Abstract
Mutations in GBA1 encoding the lysosomal enzyme β-glucocerebrosidase (GCase) are among the most prevalent genetic susceptibility factors for Parkinson’s disease (PD), with 10–30% of carriers developing the disease. To identify genetic modifiers contributing to the incomplete penetrance, we examined the effect of 1634 human transcription factors (TFs) on GCase activity in lysates of an engineered human glioblastoma line homozygous for the pathogenic GBA1 L444P variant. Using an arrayed CRISPR activation library, we uncovered 11 TFs as regulators of GCase activity. Among these, activation of MITF and TFEC increased lysosomal GCase activity in live cells, while activation of ONECUT2 and USF2 decreased it. While MITF, TFEC, and USF2 affected GBA1 transcription, ONECUT2 might control GCase trafficking. The effects of MITF, TFEC, and USF2 on lysosomal GCase activity were reproducible in iPSC-derived neurons from PD patients. Our study provides a systematic approach to identifying modulators of GCase activity and deepens our understanding of the mechanisms regulating GCase.
Subject terms: Cellular neuroscience, Molecular biology
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder characterized by motor symptoms comprising bradykinesia, rigidity, postural instability, and tremor, as well as non-motor features, including depression and cognitive impairment1. Mutations in GBA1 have emerged as a major genetic susceptibility factor for PD development2. GBA1 encodes the lysosomal enzyme β-glucocerebrosidase (GCase), which catalyzes the hydrolysis of glucosylceramide (GlcCer) and glucosylsphingosine (GlcSph) into ceramide, sphingosine, and glucose. Thus, GCase is crucially involved in the metabolism of glycosphingolipids (GSL), which play central roles in growth regulation, cell migration, apoptosis, and inflammatory responses, among other processes3.
The association between GBA1 mutations and PD was first identified in individuals with Gaucher disease (GD), the most common lysosomal storage disorder, caused by biallelic mutations in GBA1. It then emerged that not only individuals with GD but also their relatives carrying heterozygous GBA1 mutations have an elevated risk of developing PD. With a 5–20% prevalence in PD patients, GBA1 variants are a major genetic susceptibility factor for PD2,4–8. In addition, individuals with PD associated with GBA1 mutations exhibit an earlier age of disease onset and show greater cognitive impairment compared to non-mutation carriers2,5. Further strengthening the link between GCase and PD, the enzymatic activity of GCase is reduced in PD patients even in the absence of mutations within GBA19, suggesting a key role for GBA1 in the development and progression of PD.
One striking observation is that only about 10–30% of individuals with disease-associated mutations in GBA1 will develop PD8. This modest penetrance, the clustering of PD in some families carrying GBA1 variants, and phenotypic heterogeneity in GD patients, suggest the involvement of genetic modifiers influencing GCase activity, PD risk, and severity4. Genome-wide association studies (GWAS) have identified BIN1, SNCA, TMEM175, and CTSB as candidate loci influencing PD risk and progression in the context of GBA1 mutations8,10. While there is evidence that TMEM175 affects GCase activity in both cell models and humans11,12, the impact of BIN1 and CTSB on GCase function awaits experimental confirmation. Moreover, these candidate loci likely represent only a fraction of the genetic modifiers associated with disease risk, as low-prevalence single nucleotide variants (SNVs) may go undetected in GWAS13.
As transcription factors (TFs) serve as “master regulators” orchestrating entire pathways and the expression of numerous other genes, they present an attractive target for identifying comprehensive networks regulating a process of interest such as GCase activity14. Like many other lysosomal genes, the GBA1 promoter contains two Coordinated Lysosomal Expression and Regulation (CLEAR) motifs, DNA sequences recognized by Transcription Factor EB (TFEB). This nuclear-lysosomal axis enables cells to fine-tune their metabolism in response to environmental changes, such as nutrient availability15,16. Interestingly, the presence of evolutionarily conserved sequences within the 5′ and 3′ non-coding regions of GBA1 suggests the existence of additional TF binding sites that may control GCase expression, potentially by other unidentified TFs17. Yet, beyond TFEB, the transcriptional regulation of GCase remains poorly understood and no TF has appeared in GWAS studies focused on PD susceptibility.
CRISPR-based screens offer a powerful means of systematically identifying genes regulating biological processes such as GCase activity. Unlike GWAS, which are correlative, CRISPR screens enable the discovery of causal relationships18–20. Toward this goal, we recently generated an arrayed CRISPR activation (CRISPRa) library termed T.gonfio that targets each protein-coding gene with four non-overlapping guide RNAs21. Using glioblastoma cells base-edited to harbor a pathogenic GBA1 mutation, we individually activated 1634 human TFs. By assaying GCase in cell lysates, we identified 29 TFs modulating GCase activity including MITF and TFEC, two members of the MiT/TFE family of TFs to which TFEB and TFE3 also belong. MITF and TFEC increased whereas USF2 and ONECUT2 decreased lysosomal GCase activity, respectively. ONECUT2 activation did not regulate GBA1 transcript levels but resulted in the differential expression of genes associated with vesicle trafficking. These findings expand the transcriptional landscape of GBA1 regulation, deepening our understanding of the mechanisms involved in modulating GCase activity in the context of GBA1 mutations.
