Abstract
A human rotavirus (isolate M) with an atypical electropherotype with 14 apparent bands of double-stranded RNA was isolated from a chronically infected immunodeficient child. MA-104 cell culture adaptation showed that the M isolate was a mixture of viruses containing standard genes (M0) or rearranged genes: M1 (containing a rearranged gene 7) and M2 (containing rearranged genes 7 and 11). The rearranged gene 7 of virus M1 (gene 7R) was very unusual because it contained two complete open reading frames (ORF). Moreover, serial propagation of virus M1 in cell culture indicated that gene 7R rapidly evolved, leading to a virus with a deleted gene 7R (gene 7RΔ). Gene 7RΔ coded for a modified NSP3 protein (NSP3m) of 599 amino acids (aa) containing a repetition of aa 8 to 296. The virus M3 (containing gene 7RΔ) was not defective in cell culture and actually produced NSP3m. The rearranged gene 11 (gene 11R) had a more usual pattern, with a partial duplication leading to a normal ORF followed by a long 3′ untranslated region. The rearrangement in gene 11R was almost identical to some of those previously described, suggesting that there is a hot spot for gene rearrangements at a specific location on the sequence. It has been suggested that in some cases the existence of short direct repeats could favor the occurrence of rearrangement at a specific site. The computer modeling of gene 7 and 11 mRNAs led us to propose a new mechanism for gene rearrangements in which secondary structures, besides short direct repeats, might facilitate and direct the transfer of the RNA polymerase from the 5′ to the 3′ end of the plus-strand RNA template during the replication step.
Group A rotaviruses are the main cause of viral gastroenteritis in infants and in the young of many animal species. Their genome consists of 11 segments of double-stranded RNA (dsRNA) which can be separated by polyacrylamide gel electrophoresis (PAGE). Electropherotype profiles of rotavirus dsRNA typically show four size classes of segments according to their molecular weight (10). Variations in the mobility of individual RNA segments allow a genetic characterization of rotavirus strains. However, group A rotaviruses showing unusual electropherotypes in which segments of standard size are replaced by rearranged forms of larger size have been described. Such viruses with a rearranged genome (for a review, see reference 9) were first isolated from chronically infected immunodeficient children (30) and later recovered either from asymptomatically infected immunocompetent children (5) or from animals (4, 33, 41). Rotaviruses with genome rearrangements were also generated in vitro by serial passage at a high multiplicity of infection of animal (16, 38), or human (19, 27) strains. Rotaviruses carrying rearranged genes are generally not defective, and the rearranged segments can reassort in vitro and replace their normal counterparts structurally and functionally (1, 6, 14). Gene rearrangements in human rotaviruses recovered from stool samples mostly involve segment 11 and less frequently involve segments 6, 8, 9, and 10. It is not known whether the rearrangements in segment 11 occur more frequently or if viruses with a rearrangement in segment 11 have some selective advantage so that they are detected more easily (10). Gene rearrangements generated in vitro have also been reported for segment 5 of bovine (16, 42) and segment 7 of human (19, 27) rotaviruses.
Nucleotide sequences of rearranged genes from several group A rotavirus strains have been described (3, 12, 13, 15, 25, 27, 28, 36, 38, 42). In most cases, the rearrangement resulted from a partial head-to-tail duplication of the gene: the sequence included a normal 5′ untranslated region (UTR) followed by a normal open reading frame (ORF). The duplication started from various positions after the stop codon and extended up to the 3′ end, leading to a long 3′ UTR (9). Thus, the rearranged gene expressed a normal protein product (3, 27, 38). However, Tian et al. described two bovine rotavirus variants with rearrangements in the gene 5 that modified the ORF (42). The resulting viruses retained their capacity to grow in cell culture, although they expressed modified NSP1 proteins (15, 42). So far, no mosaic structures due to an intermolecular recombination have been described in rearranged genes.
Thus, genome rearrangements have been proposed to play a part in the evolution of rotaviruses (beside point mutations and gene reassortments) and to contribute to their diversity (9, 39). Moreover, it has been suggested that rearranged segments containing a partial duplication of the ORF might be more efficient templates for dsRNA synthesis than are their homologous wild-type counterparts and thus may be preferentially selected during viral replication (29). The mechanism by which genome rearrangements occur in rotavirus genes has yet not been defined, and different models have been proposed (see reference 9 for a review). Current hypotheses suggest that the RNA-dependant RNA polymerase of the virus may jump back on its template during either the transcription (plus-strand synthesis) (20) or the replication (minus-strand synthesis) (9) step. Direct repeats that might favor the polymerase switch have been found close to the rearrangement site in some cases (3, 13, 20, 38) but not in others (25, 36).
In this paper we report the analysis of two rearranged genes (gene 7 and gene 11) in a group A human rotavirus isolated from an immunodeficient child. The rearrangement in gene 7 was very unusual because it contained two complete ORFs. The rearranged gene 7 underwent further evolution in vitro, with a change in the ORF leading to the expression of a modified NSP3 protein. Furthermore, the comparison of the two rearranged genes to their normal homologues and the computer modeling of their mRNAs led us to propose a mechanism for rearrangements in rotavirus genes based on the existence of secondary structures between the 3′ and 5′ ends of the plus-strand RNAs. Similarly to the model of picornaviruses in which regions of high local secondary structure such as hairpins or stem-loops have been proposed as hot spots for RNA recombination (22, 35, 43, 46), secondary structures in rotavirus mRNAs might correspond to hot spots for genome rearrangements.
MATERIALS AND METHODS
Viruses and cells.
The M isolate was a group A human rotavirus of genotype G1 isolated in Madagascar from the stool of a 6-month-old infant with severe combined immunodeficiency syndrome. Virus propagation on cell culture (confluent monolayers of MA-104 cells) was performed as previously described (11).
The different viruses present in the stool specimen were separated by cell culture under conditions of limiting dilution. Briefly, after one passage, serial dilutions of the virus yield (24 replicates for each dilution) were propagated in 24-well culture plates. After 72 h of cell culture, rotavirus antigens were detected by indirect immunofluorescence assay using a polyclonal goat antibody specific for group A rotaviruses as previously described (11). The limiting dilution for the viral growth was defined when fewer than 25% of the inoculated wells were positive for rotavirus culture. The initial virus yield was then propagated at the defined limiting dilution in 24-well culture plates. Cell lysates from each well were further propagated in 25-cm2 flasks prior to RNA analysis by PAGE.
Virus stocks of viruses M0, M1, and M3 (three cell culture-adapted viruses derived from the M isolate) were subjected to titer determination in 24-well culture plates. The infected MA104 cells were fixed 24 h postinfection and stained for determination of the fluorescence-forming units (FFU) using a polyclonal goat antibody specific for group A rotaviruses. The average titers were 3 × 105 FFU/ml for M0 and M1 and 2 × 105 FFU/ml for M3.
Serial cell culture propagation of viruses was not performed at high multiplicity of infection (MOI) but using dilutions (1:10) of the harvested cell culture fluid for adsorption on MA-104 cells monolayers. This corresponded approximately to a MOI of 0.01 FFU/cell.
Nucleic acid analysis.
