Abstract
A DNA-binding domain (DBD) was identified on simian virus 40 (SV40) major capsid protein Vp1, and the domain's function in the SV40 life cycle was examined. The DBD was mapped by assaying various recombinant Vp1 proteins for DNA binding in vitro. The carboxy-terminal 58-residue truncated Vp1ΔC58 pentamer bound DNA with a Kd of 1.8 × 10−9 M in terms of the protein pentamer, while full-length Vp1 and carboxy-terminal-17-truncated Vp1ΔC17 had comparable apparent Kds of 5.3 × 10−9 to 7.3 × 10−9 M in terms of the protein monomers. Previously identified on Vp1 was a nuclear localization signal (NLS) consisting of two N-terminal basic clusters, NLS1 (4-KRK-6) and NLS2 (15-KKPK-18). Vp1ΔC58 pentamers harboring multiple-point mutations in NLS1 (NLSm1), NLS2 (NLSm2), or both basic clusters (NLSm1 · 2) had progressively decreased DNA-binding activity, down to 0.7% of the Vp1ΔC58 level for NLSm1 · 2 Vp1. These data, along with those of N-terminally truncated proteins, placed the DBD in overlap with the bipartite NLS. The role of the Vp1 DBD during infection was investigated by taking advantage of NLS phenotypic complementation (N. Ishii, A. Nakanishi, M. Yamada, M. H. Macalalad, and H. Kasamatsu, J. Virol. 68:8209–8216, 1994), in which an NLS-defective Vp1 could localize to the nucleus in the presence of wild-type minor capsid proteins Vp2 and Vp3. This approach made it possible to dissect the role of the bifunctional Vp1 NLS-DBD in virion assembly in the nucleus. Mutants of the viable nonoverlaping SV40 (NO-SV40) DNA NLSm1, NLSm2, and NLSm1 · 2 replicated normally following transfection into host cells and produced capsid proteins at normal levels. All mutant Vp1s were able to interact with Vp3 in vitro. The mutants NLSm1 and NLSm1 · 2 were nonviable, and the mutant Vp1s unexpectedly failed to localize to the nucleus though Vp2 and Vp3 did, suggesting that the mutated NLS1 acted as a dominant signal for the cytoplasmic localization of Vp1. Mutant NLSm2, for which the mutant Vp1's nuclear localization defect was complemented by Vp2 and Vp3, displayed a 5,000-fold reduced viability. Analysis of NLSm2 DNA-transfected cell lysate revealed a 10-fold reduction in the level of DNase I-protected viral DNA, and yet virion-like particles were found among the DNase I-resistant material. Collective results support a role for Vp1 NLS2-DBD2 in the assembly of virion particles. The results also suggest that this determinant can function in the infection of new cells.
The virion particle of simian virus 40 (SV40), like those of other papovaviruses, packages the double-stranded circular viral DNA in an icosahedrally symmetric capsid. The SV40 capsid is composed of 72 pentamers of the major capsid protein Vp1, and the pentameric units are tied together by the carboxy-terminal arms which extend into neighboring pentamers (20, 27). The amino-terminal 15 amino acids of Vp1 are not visible in the crystal structure due to disorder but probably extend into the virion core to interact with the viral minichromosome (20), which consists of the viral DNA and the four cellular core histones. A lower-resolution view of murine polyomavirus virions identifies molecules of the minor capsid proteins Vp2 and Vp3 (Vp2/3) as prongs that extend from the minichromosome core into the axial cavities of Vp1 pentamers (10). Though much is known about the structure of the virion, how virions are assembled in the cell nucleus during productive infection is still not well defined. Presumed to be important for the assembly is the interaction of the capsid proteins with DNA and with histones. Since all capsid proteins of SV40 bind DNA (4, 26), but only Vp1 of polyomavirus does (2, 22), the interactions involved in the assembly process can be different even among members of the papovavirus family. Defining a functional DNA-binding domain (DBD) for the interaction of each capsid protein with viral DNA may unravel the unique process for the assembly of individual viruses.
SV40 assembly is thought to consist of two phases. In the “subvirion” assembly phase, pentamerized Vp1 associates with Vp2/3 in the cytoplasm soon after the proteins' synthesis, and they are transported to the nucleus as such subvirion complexes (9, 21). All of the SV40 Vp1, Vp2, and Vp3 harbor nuclear localization signals (NLSs) (3, 13). Consistent with the capsid proteins' interaction prior to nuclear transport, wild-type Vp2/3 could rescue the nuclear localization of an NLS-defective Vp1 and vice versa (13). This phenotypic complementation would prove useful in our functional dissection of the Vp1 DBD (see below). The second phase of “virion” assembly begins once the capsid proteins enter the nucleus, the site of viral DNA replication and packaging. In a stepwise model for virion formation, the capsid proteins are sequentially added to and arranged on the viral minichromosome, resulting in the condensation and packaging of the viral DNA and the formation of the capsid (1, 5, 15). All three SV40 capsid proteins can bind DNA nonspecifically (4, 26). The cooperative binding of Vp3 and the transcription factor Sp1 to ses, the encapsidation signal of SV40 DNA, has been proposed to provide packaging specificity by nucleating the capsid proteins' addition to the viral minichromosome (6, 11, 23). The DNA-binding and protein-interactive functions of individual capsid proteins may collectively contribute to the packaging or virion assembly process. In this study, we mapped the DBD of SV40 Vp1 and found that it overlaps with the previously identified, amino-terminal bipartite NLS (14), which comprises two clusters of basic residues, 4-lysine-arginine-lysine-6 and 15-lysine-lysine-proline-lysine-18. We also examined whether this DBD is important for the formation of infectious virions in the viral life cycle. Despite the overlap of the DBD with the NLS, the application of phenotypic complementation has allowed us to identify at least the second basic cluster, represented by mutant NLSm2, as a determinant for nuclear virion assembly. Our results are consistent with an important role of the Vp1 DNA-binding activity in the proper packaging of virions.
MATERIALS AND METHODS
Construction of plasmids.
Subcloning was performed using standard techniques (25). All mutagenized nucleotides were confirmed by double-stranded DNA sequencing, as were the absence of unwanted mutations within the Vp1 gene. In the DNA sequences below, mutated SV40 nucleotides are given in lowercase letters, and relevant restriction sites are underlined. Vp1 amino acids are numbered from the alanine of the second codon.
