Abstract
The outbreaks of severe acute respiratory syndrome coronavirus (SARS-CoV-1), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 highlight the need for countermeasures to prevent future coronavirus pandemics. Given the unpredictable nature of spillover events, preparing antibodies with broad coronavirus-neutralizing activity is an ideal proactive strategy. Here, we investigated whether SARS-CoV-2 infection and vaccination could provide cross-neutralizing antibodies (nAbs) against zoonotic sarbecoviruses. We evaluated the cross-neutralizing profiles of plasma and monoclonal antibodies constructed from B cells from coronavirus disease 2019 (COVID-19) convalescents and vaccine recipients; against sarbecoviruses originating from bats, civets, and pangolins; and against SARS-CoV-1 and SARS-CoV-2. We found that the majority of individuals with natural infection and vaccination elicited broad nAb responses to most tested sarbecoviruses, particularly to clade 1b viruses, but exhibited very low cross-neutralization to SARS-CoV-1 in both natural infection and vaccination, and vaccination boosters significantly augmented the magnitude and breadth of nAbs to sarbecoviruses. Of the nAbs, several exhibited neutralization activity against multiple sarbecoviruses by targeting the spike receptor-binding domain (RBD) and competing with angiotensin-converting enzyme 2 (ACE2) binding. SCM12-61 demonstrated exceptional potency, with half-maximal inhibitory concentration (IC50) values of 0.001–0.091 μg/mL against tested sarbecoviruses; while VSM9-12 exhibited remarkable cross-neutralizing breadth against sarbecoviruses and SARS-CoV-2 Omicron subvariants, highlighting the potential of these two nAbs in combating sarbecoviruses and SARS-CoV-2 Omicron subvariants. Collectively, our findings suggest that vaccination with an ancestral SARS-CoV-2 vaccine, in combination with broad nAbs against sarbecoviruses, may provide a countermeasure for preventing further sarbecovirus outbreaks in humans.
Subject terms: Epidemiology, SARS-CoV-2, Infection
Introduction
In the past two decades, two pathogenic bat-origin sarbecoviruses, severe acute respiratory syndrome (SARS-CoV-1) and SARS-CoV-2, have caused global pandemics, resulting in significant public health and socioeconomic burdens1–5. Bats are the reservoir hosts of three of 10 virus groups of pandemic concern, including henipaviruses (Nipah virus and Hendra virus), filoviruses (Ebola virus and Marburg virus), and coronaviruses6, as designated by the World Health Organization. More than 4000 coronavirus sequences from 14 bat families have been identified, yet the diversity of bat coronaviruses is probably much greater7. Early cases of both SARS-CoV-1 and SARS-CoV-2 were identified in animal handlers at wild animal markets in the Guangdong1,2 and Hubei Provinces3–5, respectively. Later, the close relatives of SARS-CoV-1 and SARS-CoV-2 were identified in civets8, racoon dogs8, and pangolins9,10. Subsequently, numerous SARS-CoV-related viruses (referred to as SARSr-CoVs), which are phylogenetically linked to both SARS-CoV-1 and SARS-CoV-2, have been discovered in bats from different provinces in China as well as from European, African, and Southeast Asian countries11–20. These animal coronaviruses exhibit high degrees of similarity to either SARS-CoV-1 or SARS-CoV-2, and they use the same receptor for cell entry and replicate efficiently in primary human airway cells18–23. Serological evidence of human exposure to bat coronaviruses in rural China suggests that spillovers from bats may have occurred but have remained undetected thus far24,25. The high prevalence, close coexistence, and frequent recombination events of bat- or other-origin SARSr-CoVs pose a significant risk for the emergence and outbreak of novel variants26,27, thereby emphasizing the urgent need for pan-sarbecovirus vaccines and countermeasures.
Virus neutralization represents the best reliable protection countermeasure currently available. Previous reports have demonstrated that antibodies from patients infected with SARS-CoV-2 had cross-reactivity to the spike protein of SARS-CoV-1, and vice versa. However, these cross-reactive antibody responses typically manifest as non-neutralizing antibodies28,29. Although seasonal human coronavirus antibodies are boosted following SARS-CoV-2 infection, these antibodies do not confer protection against SARS-CoV-2 infections or reduce the likelihood of hospitalization30,31. In contrast, vaccination substantially increases the magnitude and breadth of neutralizing antibodies in patients previously infected with SARS-CoV-1 and SARS-CoV-2 or variants of concern (VOCs), thereby exhibiting potent neutralization effect to certain zoonotic sarbecoviruses32,33. Recently, a number of broad neutralizing antibodies (nAbs) against multiple sarbecoviruses have been isolated from coronavirus disease 2019 (COVID-19) convalescents or vaccine recipients, thus providing potential countermeasures for future sarbecovirus pandemics34–43.
However, the extent to which SARS-CoV-2 infection and vaccination elicit broad nAb responses against sarbecoviruses originating from animal reservoirs remain poorly understood. Here, we systemically assessed the antibody response profiles to sarbecoviruses in COVID-19 convalescent individuals and vaccination recipients, from which we isolated potent broad nAbs against sarbecoviruses. These findings provide novel insights for guiding the design of pan-sarbecovirus vaccines to prevent future outbreaks.
Results
SARS-CoV-2 infection and vaccination elicited broad cross-neutralizing antibody responses against zoonotic sarbecoviruses
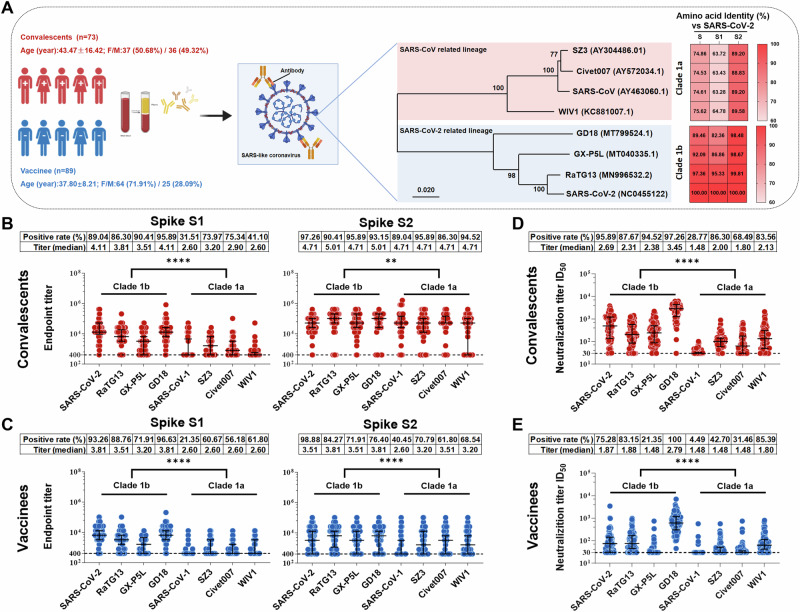
To test whether SARS-CoV-2 infection and vaccination elicit spike-specific antibodies with cross-reactivity to zoonotic sarbecoviruses, blood samples were collected from 73 COVID-19 convalescents 2 months after ancestral SARS-CoV-2 (GenBank: NC_045512.2) infection and from 89 donors 2 weeks after receiving a standard two-dose inactivated vaccine regimen (Fig. 1A, Supplementary Tables 1, 2). Blood samples from pre-pandemic healthy donors were collected as negative controls. Plasma immunoglobulin G (IgG) endpoint titers to spike S1 and S2 protein subunits of eight sarbecoviruses (SARS-CoV-1, SARS-CoV-2, RaTG13, GXP5L, GD18, SZ3, Civet007, and WIV1) were determined by enzyme-linked immunosorbent assay (ELISA). The results showed that COVID-19 convalescent plasma exhibited higher frequencies (60.41–86.3%) and titers (median: 3.51–4.11) of antibody responses to spike S1 protein subunits of clade 1b sarbecoviruses (SARS-CoV-2, RaTG13, GX-P5L, and GD18), while lower frequencies (31.51–75.34%) and titers (median: 2.6–3.2) were found to spike S1 protein subunits of clade 1a sarbecoviruses (SARS-CoV-1, SZ3, Civet 007, and WIV1) (Fig. 1B, left panel). The higher degree of homology between the spike S1 subunits of clade 1b sarbecoviruses and SARS-CoV-2, in comparison to that of clade 1a sarbecoviruses, could potentially explain these findings (Fig. 1A). In contrast to the antibody responses to spike S1 protein subunits in COVID-19 convalescent plasma, the frequencies and titers of convalescent plasma antibody responses to spike S2 protein subunits of both clades 1a and 1b sarbecoviruses showed statistically significant differences, but not as pronounced as those observed for S1 (Fig. 1B, right panel). These results may be attributed to the significant homology shared among sarbecovirus S2 protein subunits (Fig. 1A). Similar antibody response patterns were observed in inactivated vaccine recipients; however, the overall frequencies and titers were relatively lower compared to those of COVID-19 convalescents (Fig. 1B, C). There were no or very low antibody responses to sarbecovirus spike S1 and S2 protein subunits in pre-pandemic healthy donors and in the pre-vaccination donors (Supplementary Fig. 1). These results suggest that SARS-CoV-2 infection and vaccination generated spike-specific cross-reactive antibody responses to zoonotic sarbecoviruses.
