Abstract
Lipid nanoparticles (LNPs) play a crucial role in addressing genetic disorders, and cancer, and combating pandemics such as COVID-19 and its variants. Yet, the ability of LNPs to effectively encapsulate large-size DNA molecules remains elusive. This is a significant limitation, as the successful delivery of large-size DNA holds immense potential for gene therapy. To address this gap, the present study focuses on the design of PEGylated LNPs, incorporating large-sized DNA, departing from traditional RNA and ionizable lipids. The resultant LNPs demonstrate a unique particle morphology. These particles were further engineered with a DNA coating and plasma proteins. This multicomponent bionanoconstruct exhibits enhanced transfection efficiency and safety in controlled laboratory settings and improved immune system evasion in in vivo tests. These findings provide valuable insights for the design and development of bionanoarchitectures for large-size DNA delivery, opening new avenues for transformative gene therapies.
Subject terms: Drug delivery, Drug delivery, Drug delivery
Encapsulation of large-size DNA molecules into lipid nanoparticles (LNPs) remains challenging. Here, the authors engineer PEGylated LNPs with DNA and plasma proteins to improve the delivery of large DNA molecules, enhancing the gene therapy potential.
Introduction
The advent of lipid nanoparticles (LNPs) has revolutionized the field of gene therapy, offering a promising avenue for delivering therapeutic nucleic acids to target cells1. They hold immense potential in correcting genetic defects2, modulating gene expression, and delivering therapeutic factors for various applications, including genetic disorders3, cancer4, infectious diseases5, and aging-related diseases6. Initially developed for the delivery of small interfering RNAs (siRNAs)3, LNPs have demonstrated remarkable success in clinical applications, most notably in the development of mRNA vaccines against COVID-19 such as mRNA-1273/SpikeVax developed by Moderna, and BNT162b2/Comirnaty from BioNTech/Pfizer. These breakthroughs have played a substantial role in advancing mRNA technology, ultimately leading to the Nobel Prize in Medicine in 2023 (https://www.nobelprize.org/prizes/medicine/2023/advanced-information/).
However, in contrast to the remarkable achievements in siRNA and mRNA delivery, the ability of LNPs to effectively encapsulate large-size DNA molecules is less explored, thus limiting the potentially transformative opportunities for gene therapy of a wide range of genetic disorders. In fact, by introducing functional genes or gene fragments into target cells, large-size DNA can correct genetic defects, restore protein function, and potentially cure diseases that are currently intractable with conventional therapies. As an instance, previous works demonstrated an effective design of DNA/LNPs for liver targeting in vivo7, and for vaccine development8. Additionally, large-size DNA delivery opens doors to gene editing strategies, enabling the precise modification of the human genome to treat genetic diseases and combat cancer.
At the very bottom of this bottleneck, the typical size of plasmid DNAs used in molecular biology experiments (i.e. a few kilobases) poses unique challenges to efficient encapsulation and delivery compared to smaller nucleic acids such as siRNAs and mRNAs9. Its larger size and complex structure, particularly the presence of supercoiled DNA10, necessitate the development of specialized LNP formulations11–13 that can efficiently encapsulate and protect the DNA cargo while maintaining suitable physical-chemical properties and facilitating its entry into target cells.
In this study, we design PEGylated LNPs by incorporating plasmid DNA and cationic lipids, breaking away from traditional RNA and ionizable lipids14,15. Our aim was to create a robust LNP platform for large DNA delivery, overcoming associated challenges in this therapeutic approach. Using microfluidics, we encapsulated large DNA within cationic PEGylated LNPs, resulting in unique particle morphology with alternating lipid bilayers and DNA monolayers. Further engineering involved additional DNA coating and external envelopment by plasma proteins. This strategy was based on prior findings demonstrating the peculiar ability of DNA to reverse the charge of cationic lipids16 and facilitate the adsorption of an opsonin-deficient protein corona17.
The resulting coronated LNPs outperformed PEGylated LNPs in terms of stealth properties in vivo. Throughout our research, we employed both PEGylated LNPs and DNA-decorated PEGylated LNPs as control systems. In a restricted set of experiments, we also synthesized plain (i.e., unPEGylated) LNPs. This systematic approach allowed us to identify the distinct roles of each structural component in the particle structure across in vitro, ex vivo, and in vivo settings.
Overall, our results suggest that the stealth property exhibited by coronated LNPs is not only attributable to PEG18 or the protein corona19–21. Instead, it is likely conferred by a delicate interplay between PEG, surface-bound DNA, and plasma proteins which plays a crucial role in diminishing their attractiveness to immune cells, thereby reducing capture in the bloodstream. The broader implications of these findings within are thoroughly explored, and potential avenues for further research are delineated.
Results and discussion
Synthesis and physical-chemical characterization of LNPs
In this study, we have developed a multifaceted PEGylated LNP system for the delivery of plasmid DNA (hereafter simply indicated as DNA)22. The lipid composition was chosen through a rigorous multi-step screening process, as detailed in ref. 16, to facilitate the efficient escape of DNA from endosomes. This formulation comprises 50% cationic lipids, primarily aimed at efficiently encapsulating DNA, along with 48.5% zwitterionic helper lipids to boost the structural stability of the LNPs. Additionally, 1.5% PEGylated lipids have been incorporated to enhance the overall stability of the particles. Clearly, LNPs in this study lack ionizable lipids, a component that has gained significance in LNPs for its pH-sensitive behavior23. Ionizable lipids are positively charged under acidic pH conditions, facilitating the compacting of nucleic acids. Post-administration, they transition to a neutral charge, minimizing protein binding. Our rationale for excluding ionizable lipids is linked to the difficulty in controlling their protonation within acidic endosomes. In these compartments, they need to become positively charged to destabilize the membrane for the release of nucleic acids into the cytoplasm. However, achieving reliable protonation and deprotonation within cells poses challenges, potentially impacting endosomal escape and overall delivery efficiency. Furthermore, a comparison between our cationic LNP and a formulation including ionizable lipids (i.e., the formulation used for the Pfizer/BioNTech COVID-19 vaccine) revealed the superiority of our system, both plain and DNA-decorated as reported in Supplementary Fig. 1 in the Supplementary Information (SI). This suggests that the strategy adopted in this work is not well adaptable to LNPs incorporating ionizable lipids, likely due to the neutral/negative charge of these LNPs in the absence of acid pH, which represents a strength of these formulations showing reduced toxicity in the in vivo applications.
Thorough physical-chemical characterization employing dynamic light scattering (DLS) and microelectrophoresis reveals the monodispersity of the LNP formulation, substantiated by a low polydispersity index (PdI) of 0.16. These LNPs exhibit an average size of approximately 180 nm and possess a positively charged zeta potential of ~ 15 mV. This formulation served as the foundational reference point for the development of additional systems, specifically LNPs coated with further DNA. In accordance with prior literature17, it is established that DNA coating plays a structural role by facilitating the binding of an opsonin-deficient protein corona, thereby enabling effective immune evasion.
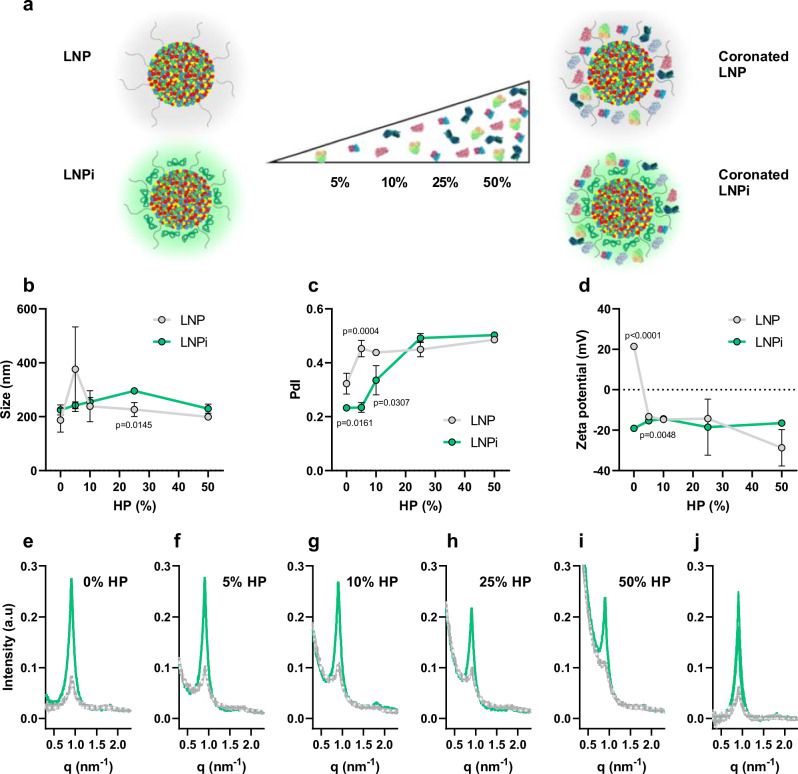
To generate DNA-decorated LNPs, the initial formulation was exposed to DNA solutions at different concentrations as schematically shown in Fig. 1a: 0.1 mg/ml, 0.2 mg/ml, and 0.3 mg/ml. These varying DNA concentrations result in LNPs with distinct DNA coating levels, categorized as low (l), intermediate (i), and high (h) which are hereafter referred to as LNPl, LNPi, and LNPh. Similar to the uncoated systems, the DNA-decorated LNPs were subjected to size and zeta potential analysis. The results are detailed in Table 1 and summarized in Fig. 1b–d. Elevating the DNA coating results in a reduction in zeta potential, with only minimal changes in size. These results affirm that LNPs can be coated with adjustable quantities of DNA to reverse the zeta potential without impacting their size. LNPl exhibits an almost neutral zeta potential (i.e., −7 ± 4). Worth noting, for LNPi and LNPh, the DNA coating significantly alters the charge from positive to negative, suggesting complete coverage of the particle surface. The PdI remains low for LNPl and LNPi, with a slight increase noted for LNPh (PdI = 0.26). The slight increase in size from 180 nm to 190 nm upon DNA addition can be attributed to the surface decoration of LNP by the DNA. The additional layer of DNA around the nanoparticles contributes to the overall increase in diameter without significantly altering the size distribution, which explains why the PdI remains nearly unchanged.
Fig. 1. Generation of DNA-decorated LNPs.
a To create DNA-decorated LNPs, the original formulation was incubated with DNA solutions at varying concentrations: 0.1 mg/ml, 0.2 mg/ml, and 0.3 mg/ml. These different levels of coating DNA in the LNPs were classified as low (l), intermediate (i), and high (h), leading to the formation of DNA-decorated LNPs referred to as LNPl, LNPi, and LNPh, respectively. Physical-chemical characterization of pristine LNPs and DNA-decorated LNPs, including (b) size, (c) polydispersity index (PdI), and (d) zeta potential. Data are presented as mean values ± standard deviation (n = 3 repeated measurements). Statistical analysis was performed by one-way ANOVA test, followed by Tukey’s multiple-comparison test. P-values < 0.05 are reported. Source data are provided as a Source Data file.
Table 1.
Size, polydispersity index (PdI), Zeta potential and Zeta deviation of LNPs and DNA-decorated LNPs (n = 3 repeated measurements)
| Size (nm) | PdI | Zeta potential (mV) | Zeta deviation (mV) | |
|---|---|---|---|---|
| LNP | 178 ± 5 | 0.161 ± 0.013 | 15.4 ± 0.2 | 5.2 ± 0.4 |
| LNPl | 192 ± 5 | 0,147 ± 0,014 | −7.4 ± 3.8 | 8.7 ± 2.3 |
| LNPi | 189 ± 8 | 0.151 ± 0.028 | −23.4 ± 1.1 | 6.1 ± 0.3 |
| LNPh | 178 ± 6 | 0.257 ± 0.010 | −30.5 ± 0.9 | 7.8 ± 0.2 |
A relevant question concerns whether the DNA, added to the bulk solution, penetrates into the preformed LNP, potentially altering its internal structure, as suggested by previous evidence by some of us on lipoplexes24. To address this question, we conducted experiments using synchrotron small-angle X-ray scattering (SAXS). In Fig. 2a–d, we present the SAXS patterns of both LNPs and DNA-decorated LNPs. The data on LNPs align with prior research25, indicating that DNA-loaded LNPs consist of internally unoriented scattering domains, resulting in a broad first-order Bragg peak and a nearly indistinguishable second-order Bragg peak.