Rotavirus genomic dsRNA was extracted either from stool suspensions (∼10% [wt/vol] in 150 mM NaCl) clarified by low-speed centrifugation or from cell culture lysate, using RNA-PLUS (Bioprobe Systems, Montreuil, France). Prior to reverse transcription PCR (RT-PCR), dsRNA segments were purified after electrophoresis in 1% low-melting-point agarose (Sigma-Aldrich Chimie S.a.r.l, St. Quentin Fallavier, France) using a modified freeze-squeeze extraction procedure (40). Briefly, the gel slice containing dsRNA was excised, crushed with a scalpel, vortexed for ∼10 s with 400 μl of phenol and frozen at −80°C for 30 min. After a rapid thawing, the supernatant was extracted with phenol-chloroform and precipitated in ethanol by standard procedures.
Rotavirus RNA genomic profiles were determined by PAGE in 10% polyacrylamide gels (1.5 mm thick) for 16 h at 20 mA at room temperature. RNA segments were made visible by ethidium bromide staining of the gel.
The genomic dsRNA segments were characterized by Northern blotting after electrophoresis in 1% agarose, using bovine rotavirus cDNA probes specific for gene 7, 8, or 9, labeled by random priming with the DIG DNA labeling and detection kit (Roche Diagnostics, Meylan, France).
Sequencing strategy and nucleotide sequence analysis.
RT-PCR amplification of genes 7, 7R, 7RΔ, 11, and 11R was performed on purified dsRNA segments using primers described in Table 1.
TABLE 1.
Primers used in RT-PCRs
| Gene (virus) | Primer
|
Use/amplification product | ||
|---|---|---|---|---|
| Name | Nucleotide sequence (5′-3′) | Location (F/R)a | ||
| 7 (M0) | P1b | CCCGTCGACGAATTCTTT(DIG) | External | Ligation to the 3′ end of the dsRNA |
| P2b | AAAGAATTCGTCGACGGG | External (F/R) | Single primer amplification/full-length cDNA | |
| 7R (M1) | A3 | GGACTTTTGAAGCAATGCAAC | 926–946 (F) | Junction zone of the rearrangement |
| A4c | CAGCAACAACTGCAGCTTCAA | 1039–1059 (R) | ||
| ZJ1c | CCACTGAAAAGCATTAATCA | 961–981 (R) | Used with P2 as forward primer/first half of gene 7R | |
| A1 | ATGCTCAAGATGGAGTCTACTCAGC | 984–1008 (F) | Used with P2 as reverse primer/second half of gene 7R | |
| 7RΔ (M3) | DPZJ3 | GAGTGGTATCTAAGGTCTATGG | 761–782 (F) | Junction zone of the rearrangement |
| RPZJ3c | CTAAGTTTATTCACGTCTTCATC | 1184–1206 (R) | ||
| DPE1 | GGCTTTTAATGCTTTTCAGTG | 1–21 (F) | First half of gene 7RΔ | |
| RPME1c | TAACATTAAAAATGTTCTGTCATAATC | 896–982 (R) | ||
| DPMB1c | GTAAGCTCTATTATTAATACTTCTTTTGAATCT | 923–955 (F) | Second half of gene 7RΔ | |
| RPB3 | GGTCACATAACGCCCCTATAGC | 1920–1941 (R) | ||
| 11 (M0) | ER1 | GAGTACAGGTCCGCTCGAATTCTTT(DIG) | External | Ligation to the 3′ end of the dsRNA |
| ER2 | AAAGAATTCGAGCGGACCTGTACTC | External (F/R) | Single primer amplification/full-length cDNA | |
| 11R (M2) | S11RA | CAGATAGTGATGACGGTAAATGTA | 509–532 (F) | Junction zone of the rearrangement |
| AS11RFc | TGAATCCATAGACACGCCAGCATC | 850–873 (R) | ||
| ZJ11ASc | GAAATTGAGGGAAGACTCGTCTACA | 611–635 (R) | Used with ER2 as forward primer/first half of gene 11R | |
| ZJ11S3c | ATGCAATTGATAGAAGATTTGTAGACG | 592–618 (F) | Used with ER2 as reverse primer/second half of gene 11R | |
Position on the sequence of the respective gene (F, forward primer; R, reverse primer)
Primers described by Lambden et al. (23).
Primer used for the RT step.
Full-length cDNA, including the 3′ and 5′ ends of genes 7 and 11 of virus M0 were obtained by the single-primer amplification strategy described by Lambden et al. (23) with some modifications. Briefly, oligonucleotides P1 (23) (for gene 7) and ER1 (for gene 11) were labeled at their 3′ ends with digoxigenin-11-ddUTP (Roche Diagnostics) as specified by the manufacturer and ligated to both 3′ ends of the dsRNA genome segments. The ligation was catalyzed by T4 RNA ligase (Roche Diagnostics). Briefly, dsRNA and 3′-end-labeled primers were added to a reaction mixture containing 2 U of T4 RNA ligase and ligation buffer and incubated for 20 h at 37°C. It was critical to maintain a ratio of 100 labeled primer molecules to 1 RNA 3′ OH extremity. After ligation, the enzyme was inactivated by heating for 2 min at 100°C and eliminated by ultrafiltration on polyvinyldiene difluoride membranes (UFC3-IPH columns; Millipore, St Quentin, France). Unligated primer molecules were removed by ultrafiltration on cellulose membranes (PLTK columns; Millipore). The efficiency of the reaction was monitored by Northern blotting followed by detection of the digoxigenin-labeled dsRNA (DIG luminescent detection kit; Roche Diagnostics). Primer-tailed dsRNAs were reverse transcribed and amplified using primer P2 (for gene 7) or ER2 (for gene 11).
Attempts to obtain full-length cDNA copies of the rearranged genes 7R and 11R by the single-primer amplification strategy always resulted in PCR products smaller than the expected size (data not shown). Such a phenomenon had previously been observed in another study of a rearranged gene (3) and might be due to an intermolecular base pairing between the duplicated sequences of the rearranged gene, leading to mispriming and incorrect elongation during the RT-PCR. To overcome this problem, the junction regions of the rearrangements of genes 7R, 7RΔ, and 11R were first amplified and sequenced, allowing the determination of new direct or reverse primers, specific for these regions. These primers used with the external primers (P2 or ER2) allowed us to sequence separately, after RT-PCR, the first and second halves of the rearranged genes, including the 3′ and 5′ ends. For gene 7RΔ, primers specific for the 5′ and 3′ termini were used instead of external primers (Table 1).
RT-PCR of the purified dsRNA gene segments was performed as follows: 1 to 5 μl of the dsRNA extract was reverse transcribed in a 50-μl reaction mixture containing 20 μM EDTA, 10 mM dithiothreitol, 0.5 mM (each) deoxynucleoside triphosphate, 0.1 μM primer, 10 U of RNase inhibitor (Life Technologies, Cergy, France), 200 U of SuperScript II (Life Technologies), and SuperScript buffer. After a 45-min incubation at 45°C the reaction was stopped by adding 1 μl of 0.5 M EDTA. Then 150 μl of H2O was added, and 5 μl of cDNA was amplified by using direct and reverse primers described in Table 1. PCR was performed in a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 to 2 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphate, 0.25 μM (each) primer, and 1.25 U of AmpliTaq DNA polymerase (Perkin-Elmer, Villebon, France). Amplification was performed in a 9700 Perkin Elmer thermocycler under PCR conditions of 35 cycles of 94°C for 30 s, 50 to 60°C for 1 min, and 72°C for 1 min. Concentrations of MgCl2 and annealing temperatures were adapted depending on the primers. PCR products were analyzed after electrophoresis in 1.5% agarose gels.