A series of pQE-Vp1 plasmids were used to express Vp1 as carboxy-terminally (histidine)6 (H6)-tagged proteins (see Fig. 1E for diagrams). First, pQE-XbaMCS was made from pQE60 (Qiagen) by knocking out the XbaI site and inserting a linker through NcoI and BgIII sites to introduce XbaI and BamHI sites in the multicloning region. To express the first 344 Vp1 residues (Vp1ΔC17), pQE-Vp1-ΔC17 was constructed by subcloning the 1,040-bp XbaI-to-BamHI fragments of pSV-Vp1 (13) into pQE-XbaMCS. To express the first 303 Vp1 residues (Vp1ΔC58) of which cysteines 104 and 254 are changed into alanines (to minimize possible oxidative cross-linking of Vp1 during protein isolation), pQE-Vp1-2CA-ΔC58 was constructed by inserting the 953-bp XbaI-to-BamHI fragment of pBS-Vp1-2CA-ΔC58 into pQE-XbaMCS. pBS-Vp1-2CA-ΔC58 was made by inserting into pBS-Vp1-ΔC58 (19) the 495-bp XbaI-to-PstI fragment from pBS-Vp1-C104A (19) and the 458-bp PstI-to-BamHI fragment from pBS-Vp1-C254A (19). To express the N-terminal mutant counterparts of ΔC17 or ΔC58 Vp1s, pQE-Vp1-ΔC17 or pQE-Vp1-2CA-ΔC58 was inserted with the 206-bp or smaller XbaI-to-AflII fragments from pSV-Vp1 -p567, -p81 (14), -p25 (14), -d20, and -d50 to yield the N-terminal mutant counterparts NLSm1, NLSm2, NLSm1 · 2 (see Table 1), ΔN(2–21), and ΔN(2–51) (see Fig. 1E). pSV-Vp1 -p567, -d20, and -d50 were made from pSV-Vp1 by site-directed mutagenesis using the SacI antisense primer (5′-CAAGAATTCGAGCTCGCCCAACTTG-3′) and either the p567-XbaI sense primer (5′-CAGGTCCATGGTCTAGA ATGAAGATGGCCCCAACAAAcgGAAAcGGAAGTTGTCCAGGGGCAG CTCCCAA-3′), the d20-XbaI sense primer (5′-CAGGTCCATGGTCTAGAATGAAGATGGCCCAAGTGCCAAAGCTCGTCAT-3′), or the d50-XbaI sense primer (5′-CAGGTCCATGGTCTAGAATGAAGATGGCCAATCCTCAAATGGGCAATCC-3′).
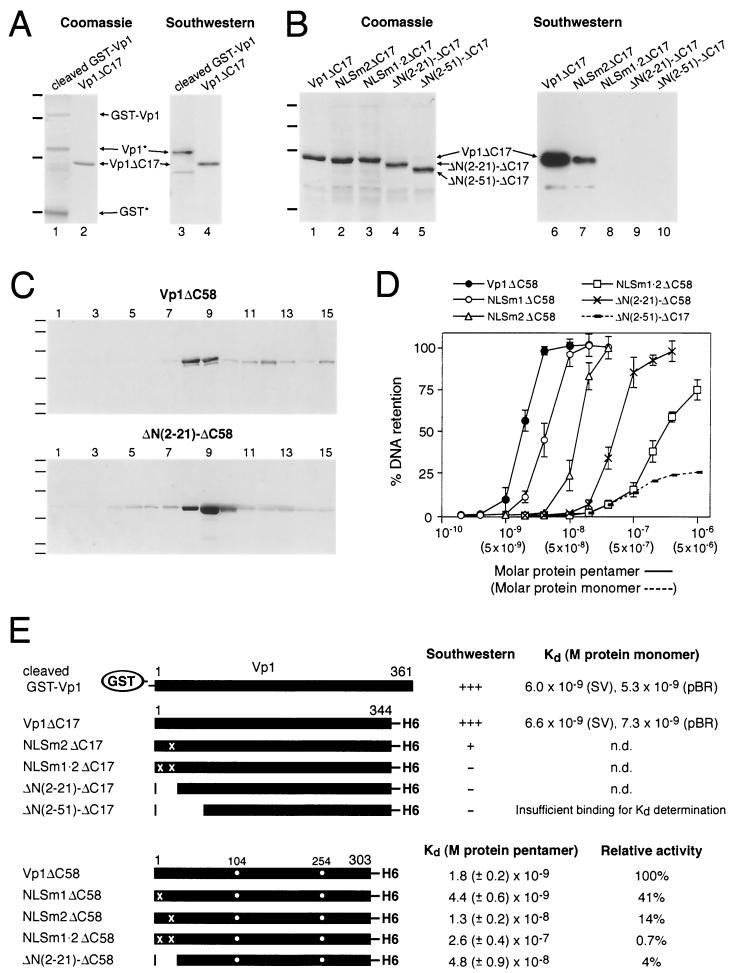
FIG. 1.
DNA binding by recombinant Vp1 proteins. (A and B) Southwestern blots. Protein samples were resolved on SDS–10% polyacrylamide gels and either stained with Coomassie blue (A, lanes 1 and 2; B, lanes 1 to 5) or electrotransferred to nitrocellulose and probed with nick-translated 32P-labeled SV40 DNA (10 to 12 ng/ml) (A, lanes 3 and 4; B, lanes 6 to 10). In panel A, 0.5 μg of Vp1 (Vp1∗, lanes 1 and 3) as a cleavage product of GST-Vp1, or 0.5 μg of Vp1ΔC17 (lanes 2 and 4), were used. Intact GST-Vp1 (GST-Vp1) and the GST moiety (GST∗) were also present in the samples of lanes 1 and 3. In panel B, 2 μg of Vp1ΔC17 (lanes 1 and 6) or of the following N-terminal mutant ΔC17 proteins were used: NLSm2 (lanes 2 and 7), NLSm1 · 2 (lanes 3 and 8), ΔN(2–21) (lanes 4 and 9), and ΔN(2–51) (lanes 5 and 10). Four bars to the left of each Coomassie gel mark the positions (from top to bottom) for molecular mass standards of 110, 74, 45, and 26 kilodaltons. (C) Isolation of pentameric Vp1ΔC58 and ΔN(2–21)-ΔC58. Sedimentation through sucrose gradients was performed as described in Materials and Methods, and an aliquot from each of the 15 fractions was analyzed by SDS-PAGE and Coomassie blue staining. In the profiles shown, twice as much protein was sedimented for ΔN(2–21)-ΔC58 (lower panel) than for Vp1ΔC58 (upper panel). Six bars to the left of each gel mark the positions for six molecular mass standards of 100, 71, 44, 28, 19, and 14 kilodaltons. Pentamers were found in fractions 8 and 9 for Vp1ΔC58 and in fractions 8 through 10 for ΔN(2–21)-ΔC58. NLSm1ΔC58, NLSm2ΔC58, and NLSm1 · 2ΔC58 gave sedimentation profiles similar to that of Vp1ΔC58. (D) Solution-phase DNA binding. Filter-binding assays were performed by incubating various concentrations of each protein with 32P-labeled DNA, and the percentages of the input radiolabel that was retained on nitrocellulose membrane upon filtration were determined. Average values from three experiments are shown with error bars for the binding of a 326-bp PCR-derived SV40 fragment by pentameric Vp1ΔC58 or its N-terminal mutant derivatives. Values from one experiment are shown for the binding of nick-translated SV40 DNA by ΔN(2–51)-ΔC17 whose monomeric concentrations are given in parentheses. Dissociation constants (Kds) were determined as molar protein concentrations at 50% DNA retention. (E) Summary of DNA-binding activities for recombinant Vp1s. For GST-Vp1-derived Vp1 and for Vp1ΔC17, the apparent Kd was given in protein monomer concentration for the binding of nick-translated SV40 (SV) or pBR322 (pBR) DNA. For ΔC58 proteins, an average Kd along with the standard deviation was given in the protein pentamer concentration for the binding of a 326-bp SV40 fragment. The relative activity is the reciprocal of Kd made relative to that of the Vp1ΔC58 Kd, which was taken to be 100%. An “X” on the schematic protein diagram represents the mutation of an N-terminal basic cluster; a dot beneath residues 104 and 254 indicates their mutation from cysteines into alanines, n.d., not done.