Fig. 1. SARS-CoV-2 infection and vaccination elicit broad cross-nAb responses against zoonotic sarbecoviruses.
A Schematic diagram of participant demographics and baseline characteristics for cross-sectional blood samples, including the number, age, and percentage of sexes of individuals (left panel), as well as an amino acid (aa) alignment of spike proteins for selected sarbecoviruses, comparing identities against SARS-CoV-2. GenBank accession numbers are given for the selected viruses. (right panel). This figure was created with Biorender.com. B, C Endpoint titers of serum IgG antibodies from 73 COVID-19 convalescents (B) and 89 vaccinees (C), determined by antibody binding to sarbecovirus spike S1 (left) and S2 (right) protein subunits. The dashed lines represent the cut-off values (endpoint titer = 400), and a value of endpoint titer > 400 was considered positive for binding. D, E Neutralization titers of plasma samples from 73 COVID-19 convalescents (D) and 89 vaccinees (E) against sarbecovirus pseudotyped viruses. The dashed lines represent the cut-off values (ID50 = 30), and a value of ID50 > 30 was considered positive for neutralization. Data are representative of technical triplicates. In (B–E), data are presented as medians ± interquartile ranges (IQRs, 25–75%), and the error bars indicate medians with IQRs. The positive rates and mean titers are shown. The Mann–Whitney U test was employed to examine the differences in endpoint and neutralization titers against sarbecoviruses clade 1a and clade 1b for COVID-19 convalescents and vaccinees. Significance levels are denoted as follows: *, P < 0.05; **, P < 0.001; ***, P < 0.001; ****, P < 0.0001.
To assess the cross-neutralizing activities of antibodies elicited by SARS-CoV-2 infection and vaccination, we constructed pseudotyped sarbecoviruses for neutralization assays. Consistent with cross-binding reactivities, COVID-19 convalescent plasma exhibited higher frequencies (87.67–97.26%) and titers (median: 2.31–3.45) of cross-nAb responses against clade 1b pseudotyped sarbecoviruses, while significantly lower frequencies (28.77–86.30%) and titers (median: 1.48–2.13) were observed for clade 1a pseudotyped sarbecoviruses (Fig. 1D). As expected, vaccination of inactivated vaccines elicited relatively lower frequencies and titers of cross-nAb responses to both 1a and 1b pseudotyped sarbecoviruses compared with SARS-CoV-2 infection (Fig. 1D, E). However, only a minority of samples exhibited very weak neutralizing activity against SARS-CoV-1 or SARS-CoV-2 in pre-pandemic healthy donors and pre-vaccination individuals (Supplementary Fig. 2). Taken together, these findings indicated that majority of individuals with natural infection and vaccination elicited broad nAb responses to most tested sarbecoviruses, particularly to clade 1b viruses, but exhibited very low cross-neutralization to SARS-CoV-1 in both natural infection and vaccination.
The breadth of the cross-neutralizing response to sarbecoviruses is associated with the potency of antibody responses elicited by SARS-CoV-2 infection and vaccination
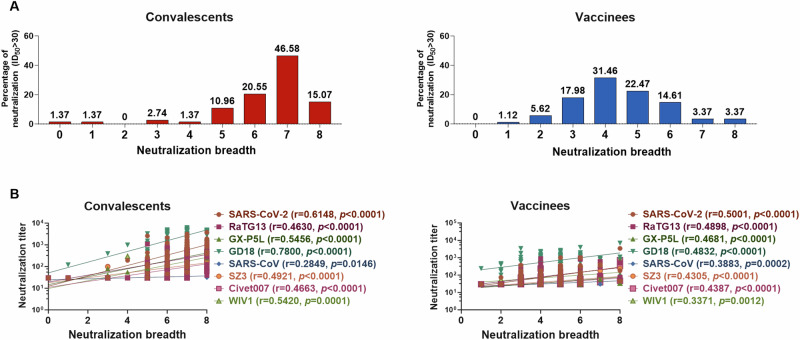
To further characterize the broad nAb responses induced by SARS-CoV-2 infection and vaccination, we categorized the neutralization breadth to eight pseudotyped sarbecoviruses on a scale ranging from 0 to 8 for each plasma sample from COVID-19 convalescents and vaccinees (Supplementary Tables 3, 4). We found that 10.96% (8/73), 20.55% (15/73), 46.58% (34/73), and 15.07% (11/73) of convalescent plasma samples exhibited cross-neutralization capacity to five, six, seven, and eight pseudotyped sarbecoviruses, respectively (Fig. 2A, left panel). In contrast, only 22.47% (20/89), 14.61% (13/89), 3.37% (3/89), and 3.37% (3/89) of vaccinees exhibited cross-neutralization capacity to five, six, seven, and eight sarbecoviruses, respectively (Fig. 2A, right panel). The majority of plasma samples from convalescents (61.65%, 45/73) could neutralize up to seven sarbecoviruses, while plasma samples from vaccinees (43.82%, 39/89) demonstrated neutralization against more than four sarbecoviruses (Fig. 2A). Then, we investigated whether the antibody titers correlated with the breath of antibodies. Intriguingly, the nAb titer to each sarbecovirus was positively associated with the nAb breadth for all plasma samples from COVID-19 convalescents and vaccine recipients (Fig. 2B). These results indicate that nAb breadth elicited by SARS-CoV-2 infection and vaccination correlated with the antibody titer magnitude.
Fig. 2. Breadth of the cross-neutralization response against sarbecovirus is associated with the potency of the antibody response.
A The neutralization breadths of convalescent (left panel) and vaccinee (right panel) plasma samples against eight sarbecoviruses are defined by the range of breadth from 0 to 8. B Correlations between neutralization breadths and neutralization titers against sarbecoviruses in convalescents (left) and vaccinees (right). Spearman’s rank correlation coefficient was employed to assess the association between the neutralization breadth and neutralization titers. P < 0.05 was considered statistically significant using a two-tailed test.
