Abstract
Age-related cataracts is a highly prevalent eye disorder that results in the clouding of the crystalline lens and is one of the leading causes of visual impairment and blindness. The disease is influenced by multiple factors including genetics, prolonged exposure to ultraviolet radiation, and a history of diabetes. However, the extent to which each of these factors contributes to the development of cataracts remains unclear. Our study identified 101 independent genome-wide significant loci, 57 of which are novel. We identified multiple genes and biological pathways associated with the cataracts, including four drug-gene interactions. Our results suggest a causal association between type 1 diabetes and cataracts. Also, we highlighted a surrogate measure of UV light exposure as a marker of cataract risk in adults.
Subject terms: Genome-wide association studies, Computational biology and bioinformatics, Genetics research
Here, the authors expand knowledge of the genetic architecture of age-related cataracts, exploring associated pathways and drug-gene interactions, and clarify the roles of type 1 diabetes and UV exposure in cataract etiology.
Introduction
Cataract is a common ocular condition characterized by opacification of the crystalline lens, leading to vision loss and potential blindness. It is the leading cause of blindness globally among adults aged 50 and over1. The prevalence of cataracts increases with age, with ~18% of individuals over the age of 60 affected by the condition2. In certain regions such as Africa, the prevalence of cataracts is even higher, with estimates suggesting that up to 40% of individuals over the age of 60 in these regions may develop the condition3. Age-related cataract, also known as senile cataract, is the most common type, accounting for 90% of all cataracts.
Genome-wide association studies (GWAS) have identified multiple genetic loci associated with a higher risk of developing cataracts4. Genes such as FOXE3 and PAX6 have been previously linked to cataracts in mice and humans. In addition to these genetic risk factors, several environmental and lifestyle risk factors are linked to the development of cataracts, including UV light exposure5,6 and a history of diabetes7. Some studies suggest that systemic diseases such as diabetes may increase the risk of cataracts and influence a younger age at cataract onset8. However, it remains unclear to what extent these factors contribute to the development of cataracts and whether a genetic propensity for cataracts is associated with lifestyle factors earlier in life.
The identification of novel genetic variants associated with cataracts could have significant medical implications for the prevention of cataracts and visual impairment, understanding the disease’s etiology, and identifying modifiable lifestyle factors that contribute to the development of the disease. In addition, drug-gene interactions can further elucidate the relationship between genes encoding proteins influenced by specific drug targets, shedding light on potential contributors to cataract development and deepening our understanding of how drugs interact with these genes. Moreover, a deeper understanding of the underlying genetic architecture could provide a way to screen individuals at high risk of developing cataracts, using methods such as polygenic risk scores (PRS). This insight could also shed light on the relationship between genetic risk for cataracts and potential contributing factors that may increase the likelihood of developing cataracts later in life.
In this study, we present the largest GWAS meta-analysis of cataracts using data from 121,725 cases and 821,856 controls of multiple ancestry. We employed gene enrichment techniques to identify genes and biological pathways associated with cataracts, and further tested the association between cataract-associated loci and gene expression through expression quantitative trait loci (eQTL) of peripheral blood and explored drug-gene interactions. In addition, we investigated the potential causal association between type 1 diabetes and cataracts using a Mendelian randomization (MR) framework to gain further insights into the relationship between diabetes and cataracts. Previous research has already established a causal association between type 2 diabetes and cataracts9. Furthermore, we estimated PRS to assess the association between cataract risk and UVR-related lesions in both young and older adults.
Results
Meta-analysis
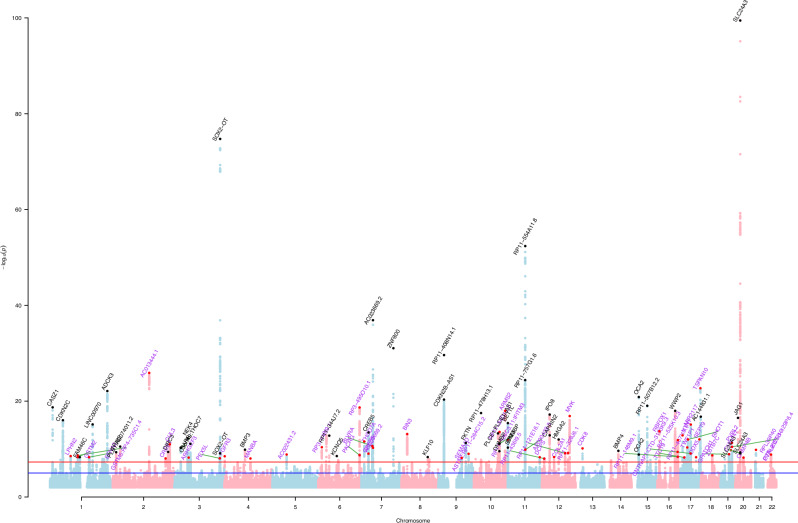
We identified 101 independent genome-wide significant loci associated with cataracts (Supplementary Data 1). Of these, 44 loci had been previously reported, and 57 were unreported; as per Fig. 1 and Supplementary Data 1 and 8. The results of our meta-analysis, which combines data from five cohorts, showed a genetic inflation factor of 1.14 and an LD-score intercept of 1.04 (se = 0.009; see Q-Q plot in Supplementary Fig. 1) and a single nucleotide polymorphism (SNP)-based heritability of 4.5%. The LD-score intercept suggests that the inflation is likely due to the polygenicity of cataracts rather than confounding factors. Prior to the meta-analysis, we assessed the genetic correlation between studies using LD score (LDSC) regression, which showed a consistent correlation between Choquet et al.4 meta-analysis and FinnGen (rg = 0.94, se = 0.04, p = 1.03 × 10−92). The univariate LDSC results for The Mass General Brigham Biobank (MGBB) study were too noisy to allow us to reliably estimate a genetic correlation. We then proceeded to correlate the effect estimates of the genome-wide significant independent loci, weighting variants by the square of the standard error of their effect estimates (also known as inverse variance weighted (IVW)). Risk-increasing alleles in the larger study were selected, and the other study was aligned to match the same allele. The weighted correlation between the previous meta-analysis, and FinnGen was high (r = 0.84), in comparison with the other two studies. The correlation between MGBB and the previous meta-analysis, and between MGBB and FinnGen, was 0.1 and 0.19, respectively, as shown in Supplementary Figs. 3–5. This is consistent with the results of LDSC, where the previous meta-analysis and FinnGen maintained a high correlation throughout the genome. Given the smaller sample size of MGBB (~3k cases) in comparison with the previous meta-analysis (~68k cases) and FinnGen (~51k cases), larger confidence intervals and lower statistical power were expected. Nevertheless, the correlation still maintains a consistent positive correlation of effects between studies. We further tried a ‘leave-one-out’ approach, where we excluded MGBB from the meta-analysis. The results did not affect the number of loci associated with cataracts, leading us to conclude that including MGBB in the meta-analysis does not significantly affect the results. However, given the consistency in the direction of the effect correlations, and a slight improvement in the effect estimates we decided to include it in the meta-analysis.