Sequencing was carried out by the dideoxynucleotide chain terminator method on an ABI Prism 377 automatic sequencer (Applied Biosystems), using the ABI PRISM DyeTerminator cycle-sequencing ready reaction kit. Nucleotide sequences were determined directly from the PCR products, except for gene 7, which was cloned in the plasmid vector pBluescript. In all cases the sequence was determined on both strands, either from two independent RT-PCR products or from plasmid DNA of three independent clones.
Secondary structure of mRNAs.
The mfold program version 3.1 (24, 47) predicts the possible secondary structures for RNA sequences. It was made available by Michael Zuker on his home page, presently located at http://bioinfo.math.rpi.edu/∼zukerm/. The program was established using the rules set up by Serra and Turner (37) and Walter et al. (44) concerning the minimal computed free energies.
Protein analysis.
Virus-infected confluent monolayers of MA-104 cells were washed with 2 ml of phosphate-buffered saline and scraped. Cell lysis was performed either for 1 h at 4°C with a buffer containing 50 mM HEPES (pH 7.0), 250 mM NaCl, 5 mM EDTA, 2 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 5 mM dithiothreitol, 0.1% NP-40, 7.5 μg of aprotinin per ml, 1 μg of leupeptin per ml, and 100 μg of phenylmethylsulfonyl fluoride per ml or for 5 min at room temperature with a buffer containing 50 mM Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate (SDS), and 2% β-mercaptoethanol. A 40-μg portion protein extract was boiled in 62.5 mM Tris-HCl (pH 6.8)–2% SDS–5% β-mercaptoethanol–25% glycerol and loaded on a 12% acrylamide gel for SDS-PAGE. Proteins were blotted on polyvinyldiene difluoride (Hybond-P; Amersham Pharmacia Biotech, Orsay, France) membranes by transverse electrophoresis in 250 mM Tris–192 mM glycine–20% methanol. After a 2-h saturation in 20 mM Tris-HCl (pH 7.6)–137 mM NaCl–0.5% Tween 20 (TBS-T) with 10% skim milk, the membranes were incubated for 90 min at room temperature with a 1:5 dilution of monoclonal mouse anti-NSP3 ID3 antibody (2), washed three times with TBS-T, and then incubated for 1 h at room temperature with a 1:10 000 dilution of a horseradish peroxidase-coupled goat anti-mouse immunoglobulin G (Jackson Immunoresearch, Interchem, Asnières, France). Finally, the membranes were washed four times with TBS-T and the proteins were revealed by enhanced chemiluminescence (ECL Western blotting detection kit; Amersham).
Nucleotide sequence accession numbers.
The full-length sequences of genes 11, 11R, 7, 7R, and 7RΔ are available under GenBank accession no. AF338244 through AF338248.
RESULTS
The human rotavirus M isolate is a mixture of viruses with standard or rearranged genes.
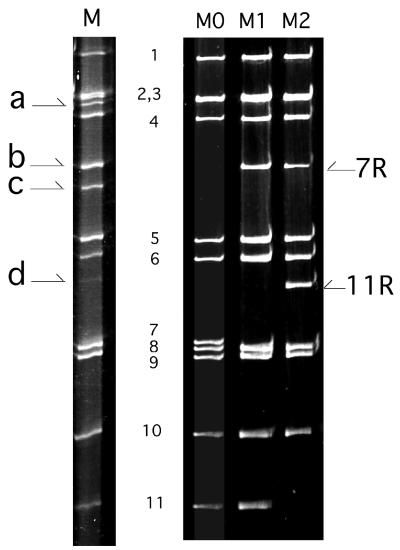
The RNA profile of the human rotavirus M isolate is shown in Fig. 1. The electropherotype displayed an unusual pattern of 14 apparent bands of dsRNA including 10 segments with standard size (segments 1 to 6 and 8 to 11) and 4 extra segments located between segments 3 and 4 (segment a), 4 and 5 (segments b and c), and 6 and 8 (segment d). In addition, segment 7 was missing from the profile and the intensity of segments 6 and 11 was lowered. Such an RNA profile, with additional high-molecular-weight segments replacing standard-size segments, strongly suggested that the isolate M genome contained rearranged genes. Moreover, this profile with 14 apparent bands of dsRNA suggested that the M isolate was a mixture of several subpopulations of viruses differing in genotype, as reported for other human rotavirus isolates with genome rearrangements (17).
FIG. 1.
RNA profiles of the M isolate and of viruses M0, M1, and M2. Lanes: M, RNA profile of the M isolate obtained from the original stool sample; M0, M1, and M2, RNA profiles of viruses M0, M1, and M2 recovered in MA-104 cell culture performed under limiting-dilution conditions. Numbers indicate the rotavirus gene segments of standard size. a, b, c, and d represent the extra segments in isolate M. 7R and 11R indicate rearranged genes 7 and 11, respectively.
Attempts to separate the various viruses contained in the M isolate by plaque purification in MA-104 cell culture directly from the stool sample were unsuccessful. Alternatively, the M isolate was grown in MA-104 cell culture from the stool supernatant under conditions of limiting dilution. This allowed us to recover cell culture-adapted viruses, all of which contained 11 genomic segments. These viruses were divided into three species named M0, M1, and M2 according to their electropherotypes (Fig. 1). M0 had a typical electropherotype of group A rotaviruses, with 11 segments located at their usual position. In M1 and M2, segment b replaced segment 7. In addition, in virus M2, segment d replaced segment 11. Northern blot analysis indicated that segment b from M1 and M2 hybridized with a cDNA probe specific for gene 7 and thus represented a rearranged gene 7. Similarly, segment d in virus M2 hybridized with a probe specific for gene 11 and represented a rearranged gene 11 (data not shown). Segments b and d were subsequently referred to as genes 7R and 11R, respectively. Although no viruses containing segments a or c could be recovered in cell culture, the existence of these RNA bands in the RNA profile of the M isolate might indicate that the M isolate contained more virus species with different genome rearrangements than were isolated. Unfortunately, the gene origin of RNA bands a and c could not be determined because there was not enough of the origin stool sample to perform Northern blot experiments on the RNA extracted from the M isolate.
These results indicated that the M isolate contained a mixture of at least three nondefective viruses, two of them containing one or two rearranged genes. This allowed us to compare the nucleotide sequences of rearranged genes with those of the standard genes from which they were derived.
The rearranged segment 7R of virus M1 contains two complete copies of the NSP3 ORF.
The complete nucleotide sequence of gene 7 from virus M0 was first determined as a reference. Gene 7 of virus M0 had 96.7% similarity to gene 7 of the human rotavirus strain Wa. However, as opposed to most rotavirus genes 7 deposited in GenBank, which contain two in-frame methionine codons (at nucleotides [nt] 26 and 35), gene 7 of M0 had a CTG in place of the first ATG. Thus, gene 7 of virus M0 contained 1,074 bp with a 5′ UTR of 34 bp and a 3′ UTR of 107 bp. The ORF ranged from nt 35 to 967 and coded for a NSP3 protein of 310 amino acids (aa).
