 1996;3:287–297. [PubMed] [Google Scholar]
- 11.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 12.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong J S, Engler J A. Domains required for assembly of adenovirus type 2 fiber trimers. J Virol. 1996;70:7071–7078. doi: 10.1128/jvi.70.10.7071-7078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong S S, Boulanger P. Protein ligands of the human adenovirus type 2 outer capsid identified by biopanning of a phage-displayed peptide library on separate domains of wild-type and mutant penton capsomers. EMBO J. 1995;14:4714–4727. doi: 10.1002/j.1460-2075.1995.tb00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong S S, Galaup A, Peytavi R, Chazal N, Boulanger P. Enhancement of adenovirus-mediated gene delivery by use of an oligopeptide with dual binding specificity. Hum Gene Ther. 1999;10:2577–2586. doi: 10.1089/10430349950016627. [DOI] [PubMed] [Google Scholar]
- 16.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC class I α2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1997;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoppe H J, Barlow P N, Reid K B. A parallel three stranded alpha-helical bundle at the nucleation site of collagen triple-helix formation. FEBS Lett. 1994;344:191–195. doi: 10.1016/0014-5793(94)00383-1. [DOI] [PubMed] [Google Scholar]
- 18.Huvent I, Hong S S, Fournier C, Gay B, Tournier J, Carrière C, Courcoul M, Vigne R, Spire B, Boulanger P. Interaction and co-encapsidation of human immunodeficiency virus type 1 Gag and Vif recombinant proteins. J Gen Virol. 1998;79:1069–1081. doi: 10.1099/0022-1317-79-5-1069. [DOI] [PubMed] [Google Scholar]
- 19.Karayan L, Gay B, Gerfaux J, Boulanger P. Oligomerization of recombinant penton base of adenovirus type 2 and its assembly with fiber in baculovirus-infected cells. Virology. 1994;202:782–795. doi: 10.1006/viro.1994.1400. [DOI] [PubMed] [Google Scholar]
- 20.Karayan L, Hong S S, Gay B, Tournier J, Dupuy d'Angeac A, Boulanger P. Structural and functional determinants in adenovirus type 2 penton base recombinant protein. J Virol. 1997;71:8678–8689. doi: 10.1128/jvi.71.11.8678-8689.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirby I, Davison E, Beavil A J, Soh C P, Wickham T J, Roelvink P W, Kovesdi I, Sutton B J, Santis G. Mutations in the DG loop of adenovirus type 5 fiber knob protein abolish high-affinity binding to its cellular receptor CAR. J Virol. 1999;73:9508–9514. doi: 10.1128/jvi.73.11.9508-9514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legrand V, Spehner D, Schlesinger Y, Settelen N, Pavirani A, Mehtali M. Fiberless recombinant adenoviruses: virus maturation and infectivity in the absence of fiber. J Virol. 1999;73:907–919. doi: 10.1128/jvi.73.2.907-919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louis N, Fender P, Barge A, Kitts P, Chroboczek J. Cell-binding domain of adenovirus serotype 2 fiber. J Virol. 1994;68:4104–4106. doi: 10.1128/jvi.68.6.4104-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michael S I, Hong J S, Curiel D T, Engler J A. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 1995;2:660–668. [PubMed] [Google Scholar]
- 26.Mullis K G, Haltiwanger R S, Hart G W, Marchase R B, Engler J A. Relative accessibility of N-acetylglucosamine in trimers of the adenovirus types 2 and 5 fiber proteins. J Virol. 1990;64:5317–5323. doi: 10.1128/jvi.64.11.5317-5323.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novelli A, Boulanger P. Deletion analysis of functional domains in baculovirus-expressed adenovirus type 2 fiber. Virology. 1991;185:365–376. doi: 10.1016/0042-6822(91)90784-9. [DOI] [PubMed] [Google Scholar]