Fig. 1. Manhattan plot showing results for the GWAS meta-analysis of cataracts; p values derived from logistic regression models are two-sided.
Each dot represents a single nucleotide polymorphism (SNP) and the red line represents the threshold for multiple testing correction (p < 5 × 10−8) and the blue line (p < 5 × 10−6). Novel loci are represented in the plot as red dots and are highlighted in purple, whereas previously known loci are presented in black.
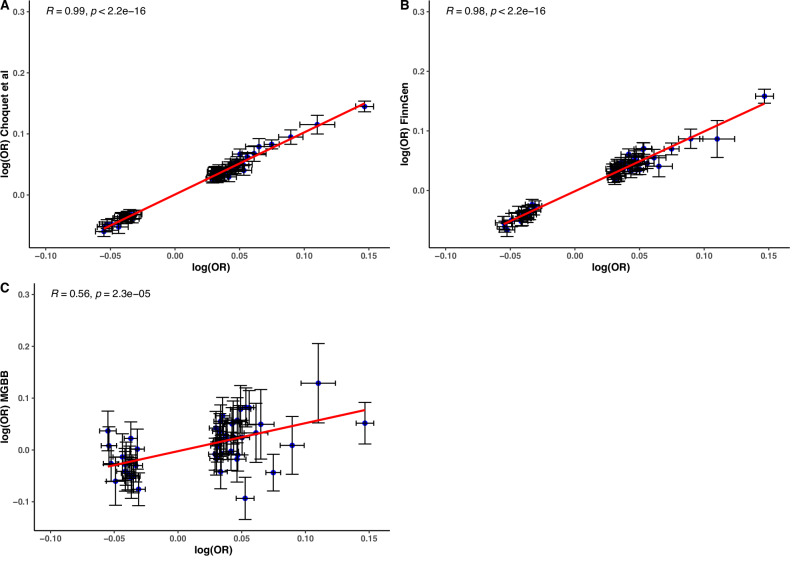
After the meta-analysis, we correlated the effect estimates between the meta-analysis and previous cataract GWAS, using independent and genome-wide significant loci, to ensure consistency in the magnitude and direction of the effect. Results showed a high correlation of the effect between our meta-analysis, FinnGen (r = 0.98, p < 2.2 × 10−16), Choquet et al.4 (r = 0.99, p < 2.2 × 10−16), and a lower correlation with MGBB (r = 0.56, p = 2.3 × 10−5) with a wider 95% CIs due to the smaller sample size in that study (Fig. 2).
Fig. 2. Correlation of allele effect estimates (log(OR)) using a two-sided linear model between the meta-analysis and previous cataract GWAS, based on independent and genome-wide significant loci (N = 101).
Effects estimates (center points) are presented as logarithms of odd ratios (log(OR)) and black crosses represent a 95% confidence interval. The studies included are (A) The previous meta-analysis for cataracts (Choquet et al.; p < 2.2 × 10−16), (B) FinnGen (p < 2.2 × 10−16), and (C) The Mass General Brigham Biobank (MGBB; p = 2.3 x 10−5).
Functional annotation and eQTL
A gene enrichment analysis using MAGMA on 19,491 genes identified 134 genes associated with cataracts, after a Bonferroni correction for multiple testing (p < 2.56 × 10−6) (Supplementary Data 2). We observed associations of previously reported genes such as BLVRA, EFNA1, KRTCAP2, and CDKN2B. We also identified two biological processes that were significantly enriched for gene ontology through gene-set analysis: pericyte cell differentiation and negative regulation of lipid biosynthetic process. These gene sets are shared with multiple disorders in GWAS catalog such as Refractive error, Type 2 diabetes, Schizophrenia, Parkinson, and Alzheimer’s disease. Results from summary-data-based Mendelian randomization (SMR) using peripheral blood eQTL data confirmed that gene expression of 30 genes out of the 134 genes identified by MAGMA is likely associated with cataract (p < 3.7 × 10−4), however only 20 genes maintain a consistent effect between eQTL of peripheral blood and the GWAS results (p heidi >0.05; Supplementary Data 3).
Drug-gene interaction
Drug-gene interactions among 20 genes associated with cataracts identified four genes (GNL3, JAG1, METTL21A, and CREB1) that interact with eight drugs. Three of these genes (CREB1, METTL21A, and GNL3) have not previously been associated at a genome-wide significant level with cataracts. GNL3 exhibited interactions with epirubicin, cyclophosphamide, and fluorouracil. Meanwhile, JAG1 was found to interact with hydrocortisone. Interactions between METTL21A and citalopram were also observed. Similarly, CREB1 displayed interactions with citalopram, lithium, nicotine, and alcohol.