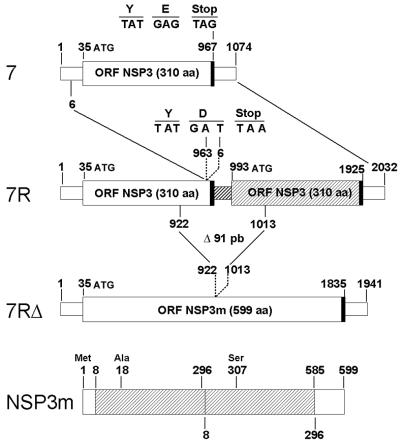
As compared to gene 7 of virus M0, gene 7R of virus M1 consisted of an almost full-length duplication of the gene (Fig. 2). The rearrangement took place after nt 963, reinitiating the sequence within the 5′ UTR at nt 6. The rearrangement not only led to the modification of the last nucleotide of the first ORF, changing the last amino acid of the protein (E-310-D), but also recreated a new in-frame stop codon TAA at nt 965. Therefore, the rearrangement of gene 7R was very unusual, since it resulted in a rearranged gene containing two complete ORFs. Except for codon 310 and for a single silent point mutation at nt 259 in the first part of the gene 7R, both ORFs were identical.
FIG. 2.
Schematic diagram of the standard and rearranged genes 7 and of the NSP3m protein. The ORFs are indicated by large boxes, and the UTRs are indicated by small boxes. Thick lines indicate the stop codons. The nucleotide positions corresponding to the gene rearrangement (gene 7R) and to the 91-bp deletion of gene 7RΔ are indicated. The nucleotide sequence of the junction region of the rearrangement is detailed. In gene 7R, the duplicated part of the gene is shaded. In NSP3m, residues are numbered referring to the first methionine codon (at nt 35) and the repeated residues (aa 8 to 296) are indicated by a cross-hatched box.
Gene 7R of virus M1 serially propagated in cell culture undergoes a further change leading to a modified NSP3 ORF.
Although gene 7R contained two complete ORFs, it was not possible to ascertain whether both ORFs were used for protein translation. Considering the hypothesis that an untranslated ORF was likely to be modified during further replication cycles, we investigated the evolution of gene 7R during serial propagation of virus M1 in cell culture as an indirect means of determining the functionality of the second ORF.
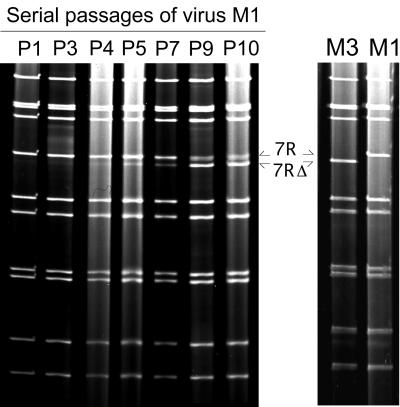
Virus M1 was serially propagated in cell culture at low MOI (≤0.01 FFU/cell), and the RNA profiles corresponding to each passage were analyzed (Fig. 3). After four passages, a 12th segment (referred to as 7RΔ) was detected below segment 7R. During further passages, the relative concentration of segment 7RΔ increased while that of segment 7R decreased. At passage 10, a subculture performed under limiting-dilution conditions allowed us to recover virus M3 containing segment 7RΔ and not segment 7R (Fig. 3). Nucleotide sequence analysis of segment 7RΔ showed that it was derived from gene 7R after a 91-bp deletion. The deletion started at nt 922 (within the first ORF of gene 7R) and ended at nucleotide 1013 (within the second ORF), thus connecting the duplicated sequence of gene 7R in frame with the first ORF (Fig. 2). Gene 7RΔ displayed an ORF of 1,941 bp coding for a modified NSP3 protein (NSP3m) of 599 aa (instead of 310). NSP3m consisted of the first 296 amino-terminal residues of NSP3 followed by a large repetition of 289 residues (aa 8 to 296) and the last 14 carboxy-terminal residues of NSP3 (aa 297 to 310). Thus, all but the first 7 and the last 14 aa of NSP3 were duplicated in the NSP3m protein. A single change in the amino acid sequence was detected in the repeated part of the protein: A (aa 18) changed to S (aa 307). As compared to gene 7R, the duplicated part of gene 7RΔ also contained an additional silent point mutation at nt 1207 (A changed to G).
FIG. 3.
Serial propagation of virus M1 in cell culture. (Left) Evolution of the virus M1 RNA profile during serial passages in MA-104 cell culture. Pn indicates the number of passages (n) performed prior to the analysis of the electropherotype. At P4, segment 7RΔ appeared below segment 7R (arrows), increased in relative concentration along with further passages, and became a majority at P9. (Right) RNA profiles of the viruses obtained after subculture of the P10 viral mixture, allowing us to recover virus M3 containing segment 7RΔ and virus M1 containing segment 7R.
Taken together, these results indicated that the duplicated coding sequence contained in gene 7R was not maintained during further replication of virus M1 in cell culture and the change in gene 7R involving both ORFs led to a rearranged gene 7 with a single ORF potentially coding for a modified NSP3 protein.
NSP3 expression in cells infected with M0, M1, or M3.
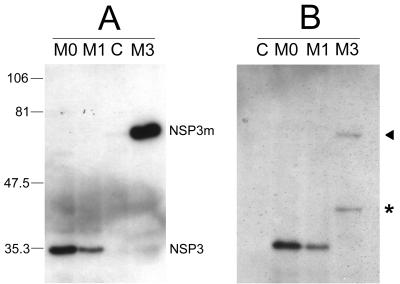
To examine the effects of gene 7 rearrangements on the expression of NSP3, MA-104 cells were infected with virus M0 (gene 7), M1 (gene 7R), or M3 (gene 7RΔ) and cell lysates were analyzed by Western blotting using an NSP3-specific monoclonal antibody (Fig. 4). As expected, the NSP3 protein produced by virus M1 had an apparent molecular mass close to 34 kDa, which was identical to that of the NSP3 protein of virus M0. However, when equivalent amounts of total proteins (40 μg) extracted from cells infected with virus M0 or M1 at the same MOI (0.1 FFU/cell) were analyzed, the signal intensity of NSP3 detected by Western blotting was reproducibly lower for M1 than for M0 (Fig. 4, compare lanes M0 and M1). This could indicate that virus M1 synthesized NSP3 in smaller amounts than did virus M0 and might favor the hypothesis that the second ORF of gene 7R was not used for protein synthesis and even lowered the expression of the first ORF. The modified protein NSP3m from virus M3 was clearly detected in Western blots when an SDS-containing buffer was used for cell lysis and protein extraction (Fig. 4A). Under these conditions, NSP3m had an apparent molecular mass of approximately 68 kDa, compared to 34 kDa for NSP3. Interestingly, when a buffer without SDS was used for cell lysis, the NSP3m protein was only faintly detected and two bands were observed, one at 68 kDa and an additional band at approximately 40 kDa (Fig. 4B). This was not the case for the NSP3 proteins of M0 or M1, which were similarly detected independent of the buffer used (compare Fig. 4A and B). The significance of the 40-kDa band observed for NSP3m remains to be established. This band might have been generated by cleavage during the extraction procedure, since the duplicated protein which contains two coiled-coil structures might be less stable and thus more accessible to proteases. However, no distinct bands corresponding to smaller cleavage products were detected on the blot, and the possibility of an artifact should not be excluded. On the other hand, the epitope recognized by the monoclonal antibody ID3, located between aa 206 and 290 (unpublished data), might not be contained in smaller cleavage products. Taken together, these results indicated that virus M3 actually produced the modified protein NSP3m and suggested that NSP3 and NSP3m might behave differently in MA-104 cells.