Fig. 2. Structure and morphology of LNP and LNPi.
Synchrotron SAXS curves of (a) LNP, (b) LNPl, (c) LNPi, and (d) LNPh (n = 10 repeated measurements). e Baseline-subtracted synchrotron SAXS curves of all the investigated samples, and corresponding structural parameters, i.e. (f) d-spacing, (g) domain size, and (h) number of repeating units (n = 10 repeated measurements). i Representative agarose gel image and (j) corresponding densitometric analysis. k–m Additional DNA in LNPs as measured by synchrotron SAXS n = 10 repeated measurements), electrophoresis (n = 4 repeated measurements), and fluorescence (n = 3 repeated measurements). n Representative TEM images of LNP. o Representative TEM images of LNPi. The frames of the TEM images correspond to the color code used throughout the manuscript to denote LNP (gray) and LNPi (light green). Data are presented as mean values ± standard deviation. Statistical analysis was performed by one-way ANOVA test, followed by Tukey’s multiple-comparison test. P-values < 0.05 are reported. Source data are provided as a Source Data file.
Of note, when DNA is added to preformed LNPs, the first-order Bragg peak becomes more pronounced, in a dose-dependent manner (Fig. 2e), confirming that exposure of LNPs to DNA leads not only to particle-surface coating but also to the penetration of DNA into the particle core. The LNP nanostructure maintains an unchanged lamellar d-spacing of approximately 6.98 nm but encompasses larger coherently scattering regions Fig. 2f. Analysis of the first Bragg peak’s half-width indicates that these scattering regions are composed of repeating units, specifically n = 4 for LNPs and 5 < n < 6 for DNA-decorated LNPs (Fig. 2h). DNA penetration within preformed LNPs facilitates in principle the delivery of a larger quantity of DNA copies from a single LNP. This is confirmed by electrophoretic measurements and fluorescence assays (Fig. 2i–m) which indicate that a specific amount of DNA adsorbed or entered LNPs. Of note, the loading efficiency of DNA in LNP systems remains consistent regardless of the buffer used (PBS, TE buffer, water, and HEPES), as demonstrated in Supplementary Fig. 2 in the SI. This consistency across different buffers reinforces the reliability of our approach and provides confidence in the reproducibility of our results under varying experimental conditions.
Taken together, the experimental tests conducted on LNP formulations (i.e., SAXS, agarose gel electrophoresis, and fluorescent DNA labeling) suggest the intermediate-coating condition as the most favorable one, as it results in systems with small size, positive surface charge, and minimal amounts of unbound DNA. Consequently, we chose LNPi for further investigation based on these findings.
To gain a deeper understanding of the structure of LNPs and LNPi, we conducted transmission electron microscopy (TEM) experiments. The representative images presented in Fig. 2n, o and Supplementary Fig. 3 in the SI illustrate the variety of morphologies observed in the measurement samples. LNPs predominantly appear as individual particles with sizes ranging between 50 nm and 100 nm, characterized by a multilamellar nanostructure closely resembling that of lipoplexes. These observations let us conclude that this structure is presumed to consist of lipid bilayers alternating with aqueous channels occupied by DNA26,27. We posit that the multilamellar structure of LNPs is induced by the size of the DNA, preventing the formation of a core-shell structure commonly reported in the literature for LNPs loaded with siRNA28,29. In addition, some multi-lamellar structures were also observed (Supplementary Fig. 3c).
We observe that LNPs appeared smaller in TEM images compared to DLS measurements. This discrepancy may be attributed to sample shrinkage during TEM preparation, which involves drying, and the sensitivity of DLS to larger and non-spherical particles, skewing the average size toward larger values30. However, the number distribution from DLS showed slightly lower values compared to the intensity distribution, aligning more closely with TEM results (Supplementary Fig. 4). Consistently, nanoparticle tracking analysis (NTA) revealed a lower mean size (Supplementary Table 1), with size distribution closer to TEM observations (mode around 70 nm, average values within 100-130 nm), as shown in Supplementary Fig. 5.
The presence of both individual particles (Supplementary Fig. 3a, b) and multi-lamellar structures (Supplementary Fig. 3c) in the TEM images suggests that LNPs have a heterogeneous morphology. This heterogeneity is maintained even after DNA decoration. Indeed, TEM clarifies that LNPi comprises both single particles (Supplementary Fig. 3d) and particles made of distinct subunits (Fig. 2o and Supplementary Fig. 3e, f). The appearance of polylobate particles in LNPi can be attributed to several factors. One potential explanation is the decoration of the multilamellar structure found in LNPs. Alternatively, DNA molecules can adhere to the surface of individual LNPs, causing them to form composite particles made up of distinct subunits. This phenomenon is supported by prior literature31, which suggests that DNA can act as a bridging molecule, linking some LNPs together or inducing shape changes that result in multi-lobe appearances. The fact that the PdI remains unchanged suggests that, despite possible structural changes and aggregation, the overall size distribution of the LNPs does not vary significantly. This implies that the DNA binds uniformly across the LNP population, maintaining a relatively homogeneous sample in terms of particle size.
In summary, combined results from DLS, synchrotron SAXS, and TEM teach us that the additional DNA partly enters preformed particles, effectively increasing the amount of DNA transported by each individual particle32. Simultaneously, it promotes association of individual particles, generating multipart particles with size below 200 nm, which is an accepted size threshold to avoid massive clearance by immune system cells33,34.
Elucidating the role of surface-bound DNA in LNP-mediated gene delivery
At this point, to evaluate the influence of DNA decoration on the transfection efficiency (TE) of LNPs, a series of experiments were executed utilizing the HEK-293 cellular model. HEK-293, composed of immortalized human embryonic kidney cells, represents one of the most extensively employed cell lines in research. To this end, we developed a variant of LNPi wherein the internal DNA was non-functional, i.e. it lacked the ability to code for a protein, as illustrated in Fig. 3a and hereafter indicated as LNPi nf-DNA.
Fig. 3. The influence of surface-bound DNA on transfection efficiency (TE) and cell viability.
a Schematic representation of LNPs loaded with functional DNA (f-DNA) in green, and non-functional DNA (nf-DNA) in yellow. These two versions of LNPs were respectively decorated with f-DNA and nf-DNA, resulting in the formation of LNPi and LNPi nf-DNA. b TE was assessed in HEK-293 cells through the luciferase assay, quantified as relative light units (RLU) per milligram of proteins (n = 6 independent measurements), and (c) through the green fluorescence protein assay (n = 2 independent measurements), measured as mean fluorescence intensity (MFI). d Cell viability data are presented as a percentage of viable cells (n = 6 independent measurements). Statistical analysis was performed by one-way ANOVA test, followed by Tukey’s multiple-comparison test. P-values < 0.05 are reported. Color code: LNP (gray); LNPi (green); LNPi nf-DNA (yellow). Data are presented as mean values ± standard deviation. Source data are provided as a Source Data file.
Preliminary assessments validated the comparability of the physical-chemical characteristics between LNPi and LNPi nf-DNA as detailed in Supplementary Table 2 of the SI. This parity in physical-chemical attributes enabled a direct comparison of the impact of external DNA on TE, effectively minimizing the influence of potential confounding factors.
To extend the applicability of our findings, we employed two distinct transfection assays: the luciferase assay and the green fluorescent protein (GFP) assay. The former provides data on the overall quantity of protein generated through gene expression in the cellular population (Fig. 3b), whereas the latter gives information about the fluorescence intensity from each single cell successfully transfected (Fig. 3c). Noteworthy, Luciferase assays show that the TE of LNPi surpasses that of LNP. This implies that the inherently negative charge of LNPi (which conventionally impedes interactions with negatively charged cell membranes) does not inhibit TE. Results from LNPi nf-DNA also suggest that external DNA is effectively involved in the cell transfection. The mean fluorescence intensity (MFI) of cells due to GFP expression follows a similar trend to that of Luciferase, albeit without statistical significance. As depicted in Fig. 3d, it is evident that this treatment exhibited no detrimental impact on the cell viability of the HEK-293 cells utilized in the experiment.
Having established that surface-bound DNA actively participates in the transfection process, we exclusively employed LNPi in our subsequent experiments. In a subsequent investigation, we explored whether the preparation of DNA-decorated LNPs using either bulk mixing or microfluidic mixing could influence the synthetic identity and functionality of LNPi. The results, as depicted in Supplementary Fig. 6 in the SI, reveal that the method of decoration employed does not have a significant impact on size, zeta potential, TE, and cell viability. Consequently, in the subsequent discussions, LNPi were prepared using bulk mixing.
Generation and physical-chemical characterization of coronated LNPi
Following the introduction of the protein corona paradigm by Dawson et al. 35, subsequent investigations have clarified that, within a physiological milieu, NPs undergo the acquisition of a protein corona. This corona alters their synthetic identity, thereby fostering the emergence of an entirely different biological identity20,36,37. A growing body of evidence indicates that the protein corona is instrumental in governing the recognition of nanoparticles by cells19, thereby exerting pivotal roles in modulating both the mobility and toxicity of the NPs. Equipping nanoparticles with an artificial protein corona ensures stability in blood and other bodily fluids, in contrast to bare NPs that tend to undergo corona formation upon exposure. This stability is crucial as it enables controlled interactions with cells, emphasizing the capacity of the artificial protein corona to maintain its integrity and functionality throughout circulation, facilitating targeted and controlled cellular engagement.
Supported by a body of prior research38–40, we endeavored LNPi with an artificial protein corona. We investigated the formation of coronated LNPi by exposing LNPi to plasma proteins as a function of human plasma (HP) concentration: the rationale behind this strategy lies in the fact that, even with functionalized NPs, the outermost layer is still susceptible to some degree of plasma protein adsorption leading to undesired masking of targeting ligands41,42. In contrast, an optimized artificial corona in HP helps in achieving the desired targeting or stealth properties43,44. To commence, LNP and LNPi were subjected to a 1-hour exposure to HP at varying concentrations (5%, 10%, 25%, and 50% vol/vol) to allow the formation of artificial protein coronas (schematically displayed in Fig. 4a) and then examined for attributes including size, PdI, zeta potential, and nanostructure.
Fig. 4. Physical-chemical characterization of coronated LNP and LNPi.
a Scheme of formation of coronated LNP and LNPi and corresponding characterization in terms of (b) size, (c) PdI, and (d) zeta potential as functions of HP concentration (% vol) (n = 3 repeated measurements). Statistical significance is evaluated by unpaired two-tailed Student’s t-test at each HP concentration. P-values < 0.05 are reported. Data are presented as mean values ± standard deviation. e–i SAXS pattern of DNA coated and plain LNPs incubated 1 h with increasing concentrations of HP (n = 10 repeated measurements). j superimposed SAXS pattern of all the particles after baseline-subtraction. Source data are provided as a Source Data file.
For LNP, an increase in particle size is observed at 5% HP, followed by a reduction, which ultimately stabilizes at values ranging from 180 to 220 nm (Fig. 4b). Although size differences at 5%HP are not statistically significant due to large experimental errors, the adsorption of plasma proteins induces a notable increase in the PdI (Fig. 4c), indicative of a certain degree of aggregation of coronated LNP that affects the uniformity of particle dispersion. By contrast, the size of coronated LNPi exhibits minimal alteration due to protein adsorption. However, PdI analysis reveals that homogeneity is maintained up to an HP concentration of 10%. Of note, examination of zeta potential trends (Fig. 4d) reveals that, irrespective of the initial surface charge of the system (positive for LNP, negative for LNPi), exposure to HP leads to a convergence towards a common negative zeta potential value, at approximately -20 mV, in keeping with previous literature37. Synchrotron SAXS experiments, as presented in Fig. 4e–i, do not display discernible alterations in the position and width of the first-order Bragg peak as a function of HP concentration in LNP as compared to LNPi, suggesting that the internal nanostructure of both the systems remain unaffected by the presence of plasma proteins on the particle surface. Such a conclusion is strongly supported by the graph in Fig. 4j, where all 10 baseline subtracted SAXS curves are shown, i.e. those corresponding to two systems (i.e., LNP and LNPi) under 5 incubation conditions. The distinctive stability observed in LNP and LNPi is likely attributable to the presence of DNA in the particle interior. Previous studies have indicated that the interaction of plasma proteins with empty cationic liposomes can impact the internal structure of the nanoparticle45.