TABLE 1.
Multiple-point mutants of the Vp1 N terminus
 |
Underlining indicates the two constituent basic clusters of the bipartite NLS, NLS1, and NLS2. Dashes are used to denote the same amino acids as wild-type Vp1.
pSG5-Vp1ΔC58-GFP was used for the transient mammalian cell expression of fluorescently tagged Vp1 fusion proteins. The proteins consist of the first 303 Vp1 amino acids connected to the red-shifted green fluorescsent protein (GFP) by a flexible linker, which is made up of three repeats of (glycine)4-serine [(G4S)3]. First, pSG5-XNAB was made from pSG5 (Stratagene) by knocking out the XbaI site and inserting into EcoRI and BamHI sites a linker that introduces unique XbaI, NotI, AgeI, and BsrGI sites. pSG5-Vp1ΔC58-GFP was then constructed by inserting three fragments into pSG5-XNAB: the 925-bp XbaI-to-NotI Vp1 fragment from pBS-Vp1-ΔC58, the 85-bp NotI-to-AgeI (G4S)3 fragment from pETC-64M5-NotAge, and the 722-bp AgeI-to-BsrGI GFP fragment from pEGFP-1 (Clontech). pETC-64M5-NotAge was made from pETC-64M5LH15His (17) by inserting a NotI linker through KpnI and NheI sites and inserting an AgeI linker through EcoRI and SacI sites.
The NLSm1, NLSm2, or NLSm1 · 2 derivatives of pBS-Vp1 (19), of pSG5-Vp1ΔC58-GFP, and of the viral plasmid NO-pSV40 (13) were obtained by substituting the 206-bp XbaI-to-AflII fragments from pSV-Vp1 -p567, -p567, -p81, or -p25, respectively. NO-SV40 DNAs were prepared from respective NO-pSV40 plasmids by BamHI digestion and ligation as previously described (13). pBS-Vp1 plasmids were used for the in vitro synthesis of Vp1 proteins from T7 promoter-driven coding sequences.
Preparation of recombinant proteins.
GST (glutathione S-transferase)-Vp1 was expressed from pGEX-Vp1, purified, and cleaved with factor Xa as described previously (7).
For H6-tagged Vp1s, XL1-Blue Escherichia coli cells harboring individual pQE-Vp1 plasmids were induced at the early log phase with 0.2 to 0.5 mM IPTG for 6 h at 30°C or for 24 h at 20°C. H6 proteins were purified using the Talon metal affinity resin (Clontech) according to the supplier's protocols as follows. Vp1ΔC17 and N-terminal-mutant ΔC17 proteins were purified according to the denaturing-buffer protocol, and the eluted protein in 20 mM Tris-Cl (pH 8.0)–100 mM NaCl–8 M urea–100 mM imidazole was dialyzed against changes of 20 mM Tris-Cl (pH 8.0)–10 mM NaCl–0.1 mM β-mercaptoethanol in which the concentration of urea was decreased from 8 M to none. The dialysate was removed of insoluble materials by a 15-min centrifugation at 15,000 × g and concentrated using a Microcon-30 concentrator (Amicon). Except for ΔN(2–51)-ΔC58, which failed to express to useful quantities in bacteria, Vp1ΔC58 and N-terminal-mutant ΔC58 proteins were prepared as follows. Each protein was purified from lysozyme- and DNase I-treated, sonicated bacterial lysate according to the native-buffer protocol, and the pentamer fraction was isolated by sedimenting 150 to 300 μg of the eluted protein, in 20 mM Tris-Cl (pH 8.0)–100 mM NaCl–100 mM imidazole, through a 10.5-ml, 5 to 20% continuous sucrose gradient in the same buffer without imidazole for 23 h at 35,000 rpm at 4°C in an SW41 rotor. Fifteen fractions were collected from the bottom of the gradient, and two to three peak pentamer fractions (see Fig. 1C), containing 50 to 75% of the total input protein, were pooled for use.
All protein preparations were quantitated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining and were stored in aliquots at 0.1 to 1 mg/ml at −70°C.
DNA-binding assays.
Southwestern blots were performed as previously described (without dithiothreitol in the blotting buffer) (4) using nick-translated, 32P-labeled SV40 DNA (4) and 0.4 to 2.0 μg of cleaved GST-Vp1 or ΔC17 Vp1s. Filter-binding assays (4) were performed using nick-translated, 32P-labeled SV40 or pBR322 DNA for cleaved GST-Vp1 and ΔC17 Vp1s, or using a 326-bp, 32P-labeled PCR fragment of SV40 DNA (see below) for ΔC58 pentamers. Each binding experiment used at least seven protein concentrations, and an apparent Kd was determined from the binding curve as the protein monomer or pentamer concentration at which 50% of the DNA probe was retained on the filter. An average Kd value from three experiments was tabulated along with the standard deviation.
The 326-bp labeled fragment, corresponding to SV40 nucleotides 1670 to 1996, was amplified by PCR in a 20-μl reaction containing 1 ng of NO-pSV40 as template; 10 pmol each of sense and antisense primers; SV40 nucleotides 1670 to 1689 and 1996 to 1976; 120 μCi of [α-32P]dATP (ICN; specific activity, 3,000 Ci/mmol); 150 nM each of dCTP, dGTP, and dTTP; 37 nM cold dATP; and 5 U of Taq DNA polymerase (Gibco-BRL) in 1× Mg2+-free PCR buffer (Gibco-BRL) supplemented with 2 mM MgCl2. The reaction was brought to 94°C for 1 min and cycled 35 times through 94°C for 45 s, 60°C for 45 s, and 72°C for 2 min, and the approximately 1 μg of PCR product was purified using the Qiaquick PCR Purification Kit (Qiagen).
Immunofluorescence and plaque assays.
TC7 cells were nuclearly microinjected with NO-SV40 DNAs and analyzed for the localization of virally encoded Vp1 and Vp2/3 by indirect immunofluorescence microscopy (3, 7) or analyzed for plaque formation (29) with the modification that a total of five agar overlays were performed, and cells were visualized on day 25. Plaque assays were also performed by transfecting CV-1 cells with NO-SV40 DNAs and infecting the harvested cell lysates onto new cells as described previously (19). To examine the localization of wild-type or mutant Vp1s expressed alone, cells were transfected as before (19) with pSG5-Vp1ΔC58-GFP DNAs, fixed at 24 h posttransfection with 3.7% formaldehyde in phosphate-buffered saline, and processed for anti-Vp1 immunofluorescence as before (3, 7). The autofluorescence of GFP was also recorded.
Analyses for viral DNA replication, capsid protein production, and viral DNA packaging.
To examine viral DNA replication, CV-1 cells on 60-mm dishes were transfected with NO-SV40 DNAs as described above and harvested at various hours posttransfection for the extraction of total viral DNAs by the Hirt method (12). One-tenth of each DNA preparation was analyzed by Southern slot blot using radiolabeled SV40 DNA as a probe. Individual slots were excised from the membrane and quantitated for radioactivity in a liquid scintilation counter.