FIG. 4.
Western blot analyses of NSP3 and NSP3m. MA-104 cells infected by viruses M0, M1, and M3 were recovered 24 h postinfection, and whole-cell lysates were analyzed by Western blotting with the monoclonal antibody ID3 specific for NSP3. C indicates mock-infected cells as a control. Numbers indicate molecular mass markers in kilodaltons. The position of viral proteins NSP3 and NSP3m are indicated. (A) Western blotting was performed after cell lysis with a 2% SDS-containing buffer. The apparent molecular mass of NSP3m (virus M3) was approximately double the molecular mass of NSP3 (viruses M0 and M1). (B) Western blot analysis was performed after cell lysis without SDS. Unlike NSP3, NSP3m was only faintly detected (arrow) and an additional band of 40 kDa was observed (asterisk).
The gene 11 rearrangement in virus M2 suggests that gene 11 rearrangements in rotaviruses occur at a definite sequence location.
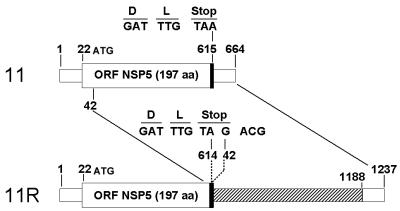
The nucleotide sequence of gene 11 from virus M0 was first determined as a reference. It consisted of 664 nt with a 594-bp ORF flanked by 5′ and 3′ UTRs of 21 and 49 nt, respectively. Gene 11 of virus M0 had 98% similarity to the gene 11 of the human rotavirus strain Wa. The rearrangement of gene 11R of virus M2 consisted of a partial duplication of gene 11 of virus M0. The rearrangement took place within the stop codon TAA at nt 614, and the sequence reinitiated within the ORF at nt 42, re-creating a new in-frame stop codon, TAG, at the same position (Fig. 5). Thus, gene 11R was 1,237 bp long and contained a normal 594-bp ORF followed by a long 3′ UTR of 622 bp. Compared to gene 11 of virus M0, no mutation was detected in gene 11R, neither in the first part nor in the duplicated part of the gene.
FIG. 5.
Schematic diagram of the standard and rearranged genes 11. The ORFs are indicated by large open boxes, and the UTRs are indicated by small open boxes. Thick lines indicate the stop codons. The nucleotide positions corresponding to the gene rearrangement (gene 11R) are indicated. The nucleotide sequence of the junction region of the rearrangement is detailed. The duplicated sequence of the gene 11R is shaded.
Strikingly, the gene 11R rearrangement was almost identical to the gene 11 rearrangements described for the human strain Z10262 (28) and the bovine strain C7/183 (36). For both these viruses, as for virus M2, the rearrangement occurred within the stop codon (at nt 614 for virus Z10262 and nt 615 for virus C7/183) and the sequence reinitiated at nt 43 (Z10262) or 41 (C7/183). Such a similarity of the gene 11 rearrangement location in three unrelated strains strongly suggested a nonrandom phenomenon. Since the rearrangements always involved the 3′ and 5′ ends of the genes, we further investigated the existence of putative secondary structures in the single-stranded RNA template for replication which might bring the 3′ and 5′ ends close together and thus favor rearrangements.
Predicted folding of gene 7 and 11 mRNAs.
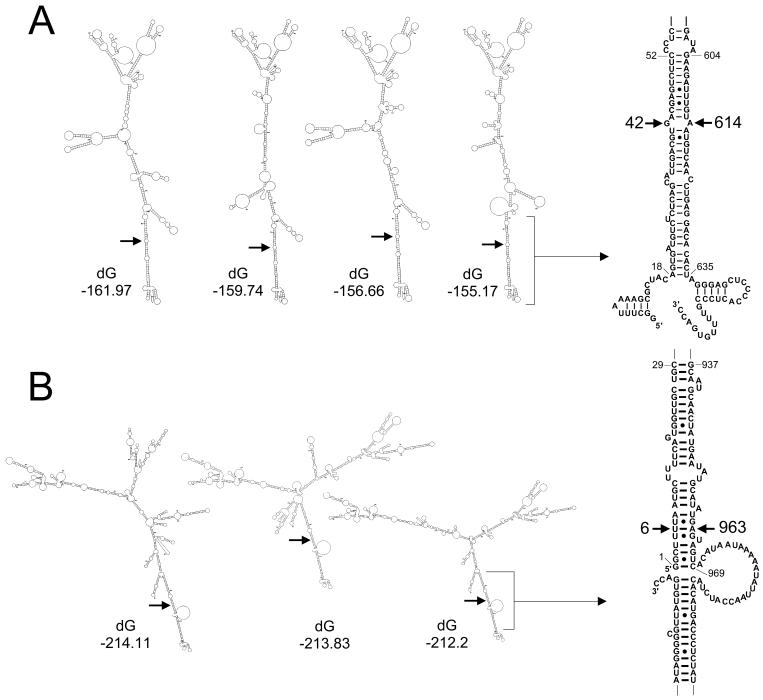
The nucleotide sequence of the single-stranded mRNA of gene 11 from virus M0 was analyzed with the mfold program based on free-energy minimization developed by Zuker et al. (47). The possible foldings obtained within 20% of the minimum free energy were analyzed. All 11 optimal secondary structures predicted by the program showed that the 3′ end (nt 604 to 635) would interact with the 5′ end (nt 52 to 18) to form a long panhandle (Fig. 6A). This base pairing placed face to face the two nucleotides (nt 42 and 614) which mark the boundary of the rearrangement of gene 11R. Similarly, for gene 7 of virus M0, 26 of the 28 most stable secondary structures predicted by the mfold program showed that the 3′ end (nt 937 to 969) would interact with the 5′ end (nt 29 to 1) of the RNA (Fig. 6B). Thus, a base pairing between nt 6 and 963, the nucleotides involved in the rearrangement of gene 7R, was predicted. Such secondary structures might favor the occurrence of rearrangement according to a mechanism that will be discussed below.
FIG. 6.
Predicted optimal secondary structures for gene 7 and 11 plus-strand RNAs. Examples of some optimal folding configurations predicted by the mfold program for genes 7 (A) and 11 (B) of virus M0 are shown. dG indicates the minimum free energy values (in kilocalories per mole). Arrows indicate the positions of the nucleotides involved in the rearrangements leading to genes 7R and 11R. On the right, the base pairing between the 3′ and 5′ ends of the RNA predicted for the most stable secondary structures is enlarged. The nucleotides involved in the gene rearrangements are indicated.
DISCUSSION
Cell culture recovery of viruses with rearranged genes.