Interpretation of SAXS results is supported by TEM images reported in Supplementary Fig. 3 in the SI.
LNP-protein corona
The integrated findings from particle size analysis, zeta potential measurements, and synchrotron SAXS collectively support a model wherein a protein corona forms around LNPs and LNPi without inducing internal structural changes. To investigate the evolving protein corona around LNP and LNPi in HP, we conducted 1D sodium dodecyl sulfate - polyacrylamide gel electrophoresis (SDS-PAGE) and bicinchoninic acid (BCA) assays, employing plain LNPs (unPEGylated) as a control. The tripartite formulation approach involving plain LNPs composed solely of lipids, LNPs comprising lipids and PEG, and LNPi featuring lipids, PEG, and DNA serves as a strategic design for discerning the distinctive contributions of each molecular species to the protein corona. This comprehensive framework facilitates a nuanced understanding of the intricate interplay among lipids, PEG, and DNA in shaping the evolving protein corona dynamics.
Representative 1D SDS-PAGE gel results in Supplementary Fig. 7a reveal that, across plasma concentrations from 5 to 50%, band intensities follow the order plain LNP » LNP ~ LNPi. This hierarchy is substantiated by the BCA assay (Supplementary Fig. 7b), emphasizing the established role of PEG in shielding LNPs against protein adsorption46.
In Fig. 5a–d, a comparison of 1D protein profiles across LNP and LNPi at varying plasma concentrations reveals distinctive coronas at each concentration. Calculating correlations between coronas (Fig. 5e) highlights the influence of DNA chains on the particle surface, altering the protein corona composition. Densitometry analysis (Fig. 5f, g) within eight major molecular weight intervals, despite gel separation limitations, offers a semiquantitative depiction of band intensity variations observed in Supplementary Fig. 7.
Fig. 5. Protein coronas of LNP and LNPi.
The coating of LNP and LNPi was characterized by densitometry analysis of one-dimensional (1D) SDS-PAGE images (n = 4 independent measurements, a representative gel image is reported in Supplementary Fig. 7 in the SI). Normalized 1D molecular weight distributions of coronated LNP and LNPi following exposure to HP = 5% (a), 10% (b), 25% (c), and 50% (d). Normalization is achieved by dividing the lane intensity profile by the integral area of the profile itself. At the highest HP concentration, acknowledged in the scientific literature for mimicking the protein excess found in physiological environments55, the coronas of LNP and LNPi predominantly differ in three molecular weight regions, as indicated by red asterisks (e) Heat map of the correlations between coronas of LNP and LNPi. Relative amounts of the most abundant proteins adsorbed in the corona of LNP (f) and LNPi (g) from plasma solutions at different concentrations (legend) after 1 h of incubation. The results are extracted as the relative intensities from four independent SDS-PAGE experiments. The asterisks in panels f and g refer to those shown in panel d. Data are presented as mean values ± standard deviation. Statistical analysis was performed by one-way ANOVA test, followed by Tukey’s multiple-comparison test. P-values < 0.05 are reported. Source data are provided as a Source Data file.
Under conditions mimicking in vivo scenarios (HP = 50%), noteworthy disparities in protein abundance between LNP and LNPi are prominent within three key molecular weight intervals. Specifically, LNPi manifests higher abundance in the 20–35 kDa and 45–58 kDa intervals, while LNP exhibits elevated abundance in the 58–75 kDa range.
Corona comparisons of LNPs and LNPi with plain LNPs reveal the highest correlation at all plasma concentrations between plain LNPs and LNPs (Supplementary Fig. 8a). This underscores the pivotal role of the shared lipid surface in shaping the protein corona, aligning with literature emphasizing lipid composition as the foremost determinant47. Conversely, lower correlations are observed between LNPi and the other two formulations, decreasing with increasing plasma concentration. This emphasizes again the substantial impact of DNA, unique to LNPi, on protein corona adsorption. We also observe that the correlation degree between the coronae of plain LNPs is significantly influenced by plasma concentration, resembling behavior found in unPEGylated cationic liposomes48. This is explained by the delicate balance in corona composition between low-affinity (LA) proteins, decreasing with increasing plasma concentration, and competitive-binding (CB) proteins, increasing with plasma concentration. In contrast, correlations clarify that LNPi coronas reach equilibrium rapidly, suggesting prompt attainment of a stable protein balance regardless of plasma concentration. LNPs exhibit an intermediate behavior.
In conclusion, investigations into protein corona formation suggest a unique protein profile for LNPi, attributable to the presence of negatively charged DNA chains on its surface. These DNA chains alter the electrostatic interactions between LNPi and plasma proteins, resulting in a distinct set of proteins adsorbed to the particle surface. Compared to LNPs, LNPi coronas reach equilibrium at lower values of protein concentration and remain stable at increasing HP percentage. This indicates that LNPi may exhibit enhanced stability in human plasma compared to LNPs, a characteristic that could improve their efficacy as drug delivery vehicles.
Understanding LNP-cell interactions
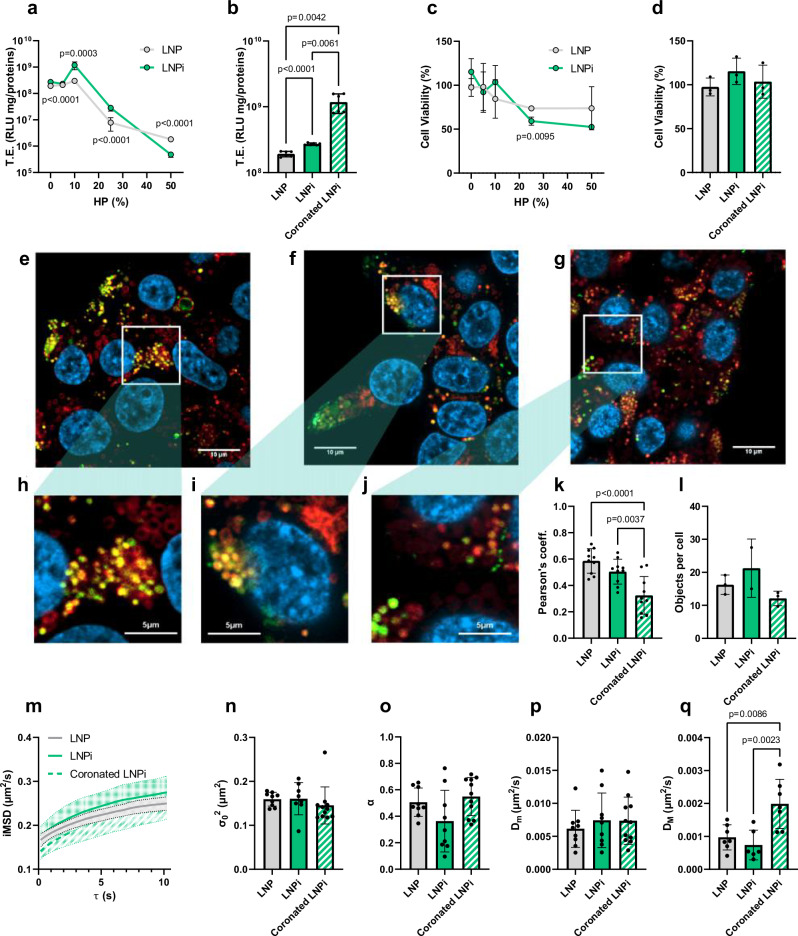
To assess the influence of protein corona on TE and cell viability, HEK-293 cells were exposed to coronated LNP and LNPi across varying percentages of HP (Fig. 6a–d). Generally, the results, as depicted in Fig. 6a–d, indicate that, regardless of HP concentration, the TE of LNPi consistently matched or exceeded that of their corresponding LNP counterparts.
Fig. 6. Investigation of LNP-cell interactions.
a TE as a function of HP concentration (% vol), (b) TE of selected conditions (i.e. both particles at 0%HP and the one exhibiting highest TE in presence of HP, n = 6 independent measurements), (c) cell viability as function of HP concentration (% vol), and (d) cell viability of selected conditions (i.e. both particles at 0%HP and the one exhibiting highest TE in presence of HP, n = 3 independent measurements). In Panels a, c, statistical significance is evaluated by two-sided Student’s t-test at each HP concentration. Statistical analysis in Panels b, d was performed by one-way ANOVA, followed by Tukey’s multiple-comparison test. P-values < 0.05 are reported. Confocal images of (e) LNP, (f) LNPi and, (g) coronated LNPi after administration to HEK-293 cells at 37 °C. Green and red channels indicate lipids, and lysosomes respectively, nuclei are stained in blue. Magnifications are displayed in (h–j). k Quantification of colocalization in terms of the Pearson’s coefficient. l number of particles per cell. m iMSD vs. time lag curves. n Offset parameter, σ02, (o) anomalous diffusion coefficient (α), (p) short-range diffusion constant (Dm), (q) long-range diffusion constant (DM). Parameters reported in panels n-q are determined through iMSD plot fitting. The datasets were made of n = 10 images per class. Statistical analysis was performed by one-way ANOVA test, followed by Tukey’s multiple-comparison test. Data are presented as mean values ± standard deviation. P-values < 0.05 are reported. Source data are provided as a Source Data file.
In particular, a significant enhancement in TE was for a HP concentration of 10%, followed by a notable decline at higher HP percentages. Furthermore, the presence of DNA did not exacerbate the cytotoxicity of LNPs at 5% and 10% HP. However, at elevated percentages (25% and 50% HP), the viability of HEK-293 cells exhibited a steady decrease, with no discernible disparities in toxicity levels observed between LNP and LNPi at the same HP concentration (Fig. 6c, d). In summary, our results demonstrate that subjecting LNPi to 10% HP (hereafter simply indicated as coronated LNPi) yields a protein-coated variant of LNPi with favorable attributes, including optimal size, negative zeta potential, and preserved nanostructure. This system displayed enhanced TE in HEK-293 cells without discernible effects on cell viability. This result contradicts prior findings49,50, which suggested that the NP-protein corona may hinder TE by directing NPs toward lysosomal degradation. To address this discrepancy, we conducted a detailed mechanistic investigation of coronated LNPi at the cellular level in the subsequent phases of the study.
To gain further insights into the cellular interactions of LNPs we performed fluorescence confocal microscopy experiments by treating Hek-293 cells with fluorescently labeled coronated LNPi. LNP and LNPi were used as a control to discern the role of PEG and DNA on particle-cell interactions.
First, we investigated the mechanism of entry of NPs into HEK-293 cells by a fluorescence-based spatiotemporal fluctuation analysis technique, allowing for direct extraction of dynamic characteristics from standard confocal imaging51–53. This information is presented graphically as mean square displacement (iMSD) versus time-delay plots (details are given in Supplementary Fig. 9 in SI). Parameters such as local diffusivity (Dm), long-range diffusion constant (DM), anomalous diffusion coefficient (α), and offset parameter (σ02) are determined through iMSD plot fitting, then visualized in a 3D space. By assessing multiple cells, the clustering of experimental points unveils the “dynamic fingerprint” of the structure at a whole-cell-population scale. In exploring the role of endocytosis in cellular uptake, we identified fingerprints of key endocytic vesicles, including caveolae, clathrin-coated vesicles, and macropinosomes (Supplementary Fig. 9b in the SI). Comparing the 3D distributions of these vesicles with those of LNP, LNPi, and coronated LNPi (Supplementary Fig. 9c), we observed consistent mechanisms of cellular internalization. Across all formulations, caveolae and clathrin-mediated endocytosis emerged as the dominant pathways, indicating minimal alteration in cellular entry modes among the three formulations.