Mendelian randomization
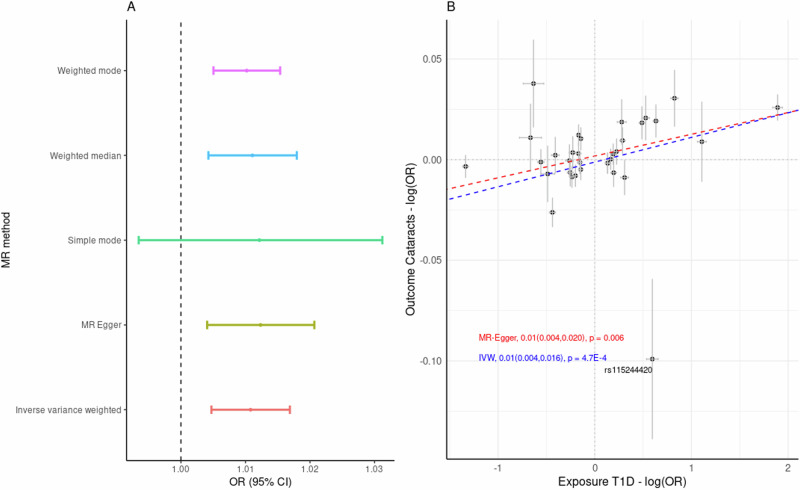
We observed a putative causal association between the genetic predisposition to type 1 diabetes and the risk of cataracts. The proportion of phenotypic variance (PVE) explained by the selected SNPs on Diabetes was ~10% (= 0.09). Analysis with MR-PRESSO identified two potential pleiotropic outliers (rs2596560 and rs9468541) and eight palindromic SNPs with intermediate allele frequencies. Once the outliers and palindromic SNPs were excluded, MR results based on IVW showed that genetic predisposition to diabetes is likely to have a causal association with higher cataract risk (OR = 1.011, 95% CI = 1.017–1.005, p = 4.7 × 10−4). Results were consistent using other MR approaches including MR Egger, Weighted median, and Weighted mode (Fig. 3, Supplementary Data 4). There was no evidence of horizontal pleiotropy (MR egger intercept p > 0.05), yet there was evidence of heterogeneity between variants (Q = 58.11, = 46.6%, p = 2.23 × 10−3). We did not find evidence of unbalanced pleiotropy, following a manual inspection of funnel plot symmetry (Supplementary Fig. 2). We further investigated the heterogeneity, as the high heterogeneity estimate could be due to the pleiotropic effects of the SNPs and contravene MR assumptions. Further investigation with MR Rucker, as a robust method to account for the heterogeneity, estimated a consistent causal association between type 1 diabetes and cataracts (OR = 1.01, 95% CI = 1.011–1.004, p = 4.7 × 10−4). We further tested the association between type 1 diabetes and cataracts by excluding the SNPs that have been associated with type 2 diabetes (p < 0.05), where the association remained significant (OR = 1.009, 95% CI = 1.002–1.015, p = 4.1 × 10−3); as per Supplementary Fig. 6. These results were consistent with the MVMR analysis when adjusting for Type 2 diabetes-related variants as a possible cofounder for the association between type 1 diabetes and cataracts (OR = 1.008, 95% CI = 1.002–1.01, p = 0.01).
Fig. 3. MR results of the putative causal association between cataracts and type 1 diabetes (N = 33).
A Forest plot based on different MR methods. Estimated effects (center point), odd ratios (OR), are presented as per unit change of the exposure with a 95% confidence interval. B Effect of variants associated with Cataracts (Outcome) and type 1 Diabetes (Exposure). Effects estimates (center points) are presented as logarithms of odd ratios (log(OR)) and gray crosses represent a 95% confidence interval. The dashed blue line shows the inverse variance weighted (IVW) fit and the red dashed line shows the MR-Egger-fit; p values derived from IVW and MR-Egger are two-sided.
Out of the 249 metabolites tested through Generalized Summary-data-based Mendelian Randomization (GSMR), 28 showed an association with cataracts, including total fatty acids, Omega 6, the ratio of phospholipids to total lipids, and cholesterol in small HDL, among others; for a complete list of nominal associations, see Supplementary Data 7. However, only the association with β-Hydroxybutyric acid was maintained after multiple testing corrections. β-Hydroxybutyric acid was highlighted as a protective factor for cataracts in our results (OR = 0.41 95% CI = 0.29–0.56, p = 7.24 × 10−8).
Polygenic risk scores
To further test the association between these genome-wide independent loci and cataracts, we conducted a PRS analysis using data from 389 cataract cases and 4,416 controls in the Busselton Healthy Aging Study (BHAS) (Fig. 4). We used a pruning and threshold approach for the PRS analysis, where the genome-wide loci were selected to evaluate the PVE explained by the genetic risk of cataracts. We found that the genetic risk of cataracts was associated with cataracts in BHAS, an independent cohort, confirming that these loci are likely associated with cataracts (AUC = 0.54, R-squared = 0.001, p = 0.01). Participants in the highest PRS decile seem more likely to have cataracts (odds ratio = 1.12, 95% CI = 0.81–1.56) compared to the rest of the cohort, however, results were not statistically significant. The PRS demonstrated similar predictive efficacy when contrasted with the PRS derived from the previous meta-analysis (AUC = 0.53, R-squared = 0.001, p = 0.02). DeLong’s test was performed to compare the ROC curves of the two models (p = 0.918).
Fig. 4.
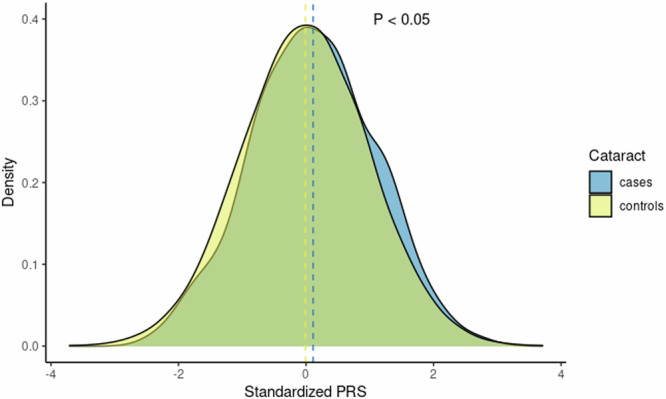
Distribution of Polygenic Risk Scores for Cataracts in Cases and Controls. Standardized PRS for cataracts are based on 389 cases and 4416 controls from the BHAS cohort (p = 0.01). The PRS distribution for cases is shown in blue, while the distribution for controls is shown in yellow.