The RNA profile of the M isolate obtained from the original stool sample contained 14 gene segments, with 4 segments migrating at unusual positions. As previously reported (17, 30), such an electropherotype corresponded to a mixture of virus subpopulations, with some viruses containing rearranged genes. As reported by others (17), plaque purification of the viruses directly from the fecal extracts was unsuccessful. However, virus culture under limiting-dilution conditions allowed us to recover viruses with a standard genome (M0) and viruses with rearranged genes: M1 (containing a rearranged gene 7) and M2 (containing rearranged genes 7 and 11). Viruses containing the slowly migrating segments a or c that were observed on the original electropherotype could not be recovered in cell culture, although 192 RNA profiles were analyzed. This could hardly be explained by the hypothesis that these viruses were in a minority in the stool sample compared to M0, M1, and M2 (Fig. 1, lane M). Indeed, M0 was readily recovered in cell culture although it was only a minor component of the stool sample as indicated by the absence of a visible segment 7 on the original electropherotype. This could indicate that virus M0 had a selective advantage due to the absence of rearranged genes while the viruses containing segments a or c had a selective disadvantage, probably because of the rearrangements themselves.
Standard and rearranged genes 7 in viruses M0, M1, and M3.
As opposed to most rotavirus genes 7 deposited in GenBank (34), both genes 7 of M0 and 7R of M1 had a CTG instead of an ATG at nt 26, indicating that the methionine codon at nt 35 was probably used as the initiating codon for NSP3. Consistent with this hypothesis, the methionine codon at nt 35 is in a better environment than the one at nt 26 for the initiation of translation according to Kozak's rules (21). In addition, Piron et al. have shown that the RNA-binding capacity of NSP3 was conserved when the protein started at the second methionine (31).
Gene 7 rearrangements had never been described before for human rotaviruses isolated in vivo, although in vitro-generated human rotaviruses with rearranged genes 7 have been reported (19, 27). The rearranged gene 7R of virus M1 had an unusual pattern, with two complete ORFs that were identical except for the last codon of the first ORF (which was changed because of the rearrangement) and for a silent point mutation in the second ORF. To our knowledge, this is the first example of a rotavirus RNA segment containing two copies of the same gene. Whether both ORFs were used by the virus M1 for the translation of NSP3 remains to be established. However, in the absence of an internal ribosome entry site on the mRNA, the second ORF was most probably not translated. Consistent with this hypothesis, virus M1 did not overexpress the NSP3 protein, as indicated by Western blot analyses.
After serial propagation of virus M1 in cell culture, gene 7R rapidly underwent a further change, leading to the deleted gene 7RΔ coding for the modified protein NSP3m. Virus M3 expressing the modified NSP3m protein was not defective and multiplied well in cell culture. A fluorescent-focus assay indicated that virus M3 replicated as efficiently as virus M0 did (results not shown). Moreover, when virus M1 was serially propagated in cell culture, virus M3 emerged and became predominant, as indicated in PAGE by the increasing relative amount of segment 7RΔ over segment 7R (Fig. 3). This could indicate that virus M3 had a selective advantage over virus M1, perhaps related to the NSP3m protein. The nonstructural protein NSP3 plays an important role in the multiplication cycle of rotaviruses. Three domains of activities have been mapped (31, 32): the N-terminal part of the protein (aa 4 to 150) which binds specifically to the 3′ end of the viral mRNAs; the middle part of the protein (aa 150 to 240), which contains a possible coiled-coil domain and is required for dimerization; and the C-terminal domain (aa 206 to 313), which is responsible for interaction of NSP3 with the translation initiation factor eIF-4GI. NSP3m contained a repetition which included the entire dimerization domain and most of the RNA-binding and the eIF-4GI-binding domains. This might have resulted in a change of some of the biological properties of the protein. For example, the fact that as opposed to NSP3, NSP3m was detected in Western blots depending on the presence of SDS in the lysis buffer might indicate a stronger interaction of NSP3m with some cellular components. This deserves further investigations, since the association of NSP3 with the cytoskeleton has been previously suggested (26).
Gene rearrangements are thought to participate in the genetic variability of rotaviruses (9). So far, the only case of a protein modification induced by a gene rearrangement was reported for two in vitro-generated variants of a bovine rotavirus strain with rearrangements in gene 5 (15, 42). For one variant (brvE), the rearranged gene contained two in-frame partial ORFs, leading to an extended NSP1 protein (728 aa instead of 491 aa) (42). For the other variant (brvA), the rearrangement was associated with several point mutations, one of which resulted in a new in-frame stop codon in the first part of the ORF, truncating the protein to its first 258 aa despite partial duplication of the gene (15). The rearranged gene 7RΔ reported here is another example of a gene with extended ORF and confirms that replication-competent viruses with modified proteins can be generated by rearrangement events. However, rotaviruses with a rearranged gene coding for a modified protein have never been isolated in vivo. In the present case, virus M3 producing a modified NSP3 protein was obtained in vitro during cell passages of virus M1, and it would have been of interest to know whether virus M1 would have evolved similarly in vivo.
Possible mechanisms for rotavirus gene rearrangement.
The molecular mechanism leading to the rearrangement of rotavirus genes is not known. A current hypothesis suggests that either during transcription (plus-strand RNA synthesis) or replication (minus-strand RNA synthesis), the viral RNA polymerase could interrupt the RNA synthesis, fall back on its template, and reinitiate RNA synthesis (9). Direct repeats close to the duplication site have been proposed as a way for the polymerase to fall back on its template (9, 20). However, direct repeats do not seem to be an absolute requirement for genome rearrangements (9). They were found in some rearranged genes (3, 13, 20, 38) but not in others (25, 36), and, in particular, no direct repeat could be found to explain the rearrangements of genes 7 and 11 reported in this study.
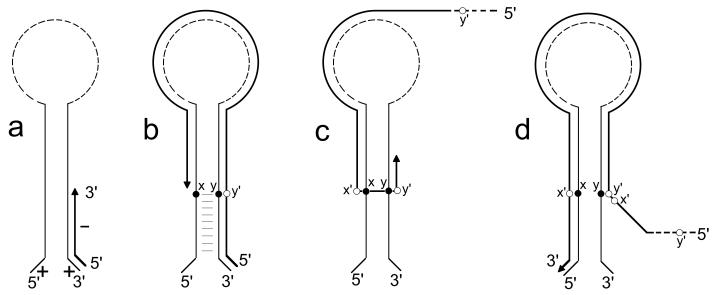
The fact that for both genes 7 and 11 the 5′ and 3′ ends of the mRNAs were predicted to form a long panhandle, resulting in placing face to face the two nucleotides involved in the rearrangement, suggested that hot spots for gene rearrangement might also be related to such secondary structures. Although the actual existence of secondary structures predicted by computer modeling remains to be established, previous studies support an interaction between the 3′ and 5′ ends of rotavirus mRNAs. The formation of a panhandle resulting from an interaction between the 5′ and 3′ ends and the single-stranded nature of at least the two 3′-terminal CC residues have been reported as structural constraints required for efficient minus-strand RNA synthesis (7, 8, 45). The predicted secondary structures of gene 7 and 11 mRNAs reported here were consistent with these constraints. This led us to propose a mechanism for gene rearrangement in which secondary structures might facilitate and direct the transfer of the RNA polymerase from the 5′ to the neighboring 3′ end of the template during the replication step (Fig. 7).