As escaping the endosome poses the primary challenge to efficient transfection54, we conducted a detailed analysis of the extent of colocalization between LNP, LNPi, and coronated LNPi with lysosomes within HEK-293 cells. The representative confocal images (Fig. 6e–g) illustrate this process, where the blue signal corresponds to cell nuclei, the green signal corresponds to fluorescently labeled particles, the red signal indicates lysosomal structure.The overlapping yellow pixels show regions of green-red colocalization. The distinct presence of these spot-like structures serves as compelling evidence of lysosomal accumulation. Notably, we observed that a significant portion of intact, fluorescently labeled LNP and LNPi exhibit substantial colocalization with lysosomes (Fig. 6h, i), while the degree of colocalization between coronated LNPi and lysosomes was lower (Fig. 6j). The visual assessment of cells was further supported by Pearson’s coefficient, as depicted in Fig. 6k. This coefficient quantifies the overlap between green and red pixels, providing an estimate of the proportion of LNPs directed towards lysosomes. This difference in the degree of colocalization cannot be explained by a different number of internalized particles per cell, which was found to be not significantly different for the three LNP formulations (Fig. 6l). This observation suggests a potentially reduced propensity for lysosomal sequestration in the case of coronated LNPi.
Fitting iMSD plots (Fig. 6m) allowed us to compare LNP, LNPi and coronated LNPi in terms of offset parameter (Fig. 6n), anomalous diffusion coefficient (Fig. 6o), local diffusivity (Fig. 6p), and long-range diffusion constant (Fig. 6q). The only statistically significant difference concerns DM. This result suggests that coronated LNPi can explore a larger region in the intracellular space. This is consistent with a reduced confinement of coronated LNPi within lysosomes and is therefore in good agreement with the confocal microscopy results presented in Fig. 6, panels e-l.
Collectively, the observed variation in lysosomal colocalization among nanoparticle formulations that employ the same endocytic mechanism could not be correlated to the physical-chemical properties of nanoparticles that control TE such as size, surface charge, and nanostructure. This observation suggests that this discrepancy may be attributed to differences in the surface properties of the nanoparticles50. Our hypothesis suggests that the artificial protein corona adorning the coronated LNPi could function as a protective shield, inhibiting premature lysosomal trafficking. Such surface alterations could potentially enhance the efficacy of gene delivery systems by prolonging their presence and diffusion within the cytoplasm, thus increasing the likelihood of successful genetic material transfer. To validate this hypothesis, understanding the composition of the protein corona is crucial. Having this information would also be valuable in understanding how immune cells capture the particles in animal models. This was thoroughly investigated in the subsequent section of our study.
The protein corona of coronated LNPi in biologically relevant conditions
All the efforts made up to this point in the work have allowed us to unequivocally identify an ideal candidate for the delivery of large-size DNA in vivo. This candidate is coronated LNPi obtained by exposing LNPi to HP = 10%. This candidate exhibits the best combination of physical-chemical properties (i.e. small size and PdI), high TE (i.e. about 109 RLU/mg proteins) and nanosafety (i.e. no cytotoxicity is detected). These properties arise from the combined presence of DNA and the artificial protein corona with which LNPi is equipped. However, it is known that the protein corona can evolve in an excess of proteins.
Therefore, to exploit artificial protein coronas in controlled laboratory settings and animal studies, it is imperative to ascertain its stability upon exposure to elevated plasma concentrations55. This was evaluated by investigating the dynamic changes of coronated LNPi using 1D SDS-PAGE, with LNP employed as a control. As illustrated in Supplementary Fig. 10 in the SI, the results reveal a notable alteration in the protein corona of LNP in response to varying plasma concentrations, whereas the biocoronated LNPi maintains stability under biologically relevant conditions at HP = 50%. These findings corroborate the preceding sections, emphasizing the distinctive ability of DNA to facilitate prompt corona adsorption, which remains resilient upon introduction into a protein-enriched milieu.
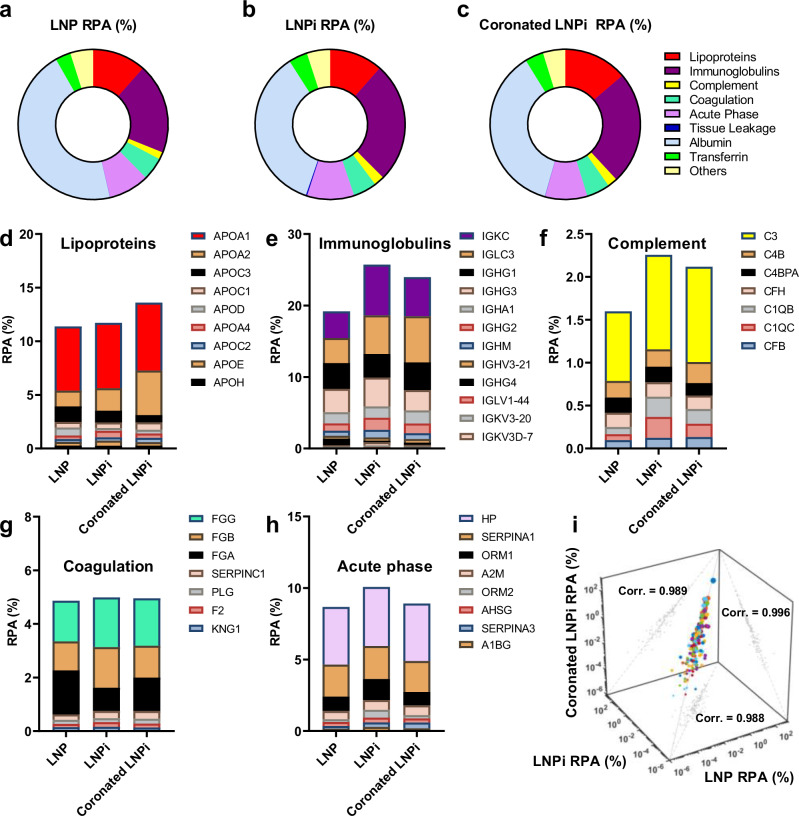
Subsequently, the coronas formed by LNP, LNPi, and coronated LNPi when exposed to HP = 50% were characterized using liquid chromatography-tandem mass spectrometry (LC-MS/MS). The full list of identified proteins is documented in Supplementary Table 3 in the SI. Figure 7a–c categorizes plasma proteins based on their biological functions56. In Fig. 7d–h we display the most abundant proteins for the five most represented protein classes: lipoproteins, immunoglobulins, complement proteins, coagulation proteins, and acute phase proteins.
Fig. 7. Correlation analysis of protein corona composition under physiologically relevant conditions.
Relative protein abundance (RPA) of protein classes in the protein corona (n = 3 repeated measurements) of (a) LNP, (b) LNPi, and (c) coronated LNPi following exposure to HP (50% vol)). RPA of the five most represented protein classes: (d) lipoproteins, (e) immunoglobulins, (f) complement proteins, (g) coagulation proteins, (h) acute phase proteins. i Scatterplot and correlation analysis of the RPAs in the coronas of the three investigated systems. Source data are provided as a Source Data file.
A good similarity in corona compositions is underscored by a correlation analysis (Fig. 7i), yielding correlation coefficients approaching unity. Particularly noteworthy is the highest correlation coefficient (0.996) observed for the pairwise comparison of LNPi and coronated LNPi, both sharing a common component — DNA chains on the particle surface. This substantiates the finding that DNA fosters the formation of resilient coronas at low plasma concentrations, with these coronas exhibiting stability even after exposure to elevated plasma concentrations. Variations in protein abundance were observed in specific molecular weight intervals, namely, the 20-35 kDa range involving Alpha-1-acid glycoprotein 1 (ORM1), the 45-58 kDa range comprising Fibrinogen chains, and the 58-75 kDa range containing human serum albumin. These proteins serve multiple biological functions, and at various levels, they have anti-inflammatory and protecting properties against macrophage activation57–62. The quantitative changes in these proteins among different formulations exhibited fold changes that consistently aligned with the trends depicted in Fig. 5. This indicates a coherent pattern of alterations in protein abundance across distinct molecular weight intervals, providing a comprehensive understanding of the formulation-specific impacts on the protein corona composition.
Sequestration of LNPs by immune cells in vitro, ex vivo, and in vivo
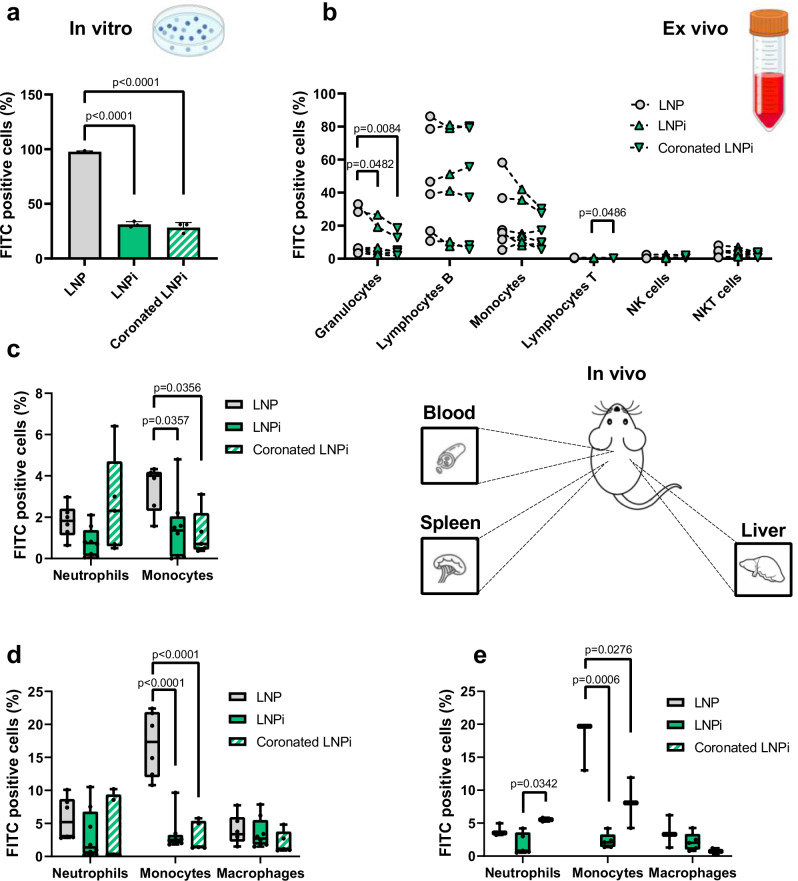
To investigate the sequestration of LNP, LNPi, and coronated LNPi in vitro, we assessed their uptake by monocytic leukemia cells (THP1) utilizing fluorescently labeled LNPs. This cell line has emerged as a standard model for assessing the modulation of monocyte and macrophage functions63. Following kinetic experiments on THP1 treated for 5 min, 10 min, 30 min, and 1 h with pristine LNP we chose a 10-minute incubation, yielding sufficient fluorescent signal and a percentage of FITC positive cells not exceeding hundred percent. Results are given in Fig. 8a as a percentage of FITC-positive cells (experimental details can be found in the Materials and Methods section). The avid capture of LNPs by THP1 cells is a noteworthy finding. It is particularly intriguing given that these particles are PEGylated, a technique traditionally regarded as the gold standard for reducing immune cell capture. This result reaffirms a potential avenue for further investigation into the interplay between surface modifications and immune cell interactions. On the other hand, a striking 100% reduction in cell uptake is observed with LNPi and coronated LNPi. This compelling result strongly indicates that the shared negative surface charge of both formulations significantly contributes to diminishing THP1 cell uptake. This phenomenon endows LNPs with inherent stealth properties, effectively evading cellular recognition. The capacity to evade internalization by THP-1 cells may lead to an extended circulation period, potentially enhancing their capability to reach therapeutic targets. Next, we investigated their capture by various distinct subpopulations of leukocytes obtained from peripheral blood mononuclear cells (PBMCs) derived from healthy donors. PBMCs have been widely used as an in vitro model for the study of immune system functionalities64. The internalization by circulating leukocytes is a critical component of the immune response, serving as an effective surveillance mechanism against foreign pathogens. When NPs are introduced into the bloodstream, sequestration by leukocytes is a barrier to efficient delivery65, thereby potentially causing a substantial impact on the particle distribution within the body. As depicted in Fig. 8b, our observations revealed the following: (i) various subpopulations of leukocytes, including granulocytes, B lymphocytes, and monocytes, exhibited a marked propensity for internalizing LNP, LNPi, and coronated LNPi; (ii) within each subpopulation of leukocytes, the cellular uptake followed the order of LNP>LNPi>coronated LNPi; (iii) it’s noteworthy to mention that the statistical significance was influenced by considerable variability among different donors.