Booster vaccination significantly augments cross-nAb potency and breadth against sarbecoviruses
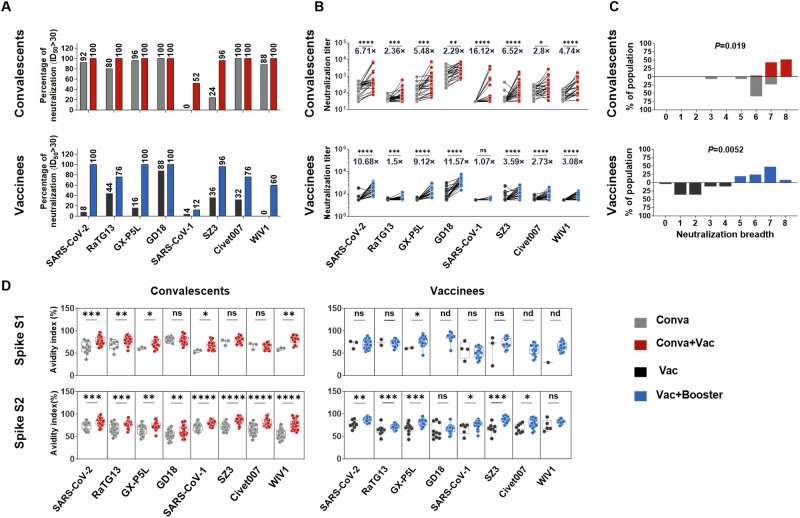
Protective antibodies are usually maintained for 6 months following primary vaccination44,45. A booster dose has been shown to significantly enhance neutralization titers to ancestral SARS-CoV-2 and its variants46,47. Here, we investigated whether administering a third booster vaccine in individuals who had already received two-dose inactivated vaccines or vaccinating COVID-19 convalescents could enhance the potency and breadth of antibody neutralization against sarbecoviruses. To this end, we compared the nAb titer, breadth, and avidity against sarbecoviruses before and after vaccination in 25 COVID-19 convalescents and before and after a third booster vaccine in 25 twice-vaccinated individuals (Supplementary Tables 5, 6). The results revealed that plasma from COVID-19 convalescent individuals persistently displayed high nAb response frequencies (80–100%) against sarbecoviruses with the exceptions of SARS-CoV-1 and SZ3 after 1 year. Furthermore, the neutralizing frequencies against sarbecoviruses were enhanced in these convalescent individuals after vaccination (Fig. 3A, top panel). In contrast, individuals who received two doses of inactivated vaccine demonstrated low frequencies (0–44%) of nAb responses to sarbecoviruses after 8 months, with the exception of GD18. However, a third booster dose increased the frequencies to 60% or greater, except for SARS-CoV-1 (Fig. 3A, bottom panel). These results suggest that administration of vaccines in COVID-19 convalescents and a third booster dose in vaccinees efficiently increased the nAb response frequencies to sarbecoviruses.
Fig. 3. Administration of a vaccine booster significantly augments cross-nAb potency and breadth against sarbecovirus.
A Percentage of plasma samples with neutralizing effects against sarbecovirus-pseudotyped virus. The plasma samples were obtained from 25 COVID-19 convalescents before and after vaccination (upper panel) and 25 vaccinees before and after receiving a third booster dose (lower panel). A value of ID50 > 30 was considered positive for neutralization. B Neutralization titers against sarbecovirus-pseudotyped virus for 25 COVID-19 convalescents before and after vaccination (upper panel) and for 25 vaccinees before and after receiving a third booster dose (lower panel). Paired t-tests were employed to examine the differences in neutralization titers pre- and post-vaccination for COVID-19 convalescents and pre- and post-booster vaccination for vaccinees. Significance levels are denoted as follows: *P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. A two-tailed test with P < 0.05 was considered statistically significant. ns, no significant. C Neutralization breadth for COVID-19 convalescents before and after vaccination (upper panel) and for vaccinees before and after receiving a third booster dose (lower panel) against sarbecoviruses. D Avidity indices of plasma sarbecovirus spike S1-specific IgG (upper panel) and S2-specific IgG (lower panel) for 25 COVID-19 convalescents before and after vaccination (left panel) and for 25 vaccinees before and after receiving a third booster dose (right panel). The convalescent plasma before vaccine administration was obtained at 248-387 days after illness onset (median=377; IQR = 371–382), while the convalescent plasma after vaccine administration was obtained at 15-201 days after third dose administration (median=118; IQR = 22-151). The plasma collected from vaccine recipients before receiving the booster dose was obtained at 271–307 days after a two-dose standard regimen (median=276; IQR = 275–278), while the plasma collected after receiving the booster dose was obtained at 14–92 days after a third booster dose (median=20; IQR = 18–24). Data are representative of technical triplicates. Data are presented as medians ± IQRs (25–75%), with error bars signifying the medians with IQRs. For C–D, the Mann–Whitney U test was employed to examine the differences pre- and post-vaccination for COVID-19 convalescents and pre- and post-booster vaccination for vaccinees. Significance levels are denoted as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. A two-tailed test with P < 0.05 was considered statistically significant. ns, no significant. nd, not detection. Conva, COVID-19 convalescents. Conva+Vac, COVID-19 convalescents after vaccination. Vac, individuals who received a two-dose vaccination. Vac+Booster, individuals who received a three-dose vaccination.
After receiving vaccines, the average neutralization titers of COVID-19 convalescents increased by 6.71-, 2.36-, 5.48-, 2.29-, 16.12-, 6.52-, 2.8- and 4.74-fold to pseudotyped SARS-CoV-2, RaTG13, GX-P5L, GD18, SARS-CoV-1, SZ3, Civet007 and WIV1, respectively (Fig. 3B, top panel). After the third vaccine booster, the average neutralization titers of vaccinees increased by 10.68-, 1.5-, 9.12-, 11.57-, 1.07-, 3.59-, 2.73- and 3.08-fold to pseudotyped SARS-CoV-2, RaTG13, GX-P5L, GD18, SARS-CoV-1, SZ3, Civet007 and WIV1, respectively (Fig. 3B, bottom panel). Vaccination in convalescents showed the highest enhancement of neutralization titers against SARS-CoV-1, while administering a third vaccine to vaccinees resulted in maximum promotion against GD18 (Fig. 3B). The assessment of plasma from convalescent individuals pre- and post-vaccination revealed an increase in neutralizing breadth, from neutralizing 6–7 sarbecoviruses (84%) prior to vaccination to neutralizing 7–8 sarbecoviruses (96%) after vaccination. Similarly, receipt of a third booster dose in vaccinees resulted in an enhancement in breadth, from neutralizing 1–2 sarbecoviruses (72%) prior to boost to neutralizing 5–7 sarbecoviruses (92%) post-boost (Fig. 3C). These results indicate that convalescents after vaccination and vaccinees who received a third booster dose exhibited significantly augmented breadths of cross-neutralization against sarbecoviruses.
Antibody avidity reflects the quality of the antibody response. Herein, we measured the avidity indices of spike-specific antibodies to sarbecovirus spike antigens S1 and S2 in convalescents before and after vaccination and in vaccinees before and after receiving a third booster dose. The data revealed that the majority of S1 antibodies were close to baseline in both convalescents before vaccination and vaccinees before receiving a third booster dose. S2 antibodies were also present at low levels in vaccinees (Supplementary Fig. 3). The results showed that vaccination in convalescents and receipt of a third booster in vaccinees further promoted antibody avidity against sarbecovirus spike antigens (Fig. 3D). Thus, administration of vaccines in COVID-19 convalescents, and receipt of a third dose of booster in vaccinees, efficiently increased the preexisting antibody responses to sarbecoviruses by enhancing nAb titer, breadth, and avidity.