Results
A cellular model system to study L444P mutant GCase activity
Large-scale genetic screens require scalable, easy-to-manipulate model systems. We selected the human glioblastoma line LN-229, which possesses intermediate endogenous GCase activity and expression levels22 compared to other cancer cell lines. As the detection of enhancers of mutant GCase might be clinically most relevant, we aimed to generate an LN-229 line harboring the pathogenic GBA1 L444P variant (NM_000157.4:c.1448T > C) in all alleles (Fig. 1a) using the adenine base editor ABE8e23. The L444P mutation results in misfolding of GCase and recognition of the protein by the endoplasmic reticulum (ER) quality-control system, which ultimately leads to its proteasomal degradation and greatly reduced lysosomal levels of the variant24. Just one mutant L444P allele confers an increased risk of developing PD, whereas a homozygous mutation drastically reduces enzymatic activity and manifests as Gaucher disease (GD), which is often accompanied by severe neurological involvement (GD type II or III)25–27.
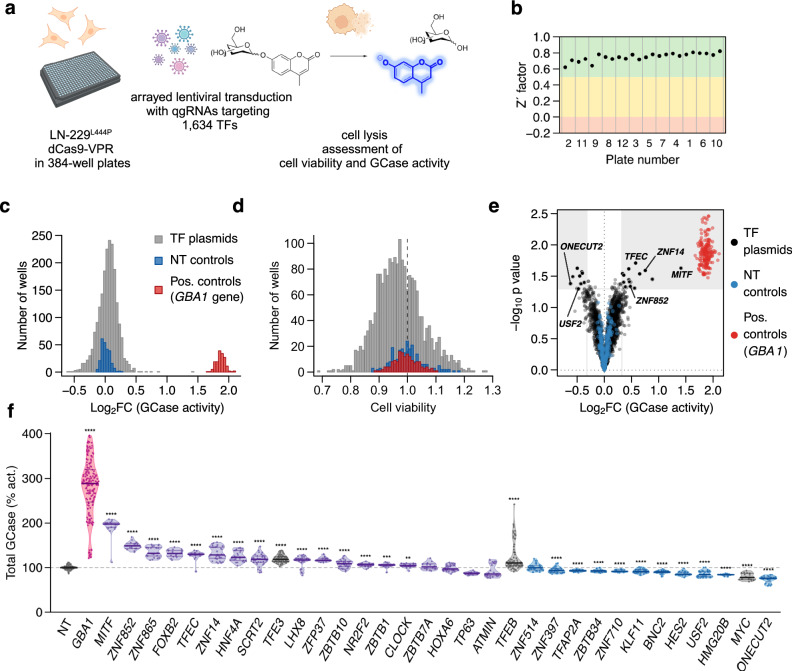
Fig. 1. A forward-genetics screen for GCase activity modulators.
a Study workflow. b Read frequencies (next-generation sequencing) of the desired T > C point mutation in the base-edited LN-229L444P (orange) and its parental LN-229WT (blue) line. c GCase enzymatic activity in cell lysates of LN-229WT (WT), LN-229L444P (L444P), and GBA1-ablated (KO) LN-229 cells. 30 wells/genotype, two independent experiments. AT3375: GCase-selective inhibitor. d. Immunoblot of cellular GCase protein in LN-229ΔGBA1 (KO), LN-229L444P (L444P), and LN-229WT (WT) after treatment with the proteasome inhibitor MG-132 (+) or with DMSO vehicle (−). N = 4 experiments, unpaired two-samples t-test. e. Lysate-based GCase assay in LN-229WT (WT) and LN-229L444P (L444P) cells after treatment with the proteasome inhibitor MG-132 or with DMSO vehicle. N = 18 wells/condition, unpaired two-samples t-test fluorescence intensity was normalized to average cell number/well. DMSO-treated WT cells were taken as reference (100%).
Next-generation sequencing using primers that selectively amplify GBA1 but not its highly homologous pseudogene GBAP128 confirmed the presence of the L444P mutation in all alleles of the polyploid LN-229 line (Fig. 1b). A single-cell derived edited clone (hereafter referred to as LN-229L444P) was used for all subsequent experiments. GCase activity in LN-229L444P lysates was reduced to 7% of the parental line expressing wild-type GCase (LN-229WT) (Fig. 1c) when measured with a lysate-based assay employing the artificial substrate 4-methylumbelliferyl-β-D-glucopyranoside (4MU-Glc). This is similar to the residual enzymatic activity reported in fibroblasts from patients with a homozygous L444P mutation27. Treatment of LN-229L444P cells with the proteasome inhibitor MG-132 increased GCase protein levels to 145% and GCase activity in cell lysates to 139% of DMSO-treated LN-229L444P control cells, while GCase protein levels and activity remained unchanged in LN-229WT cells (Fig. 1d, e and Supplementary Fig. 1a). The electrophoretic pattern differences (Fig. 1d) indicate differences in the forms of N-glycosylation found on the WT and L444P variant of GCase. ER-retained proteins, such as the L444P GCase variant, typically display only mannose rich N-glycans, which can be degraded by the endo-glycosidase EndoH. In contrast, properly folded proteins passing into the Golgi apparatus have more elaborate N-glycans that are less susceptible to EndoH. Accordingly, digestion of cell lysates expressing the L444P variant show near complete susceptibility to EndoH digestion, which contrasts with the WT enzyme (Supplementary Fig. 1b). While a decrease in overall levels of the L444P variant of GCase is often seen, here we see only a modest trend to a reduction in its levels. This difference may arise from the rate of degradation of the L444P variant being relatively slow within this cell line under the culture conditions, leading to increased levels being retained in the ER and then observed by immunoblot. Alternatively, the effect may arise from the antibody being used in this study recognizing the L444P misfolded GCase, while other antibodies not being able to do so29. In any event, the increase in levels of the L444P variant observed upon proteasome inhibition, coupled with its high susceptibility to EndoH supports the L444P variant being retained within the ER as generally reported.