Fig. 8. Capture of LNP, LNPi and coronated LNPi by immune cells in vitro, ex vivo, and in vivo.
a The percentage of FITC-positive THP1 cells was quantified after a 10-min treatment with LNP, LNPi, and coronated LNPi, with error bars representing the standard deviation of n = 3 independent measurements. Statistical analysis was performed using one-way ANOVA, followed by Tukey’s multiple-comparison test. P-values < 0.05 are reported. b Analysis of FITC-positive leukocyte subpopulations was conducted after treatment with LNPs. FACS analysis was carried out on PBMC from blood samples of n = 6 healthy donors following a 5-minute incubation with LNPs. Statistical analysis was performed using ratio-paired Student’s t-tests. c–e The distribution of LNP, LNPi, and coronated LNPi in the blood (c, n = 6, 8, 5, respectively), spleen (d, n = 6, 8, 5, respectively), and liver (e, n = 3, 5, 3, respectively) was evaluated after intravenous injection in 8–10-weeks-old C57BL/6 female mice. Uptake was expressed as a percentage of FITC-positive cells (including neutrophils, monocytes, and macrophages). Mice were sacrificed two hours post-LNP injection. Statistical analysis was conducted through one-way ANOVA followed by Tukey’s multiple-comparison test. P-values < 0.05 are reported. Boxplots are represented by minimum (lower whisker), maximum (upper whisker), sample median (central line), and the first and third quartiles (lower and upper bound of boxes). Source data are provided as a Source Data file. Sketches have been created by using Biorender.com.
As a last step, animal experiments were conducted to elucidate the stealth properties of coronated LNPi in vivo. LNP and LNPi were employed as a reference. The particles were administered to female C57BL/6 mice, aged 8 to 10 weeks, via intravenous venipuncture (i.v.). Subsequently, we assessed particle capture by the primary populations of phagocytes in the blood, namely monocytes and neutrophils. Additionally, we examined particle capture in the spleen and liver, focusing on FITC-positive monocytes, neutrophils, and macrophages. As Fig. 8c shows, the uptake of monocytes in the blood decreased from a mean of 3.4% of FITC-positive cells in mice treated with plain LNP to 1.4% and 1.2 % in mice treated with LNPi and coronated LNPi, respectively. In the spleen (Fig. 8c), LNP uptake was observed in 17% of monocytes, while for LNPi and coronated LNPi, the figures were 2.5 % and 2.1% respectively. In the liver, the proportion of monocytes exhibiting LNP uptake was 17.5%, which then decreased to 2.3% and 8% after treatment with LNPi and coronated LNPi. Similar trends were noted for resident macrophages in both organs, with lower uptake for LNPi and coronated LNPi. Collectively, in vitro, ex vivo, and in vivo investigations have demonstrated LNPi and coronated LNPi exhibit excellent ability to evade capture by immune cells, significantly superior to LNP. This unique capability is not attributed to PEG, a shared structural component among the three systems used, but to DNA, which emerges as the primary element conferring stealth properties to LNPs. The observed differences between LNPi and coronated LNPi can be interpreted through the distinct nature of their protein coronas66. When LNPi is administered in vivo, it forms an endogenous corona as it interacts with the biological fluid. This endogenous corona reflects the complexity of the physiological environment and its dynamic conditions. In contrast, coronated LNPi possesses an artificial corona, which is pre-formed in vitro under conditions of excess protein. This artificial corona, while almost identical in protein composition to an endogenous corona, is formed in a controlled environment that primarily involves plasma proteins and lacks the full spectrum of biomolecular interactions present in vivo. This may result in subtle differences in how immune cells recognize and process these nanoparticles.
Biodistribution of LNP, LNPi and coronated LNPi in vivo
After evaluating the capture of LNP, LNPi, and coronated LNPi by immune cells, the next logical step was to study their biodistribution and toxicity in vivo. This sequence was chosen because understanding how these NPs interact with immune cells can provide insights into their subsequent behavior in the body. Immune cell capture in the blood can significantly influence the biodistribution of NPs, impacting their ability to reach and accumulate in organs. Therefore, assessing biodistribution helps elucidate the relationship between NP design, immune evasion, and delivery. Figure 9 shows the biodistribution of LNP, LNPi, and coronated LNPi in key organs, including the liver, lungs, spleen, kidneys, and heart.
Fig. 9. Biodistribution of LNP, LNPi and coronated LNPi in vivo.
Nanoparticle accumulation in the liver, lungs, spleen, kidneys, and heart was estimated by fluorescence associated with fluorescently labeled nanoparticles (TexasRed). Images of organs from a representative mouse per group (n = 4/group of 8-10 weeks-old C57BL/6 female mice), 3 h after injection with (a) LNP, (b) LNPi, and (c) coronated LNPi. d Quantification of the fluorescence associated with the observed organs, reported as total flux (photons/sec). The signal from non-treated mice (N.T.) is reported as a control. For each organ, statistical analysis was performed by one-way ANOVA, followed by Tukey’s multiple-comparison test. P-values < 0.05 are reported. Data are presented as mean values ± standard deviation. Source data are provided as a Source Data file.
All formulations predominantly delivered to the liver, consistent with the protein corona composition shown in Fig. 7, where human serum albumin (HSA) and apolipoproteins, particularly APO-A1, were most abundant. APO-A1 is the main protein component of high-density lipoproteins (HDL) and plays a crucial role in cholesterol transport and metabolism. Its presence in the protein corona of nanoparticles is supposed to facilitate hepatic targeting, as the liver is a primary site for cholesterol processing and HDL metabolism. Therefore, the enrichment of APO-A1 in the protein corona of these formulations enhances their recognition and uptake by liver cells. The observed biodistribution pattern correlates with the differential clearance of the formulations by immune cells, particularly monocytes, neutrophils, and macrophages, as shown in both in vitro and in vivo experiments. In particular, LNPi demonstrated significantly lower uptake by immune cells and showed enhanced biodistribution, suggesting a reduced immune system clearance. The decreased internalization by immune cells for the DNA-coated particles, attributed to their negative surface charge and DNA-induced stealth properties, likely contributes to their increased accumulation in organs. While inducing specific organ tropism was beyond the scope of this study, the observed accumulation of each formulation in organs and their evasion from immune cell clearance highlights the importance of surface engineering. Enhanced stealth properties, as demonstrated by LNPi, offer a promising strategy for improving organ-specific accumulation. In addition to evaluating the biodistribution of LNPi and coronated LNPi, we also assessed their cytotoxicity in various organs. This complementary analysis provides a more comprehensive understanding of the safety profile of these NPs. Immunohistochemical images of all examined organs, as shown in Supplementary Fig. 13, revealed no signs of toxicity. This thorough investigation underscores the potential of LNPi and coronated LNPi as safe and effective delivery systems, enhancing our confidence in their biocompatibility and suitability for further applications. Although the primary goal was not to develop a formulation for a specific therapeutic application, LNPi shows potential for hepatic delivery, which could be beneficial for treating autoimmune diseases. Additionally, the study provides a solid foundation for future formulations that not only exhibit stealth characteristics but also possess targeted tropism influenced by their protein corona composition.
LNPs are emerging as pivotal players in the landscape of gene therapy, offering promising avenues for tackling genetic disorders, cancer, and the challenges posed by evolving pandemics like COVID-19 variants. Contrary to the notable achievements in delivering small nucleic acids such as siRNA and mRNA, the effective employment of large-size DNA molecules remains unattained. This limitation is particularly noteworthy considering the transformative potential large-size DNA delivery holds in the treatment of diverse genetic diseases. In response to this bottleneck, our study embarks on the strategic design of PEGylated LNPs, deviating from conventional approaches that predominantly involve RNA and ionizable lipids. The innovative integration of large-sized DNA, cationic lipids, and protein corona into the LNP architecture marks a difference with respect to traditional methodologies. Motivated by the unique capacity of DNA to neutralize cationic lipids and facilitate the formation of an opsonin-deficient protein corona, our LNPs undergo additional modifications post-synthesis. This includes the introduction of DNA coating and plasma proteins, culminating in the creation of a coronated LNP. Beyond its visual distinctiveness, the coronated LNP demonstrates superior TE in in vitro studies, as well as enhanced evasion of the immune system and safety in both controlled laboratory conditions and in vivo experiments. The success of the coronated LNP is underpinned by its advanced capabilities in evading lysosomal degradation and immune cell capture. This remarkable attribute stems from a nuanced interplay among key components within the LNP structure, encompassing PEGylation, the protein corona, and the embedded DNA. Our findings address a significant gap in LNP technology and contribute to advancing the field of large-size DNA delivery. By elucidating the complex dynamics influencing LNP functionality at the molecular level, we provide a basis for potential advancements in gene therapies. This emphasizes the importance of continuous exploration and improvement in this promising therapeutic approach beyond existing paradigms.
Methods
Ethical oversight
Informed and written consent in accordance with the Declaration of Helsinki was obtained from all healthy blood donors, and approval was obtained from the Ethics Committee of the Sapienza University of Rome (https://www.uniroma1.it/it/pagina/componenti-del-comitato-etico-la-ricerca-transdisciplinare). All animal experiments were approved by local ethic authorities (Ministry of Health) and conducted in accordance with Italian Governing Law (D.lgs 26/2014) - Protocol number: C1368.26 - PI: G Canettieri.
Preparation of LNPs
LNP has been developed using cationic lipids 1,2-Dioleoyl-3-trimethyl-ammonium-propane (DOTAP), (3β-[N-(N’, N′-dimethyl-aminoethane)-carbamoyl])-cholesterol (DC-Chol), zwitterionic lipids, cholesterol, dioleoyl phosphatidyl ethanol-amine (DOPE), and PEG-lipid 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)−2000 (DOPE-PEG2k). The LNP referred to as ionLNP was prepared using Ionizable lipid 6-((2-hexyldecanoyl)oxy)-N-(6-((2-hexyldecanoyl)oxy)hexyl)-N-(4-hydroxybutyl)hexan-1-aminium (ALC-0315), helper lipid 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (DSPE-PEG2k). All the lipids were purchased from Avanti Polar Lipids (Alabaster, AL). For LNP preparation, lipids were dissolved separately in absolute ethanol in a molar ratio of 0.133: 0.4: 0.133: 0,32: 0,015 for DOTAP: DC-Chol: cholesterol: DOPE: DopePEG, while ionLNP was prepared by dissolving in absolute ethanol ALC-0315, DSPC, cholesterol and DSPE-PEG in 0.463: 0.094: 0.427: 0.016 molar ratio until a concentration of 12,5 mM. The PmirGLO plasmid DNA encoding for firefly luciferase was bought from Promega (Madison, WI, USA) while pEGFP plasmid encoding for green fluorescent protein was a gift from Koen Venken (Addgene plasmid # 165830; http://n2t.net/addgene:165830; RRID: Addgene_165830). pDNAs were diluted in sodium acetate 25 mM, having pH4, to reach a final concentration of 0.2 mg/ml. NanoAssemblr® Ignite™ microfluidic platform (Vancouver, BC, Canada) was used to assemble the LNPs, the mixing of the lipid and DNA solution was performed at a total flow rate of 2 ml/min and a flow rate ratio of 3:1 (DNA/lipid). After the assembling residual ethanol, corresponding to 25% of total volume was eliminated through dialysis performed using a dialysis cassette with 3.5 kDa molecular weight cut off (MWCO) (Thermo Scientific, Rockford, AZ, USA) against phosphate saline buffer (PBS) for 19 h.