Characterization of monoclonal antibodies (mAbs) isolated from COVID-19 convalescents and vaccinees
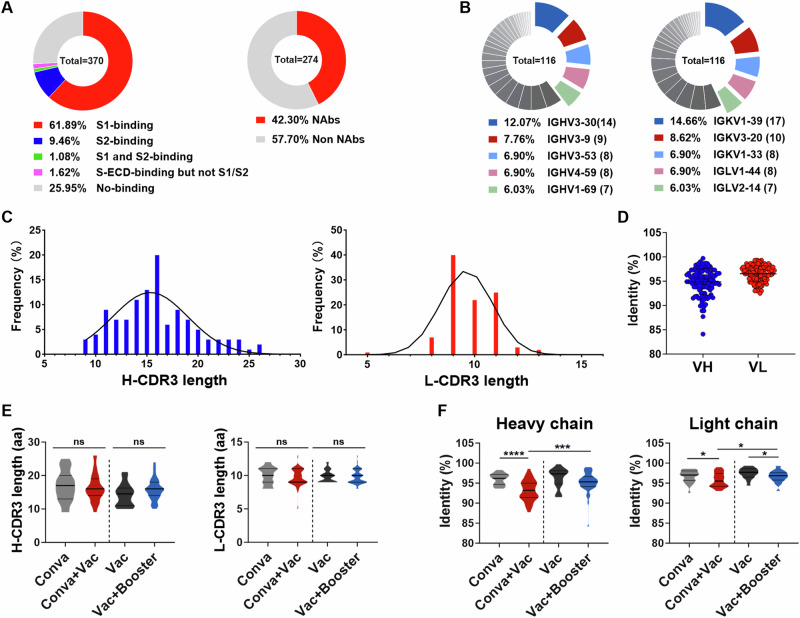
To further characterize the nAb responses elicited by SARS-CoV-2 infection and vaccination, we sorted single memory B cells specific to SARS-CoV-2 spike antigen from COVID-19 convalescents and vaccinees by fluorescence-activated cell sorting (FACS). A total of 370 mAbs were cloned and purified (Fig. 4A), of which 74.1% (274/370) exhibited binding activity to the SARS-CoV-2 spike protein. Moreover, these mAbs exhibited distinct binding domains in the spike protein, of which 229 mAbs bound to spike S1, 35 mAbs bound to spike S2, 4 mAbs bound to both spike S1 and S2, and 6 mAbs bound neither S1 nor S2 alone but did bind the intact extracellular domain of the spike protein (Fig. 4A). To assess the neutralizing activity of these mAbs, we performed a primary neutralization screening (10 μg/mL antibody concentration) using SARS-CoV-2 pseudotyped virus, which revealed that 42.3% (116/274) of mAbs displayed neutralizing activities to SARS-CoV-2 (Fig. 4A).
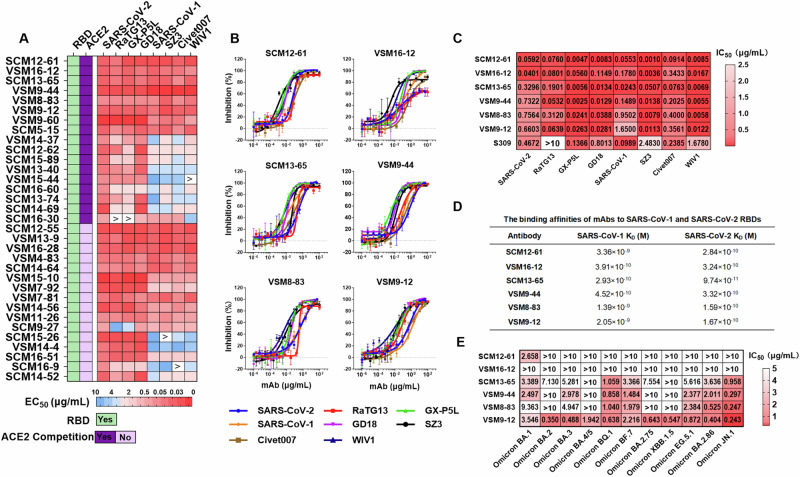
Fig. 4. Isolation and characterization of cross-neutralizing antibodies to sarbecoviruses.
A Schematic diagram of the antibody source and quantity (left). Based on spike-binding assays, 370 mAbs were categorized into five types: S1-binding mAbs (red in pie charts), S2-binding mAbs (blue), S1- and S2-binding mAbs (green), extracellular domain of the spike protein (S-ECD) but not S1- and S2-binding mAbs (pink), and no binding (gray). The percentages of neutralizing (red) and non-neutralizing (gray) mAbs were evaluated by SARS-CoV-2 pseudotyped particle assays at an antibody concentration of 10 μg/mL. B–D The gene region usage, CDR3 lengths, and SHM were analyzed for 116 nAbs using IgBLAST. Pie chart showing the germline gene region usage of these nAbs. The five gene regions most used by heavy (left) and light (right) chains are given (B). Distributions of H-CDR3 and L-CDR3 length over a total of 116 nAbs (C). Percent identities of heavy and light chains to the inferred germline among 116 nAbs. Data are presented as medians ± IQRs (25–75%), and the error bars indicate the medians with IQRs (D). E Comparison of the lengths of H-CDR3 (left) and L-CDR3 (right) for nAbs from four distinct cohorts. F Comparison of the SHM levels of heavy (left) and light (right) chains. In panels E and F, Kruskal–Wallis tests were used with post hoc Dunn’s multiple comparisons tests. Significance levels are denoted as follows: *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. A two-tailed test with P < 0.05 was considered statistically significant. ns, no significant. Conva, COVID-19 convalescents. Conva+Vac, COVID-19 convalescents after vaccination. Vac, individuals who received a two-dose vaccination. Vac+Booster, individuals who received a three-dose vaccination.
Next, we analyzed the immunoglobulin (Ig) sequences (heavy and light chains) of these SARS-CoV-2 spike-specific nAbs. The variable region sequences of the heavy and light chains exhibited a diverse distribution of gene segments. Of the heavy chains, 12.07% (14/116) utilized VH3-30 (Fig. 4B, left pie), one of the most predominant epitopes for SARS-CoV-2 nAbs, as previously reported48–50. Of the light chains, 14.66% and 8.62% employed Vκ1-39 and Vκ3-20, respectively, which are germline epitopes commonly observed for SARS-CoV-2 nAbs49,51 (Fig. 4B, right pie). The lengths of heavy and light complementarity determining region 3 (CDR3) were distributed in a defined range (Fig. 4C), ranging from 8 to 26 amino acids for heavy CDR3 (H-CDR3). Notably, 34% of nAbs exhibited CDR3 lengths between 14 and 16 amino acids, consistent with previous reports49 (Fig. 4C, left panel). For light CDR3 (L-CDR3), 87% of nAbs ranged from 9 to 11 amino acids in length (Fig. 4C, right panel). Furthermore, the identity of VH sequences to the germline ranged from 84.1% to 99.7% (median, 95.1%), while the VL sequences showed identities ranging from 92.5% to 99.3% (median, 96.8%) (Fig. 4D). The lengths of H-CDR3 and L-CDR3 were not significantly altered by the administration of vaccines to convalescents or a third booster dose to vaccinees, but there was an increase in antibody somatic hypermutation (SHM) level (Fig. 4E, F). These results are consistent with the observation that vaccination with a booster promotes the maturation of nAbs (Fig. 3D).