We investigated the association between the PRS for cataracts and measures of ocular UVR exposure in the Raine Study Gen2 and BHAS cohorts. The PRS for cataracts was nominally associated with pterygium area in young adults (the Raine Study Gen2; p = 0.029) after multiple testing corrections (p = 0.05/2 traits = 0.025), but not in older adults (BHAS; p = 0.189). Furthermore, the PRS for cataracts was not associated with CUVAF in young adults (p = 0.201) and was nominally associated in older adults (p = 0.047) (Supplementary Data 5).
Discussion
This study identified 101 independent genome-wide significant loci associated with cataracts, of which 57 are novel, expanding our understanding of the genetic basis of this condition. The PRS analysis showed an association between these genome-wide independent loci and the risk of cataracts in independent datasets, providing further evidence for an association between these loci and the disease.
We used MAGMA to perform gene enrichment analysis and found that 134 genes were associated with cataracts. Additionally, we identified two biological processes, pericyte cell differentiation and the negative regulation of the lipid biosynthetic process, that were associated with age-related cataracts. These results are consistent with previous studies that implicate lipid accumulation in disrupting the structure and function of lens proteins10,11 by affecting homeostasis and the biosynthesis process of lipids, which leads to an opacification of the lens.
Negative regulation of lipid biosynthesis has also been linked to the development of metabolic disorders such as type 2 diabetes12,13, where a dysregulation of lipid biosynthesis can lead to the accumulation of lipids such as triglycerides and cholesterol in several organ tissues14,15. Lipids are also an important component of the lens membranes and dysregulation of the lipid metabolism has been associated with the development of cataracts16. Drugs that target negative regulation of lipid biosynthesis have shown promise as potential treatments for cataracts17 by modulating lipid metabolism and reducing the accumulation of lipids in the lens, which is a hallmark of cataract formation. We further assessed blood lipid metabolites and their association with cataracts. Notably, we observed a nominal causal association between the ratio of phospholipids to total lipids and the risk of cataracts. This finding aligns with prior research that has emphasized the phospholipid-to-cholesterol ratio as a factor contributing to cataract etiology. However, among all the tested associations, only β-Hydroxybutyric acid demonstrated sustained statistical significance following correction for multiple testing, and there is no established link between β-Hydroxybutyric acid, a ketone body produced during fasting or ketosis, and the development or prevention of cataracts. Consequently, further research is necessary to elucidate the specific role of lipid metabolites in the etiology of cataracts.
Drug-gene interactions involve genes that encode proteins potentially influenced by drug-like molecules, predicted based on sequence and structural similarity to existing drug targets. These interactions can aid in the identification of potential risk factors for cataract development and in uncovering the mechanisms of interaction with drugs. Our results suggest that specific interactions between genes and drugs highlight mechanisms that may play a role in the development of cataracts. In particular, the gene CREB1 exhibits interactions with lithium, nicotine, and alcohol, indicating their potential relevance to cataract formation. Consistently, previous studies have linked lithium18, nicotine19, and alcohol consumption20 with an increased risk of developing cataracts. Citalopram, an antidepressant known as a selective serotonin reuptake inhibitor21, has been associated with an increased risk of cataract surgery22. The interaction between METTL21A and CREB1 with citalopram could highlight a mechanism that is triggered by citalopram and leads to an increased risk of cataracts.
Meanwhile, the interaction of JAG1 with hydrocortisone underlines a potential drug that could be used for the treatment of cataracts. Hydrocortisone is a corticosteroid drug that has been evaluated for its potential as an anti-cataract steroidal drug23, which is consistent with the findings of this study. The mechanism of interaction with other drugs such as epirubicin, cyclophosphamide, and fluorouracil is not well understood, so further research is needed to determine if this interaction could lead to potential treatments. This information sheds light on the complex genetic basis of cataracts and provides new insights into the underlying mechanisms behind gene-drug interaction.
We observed a potential causal association between genetic predisposition to type 1 diabetes and an increased risk for cataracts. Despite the already well-established high prevalence of cataracts among patients with diabetes, this is a noteworthy finding as it suggests that the genetic variants associated with type 1 diabetes may also increase the risk of cataracts. This is consistent with previously published research that identified a potential causal association between type 2 diabetes and cataracts9, likely pointing to a common underlying mechanism between the etiology of the two conditions. However, patients diagnosed with diabetes often undergo frequent eye examinations24. More frequent and careful examinations could lead to a higher diagnosis rate within the disease subgroups which could inflate the association between type 1 diabetes and cataracts. As such, additional research incorporating a wider sample is necessary to establish the persistence of the causal relationship.
The PRS cross-trait analyses indicated a potential association between the risk of developing cataracts and increased CUVAF area in older adults, while results in young adults were non-significant. These findings may highlight that persistent exposure to sunlight may influence the development of cataracts later in life. Thus, this implicates UVR, specifically CUVAF area, as a potential marker of cataract risk in adults.
This study contributes to our understanding of the genetic basis of cataracts and has yielded several noteworthy findings. Our analysis identified 101 loci and 134 genes associated with cataracts, doubling the number of known loci. Notably, we have identified a potential overlap between negative regulation of lipid biosynthesis and the development of cataracts, as well as drug-gene interactions that may expand the range of therapeutic options available for the treatment of cataracts. Furthermore, our findings provided evidence of a putative causal relationship between genetic predisposition to type 1 diabetes and an increased risk of cataracts. Finally, we highlight a potential association between exposure to UVR and the risk of developing age-related cataracts; however, further research is required to establish the reliability of this as a marker of cataract risk. The results of this study contribute to the understanding of biological mechanisms involved in the development of cataracts and open up potential new avenues for the treatment of cataracts.