DNA coating of LNPs
LNPs prepared at a final concentration of 0.1 mg/ml were incubated with DNA solutions (i.e. DNA in H2O for LNP and DNA in PBS 1x for ionLNP) at three different concentrations: 0.1 mg/ml, 0.2 mg/ml, and 0.3 mg/ml, to prepare low, intermediate, and high DNA-coated LNPs, respectively. The pDNAs pmirGLO (Promega, Madison, WI, USA), and pEGFP (Addgene # 165830) were used to develop functional DNA coated LNPs while pVAX1 (Thermo Fisher Scientific, Waltham, MA, USA) was utilized for the non-functional DNA coated LNPs development. The incubation was performed at room temperature for 30 min and the LNPs were stored at 4 °C.
Size and zeta potential
Size and zeta potential measurements were performed using Zetasizer Nano ZS90 (Malvern, UK). Size measurement was performed through dynamic light scattering after LNPs dilution 1:10 in H2O and results are reported as mean of Z-average (intensity weighted hydrodynamic mean size of the LNPs) ± SD of three repeated measurements. LNPs were instead diluted 1:60 in H2O for microelectrophoresis and the results are reported as mean ± standard deviation of three measurements. Nano Tracking analysis NTA was performed using NanosightPro (Malvern), LNP and LNPi were diluted 14’000 times using H2O the results are presented as mean ± SD of 10 reads.
SDS-PAGE and profile analysis
Plain LNP and DNA-coated LNPi were diluted in PBS 1X until a final concentration of 2 mg/ml lipids and incubated with different amount of human plasma (HP) purchased from Sigma Aldrich Inc. (Merk KGaA, Darmstadt, Germany) for a final volume of 5%, 10%, 25%, and 50% calculated with following formula (1):
| 1 |
Incubation with HP was performed at 37 °C for 1 h. In the study of the PC stability, particles were incubated again in 50% HP volume for 1 h at 37 °C. After the incubation, the samples were diluted in H2O to reach the same volume and centrifuged at 18620 g for 15 min at 4 °C. The supernatant was discarded, and the pellets were resuspended in 200 µl of H2O. The latter washing procedure was repeated 3 times. At the end, the pellets were resuspended in 20 µl of Laemmli Loading buffer 1X and boiled 10 min at 100 °C and then centrifuged at 4 °C, 18,620 g for 15 min. 10 µl of the so obtained supernatant was loaded on a gradient polyacrylamide gel stain-free (4 − 20 % TGX precast gels, BioRad) and run for about 150 min at 100 V. Gel images were acquired with a ChemiDoc gel imaging system (Bio-Rad, CA) and processed with custom MatLab scripts (MathWorks, Natick, MA), as reported in previous work67. All the employed scripts (which have been tested on Windows 7 and Windows 10), a user guide, and a sample image to test the codes are available as Supplementary Software 1.
Synchrotron SAXS
Synchrotron small-angle X-ray scattering (SAXS) measurements were performed at the Austrian SAXS beamline ELETTRA (Trieste, Italy)68, with a Pilatus3 1 M (Dectris, Baden, Switzerland) detector, calibrated by using silver behenate powder (d-spacing = 5.838 nm). q-range was fixed in the range 0.05–5 nm−1, with an exposure time of 20 s. Correction for background, primary beam intensity, and detector efficiency were included in the analysis. Furthermore, blank and buffer samples were measured for all the investigated systems to quantify their contributions to the SAXS patterns. In this way, the amount of non-encapsulated DNA in LNPs and the scattering contribution of free proteins for LNP-HP complexes were assessed. SAS data collection table is provided in the Supplementary Information (Supplementary Table 7).
TEM experiments
Samples for imaging were diluted to 1 mg/ml using ultrapure water and deposited onto formvar/carbon-coated copper grids (Ted Pella 01801) glow discharged for 120 s (Pelco easiGlow). Grids were stained with 2% uranyl acetate staining, washed in ultrapure water, and dried in air. Samples were examined using a JEOL JEM-F200 transmission electron microscope at 200 kV, with images captured by a GATAN Rio 16 CMOS camera.
PicoGreen assay
The amount of encapsulated pDNA (i.e., pmirGLO) and decorating DNA in LNPs was evaluated through the Quanti-iT PicoGreen dsDNA assay kit (ThermoFisher Scientific, Waltham, MA, USA). Reagents were prepared following the manufacturer’s protocol, LNPs were diluted 150-fold in TE buffer and then placed in a 96-well plate (Corning ® 96Well solid polystyrene microplate, Sigma Aldrich, St. Luis, MI, USA). Samples were diluted two-fold with fluorescent reagent Quanti-iT Pico-Green. TritonX-100 (1% v/v) was added to the wells to lyse the LNPs and measure the total DNA (i.e., the sum of encapsulated DNA and free DNA). After 5 min of incubation at room temperature, the fluorescence was measured (475 nm excitation wavelength, 500–550 nm emission wavelength) using Glomax Discovery System (Promega, Madison, WI, USA).
Cell culture
Human embryonic kidney (HEK-293) cell line purchased from American type culture collection (ATCC, Rockville, MD, USA) was maintained in Dulbecco Modified Eagle Medium (DMEM) (Gibco Life Technologies) supplemented with 2 mM L-glutamine, 100 IU/mL penicillin/Streptomycin and Fetal Bovine Serum (FBS) (Gibco Life Technologies) 10% and under humidified atmosphere at 37 °C and 5% CO2. Human monocyte cell line THP-1 cells were purchased from ATCC (ATCC® TIB-202™) and maintained in the RPMI-1640 medium (Gibco, Carlsbad, CA) supplemented with 2 mM l-glutamine, 100 IU/mL P/S, and 10% FBS.
Transfections
HEK-293 cells were seeded in a complete DMEM medium, supplemented in 24-well plates, 40,000 cells per well. After 24 h the medium was replaced by 400 µl Opti-MEM reduced serum medium (Gibco Life Technologies, Carlsbad, CA, USA), containing plain and DNA-coated LNPs for a final concentration of 1 μg encapsulated DNA/well with or without HP and incubated for 3 h at 37 °C. After 3 h the treatment was removed and replaced with 1 ml of supplemented culture medium and incubated for 48 h. After incubation the cells were washed with PBS 1X and lysed using 60 μl of passive lysis buffer (Promega) per well and incubated at room temperature for 10 min. 30 μl of cell lysate was distributed (10 μl per well) into 3 wells of a white 96-well plate and diluted with 100 μl of luciferase substrate reagent purchased from Promega and measured by Glomax Discover System (Promega, Madison, WI, USA). The remaining 30 μl were placed in 3 wells of a second 96-well plate and diluted with 200 μl of BCA reagent of BCA Assay Protein Kit (thermo Fisher Scientific, Waltham, MA, USA) prepared following the protocol and finally incubated 30 min at 37 °C, 5% CO2.At the end the protein content was evaluated using Glomax Discover System (Promega, Madison, WI, USA). TE was reported as a relative light unit (R.L.U) per milligrams of proteins.
Cell viability
10000 cells per well were seeded into 96-well plate in supplemented DMEM and incubated for 24 h at 37 °C, 5% CO2. After the incubation the medium was removed and replaced with 100 μl of Opti-MEM reduced serum medium (Gibco Life Technologies, Carlsbad, CA, USA), with LNPs for a final concentration of 0.25 μg encapsulated DNA and placed in the incubator at 37 °C, 5% CO2. After 3 h, the treatment was removed, and the cells were washed with PBS and 200 μl of supplemented DMEM was added to each well. After 48 h 100 μl of the medium was removed from each well and 50 μl per well of XTT prepared following the protocol was added. Absorbance was measured at 450 nm wavelength using Glomax Discover System (Promega, Madison, WI, USA). Cell viability was expressed as percentage in respect to untreated cells.
Confocal microscopy
Live-cell imaging was performed with a Zeiss LSM 800 confocal microscope equipped with a 63×, 1.4 N.A. oil immersion objective. HEK-293 cells were seeded on glass-bottom dishes (Willco wells # HBST-3522) 24 h before the experiment. On the day of confocal acquisitions, cells were incubated with LNP, LNPi and coronated LNPi for 3 h at 37 °C. Nuclei were stained with DAPI (ThermoFisher). To evaluate the colocalization between LNPs and lysosomes, cells were stained with LysoTracker DeepRed (ThermoFisher) 30 min before imaging. Confocal acquisitions (512 × 512 pixels, 100 nm pixel size) were taken by sequentially exciting fluorescently labeled particles at 561 nm and LysoTracker DeepRed at 640 nm. Emission was collected in the 570–645 nm range and in the 645–700 nm range, respectively. To perform image, Mean Square Displacement (iMSD) analysis, time series of 500 frames were acquired (256 × 256 pixels, 50 nm pixel size, 204.80 ms acquisition time per frame). Fluorescently labeled LNPs were acquired exciting at 561 nm and collecting the fluorescence between 570 nm and 700 nm.
iMSD analysis
iMSD analysis of the time-lapse movies was carried out with custom scripts working on MATLAB (MathWorks Inc., Natick, MA, USA), as described in detail in refs. 51,53. The source code for iMSD analysis (which has been tested on Windows 7 and Windows 10) can be found at the following link: https://springernature.figshare.com/articles/dataset/iMSD_code/6146495/1, along with the corresponding user manual, and a description of the code’s functionality. These files are also attached as Supplementary Information of this work. Briefly, the spatiotemporal correlation function (STCF) was calculated for each time-lapse series; 2D-Gaussian fitting of STCF provides the iMSD curve, describing the ensemble diffusion law of imaged objects. Each iMSD curve was then fitted to extract the structural dynamic parameters: diffusion coefficient and y-axis intercept value, with the latter related to average particle size.
Nano-liquid chromatography tandem mass spectrometry
For mass spectrometry analysis, all chemicals, reagents and organic solvents of the highest grade available were purchased from Merck Life Science (Darmstadt, Germany), unless otherwise stated. Trifluoroacetic acid (TFA) was supplied by Romil Ltd (Cambridge). Bond elut C18 EWP cartridges (50 mg) were purchased from Agilent (Santa Clara, USA). Trypsin was provided by Promega (Madison, WI, USA).
Shotgun proteomics sample preparation
Protein pellets were prepared as described in ref. 69; briefly, they were dissolved in 100 µL of 8 mol L-1 urea in 50 mmol tris(hydroxymethyl)aminomethane hydrochloride (tris-HCl) buffer (pH = 7.8). After complete solubilization, protein samples were reduced with 2 µL of 200 mmol L-1 dithiothreitol for 1 h at 37 °C and alkylated with 8 µL of 200 mmol L-1 2-iodoacetamide for 1 h in the dark at room temperature. The alkylation was quenched with further 8 µL of 200 mmol L−1 dithiothreitol. The urea concentration was then diluted to 1 mol L-1 with 50 mmol L-1 tris-HCl buffer (pH = 7.8) before overnight tryptic digestion with 2 µg of trypsin at 37 °C. The enzymatic reaction was stopped by adding TFA to reach pH 2.5. The resulting peptide mixtures were purified by solid-phase extraction on Bond elut C18 EWP cartridges, evaporated to dryness and dissolved in 100 µL of 0.1% formic acid.