One important caveat of lysate-based assays employing artificial substrates is hydrolysis by human β-glucosidases other than GCase, such as GBA2 and GBA330–33. To ascertain the specificity of our assay for GCase activity in LN-229 cells, we conducted dose-response experiments in cell lysates using three inhibitors: conduritol-β-epoxide (CBE), which inhibits glycosidases beyond GCase at higher concentrations34, the highly selective GCase inhibitor AT337532, and the GBA2-selective inhibitor miglustat35. Treatment of LN-229WT cells with CBE and AT3375 at commonly employed working concentrations (300 µM and 10 µM, respectively) reduced the fluorescent signal to <5% of that seen in DMSO-treated cells, whereas miglustat caused no decrease (Supplementary Fig. 1b-d). To further confirm specificity of the assay, we generated LN-229 cells in which GBA1 was ablated (LN-229ΔGBA1) by applying sgRNAs targeting the catalytic domain of GCase. In LN-229ΔGBA1 lysates, the turnover of 4MU-Glc was reduced to 1.5% of the levels observed in LN-229WT cells (Fig. 1c). Collectively, these data confirmed premature proteasomal degradation of the L444P mutant GCase in engineered LN-229L444P cells. Additionally, dose-response curves showed that 4MU-Glc was predominantly hydrolyzed by GCase in LN-229 cells30. Thus, we concluded that the LN-229L444P line is suitable for identifying L444P mutant GCase activity modulators using a lysate-based assay.
An arrayed CRISPRa screen identifies 11 TFs as modulators of mutant GCase activity
To identify TFs regulating GCase activity, we performed an arrayed transcriptional activation (CRISPRa) screen (Fig. 2a) using the T.gonfio library21. To achieve high on-target activity, each protein-coding gene is targeted by four non-overlapping sgRNAs referred to as quadruple-guide RNAs (qgRNAs). To maximize their versatility, qgRNA plasmids were designed to tolerate human DNA polymorphisms. In addition, each major transcription start site (TSS) is targeted individually to account for potential divergence in TSS activities among different cell types. LN-229L444P cells stably expressing dCas9-VPR were seeded and transduced with lentiviruses carrying 1889 qgRNAs targeting 1634 human TFs14 in three replicate 384-well plates, resulting in 6,729 transductions (including controls), as 171 TFs had more than one TSS. Five days post-transduction, GCase activity was measured on two replicate plates in cell lysates using 4MU-Glc as the substrate. Cell viability was assessed with the CellTiter-Glo assay on the third replicate plate (Supplementary Fig. 2a).
Fig. 2. Arrayed CRISPR activation screen of human TFs in LN-229L444P cells.
a Overview of the screening workflow. b Z’ factor of each assay plate reporting on the separation between cells exposed to qgRNA lentiviruses targeting GBA1 (positive controls) vs. nontargeting (NT) qgRNA (negative controls). Plates are ordered in ascending order of Z’ factor of the replicate plates, with plate numbers (order of pipetting) provided on the x-axis. Histograms of enzymatic activity (c) and viability (d) of LN-229L444P cells treated with TF-activating (grey), GBA1-specific (red), and nontargeting qgRNAs (blue). GCase activity was normalized to wells infected with NT controls. e Changes in GCase activity (abscissa) and ps (ordinate) after activation of designated transcription factors. f Re-testing of 29 candidates with a higher number of replicates. Grey: NT controls and TFs previously found to regulate GCase expression (TFE3, TFEB, and MYC). Purple and blue: TFs increasing and decreasing GCase activity in the primary screen, respectively. Red: positive controls (GBA1-activated cells). N ≥ 10 transductions in ≥2 independent experiments; unpaired t-test. Asterisks: ps (here and henceforth). ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
Positive (GBA1-activated) and non-targeting control (NT ctrl) qgRNAs yielded distinct signals with no overlap in fluorescence intensities, resulting in a Z′-factor >0.5 on all plates36 (Fig. 2b, c). The coefficient of variation between duplicate samples (R2) was 0.71, demonstrating a high degree of technical reproducibility (Supplementary Fig. 2d). No major changes in cell viability were observed after lentiviral transduction and activation of TFs (Fig. 2d). Except for a minor plate gradient towards lower values around well A24 (Supplementary Fig. 2b), the quality measurements confirmed the robustness of the primary screen, instilling confidence in our selection of candidate TFs for further investigation.