Methods
Cohorts
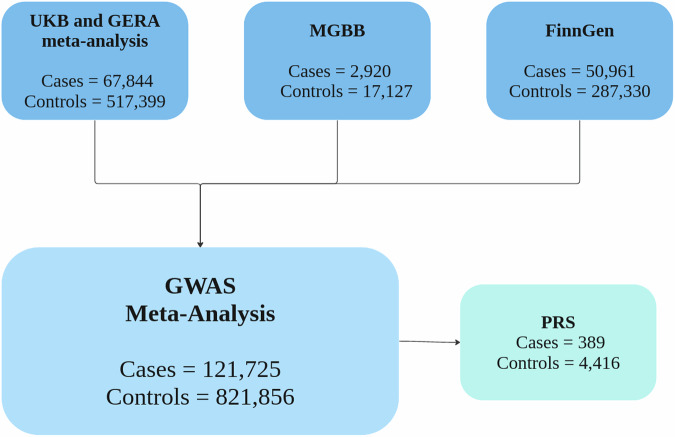
To identify risk loci for cataracts, we conducted a GWAS meta-analysis that encompasses 67,844 cases and 517,399 controls from a meta-analysis of the United Kingdom Biobank (UKB) and Genetic Epidemiology Research in Adult Health and Aging (GERA) cohorts4, 2920 cases and 17,127 controls from the MGBB and 50,961 cases and 287,330 controls from FinnGen. We leveraged data from two independent Australian cohorts, the Raine Study and BHAS25–27, to derive PRS based on the meta-analysis results and test their predictive ability to validate the associated loci (Fig. 5). A brief description of each of the cohorts included in the present study is provided below.
Fig. 5. Cohorts included in the cataract meta-analysis and polygenic risk score (PRS) analysis.
UK biobank (UKB), Adult Health and Aging (GERA), Mass General Brigham Biobank (MGBB).
The MGBB (formerly Partners HealthCare Biobank) is a long-term medical research repository allocated within the Partners HealthCare System in Boston, Massachusetts28. The Biobank is dedicated to collecting and storing biospecimens, such as blood and tissue samples, from over 130,000 participants and genomic data from over 65,000 participants; of which 47% are over 60 years of age. The Partners HealthCare Biobank operates with the highest ethical standards and follows the Declaration of Helsinki to ensure the protection of the rights and welfare of study participants. We performed a GWAS in 2920 cases and 17,127 controls using PLINK 1.90 beta. The phenotype was defined based on ICD-9/ICD-10 codes in the electronic health records of the participants.
The FinnGen Study is a biobank located in Finland that aims to encourage human genetics research and promote the discovery of novel treatments for various diseases. It is one of the largest biobanks in Europe including over 220,000 participants with genomic data and with a median age of 63 years29. The biobank is maintained by the Institute for Molecular Medicine Finland. The summary statistics used in this study included 50,961 cases and 287,330 controls and are labeled as “senile cataracts” as part of Finngen release eight (https://r8.finngen.fi/pheno/H7_CATARACTSENILE).
The current meta-analysis also includes the previous larger cataract multi-ancestry meta-analysis that encompassed GERA and UKB4, using their publicly available summary statistics. The GERA cohort contains clinical and genomic data of over 110,000 participants; 33,145 patients who have undergone cataract surgery and 64,777 controls were included in this study. The UKB is a large prospective study following the health of ~500,000 participants, including 34,699 cataract cases defined as “participants with a self-reported cataract operation (f20004 code 1435) or/and a hospital record including a diagnosis code (ICD-10: H25 or H26)”4, and 452,622 controls were included in the meta-analysis. Cataracts were clinically diagnosed based on ICD-9/ICD-10 criteria in GERA, the MGBB, and FinnGen. Detailed information regarding phenotype definition, genotyping, QC, and imputation procedures for all cohorts is provided in Supplementary Data 6.
GWAS meta-analysis
We conducted an IVW fixed-effect meta-analysis of 121,725 cases and 821,856 controls using METAL30. All variants were aligned to the positive strand on build GRC37 hg19 and those with a minor allele frequency (MAF) < 1% were removed. In cases where the MAF was not available for the study (i.e., GERA and MGBB), it was derived from a reference panel based on five thousand healthy individuals from UKBB. Linkage disequilibrium (LD) clumping was used to identify the independent genome-wide significant loci within a 1 Mb window. Clumping of the results was conducted in PLINK 1.9 with a P value cut-off of 5e-8 and r-squared >0.01 using the 1000 Human Genome Project reference panel. Employing the same configuration for the clumping analysis, we estimated the independent loci for the previous, larger meta-analysis, resulting in 44 independent loci. Loci that were shared between the previous and this study’s meta-analysis were considered known, while those unique to this study were deemed as novel. SNP heritability estimates for the GWAS meta-analysis were estimated using LDSC v1.0.131. Manhattan plots were generated using custom code32.
Functional annotation and eQTL
To provide insights into the genes and biological pathways underlying cataract etiology, we used functional annotation as implemented in FUMA (Functional Mapping and Annotation of Genetic Variants)33 v1.3.6 and mapped variants to protein-coding genes using MAGMA (Meta-Analysis Gene-set Mining of GWAS) v1.08. FUMA is a tool for functional annotation of genetic variants that integrates functional genomics data from various sources, such as GWAS and eQTL to provide functional annotation and prioritize genetic variants. MAGMA performs gene-based and gene-set analyses to identify genes and genetic pathways that are enriched for genetic variants associated with a trait34. We used a p value threshold of 5e-8 to define loci associated with cataracts in GWAS and applied a Bonferroni correction for multiple testing and corrected for the total number of genes in the gene-based analysis (p = 0.05/19,491 genes) and gene-set analysis (p = 0.05/17,017 gene-sets). Gene-based analysis was used to prioritize associations between genes and cataracts. Gene-set tests, also known as pathway analysis or enrichment analysis, were employed to assess groups of genes that are functionally related and collaborate in biological pathways that are associated with cataracts etiology process. Gene sets were obtained from Msigdb v7.0 for “Curated gene sets” and “GO terms” as part of the magma analysis and are incorporated in FUMA V.108 pipeline.
Genes identified through gene-based analysis in MAGMA were further evaluated with the integration of blood eQTL data from 2765 individuals35 of the Consortium for the Architecture of Gene Expression by using SMR. SMR is a commonly used method to interrogate the association between gene expression and complex human traits using GWAS summary statistics36. We applied a Bonferroni correction, which was set at 0.05 divided by the number of genes (N = 134), to account for multiple testing.
Drug-gene interaction
Genes that were consistent between MAGMA and SMR analyses were evaluated for their potential drug-gene interactions using vs 4.0 of the Drug-Gene Interaction Database37. The Drug-Gene Interaction Database is a carefully curated database that collects information on both established and predicted interactions between drugs and genes. It combines data from various sources to give a comprehensive overview of drug-gene interactions. Drug-gene interactions are instrumental in the development of strategies for preventing cataracts, as they provide information about potential drug exposures that could be related to therapeutic approaches for comorbid conditions or lifestyle factors that increase the likelihood of cataracts.