MAbs isolated from COVID-19 convalescents and vaccinees exhibit broad neutralizing profiles to multiple sarbecoviruses
To investigate whether the nAbs that neutralize SARS-CoV-2 have cross-neutralizing activities against pesudotyped sarbecoviruses, as observed in the plasma from COVID-19 convalescents and vaccinees (Fig. 1D), based on the neutralization capacity to SARS-CoV-2 pseudotyped virus at a concentration of 10 μg/mL and the binding breadth to sarbecovirus spike antigen at a concentration of 1 μg/mL, 33 nAbs were selected for further investigation (Supplementary Table 7–8). All of these nAbs bound to the SARS-CoV-2 spike receptor-binding domain (RBD), with half of them competing with angiotensin-converting enzyme 2 (ACE2) binding. Moreover, 39.4% (13/33) of the nAbs showed cross-binding activity to spike S1 protein subunits from eight sarbecoviruses at the nanogram level, with a half-maximal effective concentration (EC50) range of 0.015–0.1 μg/mL (Fig. 5A). For neutralization profiles, 18.18% (6/33) of the nAbs (SCM13-65, VSM8-83, VSM16-12, SCM12-61, VSM9-12, and VSM9-44) neutralized all tested sarbecoviruses at a concentration of 10 μg/mL (Supplementary Table 9). The half-maximal inhibitory concentration (IC50) values of these six antibodies against the eight sarbecoviruses ranged between 0.001 and 1.65 μg/mL (Fig. 5B, C). S309 also cross-neutralized several sarbecoviruses, but with markedly lower potency than the neutralization capacities of SCM12-61 and SCM13-65 in our assay (Fig. 5C). SCM12-61 exhibited a broad neutralizing profile to all sarbecoviruses by efficiently inhibiting cell entry with an IC50 range of 0.001–0.0914 μg/mL (Fig. 5B, C). The binding affinities to RBDs of SARS-CoV-1 and SARS-CoV-2 were also in the nanomolar to picomolar ranges, with values of 3.36 nM and 0.284 nM, respectively. SCM13-65 showed the highest affinity towards the RBD of SARS-CoV-2, followed by VSM8-83 and VSM9-12. Furthermore, SCM13-65 also exhibited the highest affinity to the SARS-CoV-1 RBD, followed by VSM16-12 and VSM9-44 (Fig. 5D). Although these six nAbs show broad neutralization activities against the tested sarbecoviruses, whether these nAbs also present neutralizing activities to SARS-CoV-2 omicron variants are not known. To this end, the neutralizing profiles of these nAbs against Omicron variants (BA.1, BA.2, BA.3, BA.4/5, BQ.1, BF.7, BF.2.75, XBB.1.5, EG.5.1, BA.2.86, JN.1) were determined. Our results revealed that most of six nAbs showed limited neutralization activity against the Omicron variants. In contrast, VSM9-12 present broad neutralizing profile all tested Omicron subvariants and sarbecoviruses (Fig. 5E), highlighting the potential of broad nAbs in combating sarbecoviruses and current emerged variants.
Fig. 5. The mAbs isolated from COVID-19 convalescents and vaccinees exhibit broad neutralizing profiles against sarbecoviruses.
A RBD-binding, ACE2 competition, and spike-binding EC50 values were determined for 33 nAbs by ELISA and are presented as a heatmap. Binding was defined as an OD450 value three-fold greater than the cut-off value (i.e., the average OD450 value of negative antibody) at an antibody concentration of 1 μg/mL; ACE2 competition was defined as an antibody binding to the RBD and inhibiting ACE2 binding by > 50%. B Neutralization curves for six nAbs are presented as the percentage of neutralization for pseudotyped sarbecoviruses. Data are representative of technical triplicates. A broad nAb S309 and an unrelated antibody HNC5 were used as the positive and negative controls, respectively. C Neutralization activity (IC50) levels of six nAbs against sarbecoviruses are presented as a heatmap, with darker colors indicating greater levels of neutralization activity. Data are derived from (B). D The binding affinities (KD) of six nAbs for SARS-CoV-1 and SARS-CoV-2 RBD proteins. mAbs were bound to the proA biosensor and then tested for binding to SARS-CoV-1 or SARS-CoV-2 RBD proteins using BLI. E Neutralization activity (IC50) levels of six nAbs against a panel of SARS-CoV-2 Omicron subvariants are presented as a heatmap, with darker colors indicating higher levels of neutralization activity.
Discussion
The existence of vast reservoirs of sarbecoviruses in wild animals, including bats, civets and pangolins, plus the potential for recombination events, indicates that sarbecoviruses are likely to spill over into humans in the future and initiate new epidemics and/or pandemics21,23,52. Recent evidence has shown that animal-associated SARS-CoV-1/2-like sarbecoviruses cause respiratory infections and mild disease in mice53,54. Thus, safe and effective countermeasures to combat potential sarbecovirus pandemics are urgently needed55. Here, we have shown that both SARS-CoV-2 infection and vaccination elicited broad nAb responses to sarbecoviruses that originated from bat, civet, and pangolin hosts. Furthermore, nAbs isolated from SARS-CoV-2 convalescents and vaccinees exhibited neutralizing potency and breadth to these animal-originated sarbecoviruses. These findings provide an option to combat future outbreaks of pre-emergent sarbecoviruses in humans. The SARS-CoV-2 vaccines based on the ancestral strain effectively reduced the severity and transmission of SARS-CoV-2 infection within a limited timeframe. However, their efficacy against variants has significantly diminished due to viral evolution and the waning of vaccine-elicited immune responses56,57. The effectiveness of antibody responses elicited by SARS-CoV-2 infection and vaccination against animal-associated high-risk sarbecoviruses remains unclear. Recent efforts have been made to evaluate the efficacy of SARS-CoV-2 vaccine-induced antibody responses to sarbecoviruses in mice and macaques58–60. Immunization of macaques with the SARS-CoV-2 RBD elicited cross-neutralizing antibody responses against both SARS-CoV-1 and SARS-CoV-258. Chimeric spike messenger RNAs (mRNAs) induced high levels of broadly protective neutralizing antibodies against high-risk sarbecoviruses59. An inactivated SARS-CoV-2 vaccine provided full protection against SARS-CoV-2 and rWIV1, as well as partial protection against rRsSHC014S infection in human ACE2 transgenic mice60. In the current study, we found that infection with the ancestral strain of SARS-CoV-2 elicited antibodies with cross-neutralizing activities to sarbecoviruses derived from bats, civets, and pangolins. The cross-neutralizing titers to clade 1b sarbecoviruses were comparable to those to SARS-CoV-2, while the titers to clade 1a sarbecoviruses were relatively lower. Immunization with inactivated vaccines in humans using a standard two-dose regimen also elicited cross-neutralizing antibody responses against tested sarbecoviruses in a similar pattern to SARS-CoV-2 infection, albeit displaying relatively lower titers. The cross-neutralizing titer and breadth were associated with the initial neutralizing titer to SARS-CoV-2 itself. Due to the continuous waning in neutralizing antibody responses, both SARS-CoV-2 infection and vaccination elicited transient cross-neutralizing antibodies against sarbecoviruses. However, waning cross-neutralizing antibody responses can be promptly enhanced by vaccination with an ancestral SARS-CoV-2 vaccine in COVID-19 convalescents and through the administration of a third vaccine dose in vaccinated recipients. These findings suggest that preexisting humoral immunity may be beneficial in preventing sarbecovirus infection when a spillover from bats or other wild animals occurs7. Once a sarbecovirus, particularly a clade 1b virus, establishes infection in humans, emergency immunization of uninfected individuals with an ancestral SARS-CoV-2 vaccine may be an option as a preventive measures against sarbecovirus transmission in the population. Thus, although ancestral SARS-CoV-2 vaccines have reduced efficacy against SARS-CoV-2 variants, they may still be valuable in preventing future pre-emergent sarbecovirus infections.
In addition to vaccination, nAbs represent an effective countermeasure to combat SARS-CoV-2 variants and to protect from future sarbecovirus spillover events. Previously, several isolated and engineered nAbs showed cross-neutralizing activity against multiple sarbecoviruses, including SARS-CoV-1, RaTG13, WIV1, and pangolin coronaviruses, and most of these broad nAbs bound to the ACE2 binding site36,40,61,62. We have identified 33 RBD-directed antibodies that potently neutralized a panel of sarbecoviruses. Half of these broad nAbs targeted the ACE2 binding site and showed high neutralization breadth and potency against tested sarbecoviruses. This discovery provides additional options for developing effective countermeasures for future pre-emergent sarbecovirus outbreaks. Although the most of nAbs against sarbecoviruses present limited neutralizing activities and breadths to SARS-CoV-2 Omicron variants, one of these antibodies, VSM9-12, exhibits broad neutralizing efficacy to all the tested Omicron variants, highlighting the potential to combating emerging, remerging, and preemerging sarbecoviruses. However, not all ACE2-dependent sarbecoviruses were sensitive to SARS-CoV-2 nAbs and vaccines63; moreover, several ACE2-independent sarbecoviruses pose broader zoonotic threats64. Among the 33 identified broad nAbs, approximately half of these broad nAbs targeted outside the RBD, thereby offering additional strategies to combat ACE2-independent or less ACE2-sensitive sarbecoviruses.