Clinical observations in GD patients undergoing enzyme replacement therapy suggest that even small increases in GCase activity (≈10%) can be clinically meaningful37. Hence, using the log2-transformed fold change (log2FC) fluorescence intensity values, we applied the following cut-off criteria for candidate selection: (a) a p < 0.05 computed from two experimental replicates and (b) a mean log2FC > 0.32 or < −0.32 for up- and downregulators of GCase activity. A log2FC of ±0.32 resulted in 4.5 standard deviations (SD) from the mean fluorescence values of the NT controls. None of the 249 NT controls met these cut-off criteria. These selection criteria resulted in the nomination of 29 TFs (18 up- and 11 down-regulators of enzymatic activity) as candidate regulators of GCase activity (Fig. 2e). To prioritize the strongest candidates and minimize false-positive hits, we retested qgRNAs targeting those 29 candidate TFs in ≥2 independent experiments; each TF was tested with ≥10 replicates. 11 of the 29 candidates (38%), namely MITF, ZNF852, ZNF865, FOXB2, TFEC, ZNF14, HNF4A, SCRT2, USF2, HMG20B, ONECUT2 exhibited changes in GCase activity equal to or greater than those observed following CRISPRa of TFE3 (median log2FC = 0.25 = log2(1.19)), a member of the microphthalmia (MiT/TFE) family of TFs previously shown to regulate GBA1 expression and GCase activity38,39. Notably, MITF transcription start site 2 (TSS2) (i.e., the more 3′, downstream TSS, hereafter referred to as MITF) and TFEC, two members of the MiT/TFE family, were among the 11 reproducible candidates (Fig. 2f). All those 11 candidates could also be reproduced in LN-229WT dCas9-VPR cells (Supplementary Fig. 3a). In summary, 29 out of 1,634 TFs met our primary cutoff criteria, with 11 demonstrating robust and reproducible effects on GCase activity in the lysate-based assay upon retesting. These 11 TFs were subjected to further validation.
MITF and TFEC increase GBA1 transcript levels
As the enzymatic activity of GCase can be regulated at multiple levels (Supplementary Fig. 3b), we sought to differentiate between those TFs regulating GCase activity by controlling gene expression and those acting post-transcriptionally. However, these processes are not mutually exclusive. We performed CRISPRa of the TFs in LN-229L444P cells and measured GBA1 transcript levels by reverse transcription quantitative PCR (RT-qPCR) and GCase protein levels through immunoblotting. We expected that most TFs would directly regulate GBA1 expression, affecting GCase protein abundance. While unchanged GBA1 RNA levels make a transcriptional regulatory effect unlikely, changes in transcript levels do not necessarily imply transcriptional regulation, as alterations in RNA stability could contribute to the observed effect. For RT-qPCR, we used primers specifically targeting GBA1 to avoid amplifying the highly homologous GBAP1 pseudogene which shares over 90% sequence identity with GBA1 (Fig. 3a)28.
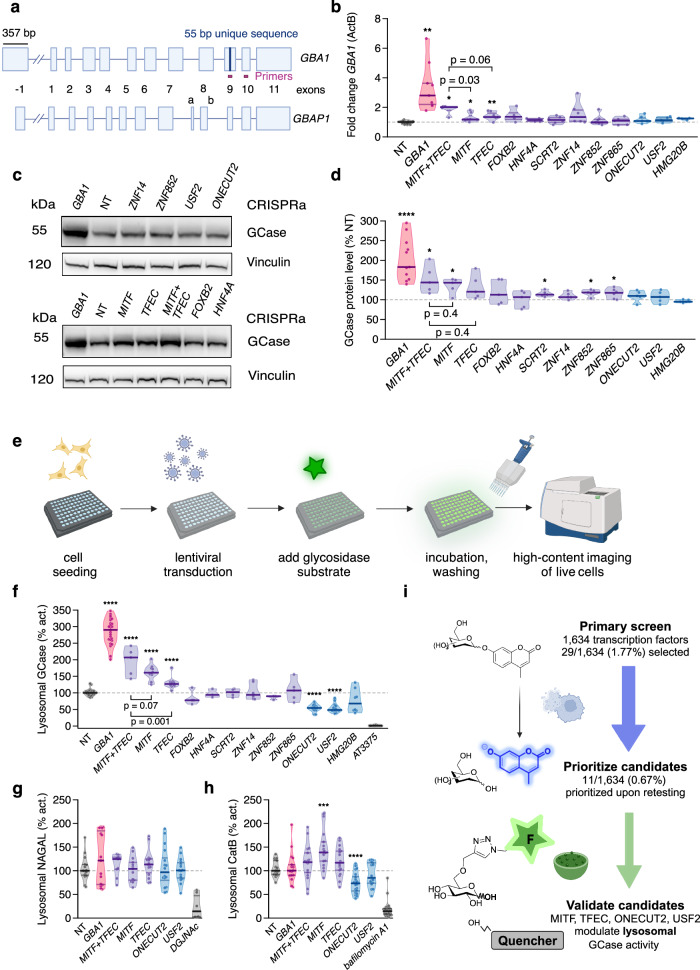
Fig. 3. GBA1 expression, GCase protein levels, and lysosomal hydrolase activities in LN-229L444P cells following activation of TFs.