Mendelian randomization
We estimated the putative causal relationship between Type 1 Diabetes38 and cataracts using the TwoSampleMR framework implemented as a package in R 4.0.2. “TwoSampleMR”39 package v 0.5.5 is an R package that enables the estimation of causal effects between an exposure and an outcome of interest using summary-level data from GWAS through two-sample MR. The package includes various methods such as IVW, weighted median, MR-Egger, MR-PRESSO, and MR Rucker. We selected independent instrumental variables based on clumping using PLINK 1.9, with the following parameters: --clump-r2 0.001, --clump-p1 5e-8, --clump kb 1000. If the IVW estimate showed evidence for a nominal causal association, we further re-assessed the MR relationship using a series of alternative MR models, including MR-Egger, weighted median, simple, and weighted mode40. We calculated the proportion of PVE explained by SNP based on the equation below and used the traits where SNPs collectively explained at least 1% of the PVE. Here, β is the effect of the variant, MAF is the minor allele frequency, SE is the standard error and N is the sample size.
| 1 |
Multivariate Mendelian randomization (MVMR) was employed to assess whether the genetic variants associated with Type 2 Diabetes41 were causally influencing the association between Type 1 diabetes and Cataracts; this analysis is included in the “TwoSampleMR” package.
Considering the association between lipid metabolites and cataracts highlighted by the gene-set analysis, we further assessed a potential causal relationship through an MR framework. We utilized summary statistics data from 249 metabolic markers, including amino acids and metabolites related to glycolysis and fatty acids. This data was generated through metabolic profiling conducted by Nightingale Health’s NMR metabolomics platform, which analyzed over 118,000 participants from the UKB42. We employed GSMR43 v1.91 to investigate the potential causal relationship between cataracts and the mentioned phenotypes. This method only requires GWAS summary statistics to estimate MR effect sizes and accounts for correlated SNP instruments by modeling LD from a pre-specified reference panel. Additionally, it uses the HEIDI-outlier statistical test to look for heterogeneous SNP outliers. We applied specific parameters to select independent instrumental variants (--clump-r2 0.001, --gwas-thresh 5e-8, and --clump kb 1000 --heidi-thresh 0.01). To control for Type-1 Error, we applied the Bonferroni correction by setting the p value threshold to 0.05/(249 metabolites) = 2e-04. To avoid sample overlap with UKB participants, we excluded GERA-UKB meta-analysis and based the MR results on a meta-analysis of FinnGen and MGBB.
Polygenic risk scores (PRS)
PRS is a statistical method that adds the number of risk alleles a person carries weighted by their effect sizes to estimate an individual’s risk for developing a particular disease. PRS can be used to predict an individual’s risk for cataracts and can also help identify individuals who may benefit from early intervention. A brief description of each of the cohorts included in the PRS analysis is provided below.
The Raine Study is a prospective multigenerational observational study from Western Australia44,45. Between 1989 and 1991, 2900 women in the first trimester of their pregnancy were recruited from metropolitan Perth, Western Australia. A total of 2868 offspring (Gen2) were born to these women, and the birth cohort has been undergoing a series of health and medical examinations since before they were born. Comprehensive eye examinations were conducted at 20 and 28 years of age25,26. The Raine cohort consists of individuals under 30 years of age, which means they do not have age-related cataracts. Therefore, only estimates for UVR-related phenotypes (as explained in the section below) were assessed in this cohort.
Blood specimens were obtained from participants during the Gen2 14- and 17-year follow-up assessments. Of the 1592 participants, samples were analyzed in 2010 using the Infinium HD Human660W-Quad Beadchip Array while samples of an additional 310 participants were analyzed in 2013 using the Infinium OmniExpress-24 BeadChip Array.
The BHAS is a long-term, population-based study of 5107 adults born between 1946 and 1964 recruited from the City of Busselton, a coastal city in Western Australia and is focused on two examination periods conducted between 2010 and 201527 and between 2016 and 2022. The BHAS involves the collection of detailed data on various aspects of health and well-being, including physical, cognitive, and mental health, as well as lifestyle and environmental factors. All follow-ups of the Raine Study and phases of the BHAS have been approved by the University of Western Australia Human Research Ethics Committee and comply with the Declaration of Helsinki.
BHAS participants underwent blood sample collection and were genotyped using the Illumina Infinium Global Screening Array. Quality control measures were implemented to ensure data accuracy, including the exclusion of data with a single SNP call rate <0.95, Hardy-Weinberg equilibrium p value less than , and MAF less than 0.01. Population outliers were identified and excluded through a principal component analysis to maintain participants with known European ancestry using data from the 1000 Human Genome Project reference. The post-quality control data was then imputed against the TOPMed reference panel. SNPs with an imputation accuracy >0.3 and MAF > 0.01 were included in further analysis.
The PRS for cataracts was generated using PLINK 2.0. We selected independent SNPs using the following parameters: --clump-r2 0.05, --clump-p1 5e-8, and --clump kb 1000. We used a subset of the UK biobank that includes 5000 healthy individuals as a linkage disequilibrium reference for the clumping process. We used a generalized linear model to assess the correlation between the scores derived based on cataract genome-wide significant SNPs (p < 5e-8) and cataracts in an independent cohort (Busselton, N cases = 389, N controls = 4416). We also conducted a comparison of the PRS results with a PRS derived from the preceding cataracts meta-analysis4, employing the same parameters.