Shotgun proteomics analysis
Peptide samples were analyzed by nanoHPLC-MS/MS as described in ref. 70, on a Ultimate 3000 nanoHPLC system coupled to Orbitrap Elite (Thermo Fisher Scientific), with minor modifications. Samples (20 µL) were injected and on-line preconcentrated on a 300 μm i.d. × 5 mm Acclaim PepMap 100 C18 μ-column (Thermo Scientific) using water/acetonitrile, 99:1 (v/v) containing 0.1% (v/v) TFA at a flow rate of 10 μL min−1. Peptides were then separated on an EASY-Spray column (15 cm × 75 μm i.d. PepMap C18, 2-μm particles, 100 Å pore size; Thermo Fisher Scientific) at 300 nL min−1 and at 35 °C. A 100 min-long multistep gradient was used for peptide separation, using water (phase A) and acetonitrile (phase B) both with 0.1% formic, as follows: 1% B for 5 min; 1–5% B in 2 min; 5-35% B in 90 min. The column was then washed at 90% B for 10 min and equilibrated at 1% B for 30 min. Full scan spectra were acquired at 30,000 resolution (full width at half maximum at m/z 400) in the range 300-2000 m/z. MS/MS spectra were acquired in top 10 data-dependent mode at 15,000 resolution rejecting singly and unassigned charge states and fragmenting precursor ions by higher-energy collisional dissociation with 35% normalized collision energy, isolation window of 2 m/z, and dynamic exclusion (repeat count of 1, repeat duration of 30 s, exclusion duration of 70 s). For each sample, three technical replicates were performed. The acquired raw MS/MS data files from Xcalibur software (version 2.2 SP1.48, Thermo Fisher Scientific) were searched against Swiss-Prot human database by MaxQuant search engine71, (v1.6.3.4)71 with the automatic setting for tryptic peptide matching and label free analysis, according to the following settings: protein sequence database: Swissprot human; protease: trypsin; cleavage site: lysine and arginine; number of missed cleavages: 2; fixed and variable modifications: carbamidomethylation on cysteine was fixed, while oxidation of methionine, acetylation of protein N-termini, and deamidation of glutamine and asparagine were set as variable modifications; mass tolerance for precursor and fragment ions: 10 and 20 ppm; minimum peptide length: 8; applied score cutoffs:false; discovery rate = 0.01 for peptide-spectrum matches, peptides and proteins identifications; minimum number of unique peptides for protein identification: 1 razor unique peptide. For data analysis, the MaxQuant output “proteinGroups.txt” was used. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE72 partner repository with the dataset identifier PXD054751.
In vitro uptake of LNPs from immune system cells
To investigate the uptake of LNPs in the THP-1 cell line, LNP, LNPi and coronated LNPi were prepared using NBD (fluorescent lipid/total lipid = 5/1000 mol/mol). THP-1 cells were cultured at a density of 500,000 cells per milliliter in 24-well dishes and treated using 1 µg of encapsulated DNA in a final volume of 1 ml. After the treatment, the cells were rinsed with cold PBS and subsequently analyzed using a FACS Canto instrument from BD Biosciences in San Jose, CA. The cells were gated based on forward versus side scatter to exclude debris, and their specific emission was then assessed. Data analysis was carried out using FlowJo software.
Ex vivo uptake of LNPs from immune system cells
For the sequestration of particles from circulating peripheral blood mononuclear cells (PBMCs) LNPs prepared using NBD (fluorescent lipid/total lipid = 5/1000 mol/mol) were incubated with healthy donors’ blood samples (120 µl vol) for 10 min at 37 °C. Informed and written consent in accordance with the Declaration of Helsinki was obtained from all healthy blood donors, and approval was obtained from the Ethics Committee of the Sapienza University of Rome (https://www.uniroma1.it/it/pagina/componenti-del-comitato-etico-la-ricerca-transdisciplinare). Blood donors were anonymous unpaid volunteers and no sex-related analysis was performed. The treatment of cells was performed using 95 µg (mass of lipids) of NBD-labeled LNPs (6 µl vol) corresponding to 0.3 µg of encapsulated DNA. Then cells were washed with PBS and red blood cells were then lysed with a buffer containing 155 mM NH4Cl, 12 mM NaHCO3, 0.1 mM EDTA. After washing by centrifugation cells were labeled with the following antibodies: anti-CD3/BV510, (clone HIT3a, cat. 564713, lot. 3109460), CD56/PE (clone NCAM16.2, cat. 345812, lot.3012189), anti-CD14/APC (clone MOP9, cat. 340436, lot. n/a), anti-CD45/PECy7 (clone HI30, cat. 557748, lot.0349447), and anti-CD19/BV450 (clone HIB19, cat. 560353, lot.3065933), all obtained from BD Bioscience and used according to the manufacturer’s instructions. The fluorescence of the internalized LNPs was assessed through immunofluorescence and FACS analysis using a FACSCanto instrument from BD Biosciences, San Jose, CA. The measurements were expressed as the percentage of positive cells, with gating on distinct leukocyte subpopulations40. The gating strategy is shown in Supplementary Fig. 11 in the SI. Briefly, residual red blood cells were eliminated from the analysis by gating on CD45+ population. Next, in order to exclude any clumps of multiple cells, cell doublets were gated out by looking at FSC-A vs FSC-H followed by FSC-A vs SSC-A gate to eliminate debris. Leukocyte populations were identified by gating on SSC-Ahigh (granulocytes); CD45 + CD14+ (monocytes; αCD45PECy7/ αCD14APC); CD45 + CD3-CD56+ (NK cells; αCD45PECy7+/αCD3BV410/ αCD56PE); CD45 + CD3 + CD56- (T lymphocytes; αCD45PECy7/ αCD3BV410/ αCD56-PE): CD45 + CD3 + CD56+ (NKT cells; αCD45PECy7/ αCD3BV410/ αCD56PE); CD45 + CD3-CD56-CD19+ (B lymphocytes; αCD45PECy7/ αCD3BV410/ αCD56PE/ αCD19/BV450). Data analysis was performed using the FlowJo program.
In vivo animal experiments
To investigate cellular uptake of nanoparticles in vivo, 200 µl of i) LNP, ii) LNPi, and iii) coronated LNPi at a final concentration of 0.05 mg/ml (10 µg of encapsulated DNA) were intravenously injected (i.v.) in 8-10 weeks old female C57BL/6 mice (Charles River, n = 8 mice per group; 200 µl/each mouse). The control group was i.v. injected with the same volume of PBS. At 2 h after injection, mice were sacrificed followed by cardiac puncture to collect blood, further added to heparin tubes. Freshly isolated cells from the spleen, liver, and blood were prepared and stained as described in ref. 17. Briefly, leukocytes were isolated from the liver through mechanical dissection and further purified using a Percoll gradient73. After lysis of red blood cells, 2 × 106 of splenocytes, liver-derived leukocytes, or 50 μL of peripheral blood mononuclear cells (PBMCs) were washed and resuspended in staining buffer (PBS without Ca2 + Mg2 + , BSA 0.5%, EDTA 2 mM, and NaN3 0.025%). After 10 mins of incubation with 0.1μg anti-CD16/32 (clone 2.4G2, Stemcell Technologies), cells were stained with anti-F480/BV421 (clone BM8, cat. 123137, lot. B330997, BioLegend), anti-CD11b/FITC (clone MI/70, cat. 110112-82, lot. 1989138 eBioscience) and anti-GR1/APC (clone R86-85C, cat.2123757, lot. 175931-82 Invitrogen) for 25 min at 4 °C. These antibodies were used according to the manufacturer’s instructions. The gating strategy is reported in Supplementary Fig. 12 in the SI. Samples were analyzed by flow cytometry using a FACSCanto II (BD Biosciences, San Jose, CA), and data analysis was performed using the FlowJo program.
For biodistribution analysis, 200 µl of (i) LNP, (ii) LNPi, and (iii) coronated LNPi labeled with Texas Red at final concentration of 0.25 mg/mL of encapsulated DNA (50 µg/mice) were intravenously injected (i.v.) in 8-10 weeks old female C57BL/6 mice (Charles River, n = 4 mice per group; 200 µl/each mouse). 3 h after injection, mice were anesthetized, and their abdomen and chest were depilated with hair removal cream. Fluorescence images were acquired by IVIS Lumina III In Vivo Imaging System (Caliper Life Science, Waltham, MA, USA). At the end of the fluorescence acquisition, organs (heart, kidney, liver, lung and spleen) were collected for ex-vivo imaging. Mice and organs were imaged with an exposure time of 1 s. Fluorescence images were quantified using Living Image software (Caliper Life Science).
To assess the toxicity of the LNPs against different tissues, heart, kidney, liver, lung, and spleen were isolated 24 h after injection and fixed immediately in 4% paraformaldehyde and paraffinized after 24 h. 4 µm section from paraffin-embedded tissues were stained with hematoxylin and eosin (H&E). All slides were examined for tissue inflammatory reactions by a pathologist in a blinded manner.
Animals were housed in the Institute’s Animal Care Facilities, which meet international standards and were checked regularly by a certified veterinarian responsible for health monitoring, animal welfare supervision, and revision of experimental protocols and procedures. The mice were kept in a room with a temperature of 18–25°, relative humidity of 40–70%, and noise below 85 decibels. The room provided an appropriate period of alternating light and dark, usually 12/12 h, to simulate the normal circadian rhythm of mice.
All animal experiments were approved by local ethic authorities (Ministry of Health) and conducted in accordance with Italian Governing Law (D.lgs 26/2014) - Protocol number: C1368.26 - PI: G Canettieri. We followed the European and national regulations for the care and use of animals to protect them for experimental and other scientific purposes (D.lgs 26/2014).
Reporting on sex: Prior investigations have revealed notable distinctions in the capture efficiency of natural killer (NK) cells between male and female mice in response to lipid nanoparticles74. Given these observed gender-specific variances, a decision was made to focus exclusively on female mice in the present study. This choice was aimed at minimizing confounding factors and ensuring a more homogenous experimental cohort. This approach aligns with a growing recognition of the impact of sex-based differences in immune responses and underscores the importance of considering such factors in experimental design to enhance the precision and interpretability of research findings.
Minimum Information Reporting in Bio−Nano Experimental Literature
The investigations conducted in this study, encompassing physical-chemical characterization, biological examinations, and comprehensive experimental particulars, adhere to the Minimum Information Reporting in Bio−Nano Experimental Literature reporting standard for bio−nano research75. Additionally, we have included a corresponding checklist of this information in Supplementary Table 4 in the SI.
Statistics and reproducibility
In bar charts, the results are reported as mean ± standard deviation. The number of replicates for each sample varied among experiments, as follows: n = 2 for cell transfection experiments by flow cytometry, n = 3 for size, PdI, zeta-potential measurements, fluorescence assay, cell viability as a function of HP%, proteomics experiments, and flow cytometry experiments in THP1 cells; n = 4 for agarose gel electrophoresis experiments, SDS-PAGE experiments, and in vivo biodistribution experiments; n = 3-8 for in vivo uptake experiments; n = 6 for transfection efficiency by luciferase assay, and ex vivo uptake experiments; n = 10 for synchrotron SAXS measurements, colocalization analysis, and iMSD analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The research leading to these results has received funding from the European Union - NextGenerationEU through the Italian Ministry of University and Research under PNRR - M4C2-I1.3 Project PE_00000019 “HEAL ITALIA” to Giulio Caracciolo. CUP: B53C22004000006.
Author contributions
D.P., A.Z., S.C., and G. Car. conceived the project. D.P., A.L., A.L.C., H.A., M.R., F.C., A.Z., S.C., G.C., and G. Car. were involved in experimental design. S.R., L.D., E.Q., E.V., M.V.G., A.M., B.N., L.P., V.D.L., S.G., L.L., C.M.M., C.N., F.M., G.P., M.G.P., and L.D.M. contributed to data collection and/or interpretation. S.R., L.D., A.Z., S.C., and G. Car. contributed to writing the paper. A.Z., and G.Car. contributed to securing funding. All the authors provided scientific input and read and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks Hélder A. Santos, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided with this paper. Mass spectrometry data are stored in a public ProteomeXchange dataset, with accession number: PXD054751 [proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD054751]. Source data are provided with this paper.
Code availability
The custom codes used for SDS-PAGE analysis are provided as Supplementary Software 1. The archive contains all the necessary scripts, a manual, and data for a representative example.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Serena Renzi, Luca Digiacomo, Daniela Pozzi.
Contributor Information
Alessandra Zingoni, Email: alessandra.zingoni@uniroma1.it.
Saula Checquolo, Email: saula.checquolo@uniroma1.it.