a GBA1 and GBAP1 genes approximately to scale. Boxes: exons; lines: introns. Purple: 55-bp sequence unique to GBA1 exon 9. Pink: GBA1 RT-PCR primers. b Fold changes GBA1 mRNA (RT-qPCR) after CRISPRa of TFs in LN-229L444P cells. -actin (ActB) was used for normalization (N = 3–8 repeats). c GCase protein levels after CRISPRa of TFs in LN-229L444P cells. Cells were harvested 5 days post-transduction. d GCase protein quantification after CRIPSRa of TFs in LN-229L444P cells (N = 5–11 experiments). e Microscopy-based assessment of lysosomal hydrolase activities in live LN-229L444P cells. f GCase activity assessed by LysoFQ-GBA1 in live LN-229L444P cells after CRISPRa (N = 3–9 experiments). g α-N-acetylgalactosaminidase (NAGAL) activity in live LN-229L444P cells after CRISPRa of TFs. DGJNAc: NAGAL inhibitor (3 experiments). h Cathepsin B activity assessed in live LN-229L444P cells after CRISPRa of TFs. Bafilomycin A1: v-ATPase inhibitor (N = 3 experiments). i Hit selection process. Solid lines in C and D: medians, 25%, and 75% quartiles. Medians of NT controls: 100%. Unpaired t-test.
CRISPRa of MITF and TFEC significantly increased GBA1 transcript levels (fold changes 1.18 and 1.35, respectively) (Fig. 3b). As GBAP1 has been suggested to regulate GBA1 levels through miR-22-3p sequestration28, we also assessed transcript levels of GBAP1 after CRISPRa of our candidates, which were unchanged (Supplementary Fig. 3c). At the protein level, CRISPRa of MITF, ZNF865, ZNF852, and SCRT2 also increased GCase protein levels (135%, 125%, 120%, and 115% of the NT control, respectively) (Fig. 3c, d). In summary, CRISPRa of MITF and TFEC altered GBA1 transcript levels, suggesting their effects on GCase activity are at least partly mediated through regulating GBA1 RNA abundance. In line with a direct transcriptional regulation of GBA1 RNA levels via promoter binding, CRISPRa of MITF increased luminescence intensity in a GBA1 promoter luciferase assay (Supplementary Fig. 3d)28. Furthermore, CRISPRa of MITF, ZNF852, ZNF865, and SCRT2 increased GCase protein levels, indicating that changes in protein abundance contribute to the observed increase in GCase activity in the lysate-based assay.
Four TFs modulate lysosomal GCase activity
While lysate-based assays report on total cellular enzyme activity, they require the disruption of the lysosomal microenvironment32. Accordingly, we aimed to corroborate the effect of our candidate TFs on GCase activity in its native lysosomal environment. To quantify lysosomal GCase activity in live LN-229L444P cells following CRISPRa of our 11 candidates, we employed the fluorescence-quenched cell-active substrate LysoFQ-GBA1, which specifically and quantitatively reports on GCase activity in lysosomes after being taken up via endocytosis32 (Fig. 3e, Supplementary Fig. 4). The fluorescence intensities observed in wells treated with the highly selective GCase inhibitor AT3375 were <5% of the values in NT-treated control wells, confirming that the fluorescent signal originated from the turnover of LysoFQ-GBA1 by GCase. Among the 11 TFs, two up-regulators (MITF and TFEC) and two down-regulators (USF2 and ONECUT2) significantly changed lysosomal GCase activity (Fig. 3f).
We then asked if the candidate TFs exclusively regulate GCase activity or exert a broader effect on lysosomal hydrolase activities, as was described for TFEB and TFE316,39. To do this, we used the bis-acetal-based (BAB) fluorescence-quenched substrate for α-N-acetylgalactosaminidase (NAGAL-BABS)40 and the Magic Red substrate for cathepsin B (CatB) in live LN-229L444P cells. No effect on lysosomal NAGAL activity was observed following the activation of our TFs. However, activation of MITF increased CatB activity while activation of ONECUT2 decreased it (Fig. 3g, h). Hence, the directionality of these effects paralleled those seen for GCase activity. Activation of TFEC and USF2 impacted CatB activity in the same direction as GCase activity measured in the lysate assay, but the effects were not significant (p = 0.076 and p = 0.1; Fig. 3h).
Overall, among the 11 modulators of GCase activity identified using the lysate-based assay, CRISPRa of MITF, TFEC, ONECUT2, and USF2 significantly altered lysosomal GCase activity in live LN-229L444P cells (Fig. 3i). Additionally, besides their impact on GCase, CRISPRa of MITF and ONECUT2 significantly affected CatB activity, indicating a regulatory influence of these TFs on lysosomal hydrolases beyond GCase (Supplementary Fig. 5a–e). The absence of effects on NAGAL activity suggests that distinct mechanisms and TFs govern different lysosomal hydrolases.