We further explored the association between the PRS of cataracts and two surrogate measurements of the UVR exposure; pterygium (Busselton N cases = 516, N controls = 4029), non-cancerous growth of the conjunctiva membrane that is highly associated with UVR exposure, and conjunctival ultraviolet autofluorescence (CUVAF; N BHAS = 4384, N the Raine Study Gen2 = 1847) a non-invasive and objective method of measuring the amount of UVR exposure at the bulbar conjunctiva46. Participants who have previously undergone pterygium removal surgery were also included in the analysis as cases25,47. Given the limited statistical power derived from the scarcity of genome-wide significant instruments for UVR exposure phenotypes, further analysis utilizing MR methods to confirm the causal relationship between UVR exposure and cataracts was not feasible.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Acknowledgements
The authors would like to acknowledge the Busselton Healthy Aging Study participants, the Raine Study participants, and their families for their ongoing participation in the study. The authors also thank the Raine Study team for study coordination and data collection, as well as the National Health & Medical Research Council (NHMRC; Australia) for their long-term contribution to funding the study over the last 30 years. The Pawsey Supercomputing Centre provided computation resources to carry out analyses required with funding from the Australian Government and the Government of Western Australia. L.M.G.M. is supported by a UQ Research Training Scholarship from The University of Queensland (UQ). M.E.R. thanks funding from the Rebecca L. Cooper Medical Research Foundation through an Al & Val Rosenstrauss Fellowship (F20231230). P.G. is supported by an NHMRC Investigator Grant (#1173390). D.A.M. is supported by an NHMRC Practitioner Fellowship. S.M. is supported by an NHMRC Senior Research Fellowship. The BHAS is supported by grants from the Government of Western Australia (Department of Jobs, Tourism, Science and Innovation and Department for Health), the Commonwealth Government (Department of Health), the City of Busselton and from private donations to the Busselton Population Medical Research Institute. In-kind support was received by the Western Australian Country Health Service and BD Biosciences. The core management of the Raine Study is funded by The University of Western Australia, Curtin University, Telethon Kids Institute, Women and Infants Research Foundation, Edith Cowan University, Murdoch University, The University of Notre Dame Australia, and the Raine Medical Research Foundation. Collection of DNA and GWAS data for Gen2 of the Raine Study was funded by grants from the NHMRC (Grant IDs 572613, 403981, 1059711) and the Canadian Institutes of Health Research (CIHR) (Grant ID MOP-82893). The eye data collection for Gen2 20- and 28-year follow-ups were funded by the NHMRC (Grant IDs 1021105 1126494, 1121979), CIHR, the Lions Eye Institute, the Australian Foundation for the Prevention of Blindness, Alcon Research Institute, Telethon Kids Institute, Australia Vision Research (formally the Ophthalmic Research Institute of Australia), and the Heart Foundation (Grant ID 102170).
Author contributions
P.G. and M.E.R. conceived the study, designed the analyses, and jointly supervised the study. S.D.T. carried out the analyses with support from S.S.Y.L. and A.I.C. S.D.T. wrote the manuscript. S.S.Y.L., G.L., and D.A.M. were involved in the collection of genetic and phenotypic data for the Raine study, while M.H. was involved in the collection of data for the Busselton Health study. S.D.T., S.S.Y.L., L.M.G.M., A.I.C., G.L., J.S.O., D.A.M., K.P.B., M.H., X.D., and S.M. advised on the study design and provided valuable input.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The meta-analysis data generated in this study have been deposited in the Zenodo database under accession code 13594388. Cataract and genotype data from the MGBB are available under restricted access for authorized researchers, as they represent sensitive patient data. Access can be obtained by applying through the following link: https://www.massgeneralbrigham.org/en/research-and-innovation/participate-in-research/biobank/for-researchers. Similarly, raw data from the Raine Study and the Busselton Study are protected by privacy laws and are only available to selected researchers. Applications can be submitted through the Raine Study [https://rainestudy.org.au/home-beta/information-for-researchers/available-data-sm/] and Busselton Study [https://bpmri.org.au/research/database-access.html] webpages. The GWAS data used in this study are available in the GWAS Catalog for the previous meta-analysis, which includes the UKB and GERA datasets, under accession code GCST90014268 [https://www.ebi.ac.uk/gwas/downloads/summary-statistics] and as part of release 8 of FinnGen [https://r8.finngen.fi/pheno/H7_CATARACTSENILE]. Supplementary data can be accessed on Figshare [10.6084/m9.figshare.22359862].
Code availability
The code used in this study is available upon request.
Competing interests
A.I.C. is currently employed by the Regeneron Genetics Center, a wholly-owned subsidiary of Regeneron Pharmaceuticals, Inc., and may own Regeneron stock or stock options. A.I.C. contributed to this work during his tenure at QIMR Berghofer Medical Research Institute and The University of Queensland. The other authors report no competing interests.
Ethics
This study adheres to the principles of the Declaration of Helsinki and follows the guidelines set forth by the International Ethical Guidelines for Biomedical Research Involving Human Subjects. We are committed to promoting diversity and inclusivity in all aspects of its design, methodology, and analysis. We have striven to ensure that the sample population represents different ancestries to enhance the generalizability and relevance of our findings. We also acknowledge the importance of diverse perspectives in the interpretation of our results and have actively sought input from researchers and clinicians from varied backgrounds.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Puya Gharahkhani, Miguel E. Rentería.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53212-6.