Giulio Caracciolo, Email: giulio.caracciolo@uniroma1.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53569-8.
References
- 1.Cullis, P. R. & Hope, M. J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther.25, 1467–1475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulkarni, J. A., Cullis, P. R. & Van Der Meel, R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Therapeutics28, 146–157 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Akinc, A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol.14, 1084–1087 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Gokita, K., Inoue, J., Ishihara, H., Kojima, K. & Inazawa, J. Therapeutic potential of LNP-mediated delivery of miR-634 for cancer therapy. Mol. Ther.-Nucleic Acids19, 330–338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thi, T. T. H. et al. Lipid-based nanoparticles in the clinic and clinical trials: from cancer nanomedicine to COVID-19 vaccines. Vaccines9, 359 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu, J., Li, T. & Zhu, J. Gene therapy strategies targeting aging-related diseases. Aging Dis.14, 398 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu, Y. et al. Multi-step screening of DNA/lipid nanoparticles and co-delivery with siRNA to enhance and prolong gene expression. Nat. Commun.13, 4282 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guimaraes, L. C. et al. Nanoparticle-based DNA vaccine protects against SARS-CoV-2 variants in female preclinical models. Nat. Commun.15, 590 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz, C. & Wagner, E. Therapeutic plasmid DNA versus siRNA delivery: common and different tasks for synthetic carriers. J. Control. Release161, 554–565 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Irobalieva, R. N. et al. Structural diversity of supercoiled DNA. Nat. Commun.6, 8440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulkarni, J. A. et al. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomed. Nanotechnol. Biol. Med.13, 1377–1387 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Algarni, A. et al. In vivo delivery of plasmid DNA by lipid nanoparticles: the influence of ionizable cationic lipids on organ-selective gene expression. Biomater. Sci.10, 2940–2952 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Scalzo, S. et al. Ionizable lipid nanoparticle-mediated delivery of plasmid DNA in cardiomyocytes. Int. J. Nanomed.30, 2865–2881 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biscans, A., Ly, S., McHugh, N., Cooper, D. A. & Khvorova, A. Engineered ionizable lipid siRNA conjugates enhance endosomal escape but induce toxicity in vivo. J. Control. Release349, 831–843 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni, J. A., Witzigmann, D., Chen, S., Cullis, P. R. & van der Meel, R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Acc. Chem. Res.52, 2435–2444 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Quagliarini, E. et al. Mechanistic insights into the superior DNA delivery efficiency of multicomponent lipid nanoparticles: an in vitro and in vivo study. ACS Appl. Mater. Interfaces14, 56666–56677 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Giulimondi, F. et al. Opsonin-deficient nucleoproteic corona endows UnPEGylated liposomes with stealth properties in vivo. ACS Nano16, 2088–2100 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, V. et al. Shielding of lipid nanoparticles for siRNA delivery: impact on physicochemical properties, cytokine induction, and efficacy. Mol. Ther.-Nucleic Acids3, e210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schöttler, S., Landfester, K. & Mailänder, V. Controlling the stealth effect of nanocarriers through understanding the protein corona. Angew. Chem. Int. Ed.55, 8806–8815 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Ke, P. C., Lin, S., Parak, W. J., Davis, T. P. & Caruso, F. A decade of the protein corona. ACS Nano11, 11773–11776 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Rampado, R., Crotti, S., Caliceti, P., Pucciarelli, S. & Agostini, M. Recent advances in understanding the protein corona of nanoparticles and in the formulation of “stealthy” nanomaterials. Front. Bioeng. Biotechnol.8, 166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui, L. et al. Efficient delivery of DNA using lipid nanoparticles. Pharmaceutics14, 1698 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam, K. et al. Unsaturated, trialkyl ionizable lipids are versatile lipid‐nanoparticle components for therapeutic and vaccine applications. Adv. Mater.35, 2209624 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Caracciolo, G., Pozzi, D., Caminiti, R. & Amenitsch, H. Formation of overcharged cationic lipid/DNA complexes. Chem. Phys. Lett.429, 250–254 (2006). [Google Scholar]
- 25.Quagliarini, E. et al. Microfluidic formulation of DNA-loaded multicomponent lipid nanoparticles for gene delivery. Pharmaceutics13, 1292 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caracciolo, G. et al. Enhanced transfection efficiency of multicomponent lipoplexes in the regime of optimal membrane charge density. J. Phys. Chem. B112, 11298–11304 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Marchini, C. et al. Tailoring lipoplex composition to the lipid composition of plasma membrane: a trojan horse for cell entry? Langmuir26, 13867–13873 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Kulkarni, J. A. et al. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano12, 4787–4795 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Viger-Gravel, J. et al. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced NMR spectroscopy. J. Phys. Chem. B122, 2073–2081 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Ribeiro, L. Nd. M., Couto, V. M., Fraceto, L. F. & De Paula, E. J. S. R. Use of nanoparticle concentration as a tool to understand the structural properties of colloids. Sci. Rep.8, 982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sternberg, B., Sorgi, F. L. & Huang, L. New structures in complex formation between DNA and cationic liposomes visualized by freeze—fracture electron microscopy. FEBS Lett.356, 361–366 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Palchetti, S. et al. Manipulation of lipoplex concentration at the cell surface boosts transfection efficiency in hard-to-transfect cells. Nanomed. Nanotechnol. Biol. Med.13, 681–691 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Oh, N. & Park, J.-H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int. J. Nanomed.9, 51–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manolova, V. et al. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol.38, 1404–1413 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Cedervall, T. et al. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl Acad. Sci. USA104, 2050–2055 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dawson, K. A. & Yan, Y. Current understanding of biological identity at the nanoscale and future prospects. Nat. Nanotechnol.16, 229–242 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Walkey, C. D. & Chan, W. C. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev.41, 2780–2799 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Simon, J. et al. Exploiting the biomolecular corona: pre-coating of nanoparticles enables controlled cellular interactions. Nanoscale10, 10731–10739 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Dal Magro, R. et al. Artificial apolipoprotein corona enables nanoparticle brain targeting. Nanomed.: Nanotechnol., Biol. Med.14, 429–438 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Giulimondi, F. et al. Interplay of protein corona and immune cells controls blood residency of liposomes. Nat. Commun.10, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salvati, A. et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol.8, 137–143 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Nienhaus, K. & Nienhaus, G. U. Mechanistic understanding of protein corona formation around nanoparticles: old puzzles and new insights. Small19, 2301663 (2023). [DOI] [PubMed] [Google Scholar]
- 43.Digiacomo, L., Pozzi, D., Palchetti, S., Zingoni, A. & Caracciolo, G. Impact of the protein corona on nanomaterial immune response and targeting ability. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol.12, e1615 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Wang, Y.-F. et al. The Yin and Yang of the protein corona on the delivery journey of nanoparticles. Nano Res.16, 715–734 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Digiacomo, L., Pozzi, D., Amenitsch, H. & Caracciolo, G. Impact of the biomolecular corona on the structure of PEGylated liposomes. Biomater. Sci.5, 1884–1888 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Viard, M. et al. Design and biological activity of novel stealth polymeric lipid nanoparticles for enhanced delivery of hydrophobic photodynamic therapy drugs. Nanomed. Nanotechnol., Biol. Med.14, 2295–2305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caracciolo, G. et al. Lipid composition: a “key factor” for the rational manipulation of the liposome–protein corona by liposome design. RSC Adv.5, 5967–5975 (2015). [Google Scholar]
- 48.Caracciolo, G. et al. Evolution of the protein corona of lipid gene vectors as a function of plasma concentration. Langmuir27, 15048–15053 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Palchetti, S. et al. A mechanistic explanation of the inhibitory role of the protein corona on liposomal gene expression. Biochim. Biophys. Acta Biomembr.1862, 183159 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Bertoli, F., Garry, D., Monopoli, M. P., Salvati, A. & Dawson, K. A. The intracellular destiny of the protein corona: a study on its cellular internalization and evolution. ACS Nano10, 10471–10479 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Digiacomo, L., Digman, M. A., Gratton, E. & Caracciolo, G. Development of an image Mean Square Displacement (iMSD)-based method as a novel approach to study the intracellular trafficking of nanoparticles. Acta Biomater.42, 189–198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Digiacomo, L. et al. Dynamic fingerprinting of sub-cellular nanostructures by image mean square displacement analysis. Sci. Rep.7, 14836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferri, G. et al. Time-lapse confocal imaging datasets to assess structural and dynamic properties of subcellular nanostructures. Sci. Data5, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cardarelli, F. et al. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci. Rep.6, 25879 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monopoli, M. P. et al. Physical− chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc.133, 2525–2534 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Tenzer, S. et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol.8, 772–781 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Fournier, T., Medjoubi-N, N. & Porquet, D. Alpha-1-acid glycoprotein. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol.1482, 157–171 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Huang, Z. & Ung, T. Effect of alpha-1-acid glycoprotein binding on pharmacokinetics and pharmacodynamics. Curr. Drug Metab.14, 226–238 (2013). [PubMed] [Google Scholar]
- 59.Deng, Z. J., Liang, M., Monteiro, M., Toth, I. & Minchin, R. F. Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Nat. Nanotechnol.6, 39–44 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Takeuchi, T. et al. Molecularly imprinted nanogels acquire stealth in situ by cloaking themselves with native dysopsonic proteins. Angew. Chem. Int. Ed.56, 7088–7092 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Hochepied, T., Berger, F. G., Baumann, H. & Libert, C. α1-Acid glycoprotein: an acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev.14, 25–34 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Wan, S. et al. The “sweet” side of the protein corona: effects of glycosylation on nanoparticle–cell interactions. ACS Nano9, 2157–2166 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Chanput, W., Mes, J. J. & Wichers, H. J. THP-1 cell line: an in vitro cell model for immune modulation approach. Int. Immunopharmacol.23, 37–45 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Reddy, M., Eirikis, E., Davis, C., Davis, H. M. & Prabhakar, U. Comparative analysis of lymphocyte activation marker expression and cytokine secretion profile in stimulated human peripheral blood mononuclear cell cultures: an in vitro model to monitor cellular immune function. J. Immunol. Methods293, 127–142 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Betker, J. L. et al. Nanoparticle uptake by circulating leukocytes: a major barrier to tumor delivery. J. Control. Release286, 85–93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caracciolo, G. Artificial protein coronas: directing nanoparticles to targets. Trends Pharmacol. Sci.45, 602–613 (2024). [DOI] [PubMed] [Google Scholar]
- 67.Digiacomo, L. et al. Optimal centrifugal isolating of liposome–protein complexes from human plasma. Nanoscale Adv.3, 3824–3834 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amenitsch, H. et al. First performance assessment of the small-angle X-ray scattering beamline at ELETTRA. J. Synchrotron Radiat.5, 506–508 (1998). [DOI] [PubMed] [Google Scholar]
- 69.Caracciolo, G. et al. Disease-specific protein corona sensor arrays may have disease detection capacity. Nanoscale Horiz.4, 1063–1076 (2019). [Google Scholar]
- 70.La Barbera, G. et al. Development of an enrichment method for endogenous phosphopeptide characterization in human serum. Anal. Bioanal. Chem.410, 1177–1185 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc.11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Deutsch, E. W. et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res.51, D1539–D1548 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi, W. et al. Isolation and purification of immune cells from the liver. Int. Immunopharmacol.85, 106632 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Vulpis, E. et al. The possible role of sex as an important factor in development and administration of lipid nanomedicine-based COVID-19 vaccine. Mol. Pharm.18, 2448–2453 (2021). [DOI] [PubMed] [Google Scholar]
- 75.Faria, M. et al. Minimum information reporting in bio–nano experimental literature. Nat. Nanotechnol.13, 777–785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Source data are provided with this paper. Mass spectrometry data are stored in a public ProteomeXchange dataset, with accession number: PXD054751 [proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD054751]. Source data are provided with this paper.
The custom codes used for SDS-PAGE analysis are provided as Supplementary Software 1. The archive contains all the necessary scripts, a manual, and data for a representative example.