To examine the steady-state levels of virally encoded capsid proteins, CV-1 cells on a 60-mm dish were transfected with a 4:1 molar mixture of each NO-SV40 DNA and pmiwZ (28), which expresses β-galactosidase under the control of a complex of Rous sarcoma virus and β-actin promoter-enhancers. The cells were harvested at 72 h posttransfection, and aliquots of cells containing equal β-galactosidase activities as determined from the β-Galactosidase Assay Kit (Stratagene) were analyzed by anti-Vp1 and anti-Vp3 Western blots using polyclonal rabbit anti-Vp1 and anti-Vp3 sera (16) and the Enhanced Chemiluminescence Western Blotting System (Amersham-Pharmacia).
To examine viral DNA packaging, NO-SV40 DNAs-transfected CV-1 cells were harvested at 72 h posttransfection and analyzed as described previously (19). Briefly, cells from each 150-mm dish were lysed by sonication in 0.5 ml of hypotonic buffer, and the total viral DNA and DNase I-resistant viral DNA were extracted from aliquots of the cell lysate. After the extracted viral DNAs were linearized with KpnI, and the proportion of DNase I-resistant to total viral DNA was determined by Southern blot and phosphorimaging. A value of 65 to 80% was obtained for wild-type NO-SV40 transfected lysate. To examine the presence of virions or virion-like particles, DNase I-treated lysates were sedimented through 5 to 32% sucrose gradient, and the resulting fractions were analyzed for total viral DNA by Southern blot as before (19), as well as analyzed for Vp1 by Western blot using polyclonal rabbit anti-Vp1 serum and 125I-labeled protein A detection as described elsewhere (21). Note that the amount of DNase I used in the above assays was several times higher than was needed to completely degrade free viral DNA and yet did not noticeably affect the integrity of wild-type virion particles (19).
Vp3 interaction assay.
The GST-Vp3 fusion protein and its preparation from the soluble fraction of the bacterial lysate by binding to glutathione affinity resin have been described (4). 35S-labeled Vp1 proteins were synthesized by in vitro transcription and translation of pBS-Vp1 plasmids as described previously (19). For the interaction assay, 5 μl of a slurry of glutathione resin bound with 10 pmol of GST-Vp3 was extensively washed and then reacted for 30 min at room temperature with 50 fmol of 35S-labeled wild-type or NLS-mutant Vp1 in 400 μl of a buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, and 10 μg of aprotinin, 10 μg of leupeptin, 10 μg of pepstatin, and 50 ng of ethidium bromide per ml). The resin was collected and extensively washed with the same buffer without ethidium bromide, and the bound proteins were analyzed by SDS-PAGE and fluorography using the Amplify reagent (Amersham-Pharmacia). In control assays, the same amount of resin-bound GST, expressed from pGEX-3X (Amersham-Pharmacia)-transformed bacteria, was used instead of resin-bound GST-Vp3.
RESULTS
DNA binding by recombinant SV40 Vp1 proteins.
We used two types of in vitro assays to examine the DNA-binding abilities of E. coli-expressed recombinant Vp1s. In the Southwestern assay, proteins resolved by SDS-PAGE were transferred to nitrocellulose and renatured on the membrane before probing with labeled DNA. GST-Vp1 was inactive in DNA binding (Fig. 1A, GST-Vp1 band in lanes 1 and 3) unless the Vp1 moiety (Fig. 1A, Vp1∗ band in lanes 1 and 3) was freed by proteolytic cleavage at its amino-terminal junction with GST. Vp1ΔC17, with a natural N terminus and a C-terminal polyhistidine tag, was active in Southwestern assay (lanes 2 and 4). To estimate the DNA binding affinities of GST-Vp1-derived Vp1 and Vp1ΔC17, a solution-phase filter-binding assay was used. Both proteins had dissociation constants (Kds) in the range of 5.3 × 10−9 to 7.3 × 10−9 M for interacting with either SV40 or pBR322 DNA (Fig. 1E). The recombinant Vp1's apparent lack of DNA-sequence specificity (Fig. 1E) is in agreement with a previous report (26).
Since Vp1ΔC17 was found mostly in inclusion bodies in bacteria, its purification required urea denaturation and subsequent renaturation steps. We sought to improve recombinant Vp1 design and preparation in the following ways. First, recombinant SV40 Vp1s can be expected to exist as pentamers or their complexes, similar to bacterially purified polyomavirus Vp1 (24). Deleting up to about 58 amino acids from the C terminus should eliminate the assembly of SV40 Vp1 pentamers into higher complexes without affecting pentamerization. Second, urea-denatured proteins may not regain proper folding and tend to aggregate upon the denaturant's removal. It would be preferable to use native buffer conditions throughout purification. Third, Vp1 pentamers may also aggregate through disulfide linkages, such as those between cysteines 104 observed in virion crystal structure (27) or perhaps artifactual ones involving cysteine 254. Eliminating these cysteines should minimize such cross-links. Hence, we expressed the C-terminal-58 truncated Vp1ΔC58 in which cysteines 104 and 254 are replaced by alanines, purified it under native buffer conditions, and isolated the pentamer fraction in a sucrose gradient (Fig. 1C). To derive a Kd that is more likely to reflect the one-to-one stoichiometric interaction between the pentamer and DNA, the filter-binding assay was performed using 326-bp, PCR-generated SV40 DNA probe rather than nick-translated DNA, which may consist of fragments that are longer or heterogeneous in length. Under these conditions the Kd measured for Vp1ΔC58 was 1.8 (±0.2) × 10−9 M in terms of the protein pentamer (Fig. 1D and E). Thus, the DNA-binding ability of Vp1 as a homogeneous, C-terminal-58 truncated pentamer was comparable to those of full-length Vp1 and Vp1ΔC17. These results indicated that the Vp1 C terminus does not mediate DNA binding and led us to investigate whether the Vp1 DBD lies in the N terminus.
The Vp1 DBD overlaps with the bipartite Vp1 NLS.
The N terminus of Vp1 is likely to harbor a DBD because of its proximity to the core of the virion (20) and by analogy to the mapped polyomavirus Vp1 DBD (22). It is also the location for the previously identified bipartite NLS (14), which consists of two clusters of basic residues. We will refer to the first cluster, 4-lysine-arginine-lysine-6, as NLS1 and to the second cluster, 15-lysine-lysine-proline-lysine-18, as NLS2 (Table 1). To test whether these basic clusters also mediate Vp1 DNA binding, sets of multiple-point mutations (NLSm1, NLSm2, and NLSm1 · 2) and N-terminal truncations [ΔN(2–21) and ΔN(2–51)] were introduced into Vp1ΔC17 and Vp1ΔC58, and DNA-binding assays were performed. In NLSm1, NLS1 is mutated into asparagine-glycine-asparagine; in NLSm2, NLS2 is changed to asparagine-asparagine-proline-asparagine; and in NLSm1 · 2, both clusters are correspondingly changed (Table 1). In ΔN(2–21) or ΔN(2–51), residues 2 to 21 or residues 2 to 51, respectively, were deleted. Among the mutant ΔC17 proteins, NLSm2 had partial activity in the Southwestern blot, while NLSm1 · 2 and both N-terminal deleion mutants were inactive (Fig. 1B), suggesting that the basic clusters were necessary for the DNA binding. For the N-terminal mutant ΔC58 proteins, the Kds were determined with purified pentamers as before. The multiple-point mutant ΔC58 proteins sedimented in the sucrose gradients with a similar profile as Vp1ΔC58 (data not shown), while the N(2–21)-ΔC58 pentamer sedimented somewhat slower than Vp1ΔC58, an expected result for the additional deletion (Fig. 1C). We found that mutants NLSm1, NLSm2, ΔN(2–21), and NLSm1 · 2 bound DNA with progressively decreased affinities; their Kds in terms of the protein pentamer, 4.4 (±0.6) × 10−9, 1.3 (±0.2) × 10−8, 4.8 (±0.9) × 10−8, and 2.6 (±0.4) × 10−7, respectively, correspond to 41, 14, 4, and 0.7% of the Vp1ΔC58 activity, respectively (Fig. 1D and E). A Kd was not determined for ΔN(2–51)-C58 because of difficulty with its bacterial expression, but its ΔC17 counterpart, ΔN(2–51)-ΔC17, was found not to have sufficient DNA-binding affinity for Kd measurement (Fig. 1D and E). These results indicate that NLS1 and NLS2—that is, NLS1-DBD1 and NLS2-DBD2—represent essential parts of the Vp1 DBD, with the latter part contributing more to DNA binding than the former part. The N-terminal 22-to-51 region also appears to harbor a small fraction of total binding activity.