References
- 1.Steinmetz, J. D. et al. Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the Right to Sight: an analysis for the Global Burden of Disease Study. Lancet Glob. Health9, e144–e160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashemi, H. et al. Global and regional prevalence of age-related cataract: a comprehensive systematic review and meta-analysis. Eye34, 1357–1370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khoza, L. B. et al. Survey on prevalence of cataract in selected communities in Limpopo Province of South Africa. Sci. Afr.8, e00352 (2020). [Google Scholar]
- 4.Choquet, H. et al. A large multiethnic GWAS meta-analysis of cataract identifies new risk loci and sex-specific effects. Nat. Commun.12, 3595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyashita, H. et al. Association between ultraviolet radiation exposure dose and cataract in Han people living in China and Taiwan: a cross-sectional study. PLoS ONE14, e0215338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts, J. E. Ultraviolet radiation as a risk factor for cataract and macular degeneration. Eye Contact Lens37, 246–249 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Kiziltoprak, H., Tekin, K., Inanc, M. & Goker, Y. S. Cataract in diabetes mellitus. World J. Diabetes10, 140–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu, W.-L., Shen, P.-C., Lee, C.-H., Su, Y.-T. & Chen, L.-M. High risk of early cataracts in young type 1 diabetes group: a nationwide cohort study. Int. J. Endocrinol.2020, 8160256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, H. et al. Mendelian randomization study reveals a population-specific putative causal effect of type 2 diabetes in risk of cataract. Int. J. Epidemiol.50, 2024–2037 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Shin, S. et al. Qki activates Srebp2-mediated cholesterol biosynthesis for maintenance of eye lens transparency. Nat. Commun.12, 3005 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vejux, A., Samadi, M. & Lizard, G. Contribution of cholesterol and oxysterols in the physiopathology of cataract: implication for the development of pharmacological treatments. J. Ophthalmol.2011, 471947 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrenko, V. et al. Type 2 diabetes disrupts circadian orchestration of lipid metabolism and membrane fluidity in human pancreatic islets. PLoS Biol.20, e3001725 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji, J. et al. Type 2 diabetes is associated with suppression of autophagy and lipid accumulation in β-cells. J. Cell. Mol. Med.23, 2890–2900 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon, H., Shaw, J. L., Haigis, M. C. & Greka, A. Lipid metabolism in sickness and in health: emerging regulators of lipotoxicity. Mol. Cell81, 3708–3730 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, K. & Czaja, M. J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ.20, 3–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borchman, D. Lipid conformational order and the etiology of cataract and dry eye. J. Lipid Res.62, 100039 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montastruc, G. et al. Lipid-lowering drugs and the risk of cataract: an observational post marketing study. Arch. Cardiovasc. Dis. Suppl.12, 190 (2020). [Google Scholar]
- 18.Chu, C.-S., Lin, C.-H., Lan, T.-H. & Chou, P.-H. Associations between use of mood stabilizers and risk of cataract: a population-based nested case-control study. J. Affect. Disord.227, 79–81 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Ye, J. et al. Smoking and risk of age-related cataract: a meta-analysis. Invest. Ophthalmol. Vis. Sci.53, 3885–3895 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Fukai, K. et al. Alcohol use patterns and risk of incident cataract surgery: a large scale case–control study in Japan. Sci. Rep.12, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-Marín, L. M. et al. The pharmacogenomics of selective serotonin reuptake inhibitors. Pharmacogenomics23, 597–607 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Erie, J. C., Brue, S. M., Chamberlain, A. M. & Hodge, D. O. Selective serotonin reuptake inhibitor use and increased risk of cataract surgery: a population-based, case-control study. Am. J. Ophthalmol.158, 192–197.e1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rana, D., Sharma, R. & Kumar, A. Comparative potential of hydrocortisone, deoxycorticosterone and dexamethasone in the prevention of cataract: an in silico and in vitro study. Orient. Pharm. Exp. Med.18, 403–422 (2018). [Google Scholar]
- 24.Hanna, S. Optometry Australia Diabetes Guidelines Working Group Optometry Australia–guidelines on the examination and management of patients with diabetes. Clin. Exp. Optom.99, 120–126 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. S.-Y. et al. Rationale and protocol for the 7- and 8-year longitudinal assessments of eye health in a cohort of young adults in the Raine Study. BMJ Open10, e033440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yazar, S. et al. Raine eye health study: design, methodology and baseline prevalence of ophthalmic disease in a birth-cohort study of young adults. Ophthalmic Genet.34, 199–208 (2013). [DOI] [PubMed] [Google Scholar]
- 27.James, A. et al. Rationale, design and methods for a community-based study of clustering and cumulative effects of chronic disease processes and their effects on ageing: the Busselton healthy ageing study. BMC Public Health13, 936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boutin, N. T. et al. The evolution of a large biobank at Mass General Brigham. J. Pers. Med.12, 1323 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature613, 508–518 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet.47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creators Diaz Torres, S. Uncovering genetic loci and biological pathways associated with age-related cataracts through GWAS meta-analysis (scripts). 10.5281/zenodo.13624099 (2023). [DOI] [PMC free article] [PubMed]
- 33.Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun.8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol.11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd-Jones, L. R. et al. The genetic architecture of gene expression in peripheral blood. Am. J. Hum. Genet.100, 371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet.48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Freshour, S. L. et al. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res.49, D1144–D1151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature594, 398–402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol.44, 512–525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai, L. et al. Genome-wide association analysis of type 2 diabetes in the EPIC-InterAct study. Sci. Data7, 393 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Julkunen, H. et al. Atlas of plasma NMR biomarkers for health and disease in 118,461 individuals from the UK Biobank. Nat. Commun.14, 604 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun.9, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKnight, C. M. et al. Birth of a cohort–the first 20 years of the Raine study. Med. J. Aust.197, 608–610 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Dontje, M. L., Eastwood, P. & Straker, L. Western Australian pregnancy cohort (Raine) Study: generation 1. BMJ Open9, e026276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lingham, G. et al. Conjunctival ultraviolet autofluorescence area decreases with age and sunglasses use. Br. J. Ophthalmol. 10.1136/bjophthalmol-2021-320284 (2021). [DOI] [PubMed]
- 47.McKnight, C. M. et al. Pterygium and conjunctival ultraviolet autofluorescence in young Australian adults: the Raine study. Clin. Exp. Ophthalmol.43, 300–307 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
The meta-analysis data generated in this study have been deposited in the Zenodo database under accession code 13594388. Cataract and genotype data from the MGBB are available under restricted access for authorized researchers, as they represent sensitive patient data. Access can be obtained by applying through the following link: https://www.massgeneralbrigham.org/en/research-and-innovation/participate-in-research/biobank/for-researchers. Similarly, raw data from the Raine Study and the Busselton Study are protected by privacy laws and are only available to selected researchers. Applications can be submitted through the Raine Study [https://rainestudy.org.au/home-beta/information-for-researchers/available-data-sm/] and Busselton Study [https://bpmri.org.au/research/database-access.html] webpages. The GWAS data used in this study are available in the GWAS Catalog for the previous meta-analysis, which includes the UKB and GERA datasets, under accession code GCST90014268 [https://www.ebi.ac.uk/gwas/downloads/summary-statistics] and as part of release 8 of FinnGen [https://r8.finngen.fi/pheno/H7_CATARACTSENILE]. Supplementary data can be accessed on Figshare [10.6084/m9.figshare.22359862].
The code used in this study is available upon request.