FIG. 7.
Possible mechanism for rearrangements of rotavirus genes. (a) During the replication step, the viral RNA polymerase initiates the minus-strand synthesis at the 3′ end of the plus-strand RNA. (b) The RNA polymerase reaches nucleotide position x, in the panhandle formed by the 5′ and 3′ ends of the RNA, and interrupts RNA synthesis. (c) Without releasing the nascent minus strand, the RNA polymerase falls back on its template at nucleotide y in the panhandle and reinitiates replication. (d) The RNA polymerase completes the replication up to the 5′ end, thus duplicating the sequence x′-y′ (complementary to x-y). Since minus-strand RNAs were predicted to fold quite differently from plus-strand RNAs (data not shown), a similar model could not be proposed to explain the occurrence of rearrangement at the transcription step. In addition, at the transcription stage, the minus-strand RNA is associated with the plus strand and is probably not subject to folding.
RNA recombination has been reported to occur by various mechanisms in a number of RNA viruses (see reference 22 for a review). Rotavirus genome rearrangements can be considered intramolecular recombination events (9) consistent with the copy choice mechanism described for poliovirus, in which the viral RNA polymerase switches templates during RNA minus-strand synthesis (18, 22). Poliovirus recombination is favored by the presence of homologous sequences in the neighborhood of the recombination site and has been reported to occur either at random (18) or at specific sites (35, 43). Indeed, regions of high local secondary structure such as hairpins and stem-loops have been reported as recombinational hot spots for poliovirus (35, 43) or foot-and-mouth disease virus (46). Similarly, secondary structures in rotavirus mRNAs might correspond to hot spots of recombination, but, as opposed to the poliovirus model, rotavirus genome rearrangements should be considered to be nonhomologous recombination events occurring within the same RNA template. What initiates the template switch within the 5′-3′ panhandle of rotavirus mRNA remains to be determined. Neighboring nucleotides other than those observed for genes 7R and 11R might be used for rearrangement, although some restrictions might exist because only the viable resulting viruses will be subsequently selected, as suggested by Lai for other RNA viruses (22). For instance, rearrangements occuring upstream of the stop codon and leading to a modified ORF might not be selected.
However, some of the rearrangements previously reported for other genes 11 (12, 13, 20) or 7 (27) for which the nucleotides involved in the rearrangement are not neighbors in the predicted secondary structures could not be directly explained by the model we propose. It is of interest that as opposed to genes 7R and 11R, these rearranged genes contain numerous mutations in the duplicated part of the sequence. Such mutations may account for a genetic drift, indicating that after genome rearrangement, the virus underwent multiple replication cycles during which deletions might also have occurred, concealing the initial location of the rearrangement. This was the case for the rearranged gene 7R, which underwent a deletion during the cell culture propagation of virus M1, leading to gene 7RΔ, in which the nucleotides involved in the initial rearrangement could no longer be identified.
In summary, the theoretical model we propose, implying the existence of secondary structures between the 3′ and 5′ ends of mRNAs, could apply to some but not all examples of gene rearrangements. However, since rearranged genes seem to be likely to evolve rapidly with time, it should be considered that in addition to the initial event of rearrangement, a rearranged gene may be the result of a multiple-step process involving different mechanisms. Analyses of additional sequences of rotavirus gene rearrangements will help us gain a better understanding of this process. The finding that the rearranged gene 7RΔ had a modified ORF confirms that gene rearrangements not only participate in the genetic variability of rotaviruses but also can lead to the synthesis of modified proteins. In the absence of a reverse genetic system for rotaviruses, the isolation of nondefective rotaviruses with rearranged genes coding for modified nonstructural proteins may be a means of specifying some of their functions.
ACKNOWLEDGMENTS
We are grateful to Anne-Marie Cassel-Béraud for providing the clinical sample containing the rotavirus M isolate. We thank Jean Cohen for helpful discussions and critical reading of the manuscript. We thank Paul Dény for constructive comments. We thank Laurence Albiges for excellent technical assistance and Christophe Goujon for advice on computer modeling.
This work was supported in part by the MESRT grant Programme de Recherches Fondamentales en Microbiologie, Maladies Infectieuses et Parasitologie “Réseau de Recherche sur les Gastro-Entérites à Rotavirus.”
REFERENCES
- 1.Allen A M, Desselberger U. Reassortment of human rotaviruses carrying rearranged genomes with bovine rotavirus. J Gen Virol. 1985;66:2703–2714. doi: 10.1099/0022-1317-66-12-2703. [DOI] [PubMed] [Google Scholar]
- 2.Aponte C, Mattion N M, Estes M K, Charpilienne A, Cohen J. Expression of two bovine rotavirus non-structural proteins (NSP2, NSP3) in the baculovirus system and production of monoclonal antibodies directed against the expressed proteins. Arch Virol. 1993;133:85–95. doi: 10.1007/BF01309746. [DOI] [PubMed] [Google Scholar]
- 3.Ballard A, McCrae M A, Desselberger U. Nucleotide sequences of normal and rearranged RNA segments 10 of human rotaviruses. J Gen Virol. 1992;73:633–638. doi: 10.1099/0022-1317-73-3-633. [DOI] [PubMed] [Google Scholar]
- 4.Bellinzoni R C, Mattion N M, Burrone O, Gonzalez A, La Torre J L, Scodeller E A. Isolation of group A swine rotaviruses displaying atypical electropherotypes. J Clin Microbiol. 1987;25:952–954. doi: 10.1128/jcm.25.5.952-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besselaar T G, Rosenblatt A, Kidd A H. Atypical rotavirus from South African neonates. Arch Virol. 1986;87:327–330. doi: 10.1007/BF01315311. [DOI] [PubMed] [Google Scholar]
- 6.Biryahwaho B, Hundley F, Desselberger U. Bovine rotavirus with rearranged genome reassorts with human rotavirus. Arch Virol. 1987;96:257–264. doi: 10.1007/BF01320965. [DOI] [PubMed] [Google Scholar]
- 7.Chen D, Patton J T. De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA. 2000;6:1455–1467. doi: 10.1017/s1355838200001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Patton J T. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J Virol. 1998;72:7387–7396. doi: 10.1128/jvi.72.9.7387-7396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desselberger U. Genome rearrangements of rotaviruses. Adv Virus Res. 1996;46:69–95. doi: 10.1016/s0065-3527(08)60070-6. [DOI] [PubMed] [Google Scholar]
- 10.Estes M. Rotaviruses and their replication. In: Fields B, Knipe D, Howley P, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1625–1655. [Google Scholar]