The NLS function of the N-terminal basic clusters was confirmed in cells transfected with pSG5-Vp1ΔC58-GFP, for which the fusion protein of Vp1 N-terminal 303 residues and GFP was expressed from the SV40 early promoter in the absence of Vp2/3 and the tumor antigens. Both the Vp1 immunofluorescence (Fig. 2) and the GFP autofluorescence (data not shown) indicated that, while wild-type Vp1ΔC58-GFP localized effectively to the nucleus (Fig. 2a), the mutant counterpart NLSm1, NLSm2, and NLSm1 · 2 proteins largely accumulated in the cytoplasm (Fig. 2b, c, and d). A mostly cytoplasmic localization pattern was also observed for full-length mutant Vp1s expressed from pSV-Vp1-p81 (NLSm2) and -p25 (NLSm1 · 2), in which the tumor antigen coding sequences are present, and from pSG5-Vp1-p25 (14). Therefore, the two basic clusters within N-terminal 21 serve as the NLS of Vp1. The collective results indicate that the DBD of Vp1 overlaps with the NLS, sharing the requirement for at least the two basic amino acid clusters.
FIG. 2.
Subcellular localization of NLS-mutant Vp1ΔC58-GFP proteins. Cells transfected with wild-type (a), NLSm1 (b), NLSm2 (c), and NLSm1.2 mutant pSG5-Vp1ΔC58-GFP were fixed and stained with guinea pig anti-Vp1, followed by rhodamine-labeled anti-guinea pig antibody. Photographs of the rhodamine fluorescence are shown. The GFP autofluorescence in each case gave an essentially identical pattern, although the intensity was less than the corresponding Vp1 immunofluorescence.
Importance of Vp1 NLS-DBD for viability.
To test the role of the overlapping Vp1 DBD and NLS in the viral life cycle, we examined the viability of nonoverlapping SV40 (NO-SV40) genomes into which NLSm1, NLSm2, and NLSm1 · 2 mutations had been introduced. NO-SV40 is a viable SV40 DNA containing all of the regulatory sequences, the early genes, and spatially separated Vp2/3 and Vp1 coding sequences (13). Two types of viability assays were performed, one by microinjection of the viral DNA and the other by infection of the viral DNA-transfected cell lysate. Mutants NLSm1 and NLSm1 · 2 failed to produce plaques in the microinjection assay, and the infection assay confirmed NLSm1 to be incapable of producing infectious particles (Table 2). Compared with wild-type NO-SV40, mutant NLSm2 formed smaller plaques in the microinjection assay and formed plaques that were 5,000 times fewer, as well as smaller, in the infection assay (Table 2).
TABLE 2.
Plaque-forming abilities of NO-SV40 Vp1 NLS-DBD mutants
| NO-SV40 | Formation of plaquesa | Titer in transfected cell lysate (PFU/ml)c | Mean plaque diam (mm)c ± SD |
|---|---|---|---|
| Wild-type | Yes | 1.8 × 108 | 5.0 ± 1.2 |
| NLSm1 | No | 0d | |
| NLSm2 | Yesb | 3.5 × 104 | 0.8 ± 0.3 |
| NLSm1.2 | No | ND |
In microinjection assay.
The average plaque diameter was ca. one-fourth that of the wild type.
Determined from infection-type plaque assay. ND, not done.
No plaques were detected in 0.1 ml of transfected cell lysate.
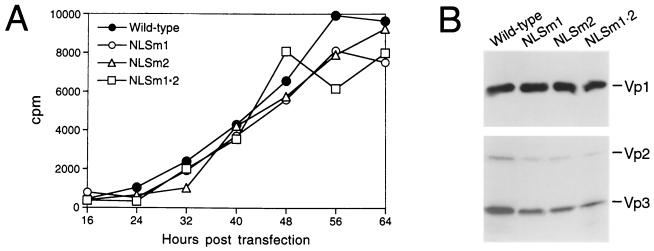
To determine which stages of the viral life cycle were affected by the Vp1 NLS mutations, we examined the state of various viral processes, beginning with viral DNA replication and capsid protein production. The amount of intracellular viral DNA, quantitated by Southern slot blot, increased steadily with increasing times posttransfection for the wild type and the three Vp1 NLS-mutant DNAs (Fig. 3A). Comparable amounts of Vp1, Vp2, or Vp3 were detected by Western blot in cell lysates from transfection with either wild-type or the three mutant viral DNAs (Fig. 3B). Thus, defects other than those in viral DNA replication or capsid protein production were responsible for the abolished or greatly reduced viabilities of the Vp1 NLS-DBD mutants.
FIG. 3.
DNA replication and capsid protein production by N-terminal Vp1 mutants. (A) Time course of viral DNA replication. Cells transfected with each NO-SV40 DNA were harvested at the indicated time points, and the total intracellular viral DNA was extracted and probed with nick-translated SV40 DNA in a Southern slot blot. The radioactivities of the individual slots were counted and plotted against time. (B) Levels of capsid proteins. Cells transfected with individual NO-SV40 DNAs were analyzed by immunoblotting with anti-Vp1 (upper panel) or anti-Vp3 (lower panel) antibodies. The amount of cells analyzed was adjusted for transfection efficiency as measured by the activity of β-galactosidase expressed from pmiwZ, which was cotransfected with each NO-SV40 DNA. Bands corresponding to Vp1, Vp2, and Vp3 are indicated at the right.
Nuclear localization defect of mutant Vp1s expressed by NO-SV40-NLSm1 and-NLSm1 · 2.
We next examined the NO-SV40 mutants for the subcellular distribution of the capsid proteins. Since mutations in either or both of NLS1 and NLS2 impaired the nuclear localization of a Vp1 fusion protein (Fig. 2), at least two different scenarios are possible when the mutant Vp1s were present along with Vp2/3 in cells introduced with the mutant viral DNAs. One is that the intact NLSs of wild-type Vp2/3 can complement the defective NLS of Vp1, as observed for NO-SV40-Vp1ΔN5 (13), and the mutant Vp1 is piggybacked to the nucleus by Vp2/3. This phenotype was seen for mutant NLSm2. Both the mutant Vp1 and the Vp2/3 were nuclearly localized (Fig. 4e and f), as were the capsid proteins expressed by wild-type NO-SV40 (Fig. 4a and b). The second scenario is that NLS complementation fails to occur, for example, because cytoplasmic Vp1-Vp2/3 interaction is somehow blocked; as a result, Vp2/3, but not the mutant Vp1, enters the nucleus. This phenotype was observed for mutants NLSm1 and NLSm1 · 2, whose mutant Vp1s remained in the cytoplasm despite the nuclear localization of Vp2/3 (Fig. 4c, d, g, and h). Thus, the major defect of nonviable mutants NLSm1 and NLSm1 · 2 appears to lie in the inability of the mutant Vp1s produced to localize to the nucleus. This defect could arise from the mutant Vp1s' lack of intrinsic Vp2/3-interactive ability or from the mutant Vp1s' inability to reach a cytoplasmic site necessary for the Vp1-Vp2/3 interaction. The former possibility is examined below.