In summary, our findings suggest that vaccination with an ancestral SARS-CoV-2 vaccine, in combination with broad nAbs effective for sarbecoviruses, may provide an option for preventing future sarbecovirus outbreaks, particularly for a clade 1b virus. However, the efficacy of this strategy needs to be further investigated in an infection model.
Methods
Study subjects
Seventy-three COVID-19 convalescents, with a median of 54 days after illness onset caused by ancestral SARS-CoV-2 infection, were recruited from Shaoyang city, Hunan Province, China (Supplementary Table 1). 25 of these convalescents received third dose administration were designated as the convalescents with vaccination group (Supplementary Table 5). Eighty-nine vaccinees who received a standard inactivated vaccine regimen (Sino Vac) were recruited from Chenzhou city, Hunan Province, China (Supplementary Table 2); 25 of these individuals received a third booster dose (Supplementary Table 6) during days 271–307 after their first vaccination. Sex- and age-matched healthy controls were recruited before the pandemic. All participants were routinely tested for SARS-CoV-2 or its variants by qualified authorities, and no exposure or infection cases were reported in these cohorts. This study was approved by the Institutional Ethical Review Board of The Central Hospital of Shaoyang, Hunan Province, China (V.1.0, 20200301) and The First People’s Hospital of Chenzhou, Hunan Province, China (V.3.0, 2021001). Informed consent was obtained from each participant, who signed written consent forms. Blood samples (20 ml) were obtained from each subject. Peripheral blood mononuclear cells (PBMCs) and plasma were isolated and stored in liquid nitrogen and in a −80 °C freezer, respectively.
Cell lines
Human embryonic kidney freestyle 293 cells (293 F) and Sf9 insect cells were used in this study for recombinant protein expression. HEK293T cells stably expressing human ACE2 (HEK293T-ACE2) were generated by lentiviral-ACE2 transduction in our laboratory for the pseudotyped virus neutralization assay65. 293 F cells were grown in serum-free FreeStyle 293 Expression Medium (Gibco, New York, USA) at 37 °C and 8% CO2 with shaking at 125 rpm. Sf9 cells were cultured in Sf-900 II SFM (Gibco, New York, USA) at 27 °C. HEK293T-ACE2 cells were maintained in DMEM (Gibco, New York, USA) with 10% fetal bovine serum (FBS; OPCEL, Hohhot, China), 1% penicillin-streptomycin, 1% minimum essential medium (MEM) nonessential amino acids, and 1% sodium pyruvate at 37 °C and 5% CO2.
Recombinant protein expression and purification
The S-ECD (extracellular domain of S protein) and RBD (receptor binding domain) proteins of SARS-CoV-2 were purchased from Sino Biological with the serial numbers 40589-V08B1 and 40592-V08B, respectively. To test the plasma and monoclonal antibody binding reactivities to distinct sarbecovirus spike proteins, the spike S1 and S2 regions from SARS-CoV-2 (GenBank accession number: YP_009724390.1), SARS-CoV-1 (Y463060.1), RaTG13 (MN996532.2), GX-P5L (MT040335.1), GD18 (MT799524.1), SZ3 (AY304486.1), Civet007 (GAY572034.1), and WIV1 (KC881007.1) were commercially synthesized (Genscript, Nanjing, China). The S1 open reading frames were cloned into the pcDNA3.1 vector and expressed in 293 F cells. The expressed S1 protein subunits were purified using a Strep-Tactin Superflow high-capacity cartridge (IBA Lifesciences, Göttingen, Germany) in an ÄKTA pure system (Cytiva, Marlborough, USA). The S2 open reading frames were cloned into the pQB vector and expressed using Sf9 cells. In brief, S2-expressing plasmids and BacIIIG bacmid vectors were co-transfected into Sf9 cells by FuGENE HD according to the manufacturer’s instructions (Promega, Wisconsin, USA). After 5 days, the cell culture supernatant containing the first generation of recombinant baculovirus (P0 virus) was harvested and stored. Working viruses expressing the target protein subunit were produced by two rounds of infection, followed by the production of recombinant spike S2 protein subunits via infection of Sf9 cells with these working viruses. The cell culture supernatants containing spike S2 protein subunits were harvested and purified using a Strep-Tactin Superflow high-capacity cartridge (IBA Lifesciences, Göttingen, Germany) in an ÄKTA pure system (Cytiva, Marlborough, USA).
ELISA
Sarbecovirus spike (S1 and S2)-specific antibody titers in COVID-19 convalescents and vaccinees were determined by ELISA. In brief, 96-well ELISA plates (JETBIOFIL, Guangdong, China) were coated with sarbecovirus spike protein subunits (S1 or S2, 200 ng/well) in phosphate-buffered saline (PBS) and incubated at 4 °C overnight. The plates were washed five times with PBS-T (0.05% Tween-20 in PBS) and then blocked with blocking buffer (2% FBS and 2% bovine serum albumin (BSA) in PBS-T) for 30 min. Two-fold serial dilutions of plasma, starting from a 1:20 dilution, were added to the 96-well plates in triplicate (100 µL/well) and incubated for 1 h at room temperature. Spike S1 and S2-specific antibodies were detected using horseradish peroxidase (HRP)-conjugated anti-human IgG (Jackson ImmunoResearch, PA, USA). Plasma samples collected from healthy subjects before the COVID-19 pandemic were used as negative controls, and SARS-CoV-2 spike RBD-specific monoclonal antibody was generated in the laboratory and used as a positive control66. The optical density at 450 nm (OD450) was measured for each reaction, and an OD450 value three-fold greater than the cut-off value (i.e., the average OD450 value of healthy controls) was considered a positive readout. The highest dilution with a positive readout was defined as the endpoint titer of the antibody response, and the data were logarithmically transformed. Plasma spike-specific antibody avidity was determined based on ELISA with an additional sodium thiocyanate (NaSCN) treatment at 100 μL/well for 15 min at room temperature. The avidity index was calculated using the formula: Avidity Index = [ODNaSCN 1 M/ODNaSCN 0 M] × 100, as previously reported67. For mAb binding, a similar assay was performed with 10-fold dilutions of mAbs, and data are presented as EC50 values.
Pseudotyped virus production and neutralization assay
Sarbecovirus and SARS-CoV-2 variants Omicron BA.1 (GISAID ID: EPI_ISL_6590782.2), BA.2 (EPI_ISL_6795834.2), BA.3 (EPI_ISL_7605589), BA.4/5 (EPI_ISL_12268493.2), BA.2.75 (GenBank accession number: OP703311.1), BF.7 (EPI_ISL_15693205), BQ.1 (EPI_ISL_15693263), XBB.1.5 (EPI_ISL_16292655), EG.5.1 (EPI_ISL_17700360), and JN.1 (EPI_ISL_18727440) pseudotyped particles were produced by co-transfecting HEK293T cells with spike-expressing plasmids and pPNL4-3 vector containing a luciferase reporter using PEI reagent (Polysciences, Pennsylvania, USA). Briefly, HEK293T cells were seeded in a 10-cm dish (Corning Costar, Massachusetts, USA) and grown to 80% confluency before transfection. The S-encoding plasmids and pPNL4-3 vector were co-transfected into HEK293T cells at a ratio of 1:3. At 48 h after transfection, the supernatant was harvested by centrifugation at 4000 × g for 15 min and then filtered through a 0.22 μm filter. All pseudotyped particles were aliquoted and stored at −80 °C until use.