- 11.Garbarg-Chenon A, Bricout F, Nicolas J C. Study of genetic reassortment between two human rotaviruses. Virology. 1984;139:358–365. doi: 10.1016/0042-6822(84)90381-7. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez S A, Mattion N M, Bellinzoni R, Burrone O R. Structure of rearranged genome segment 11 in two different rotavirus strains generated by a similar mechanism. J Gen Virol. 1989;70:1329–1336. doi: 10.1099/0022-1317-70-6-1329. [DOI] [PubMed] [Google Scholar]
- 13.Gorziglia M, Nishikawa K, Fukuhara N. Evidence of duplication and deletion in super short segment 11 of rabbit rotavirus Alabama strain. Virology. 1989;170:587–590. doi: 10.1016/0042-6822(89)90453-4. [DOI] [PubMed] [Google Scholar]
- 14.Graham A, Kudesia G, Allen A M, Desselberger U. Reassortment of human rotavirus possessing genome rearrangements with bovine rotavirus: evidence for host cell selection. J Gen Virol. 1987;68:115–122. doi: 10.1099/0022-1317-68-1-115. [DOI] [PubMed] [Google Scholar]
- 15.Hua J, Patton J T. The carboxyl-half of the rotavirus nonstructural protein NS53 (NSP1) is not required for virus replication. Virology. 1994;198:567–576. doi: 10.1006/viro.1994.1068. [DOI] [PubMed] [Google Scholar]
- 16.Hundley F, Biryahwaho B, Gow M, Desselberger U. Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology. 1985;143:88–103. doi: 10.1016/0042-6822(85)90099-6. [DOI] [PubMed] [Google Scholar]
- 17.Hundley F, McIntyre M, Clark B, Beards G, Wood D, Chrystie I, Desselberger U. Heterogeneity of genome rearrangements in rotaviruses isolated from a chronically infected immunodeficient child. J Virol. 1987;61:3365–3372. doi: 10.1128/jvi.61.11.3365-3372.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima K, Taniguchi K, Kawagishi-Kobayashi M, Matsuno S, Urasawa S. Rearrangement generated in double genes, NSP1 and NSP3, of viable progenies from a human rotavirus strain. Virus Res. 2000;67:163–171. doi: 10.1016/s0168-1702(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 20.Kojima K, Taniguchi K, Urasawa T, Urasawa S. Sequence analysis of normal and rearranged nsp5 genes from human rotavirus strains isolated in nature—implications for the occurrence of the rearrangement at the step of plus strand synthesis. Virology. 1996;224:446–452. doi: 10.1006/viro.1996.0551. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 22.Lai M M. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambden P R, Cooke S J, Caul E O, Clarke I N. Cloning of noncultivatable human rotavirus by single primer amplification. J Virol. 1992;66:1817–1822. doi: 10.1128/jvi.66.3.1817-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 25.Matsui S M, Mackow E R, Matsuno S, Paul P S, Greenberg H B. Sequence analysis of gene 11 equivalents from “short” and “super short” strains of rotavirus. J Virol. 1990;64:120–124. doi: 10.1128/jvi.64.1.120-124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattion N M, Cohen J, Aponte C, Estes M K. Characterization of an oligomerization domain and RNA-binding properties on rotavirus nonstructural protein NS34. Virology. 1992;190:68–83. doi: 10.1016/0042-6822(92)91193-x. [DOI] [PubMed] [Google Scholar]
- 27.Mendez E, Arias C F, Lopez S. Genomic rearrangements in human rotavirus strain Wa; analysis of rearranged RNA segment 7. Arch Virol. 1992;125:331–338. doi: 10.1007/BF01309651. [DOI] [PubMed] [Google Scholar]
- 28.Palombo E A, Bugg H C, Bishop R F. Characterisation of rearranged NSP5 gene of a human rotavirus. Acta Virol. 1998;42:55–59. [PubMed] [Google Scholar]
- 29.Patton J T, Chnaiderman J, Spencer E. Open reading frame in rotavirus mRNA specifically promotes synthesis of double-stranded RNA: template size also affects replication efficiency. Virology. 1999;264:167–180. doi: 10.1006/viro.1999.9989. [DOI] [PubMed] [Google Scholar]
- 30.Pedley S, Hundley F, Chrystie I, McCrae M A, Desselberger U. The genomes of rotaviruses isolated from chronically infected immunodeficient children. J Gen Virol. 1984;65:1141–1150. doi: 10.1099/0022-1317-65-7-1141. [DOI] [PubMed] [Google Scholar]
- 31.Piron M, Delaunay T, Grosclaude J, Poncet D. Identification of the RNA-binding, dimerization, and eIF4G1-binding domains of rotavirus nonstructural protein NSP3. J Virol. 1999;73:5411–5421. doi: 10.1128/jvi.73.7.5411-5421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pocock D H. Isolation and characterization of two group A rotaviruses with unusual genome profiles. J Gen Virol. 1987;68:653–660. doi: 10.1099/0022-1317-68-3-653. [DOI] [PubMed] [Google Scholar]
- 34.Rao C D, Das M, Ilango P, Lalwani R, Rao B S, Gowda K. Comparative nucleotide and amino acid sequence analysis of the sequence-specific RNA-binding rotavirus nonstructural protein NSP3. Virology. 1995;207:327–333. doi: 10.1006/viro.1995.1087. [DOI] [PubMed] [Google Scholar]
- 35.Romanova L I, Blinov V M, Tolskaya E A, Viktorova E G, Kolesnikova M S, Guseva E A, Agol V I. The primary structure of crossover regions of intertypic poliovirus recombinants: a model of recombination between RNA genomes. Virology. 1986;155:202–213. doi: 10.1016/0042-6822(86)90180-7. [DOI] [PubMed] [Google Scholar]
- 36.Scott G E, Tarlow O, McCrae M A. Detailed structural analysis of a genome rearrangement in bovine rotavirus. Virus Res. 1989;14:119–127. doi: 10.1016/0168-1702(89)90033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serra M J, Turner D H. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 38.Shen S, Burke B, Desselberger U. Rearrangement of the VP6 gene of a group A rotavirus in combination with a point mutation affecting trimer stability. J Virol. 1994;68:1682–1688. doi: 10.1128/jvi.68.3.1682-1688.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi K, Urasawa S. Diversity in rotavirus genomes. Semin Virol. 1995;6:123–131. [Google Scholar]
- 40.Tautz D, Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983;132:14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- 41.Thouless M E, DiGiacomo R F, Neuman D S. Isolation of two lapine rotaviruses: characterization of their subgroup, serotype and RNA electropherotypes. Arch Virol. 1986;89:161–170. doi: 10.1007/BF01309886. [DOI] [PubMed] [Google Scholar]
- 42.Tian Y, Tarlow O, Ballard A, Desselberger U, McCrae M A. Genomic concatemerization/deletion in rotaviruses: a new mechanism for generating rapid genetic change of potential epidemiological importance. J Virol. 1993;67:6625–6632. doi: 10.1128/jvi.67.11.6625-6632.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolskaya E A, Romanova L I, Blinov V M, Viktorova E G, Sinyakov A N, Kolesnikova M S, Agol V I. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology. 1987;161:54–61. doi: 10.1016/0042-6822(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 44.Walter A E, Turner D H, Kim J, Lyttle M H, Muller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wentz M J, Patton J T, Ramig R F. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J Virol. 1996;70:7833–7841. doi: 10.1128/jvi.70.11.7833-7841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson V, Taylor P, Desselberger U. Crossover regions in foot-and-mouth disease virus (FMDV) recombinants correspond to regions of high local secondary structure. Arch Virol. 1988;102:131–139. doi: 10.1007/BF01315570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Kluwer Academic. Dordrecht, The Netherlands: NATO ASI; 1999. pp. 11–43. [Google Scholar]