FIG. 4.
Subcellular localization of capsid proteins expressed from Vp1 N-terminal-mutant NO-SV40 DNAs. Cells were nuclearly microinjected with individual NO-SV40 DNAs, cultured for 24 h, fixed, and doubly stained with guinea pig anti-Vp1 (a, c, e, and g) and rabbit anti-Vp3 (b, d, f, and h), followed by rhodamine-labeled (a, c, e, and g) or fluorescein-labeled (b, d, f, and h) secondary antibodies. The same cells are shown in panels a and b, c and d, e and f, and g and h.
In vitro interaction of mutant Vp1s with Vp3.
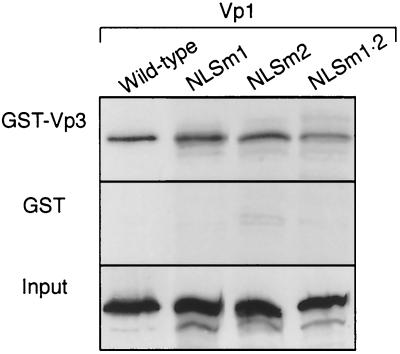
To determine if the N-terminal mutant Vp1 proteins had the intrinsic ability to associate with Vp3, an in vitro interaction assay was performed. In vitro-transcribed and-translated, 35S-labeled NLSm1, NLSm2, and NLSm1 · 2 mutant Vp1s bound resin-immobilized GST-Vp3, as did wild-type Vp1, whereas little of the Vp1s bound to GST alone (Fig. 5). Thus, the intrinsic interaction between Vp1 and Vp2/3 was not affected by the Vp1 N-terminal mutations.
FIG. 5.
Interaction of N-terminal-mutant Vp1s with Vp3. In vitro-transcribed and -translated, 35S-labeled Vp1 from each pBS-Vp1 DNA was mixed with the resin to which either GST-Vp3 (first panel) or GST (second panel) had been immobilized. The resin-bound proteins were analyzed by SDS-PAGE and fluorography. The amounts of input 35S-labeled Vp1s used for the pull-down experiments are also shown (third panel).
Nuclear virion assembly defect of NO-SV40-NLSm2.
Since all capsid proteins of mutant NLSm2 were produced in normal quantities and could localize to the nucleus, and the mutant DNA replicated normally, we tested whether the mutant could form virion-like particles. When viral DNA-transfected cell lysates were treated with DNase I, only 6.9% of the total mutant DNA remained, compared with 71% of the total wild-type DNA. Thus, mutant NLSm2 either packaged only 1/10 the amount of viral DNA as wild-type NO-SV40 or packaged the viral DNA mostly in a manner that left the DNA susceptible to DNase I digestion.
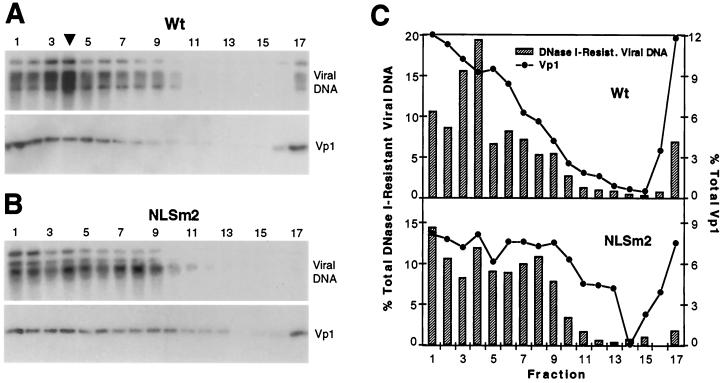
To determine if the protected mutant DNA was packaged in a particle form, the DNase I-resistant materials from wild-type and NLSm2 DNA-transfected lysates were examined by sedimentation through sucrose gradients. To adjust for the 10-fold-reduced amount of the nuclease-resistant mutant viral DNA, 10 times as much nuclease-treated NLSm2 sample as the corresponding wild-type sample was sedimented, and equal aliquots of wild-type and NLSm2 sucrose fractions were analyzed for viral DNA by Southern blot (Fig. 6). Since Vp1, whether wild type or mutant, would quantitatively remain after the nuclease treatment, it would be much more abundant in the 10-times-larger NLSm2 sample than in the wild-type sample. Accordingly, one-fifth as much of the NLSm2 fractions than the wild-type fractions was analyzed by anti-Vp1 Western blot (Fig. 6). The distribution profiles showed that wild-type viral DNA and Vp1 were present mostly in fractions 1 through 9 as well as 17 (Fig. 6A and C). About 42% of the wild-type DNA resided in fractions 3 through 5, which corresponded to the sedimentation location of purified virions (Fig. 6C). The distributions of NLSm2 DNA and the mutant Vp1 were somewhat broader than their wild-type counterparts, with comparatively more of both mutant DNA and Vp1 present in fractions 7 through 9 (Fig. 6B and C). Nonetheless, about 30% of the mutant viral DNA was present in the expected particle fractions 3 through 5 (Fig. 6C). These results indicated that mutant NLSm2 could form virion-like particles, though at a reduced level. Taken together, these results point to a reduced level of viral DNA packaging by mutant NLSm2 and hence a role for NLS2-DBD2 in the virion assembly process.
FIG. 6.
Virion particle formation by NO-SV40-NLSm2. (A and B) Sonicated lysate prepared from wild-type (Wt, A) or mutant NLSm2 (NLSm2, B) NO-SV40 DNA transfected cells was treated with DNase I, and aliquots containing equivalent amounts of DNase I-resistant viral DNAs (60 μl for wild-type raised to 600 μl with buffer and 600 μl for NLSm2) were sedimented through 5 to 32% sucrose gradients and fractionated from the bottom into 17 fractions. Five-sixths of each fraction was analyzed for viral DNA by Southern blot (upper panels). One-sixth of each wild-type fraction or one-thirtieth of each NLSm2 fraction was also analyzed for Vp1 by Western blot (lower panels). An arrowhead points to fraction 4, the peak fraction for viral DNA and Vp1 from purified virions sedimented in a parallel gradient. (C) Distribution of post-DNase I viral DNA and Vp1 in sucrose fractions. Wild-type (upper plot) and NLSm2 (lower plot). For each Southern or Western profile, the radioactivity per lane of the DNase I-resistant viral DNA or Vp1, obtained by phosphorimager quantitation of the observed band(s), was made relative to the total radioactivity of the DNA or Vp1 for all 17 fractions, which was taken to be 100%. The percentages were plotted against the fraction number.