MITF, TFEC, and USF2 alter lysosomal GCase activity in iPSC-derived forebrain neurons
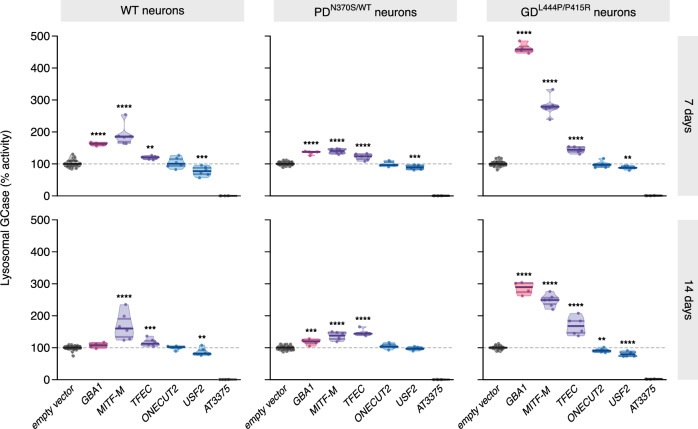
To determine whether the effect of the four candidate TFs on GCase activity extends to more clinically relevant cellular models, we introduced lentiviral overexpression vectors of these candidates into iPSC-derived forebrain neurons derived from a healthy control, from a patient with PD carrying the heterozygous GBA1N370S/WT mutation resulting in a moderate decrease in GCase activity (around 50% of WT), and from a GD patient with the compound heterozygous mutation GBA1L444P/P415R that shows drastically reduced GCase activity (around 10% of WT)32. GCase activity was assessed in live cells using LysoFQ-GBA1 at both seven and 14 days post-transduction. In line with the findings in LN-229L444P cells following CRISPRa of candidate TFs, overexpression of the MITF-M isoform resulted in the strongest increase in lysosomal GCase activity across all three neuronal lines, followed by TFEC overexpression. Overexpression of USF2 decreased lysosomal GCase activity across all three lines (to 80% and 90% of the empty vector in the WT and PD/GD lines, respectively, with 100% activity representing the activity seen in each line following transduction with empty vector). Overexpression of ONECUT2 did not affect enzymatic activity, except for a trend toward a decrease in the GBA1L444P/P415R neurons 14 days post-transduction (Fig. 4a–f, Supplementary Fig. 4f). In summary, the effect of MITF, TFEC, and USF2 on lysosomal GCase activity was replicated in iPSC-derived neurons from both GBA1 WT and GBA1 mutant individuals harboring two common pathogenic GBA1 variants (L444P and N370S), while ONECUT2 overexpression did not significantly alter lysosomal GCase activity. This discrepancy may be attributable to cell-type, isoform-and/or mutation-specific effects of ONECUT2 on enzymatic activity.
Fig. 4. Relative lysosomal GCase activity in iPSC-derived forebrain neurons.
Relative lysosomal GCase activity assessed with LysoFQ-GBA1 in live neurons derived from a healthy individual (WT neurons), from a Parkinson patient carrying a heterozygous GBA1 mutation (PDN370S/WT) or a Gaucher patient with compound heterozygous GBA1 mutations (GDL444P/P415R) after overexpression of candidate TFs at 7 (top row) or 14 days (bottom row) post-transduction. LN-229WT (WT), LN-229L444P (L444P), and GBA1-ablated (KO) LN-229 cells. 30 wells/genotype, two independent experiments. AT3375: GCase-selective inhibitor. Six replicate wells/candidate and 26 replicates for the empty vector; N = 1 experiment. Lines: medians and 25-75% quartiles. 100% Lysosomal GCase activity represents the relative activity seen in each of the three cell lines transduced with empty vector. Unpaired t-test.
USF2 is a bidirectional regulator of GCase abundance and activity
To assess whether the candidate genes regulate GCase activity bidirectionally, we ablated them using CRISPR in LN-229L444P and LN-229WT cells stably expressing Cas9. While CRISPRa of USF2 decreased GCase activity, ablating USF2 increased GCase activity in LN-229L444P in both lysate and live-cell assays (Fig. 5a, b). L444P GCase protein levels increased to 131% of the NT control following ablation of USF2 (Fig. 5c, d). The ablation efficiency was high, resulting in >75% reduction of USF2 protein levels (Fig. 5e). The effect of USF2 ablation on GCase activity and abundance was preserved in LN-229WT cells (Fig. 5f, g). USF2 exhibits functional overlap with its homolog USF1, and they bind DNA either as homo- or heterodimers41. To assess the effects of USF1, USF2, and USF1/USF2 double-knockout, transcriptomic profiling was performed in triplicates of LN-229L444P cells following CRISPR ablation. GBA1 emerged among the top five differentially expressed genes (DEGs) in the USF1/USF2 double-knockout samples (Fig. 5h, i). Transcript levels of other lysosomal hydrolases (CTSB, CTSD, GLA) and lysosomal membrane proteins (e.g., MCOLN1) were also altered. Furthermore, USF1/USF2 double-knockout significantly increased transcript levels of SCARB2 (encoding the GCase transport protein LIMP-2) and PSAP (encoding the GCase activating protein saposin C) which were previously described as modulators of GCase29 (Fig. 5i). In addition, subunits of the lysosomal v-ATPase were differentially expressed in both USF2 and USF1/2 ablated samples, consistent with phagosome acidification (GO:0090383) and lysosomal membrane (GO:0005765) being among the top five gene ontology (GO) terms identified in the overrepresentation analysis (ORA) (Fig. 5i). In summary, while CRISPRa of USF2 decreased GCase activity, its ablation increased the abundance and activity of GCase in both LN-229L444P and LN-229WT cells, establishing USF2 as a bi-directional regulator of GCase activity not restricted to the GBA1 L444P mutation. Furthermore, concurrent ablation of USF1 and USF2 enhanced transcript levels of GBA1, other lysosomal hydrolases, and lysosomal membrane proteins, reaffirming the involvement of USFs in the modulation of lysosomal gene expression, as previously documented in murine neurons41.