Neutralization assays were performed by incubating pseudotyped virus particles with mAbs or heat-inactivated plasma, and neutralization was measured by the reduction in luciferase intensity65. One day prior to infection, HEK293T-ACE2 cells were seeded at a density of 1 × 104 cells/well in a 96-well plate. The following day, serial dilutions of plasma starting with a 1:30 dilution or mAbs starting with a 100 μg/mL dilution were made in a 96-well cell culture plate for 12 dilutions, and diluted pseudotyped virus particles were added to the plate and incubated at 37 °C for 60 min. After incubation, the plasma or mAbs–virus mixture was added to the cells and centrifuged at 800 × g for 30 min. Following infection for 48 h, cells were lysed using 1× luciferase cell culture lysis buffer (Promega, Wisconsin, USA), and luciferase intensity was measured using the Luciferase Assay System (Promega, Wisconsin, USA) on a Varioskan Flash Multimode Reader (Thermo Fisher Scientific, Massachusetts, USA). Plasma from healthy controls and non-related mAb were used as negative controls. Broad nAb S309 was used as a positive control.
Spike-specific single memory B-cell sorting
Single spike-specific memory B cells were sorted from COVID-19 convalescents and vaccinees to generate mAbs. SARS-CoV-2 spike (Sino Biological, Beijing, China) was labeled with Alexa Fluor 488 and Alexa Fluor 647 (Invitrogen, California, USA) and used as probes to identify spike-specific memory B cells. PBMCs from convalescents or vaccinees were allowed to grow for 3 h after thawing and were then stained with Live/Dead reagent (Invitrogen, California, USA) at 4 °C for 30 min. Cells were washed twice with staining buffer (PBS, 1% FBS) and stained with the SARS-CoV-2 spike probes. After a 30 min incubation, cells were washed twice with staining buffer and stained with antibodies BUV373 mouse anti-human CD3 (SK7) (BD Biosciences, New Jersey, USA), PerCP cy5.5 mouse anti-human CD19 (HIB19) (BioLegend, California, USA), PE cy7 mouse anti-human CD27 (M-T271) (BioLegend, California, USA), PE/Dazzle™ 594 mouse anti-human IgD (IA6-2) (BioLegend, California, USA), and PE mouse anti-human IgG (G18-145) (BD Biosciences, New Jersey, USA), at 4 °C for an additional 30 min. After washing, the cells were resuspended for sorting in 10% FBS/Roswell Park Memorial Institute (RPMI) 1640 medium. The cells were sorted on a Beckman Coulter MoFlo XDP instrument. Spike-specific memory B cells were gated as CD3-CD19+CD27+IgD-IgG+spike-AF488+ and AF647+ B cells. Single spike-specific memory B cells were sorted into the wells of a 96-well PCR plate (Thermo Fisher Scientific, Massachusetts, USA), with each well containing 7 μL lysis buffer (1.75 μL 10× PBS, 5 mM dithiothreitol (DTT), 1% IGEPAL, 20 ng/μL random primers, and 2 U/μL RNAsin). The plate was immediately spun down and put on dry ice until further use.
Antibody gene amplification and mAb production
Total RNA from the sorted single cells was reverse-transcribed using SuperScript IV reverse transcriptase (Thermo Fisher Scientific, Massachusetts, USA), and antibody variable region genes were amplified by nested PCR68. The 96-well plate containing sorted single cells was incubated at 68 °C for 1 min and then transferred back onto ice. Then, RT reaction mix (0.2 μL of RNAsin, 40 U/μL, 2.2 μL H2O, 1.05 μL DTT, 0.5 μL dNTPs, 2.8 μL of 5× SuperScript IV buffer, and 0.25 μL SuperScript IV) was added to each well. Reverse transcription was performed at 42 °C for 5 min, 25 °C for 10 min, 50 °C for 60 min, and 94 °C for 5 min. Next, antibody variable regions (VH, Vκ, and Vλ) were amplified by nested PCR using IgG-specific primers and HotStarTaq DNA Polymerase (Qiagen, Nordrhein-Westfalen, Germany), as previously described68. The PCR products were purified and sequenced to determine the sequences of the H and κ/λ chains. The sequences of the variable regions of the mAbs were then synthesized and separately inserted into Ig heavy- or light-chain expression vectors (AbVec2.0-IGHG1, AbVec1.1-IGKC, and AbVec1.1-IGLC2-XhoI). The antibody gene usage was determined using IgBLAST.
The plasmids carrying paired heavy and light chain genes were co-transfected into 293 F cells at a 1:2 ratio using polyethylenimine (PEI, Polysciences, Pennsylvania, USA). After 5 days, the culture supernatant was harvested and filtered through a 0.22-μm filter. The expressed antibodies in supernatant were purified using HiTrap Protein A HP columns (Cytiva, Marlborough, USA) in an ÄKTA pure system (Cytiva, Marlborough, USA).
ACE2 binding assay
ACE2 binding was examined by ELISA with slight modifications. A 96-well ELISA plate coated with SARS-CoV-2 RBD protein (2 µg/ml, 100 μL/well) was blocked and incubated with mAbs (10 µg/ml, 100 μL/well) and biotin–ACE2 (Sino Biological, China) at 37 °C for 1 h. After washing, streptavidin/HRP (1:2000 dilution; Solarbio, China) was added and incubated at 37 °C for 1 h. The plate was then developed with 100 μL of 3,3’,5,5’-tetramethylbenzidine dihydrochloride (TMB), terminated by the addition of 50 μL of 1 M H2SO4, and assessed for the OD450. A positive control (biotin–ACE2 protein without mAbs) and a blank control (blocking buffer only) were included. The percentage of inhibition of ACE2 binding was calculated using the following equation:
Biolayer interferometry (BLI) assay
The binding affinities between mAbs and SARS-CoV-1/2 RBD proteins were measured by BLI assay using an Octet RH16 instrument (Sartorius, Germany). All mAbs and RBDs were diluted in assay buffer (0.1% w/v BSA and 0.02% Tween-20 in PBS). mAbs were initially bound to the proA biosensor for 90 s. After a washing step using assay buffer, the sensor-bound mAbs were exposed to wells containing serial dilutions of RBDs in assay buffer for 120 s. The sensors were then placed in assay buffer for dissociation for up to 300 s, and assay buffer without antibody was used to correct for background. The results were analyzed using Data Analysis 12.0 software to determine the Kon, Koff, and KD values for the two different RBDs.
Statistical analysis
The Kolmogorov–Smirnov test was employed to assess normality prior to conducting the comparison. Data with non-normally distributed variables were expressed as medians±IQRs (interquartile ranges). Spearman’s rank correlation coefficient was utilized to analyze the correlation between neutralization titer and neutralization breadth. For paired sample comparisons, paired t-tests were used to analyze the differences between two groups. Mann–Whitney U tests were used to analyze two independent variables. To compare the CDR3 length and SHM number, Kruskal–Wallis tests with post hoc Dunn’s multiple comparisons tests were conducted. GraphPad Prism v.9.0 and SPSS v.26 were used for all data analyses, A P value less than 0.05 was considered to be significant, denoted as *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001. ns, not significant. Unless otherwise stated, all numerical data presented in this study were obtained from at least three independent experiments.
Supplementary information
Acknowledgements
We thank all of the participants. This work was supported by the National Natural Science Foundation of China (92269115, 82061138020, 32270996, and 82102365), The Science and Technology Innovation Program of Hunan Province of China (2022RC3079), International Cooperation in Science and Technology of the Guangdong Provincial Science and Technology Program (2023A0505050091 for Y.-P.L.), the Science and Technology Planning Project of Guangdong Province (2021B1212040017 for Y.-P.L.), the SC1-PHE-CORONAVIRUS-2020: “Advancing knowledge for the clinical and public health response to the 2019-nCoV epidemic” from the European Commission (CORONADX, no. 101003562 for Y.-P.L), the Clinical Medical Innovation Technology Guide Project of the Hunan Province (2021SK50306), the scientific research project of hunan provincial health commission (B202303087545, and W20243189).