DISCUSSION
In this study, we mapped within the N-terminal 21 amino acids of SV40 Vp1, a DBD that accounted for most (96 to 99%) of the in vitro-measured Vp1 DNA-binding activity. This DBD overlaps the two basic clusters, NLS1-DBD1 and NLS2-DBD2, of the bipartite Vp1 NLS (14). Both basic clusters are required for full DNA-binding activity, though the second cluster (15-lysine-lysine-proline-lysine-18) is more important for the binding than the first cluster (4-lysine-arginine-lysine-6). The biological functions of the overlapping DBD and NLS were then dissected in vivo using mutant NO-SV40 viral genomes with the application of the NLS phenotypic complementation we have previously established (13). Nonviable multiple-point mutants NLSm1 and NLSm1 · 2 were exceptional in that their mutant Vp1s were unable to enter the nucleus despite the presence of Vp2/3, though the mutant Vp1s' ability to interact with Vp3 in vitro was not affected. For a multiple-point mutant of NLS2-DBD2, NLSm2, a greatly decreased viability was correlated with a decreased level of virion packaging but not with any defects in viral DNA replication or capsid protein production, interaction, or nuclear localization. Our results demonstrated, for the first time, that NLS2-DBD2 is important for the formation of infectious particles and that the Vp1 DNA-binding function is likely to have an essential role in this process. The N-terminal location of the DBD is consistent with the crystallographic structure of the virion in which the Vp1 N terminus is oriented toward the minichromosomal core and is disordered (20).
To obtain a Kd for homogeneous Vp1 pentamers and to favor a one-to-one molar pentamer-DNA interaction, we used the purified Vp1ΔC58 pentamer, mutated in cysteines 104 and 254 to minimize artifactual pentamer-pentamer aggregation, and a short (326-bp) DNA probe. A Kd of 1.8 × 10−9 M in terms of the protein pentamer was obtained. Two other recombinant Vp1s, cleaved GST-Vp1 and Vp1ΔC17, had apparent Kds 5.3 × 10−9 to 7.3 × 10−9 M in terms of the protein monomers, which are roughly comparable to the Vp1ΔC58 pentamer value and reinforce the reliability of our assays. Whereas the DBD of SV40 Vp1 comprises NLS1 and NLS2, that of the related polyomavirus Vp1 consists of a single basic cluster, 1-alanine-proline-lysine-arginine-lysine-5 (2, 22), with a sequence resemblance to that of NLS1-DBD1. For SV40, it is NLS2-DBD2 (represented by mutant NLSm2) that was attributed with a larger share of DNA-binding activity and a role in in vivo virion packaging. Whether mutating the polyomavirus Vp1 DBD impairs polyomavirus particle packaging would make an interesting comparison. The reported apparent Kd for DNA binding by polyomavirus Vp1 is 1-2 × 10−11 M in term of the protein monomer (22). The difference between this value and the values for SV40 Vp1 may reflect either a comparatively higher DNA-binding affinity of polyomavirus Vp1 or differences in the assay methods employed.
The DBD and the bipartite NLS of SV40 Vp1 overlap within the N-terminal 21 residues. A number of other proteins, including polyomavirus Vp1, influenza virus matrix protein M1, and proteins with bHLH/bZIP, zinc finger, and homeobox domains, are also known to harbor a DNA- or RNA-binding domain in overlap with an NLS (18). This overlapping or bifunctional domain arrangement is not surprising, since clustered basic residues typify many known NLSs and nucleic acid-binding domains (8, 18). Nevertheless, we were able to distinguish the two functions in our analysis (see below). Our data, however, do not rule out a possibility that Vp1 amino acids other than the first 21 residues also make an additional contribution to DNA binding. Such residues would conceivably be at the base of the Vp1 pentamer. This question is not addressed in this study.
The functional duality of the Vp1 N-terminal domain can potentially complicate the determination of how an individual domain function contributes to viral viability. Yet by applying NLS phenotypic complementation, we found that the phenotype of viral mutant NLSm2 was consistent with a role for the Vp1 DBD in nuclear virion assembly. This mutant was viable but had a 5,000-fold-lowered infectious titer and notably smaller plaque sizes. The pattern of capsid protein nuclear localization is consistent with the reported functional complementation of NLSs, in which an NLS-defective Vp1 (e.g., Vp1ΔN5) accumulated in the nucleus in the presence of wild-type Vp2/3 as a result of the capsid proteins' association in the cytoplasm prior to nuclear entry (13). Given this effective complementation of NLSm2 Vp1 nuclear localization, the normal levels of viral DNA replication and capsid protein production, and the normal ability of the mutant Vp1 to interact with Vp3 in vitro, the mutant's reduced viability is likely the result of the mutant Vp1's impaired DNA-binding ability. There are at least two major ways a compromised Vp1 DNA-binding function can lead to a lower viral infectivity. First, impaired Vp1 DNA binding could affect the efficiency of DNA packaging in the nucleus and reduce number of particles produced. Second, the mutant virion-like particles assembled may be less stable or less effective at cell entry or nuclear targeting steps in a new round of infection. Our NLSm2 results reflect defects possibly at both stages of the viral life cycle. The observed 10-fold reduction in the packaging of the viral DNA (Fig. 6) would certainly lower the yield of potentially infectious particles. In view of the 5,000-fold-lowered overall viability, it is also likely that the particles that did form were less infectious than wild-type particles in the next infection cycle. Further experiments would be needed to address the defect of mutant NLSm2 particles during reinfection.
Distinctly different from the apparent defect of mutant NLSm2 is the intriguing phenotype of mutants NLSm1 and NLSm1 · 2, which harbor mutations in the first basic cluster, NLS1. In contrast to NO-SV40-NLSm2 (above) and -Vp1ΔN5 (13), these two nonviable mutants produced cytoplasmically localized mutant Vp1s despite the nuclear localization of Vp2/3, indicating that the defective NLSs of the mutant Vp1s were not complemented by the presence of wild-type Vp2/3 NLS. This phenotype also indicates that Vp2/3 could enter the nucleus independently of the mutant Vp1s. Two lines of evidence support the hypothesis that the mutated NLS1 acts as a dominant signal for the cytoplasmic retention of NLSm1 and NLSm1 · 2 Vp1s. First, since all mutant Vp1 proteins were made in the mutant DNAs-transfected cells (Fig. 3B) and could interact with Vp3 in vitro (Fig. 5), the lack of complementation suggests that mutant Vp1s were prevented from interacting with Vp2/3 in vivo. Second, deleting NLS1 as in the case of NO-SV40-Vp1ΔN5 actually restored NLS complementation (13). Thus, the evidence points to the presence of an unidentified function, aside from those of nuclear localization and DNA binding, in NLS1-DBD1. That is, NLS1 may define a process in the cytoplasmic Vp1 biosynthetic pathway that precedes the association and the nuclear import of Vp1 and Vp2/3. We observed that Vp1 bearing single arginine 5-to-glycine point mutation (also part of the NLSm1 mutations) exhibited a cytoskeleton-like localization pattern when expressed from a pSG5-based plasmid (N. Ishii and N. Minami, unpublished results) but showed a diffuse cytoplasmic staining when expressed from pSV-Vp1-p1 (14), from which the tumor antigens were also expressed. The suggestive role of cytoskeletal elements in Vp1's nuclear transport and association with Vp2/3 remains to be determined.