Fig. 5. USF2 ablation increases GBA1 transcription and GCase activity/abundance.
a GCase activity in cell lysates of LN-229L444P expressing Cas9 following ablations of TFs (N = 3 experiments). b GCase activity in live LN-229L444P cells expressing Cas9 assessed with LysoFQ-GBA1 after USF2 or GBA1 ablation (N = 3). c GCase immunoblot after USF2 ablation in LN-229L444P cells expressing Cas9. d GCase protein levels after ablation of USF2 in LN-229L444P expressing Cas9 (N = 4). e Immunoblot demonstrating 75% decrease of USF2 in LN-229L444P cells expressing Cas9 after ablation. f GCase activity in LN-229WT cell lysates after ablating candidate TFs (N = 3). g Immunoblot for GCase after ablating USF2 in LN-229WT expressing Cas9. h Selected top DEGs following transcriptomic profiling after ablation of USF1 and USF2 in LN-229L444P cells. i Transcriptional profiling of select genes following ablation of USF1, USF2, or USF1/2 in LN-229L444P cells. Horizontal axes: log2 fold-change (log2FC) in gene expression. j Overrepresented gene-ontology terms. Bar length: −log10 adjusted p. ((h–j): N = 3 replicates). Median values of NT controls: 100%. Solid lines: medians, 25%, and 75% quartiles.
Transcriptomic profiling suggests a role for lysosomal pH regulation by MITF and TFEC
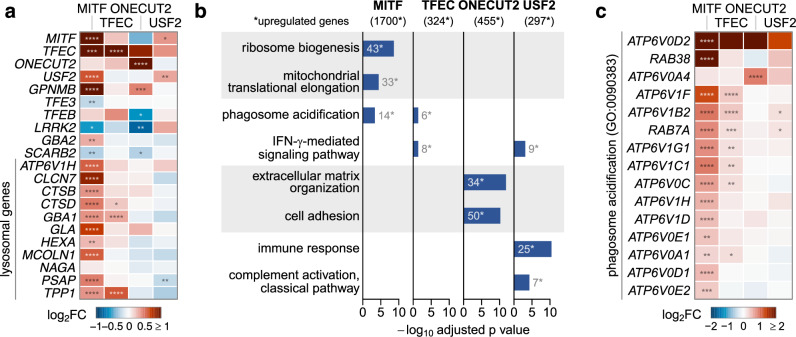
To obtain an unbiased view of the transcriptional changes induced by TF overexpression and to identify potential shared regulatory pathways, we profiled the transcriptome of LN-229L444P cells in triplicate following lentiviral transduction of the relevant CRISPRa qgRNAs. Consistent with the RT-qPCR results, GBA1 RNA levels were differentially expressed upon CRISPRa of MITF and TFEC (Supplementary Fig. 6A). CRISPRa of MITF increased the expression of multiple lysosomal genes, including CTSB, reminiscent of the effects of TFEB and TFE316. Additionally, CRISPRa of MITF resulted in the differential expression of TFEC, TFE3, and USF2 (Fig. 6a). ORA revealed shared GO terms related to proton-transporting ATPase activity (GO:0046961), proton transmembrane transporter activity (GO:0015078), and phagosome acidification (GO:0090383) in samples in which MITF and TFEC expression was activated. DEGs in those GO terms included numerous subunits of the lysosomal v-ATPase (Fig. 6b, c). Thus, MITF and TFEC may indirectly modulate GCase activity beyond regulating GBA1 expression through lysosomal pH regulation. The altered expression of TFEC, TFE3, and USF2 upon CRISPRa of MITF could indicate the existence of transcriptional networks or feedback loops in which MITF plays a dominant role.
Fig. 6. Transcriptional versus post-transcriptional mechanisms involved in GBA1 regulation.
a Transcriptional profiling of selected genes of interest following CRISPRa of candidate TFs in LN-229L444P cells. Horizontal axes: log2 fold-change (log2FC) in gene expression. b Top gene ontology terms of overrepresentation analysis (biological processes). Bar length: -log10 adjusted p. c Differentially expressed genes of GO term 0090383 (phagosome acidification) (N = 3 replicates per candidate TF).