- 28.Oliner J D, Kinzler K W, Vogelstein B. In vivo cloning of PCR products in E. coli. Nucleic Acids Res. 1993;21:5192–5197. doi: 10.1093/nar/21.22.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Reilly D, Miller L, Luckow V. Baculovirus expression vectors: a laboratory manual. New York, N.Y: Oxford University Press Inc; 1994. [Google Scholar]
- 30.Pasqualini R, Koivunen E, Ruoslahti E. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- 31.Roelvink P W, Lizonova A, Lee J G, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers B E, Douglas J T, Ahlem C, Buchsbaum D J, Frincke J, Curiel D T. Use of a novel cross-linking method to modify adenovirus tropism. Gene Ther. 1997;4:1387–1392. doi: 10.1038/sj.gt.3300541. [DOI] [PubMed] [Google Scholar]
- 33.Ruigrok R W, Barge A, Albiges-Rizo C, Dayan S. Structure of adenovirus fibre. II. Morphology of single fibres. J Mol Biol. 1990;215:589–596. doi: 10.1016/S0022-2836(05)80170-6. [DOI] [PubMed] [Google Scholar]
- 34.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 35.Santis G, Legrand V, Hong S S, Davison E, Kirby I, Imler J L, Finberg R W, Bergelson J M, Mehtali M, Boulanger P. Molecular determinants of adenovirus serotype 5 fibre binding to its cellular receptor CAR J. Gen Virol. 1999;80:1519–1527. doi: 10.1099/0022-1317-80-6-1519. [DOI] [PubMed] [Google Scholar]
- 36.Schoen G, Fender P, Chroboczek J, Hewat E A. Adenovirus 3 penton dodecahedron exhibits structural changes of the base on fibre binding. EMBO J. 1996;15:6841–6846. [PMC free article] [PubMed] [Google Scholar]
- 37.Shafren D R, Williams D T, Barry R D. A decay-accelerating factor-binding strain of coxsackievirus B3 requires the coxsackievirus-adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J Virol. 1997;71:9844–9848. doi: 10.1128/jvi.71.12.9844-9848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shayakhmetov D M, Lieber A. Dependence of adenovirus infectivity on length of the fiber shaft domain. J Virol. 2000;74:10274–10286. doi: 10.1128/jvi.74.22.10274-10286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stouten P F W, Sander C, Ruigrok R W H, Cusack S. A new triple-helical model for the shaft of the adenovirus fibre. J Mol Biol. 1992;226:1073–1084. doi: 10.1016/0022-2836(92)91053-r. [DOI] [PubMed] [Google Scholar]
- 40.Van Beusechem V W, van Rijswijk A L C T, van Es H H G, Haisma H J, Pinebo H M, Gerritsen W R. Recombinant adenovirus vectors with knobless fibers for targeted gene transfer. Gene Ther. 2000;22:1940–1946. doi: 10.1038/sj.gt.3301323. [DOI] [PubMed] [Google Scholar]
- 41.Vigne E, Mahfouz I, Dedieu J F, Brie A, Perricaudet M, Yeh P. RGD inclusion in the hexon monomer provides adenovirus type 5-based vectors with a fiber knob-independent pathway for infection. J Virol. 1999;73:5156–5161. doi: 10.1128/jvi.73.6.5156-5161.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Von Seggern D J, Chiu C Y, Fleck S K, Stewart P L, Nemerow G R. A helper-independent adenovirus vector with E1, E3, and fiber deleted: structure and infectivity of fiberless particles. J Virol. 1999;73:1601–1608. doi: 10.1128/jvi.73.2.1601-1608.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wickham T J, Carrion M E, Kovesdi I. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 1995;2:750–756. [PubMed] [Google Scholar]
- 44.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 45.Wickham T J, Tzeng E, Shears L L, 2nd, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson J M. Adenoviruses as gene-delivery vehicles. N Engl J Med. 1996;334:1185–1187. doi: 10.1056/NEJM199605023341809. [DOI] [PubMed] [Google Scholar]