In summary, our study has mapped a DBD on SV40 Vp1 and has found that the DNA-binding contribution from NLS2-DBD2 plays a role during virion assembly. This Vp1 region may also function during next-cycle infection. DBDs have now been defined for all three SV40 capsid proteins. These domains are expected to function in conjunction with other known and as-yet-unknown interactive domains during the packaging of viral DNA.
ACKNOWLEDGMENTS
P.P.L. and A.N. contributed equally to this work.
We thank Hiroshi Morioka of Hokkaido University, Sapporo, Japan, for supplying the pETC-64M5LH15His plasmid DNA. We also thank Mary A. Tran for assistance in constructing pQE-Vp1-2CA-ΔC58.
This work was supported by Public Health Service grant CA50574 from the National Institutes of Health (NIH) and by a grant from the UCLA Academic Senate. P.P.L. was supported by a predoctoral fellowship from USPHS National Research Service Award GM07185. A.M.S. and C.F.F. were supported in part by undergraduate fellowships from, respectively, NIH Minority Scientist Development program GM55052, and NIH Minority Access to Research Careers program GM08563.
REFERENCES
- 1.Blasquez V, Beecher S, Bina M. Simian virus 40 morphogenetic pathway. J Biol Chem. 1983;258:8477–8484. [PubMed] [Google Scholar]
- 2.Chang D, Cai X, Consigli R A. Characterization of the DNA binding properties of polyomavirus capsid proteins. J Virol. 1993;67:6327–6331. doi: 10.1128/jvi.67.10.6327-6331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clever J, Kasamatsu H. Simian virus 40 Vp2/3 small structural proteins harbor their own nuclear transport signal. Virology. 1991;181:78–90. doi: 10.1016/0042-6822(91)90472-n. [DOI] [PubMed] [Google Scholar]
- 4.Clever J, Dean D A, Kasamatsu H. Identification of a DNA-binding domain in simian virus 40 capsid proteins Vp2 and Vp3. J Biol Chem. 1993;268:20877–20883. [PubMed] [Google Scholar]
- 5.Coca-Prados M, Hsu M-T. Intracellular forms of simian virus-40 nucleoprotein complexes. II. Biochemical and electron microscopic analysis of simian virus 40 virion assembly. J Virol. 1979;31:199–208. doi: 10.1128/jvi.31.1.199-208.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalyot-Herman N, Ben-nun-Shaul O, Gordon-Shaag A, Oppenheim A. The simian virus 40 packaging signal ses is composed of redundant DNA elements which are partly interchangeable. J Mol Biol. 1996;259:69–80. doi: 10.1006/jmbi.1996.0302. [DOI] [PubMed] [Google Scholar]
- 7.Dean D A, Li P P, Lee L M, Kasamatsu H. Essential role of the Vp2 and Vp3 DNA-binding domain in simian virus 40 morphogenesis. J Virol. 1995;69:1115–1121. doi: 10.1128/jvi.69.2.1115-1121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 9.Gharakhanian E, Takahashi J, Clever J, Kasamatsu H. In vitro assay for protein-protein interaction: carboxyl-terminal 40 residues of simian virus 40 structural protein Vp1 contain a determinant for interaction with Vp1. Proc Natl Acad Sci USA. 1988;85:6607–6611. doi: 10.1073/pnas.85.18.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffith J P, Griffith D L, Rayment I, Murakami W T, Caspar D L D. Inside polyomavirus at 25-Å resolution. Nature. 1992;355:652–654. doi: 10.1038/355652a0. [DOI] [PubMed] [Google Scholar]
- 11.Gordon-Shaag A, Ben-nun-Shaul O, Kasamatsu H, Oppenheim A B, Oppenheim A. The SV40 capsid protein Vp3 cooperates with the cellular transcription factor Sp1 in DNA-binding and in regulating viral promoter activity. J Mol Biol. 1998;275:187–195. doi: 10.1006/jmbi.1997.1461. [DOI] [PubMed] [Google Scholar]
- 12.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 13.Ishii N, Nakanishi A, Yamada M, Macalalad M H, Kasamatsu H. Functional complementation of nuclear targeting-defective mutants of simian virus 40 structural proteins. J Virol. 1994;68:8209–8216. doi: 10.1128/jvi.68.12.8209-8216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii N, Minami N, Chen E Y, Medina A L, Chico M M, Kasamatsu H. Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. J Virol. 1996;70:1317–1322. doi: 10.1128/jvi.70.2.1317-1322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobovits E B, Aloni Y. Isolation and characterization of various forms of simian virus 40 DNA-protein complexes. Virology. 1980;102:107–118. doi: 10.1016/0042-6822(80)90074-4. [DOI] [PubMed] [Google Scholar]
- 16.Kasamatsu H, Nehorayan A. Intracellular localization of viral polypeptides during simian virus 40 infection. J Virol. 1979;32:648–660. doi: 10.1128/jvi.32.2.648-660.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi H, Morioka H, Tobisawa K, Torizawa T, Kato K, Shimada I, Nikaido O, Stewart J D, Ohtsuka E. Probing the interaction between a high-affinity single-chain Fv and a pyrimidine(6–4)pyrimidone photodimer by site-directed mutagenesis. Biochemistry. 1999;38:532–539. doi: 10.1021/bi9809167. [DOI] [PubMed] [Google Scholar]
- 18.LaCasse E C, Lefebvre Y A. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P P, Nakanishi A, Tran M A, Salazar A M, Liddington R C, Kasamatsu H. The role of SV40 Vp1 cysteines in virion infectivity. J Virol. 2000;74:11388–11393. doi: 10.1128/jvi.74.23.11388-11393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liddington R C, Yan Y, Moulai J, Sahli R, Benjamin T L, Harrison S C. Structure of simian virus 40 at a 3.8-Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 21.Lin W, Hata T, Kasamatsu H. Subcellular distribution of viral structural proteins during simian virus 40 infection. J Virol. 1984;50:363–371. doi: 10.1128/jvi.50.2.363-371.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreland R B, Montross L, Garcea R L. Characterization of the DNA-binding properties of the polyomavirus capsid protein Vp1. J Virol. 1991;65:1168–1176. doi: 10.1128/jvi.65.3.1168-1176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oppenheim A, Sandalon Z, Peleg A, Shaul O, Nicolis S, Ottolenghi S. A cis-acting DNA signal for encapsidation of simian virus 40. J Virol. 1992;66:5320–5328. doi: 10.1128/jvi.66.9.5320-5328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salunke D M, Caspar L D, Garcea R L. Self-assembly of purified polyomavirus capsid protein Vp1. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Soussi T. DNA-binding properties of the major structural protein of simian virus 40. J Virol. 1986;59:740–742. doi: 10.1128/jvi.59.3.740-742.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stehle T, Gamblin S J, Yan Y, Harrison S C. The structure of simian virus 40 refined at a 3.1-Å resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 28.Suemori H, Kadodawa Y, Goto K, Araki I, Kondoh H, Nakatsuji N. A mouse embryonic stem cell line showing pluripotency of differentiation in early embryos and ubiquitous β-galactosidase expression. Cell Differ Dev. 1990;29:181–186. doi: 10.1016/0922-3371(90)90120-l. [DOI] [PubMed] [Google Scholar]
- 29.Yamada M, Kasamatsu H. Role of nuclear pore complex in simian virus 40 nuclear targeting. J Virol. 1993;67:119–130. doi: 10.1128/jvi.67.1.119-130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]