Author contributions
X.Q., W.L., Y.-P.L., and H.C. contributed to the study design and data interpretation. Y. H., Q.W., Y.W., D.P., and R.L. contributed to patient recruitment and sample collection and processing. J.Y., J.C., S.T. X.Z., Y.T., and C.W. contributed to the serum antibody binding and avidity experiments. Z.L., X.Z., and H.C. contributed to recombinant protein expression and purification. J.Y., Y.H., and Q.W. contributed to spike-specific single memory B-cell sorting, antibody gene amplification, and mAb production. Q.W., F.C., Y.L., J.Q., and F.T. contributed to pseudotyped virus production and antibody neutralization experiments. J.Y., J.C., and S.T. contributed to the ACE2 binding assay and biolayer interferometry assay. Y. H., Q.W., T.X., F.L., and F.C. contributed to the ELISA experiments and data analysis. X.Q., Y.-P.L., Y.H., Q.W., and X.Z. drafted the manuscript. X.Q., W.L., Y.-P.L., Y.H., X.Z., and H.C. contributed to the critical revision of the manuscript for important intellectual content. X.Q., W.L., and Y.P.L. provided supervision. All authors met authorship criteria and approved the publication.
Data availability
The data generated and/or analysed in the current study are available from the corresponding authors on reasonable request.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yabin Hu, Qian Wu, Fangfang Chang, Jing Yang, Xiaoyue Zhang.
Contributor Information
Hongying Chen, Email: chenhy@nwsuaf.edu.cn.
Wenpei Liu, Email: wenpeiliu_2008@foxmail.com.
Yi-Ping Li, Email: lyiping@mail.sysu.edu.cn.
Xiaowang Qu, Email: quxiaowang@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-024-00997-8.
References
- 1.Peiris, J. S. et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet361, 1319–1325 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris, J. S., Guan, Y. & Yuen, K. Y. Severe acute respiratory syndrome. Nat. Med.10, S88–S97 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu, N. et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letko, M., Seifert, S. N., Olival, K. J., Plowright, R. K. & Munster, V. J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol18, 461–471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz-Aravena, M. et al. Ecology, evolution and spillover of coronaviruses from bats. Nat. Rev. Microbiol20, 299–314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan, Y. et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science302, 276–278 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Xiao, K. et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature583, 286–289 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Lam, T. T. et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature583, 282–285 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Lau, S. K. et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA102, 14040–14045 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, B. et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog.13, e1006698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacroix, A. et al. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect., Genet. Evol.: J. Mol. Epidemiol. Evolut. Genet. Infect. Dis.48, 10–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, W. et al. Bats are natural reservoirs of SARS-like coronaviruses. Science310, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Rihtaric, D., Hostnik, P., Steyer, A., Grom, J. & Toplak, I. Identification of SARS-like coronaviruses in horseshoe bats (Rhinolophus hipposideros) in Slovenia. Arch. Virol.155, 507–514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouilh, M. A. et al. SARS-Coronavirus ancestor’s foot-prints in South-East Asian bat colonies and the refuge theory. Infect., Genet. Evol.: J. Mol. Epidemiol. Evolut. Genet. Infect. Dis.11, 1690–1702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong, S. et al. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg. Infect. Dis.15, 482–485 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge, X. Y. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature503, 535–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou, H. et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol.30, 2196–2203.e2193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou, H. et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell184, 4380–4391.e4314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menachery, V. D. et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl Acad. Sci. USA113, 3048–3053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, X. L. et al. Isolation and Characterization of a Novel Bat Coronavirus Closely Related to the Direct Progenitor of Severe Acute Respiratory Syndrome Coronavirus. J. Virol.90, 3253–3256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menachery, V. D. et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med21, 1508–1513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, N. et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin.33, 104–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, H. et al. Human-animal interactions and bat coronavirus spillover potential among rural residents in Southern China. Biosaf. Health1, 84–90 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy, P. D. & Simon, A. E. New insights into the mechanisms of RNA recombination. Virology235, 1–9 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Rowe, C. L. et al. Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J. Virol.71, 6183–6190 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv, H. et al. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep.31, 107725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson, D. E. et al. Lack of cross-neutralization by SARS patient sera towards SARS-CoV-2. Emerg. microbes Infect.9, 900–902 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson, E. M. et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell184, 1858–1864.e1810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grobben, M. et al. Cross-reactive antibodies after SARS-CoV-2 infection and vaccination. Elife10, 10.7554/eLife.70330 (2021). [DOI] [PMC free article] [PubMed]
- 32.Tan, C. W. et al. Pan-Sarbecovirus Neutralizing Antibodies in BNT162b2-Immunized SARS-CoV-1 Survivors. N. Engl. J. Med385, 1401–1406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia, J. Z. et al. Priming conditions shape breadth of neutralizing antibody responses to sarbecoviruses. Nat. Commun.13, 6285 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wec, A. Z. et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science369, 731–736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starr, T. N. et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature597, 97–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tortorici, M. A. et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature597, 103–108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jette, C. A. et al. Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. Cell Rep.36, 109760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez, D. R. et al. A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci. Transl. Med14, eabj7125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, P. et al. A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses. Emerg. Microbes Infect.11, 147–157 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, Y. J. et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science375, 449–454 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, L. et al. An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Sci. Transl. Med14, eabn6859 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He, W. T. et al. Targeted isolation of diverse human protective broadly neutralizing antibodies against SARS-like viruses. Nat. Immunol.23, 960–970 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, X. et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2. Nat. Microbiol7, 1063–1074 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Pegu, A. et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science373, 1372–1377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Widge, A. T. et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med384, 80–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemet, I. et al. Third BNT162b2 Vaccination Neutralization of SARS-CoV-2 Omicron Infection. N. Engl. J. Med386, 492–494 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muik, A. et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science375, 678–680 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brouwer, P. J. M. et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science369, 643–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andreano, E. et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell184, 1821–1835.e1816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreer, C. et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell182, 843–854.e812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muecksch, F. et al. Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature607, 128–134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Letko, M., Marzi, A. & Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol5, 562–569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, M. Q. et al. A SARS-CoV-2-Related Virus from Malayan Pangolin Causes Lung Infection without Severe Disease in Human ACE2-Transgenic Mice. J. Virol.97, e0171922 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin, H. F. et al. Characterization of a mouse-adapted strain of bat severe acute respiratory syndrome-related coronavirus. J. Virol.97, e0079023 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burton, D. R. & Topol, E. J. Variant-proof vaccines - invest now for the next pandemic. Nature590, 386–388 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Tregoning, J. S., Flight, K. E., Higham, S. L., Wang, Z. & Pierce, B. F. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol.21, 626–636 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li, M. et al. COVID-19 vaccine development: milestones, lessons and prospects. Signal Transduct. Target Ther.7, 146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saunders, K. O. et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature594, 553–559 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez, D. R. et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science373, 991–998 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu, M. Q. et al. Inactivated SARS-CoV-2 Vaccine Shows Cross-Protection against Bat SARS-Related Coronaviruses in Human ACE2 Transgenic Mice. J. Virol.96, e0016922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinto, D. et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science373, 1109–1116 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Y. et al. Novel sarbecovirus bispecific neutralizing antibodies with exceptional breadth and potency against currently circulating SARS-CoV-2 variants and sarbecoviruses. Cell Discov.8, 36 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seifert, S. N. et al. An ACE2-dependent Sarbecovirus in Russian bats is resistant to SARS-CoV-2 vaccines. PLoS Pathog.18, e1010828 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo, H. et al. Isolation of ACE2-dependent and -independent sarbecoviruses from Chinese horseshoe bats. J. Virol.97, e0039523 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He, R. et al. SARS-CoV-2 spike-specific T(FH) cells exhibit unique responses in infected and vaccinated individuals. Signal Transduct. Target Ther.8, 393 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen, J. et al. Identification of broad neutralizing antibodies against Omicron subvariants from COVID-19 convalescents and vaccine recipients. Virol. Sin.38, 313–316 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, J. et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat. Microbiol6, 51–58 (2021). [DOI] [PubMed] [Google Scholar]
- 68.Wardemann, H. & Busse, C. E. Expression Cloning of Antibodies from Single Human B Cells. Methods Mol. Biol.1956, 105–125 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and/or analysed in the current study are available from the corresponding authors on reasonable request.