Co-activation of MITF and TFEC amplifies GBA1 expression, GCase abundance, and activity
Since members of the MiT/TFE family bind DNA as both homo- and heterodimers42, we explored whether simultaneous activation of MITF and TFEC, hereafter referred to as co-activation, would yield a stronger effect on GBA1 transcript levels, GCase abundance, and activity compared to individual activation. LN-229L444P dCas9-VPR cells were transduced with qgRNAs targeting TFEC or MITF individually or with NT controls (MOI: 4). For co-activation, cells were transduced with qgRNAs targeting TFEC and MITF (MOI: 4 each) or with NT controls (MOI: 8). GBA1 transcripts rose more strongly upon co-activation than upon individual activation of MITF and TFEC (2x, 1.2x and 1.3x, respectively) as measured by RT-qPCR and bulk RNASeq (Fig. 3b, Supplementary Fig. 6b). Moreover, co-activation of MITF and TFEC led to the most pronounced increase in GCase protein levels (mean increase of 150% for co-activation versus 130% for individual activations compared to the NT control) (Fig. 3d). Consistent with their effects on GBA1 transcript and GCase protein levels, co-activation of MITF and TFEC more substantially increased lysosomal GCase activity compared to their individual activation (173% vs. 157% and 147% of the NT control, respectively). However, the difference between co- and individual activation on lysosomal GCase activity was only significant for TFEC (p = 0.001) (Fig. 3f). To summarize, co-activation of MITF and TFEC resulted in a synergistic effect on GBA1 expression, GCase abundance and activity, with the most substantial increase detected at the RNA level.
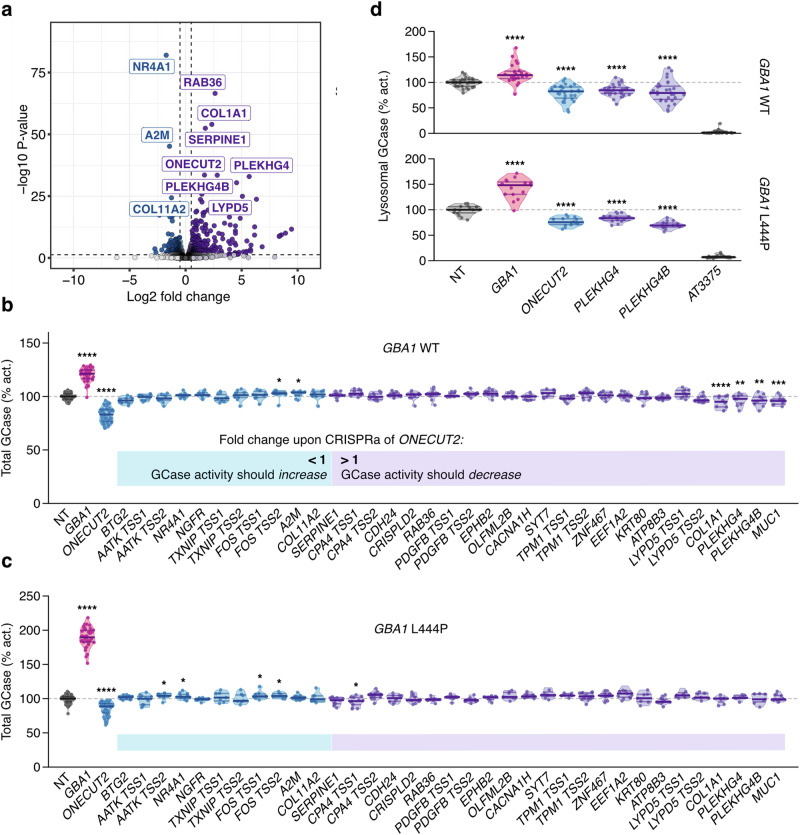
PLEKHG4 and PLEKHG4B may mediate the downstream effects of ONECUT2 on GCase activity
RT-qPCR and RNASeq showed that ONECUT2 activation did not alter GBA1 RNA levels (Fig. 3b, Supplementary Fig. 6a), indicating this TF regulates GCase posttranscriptionally. Many of the top DEGs identified after ONECUT2 activation are predicted to be involved in vesicle trafficking (Fig. 7a). Therefore, we hypothesized that ONECUT2 regulates GCase activity by influencing its trafficking to the lysosome or via lysosomal exocytosis. To identify potential mediators of these effects, we tested GCase activity upon activating each of the top 30 DEGs identified through transcriptomic profiling. LN-229L444P and LN-229WT cells stably expressing dCas9-VPR were transduced with qgRNAs targeting the top 30 DEGs, and GCase activity was assessed five days post-transduction in cell lysates with 4MUG-Glc. Although changes in total GCase activity were modest and the overlap between LN-229WT and LN-229L444P was limited, CRISPRa of PLEKHG4 and PLEKHG4B showed a significant reduction of GCase activity in LN-229WT cells (96 and 97% of the NT control) (Fig. 7b, c). Both genes encode guanine nucleotide exchange factors (GEFs) regulating cytoskeleton dynamics at the Golgi apparatus43,44. Since alterations in trafficking are likely to induce greater changes in lysosomal rather than total GCase activity, we assessed the effect of CRISPRa of PLEKHG4 and PLEKHG4B on lysosomal GCase activity using LysoFQ-GBA1. Indeed, activation of PLEKHG4 and PLEKHG4B significantly decreased lysosomal GCase activity in both LN-229WT and LN-229L444P cells (mean of 84 and 80% or 83 and 70% of the NT control, respectively) (Fig. 7d). Additionally, we evaluated alterations in protein abundance following CRISPRa of ONECUT2 by mass spectrometry analysis in LN-229L444P cells. Six of the top DEGs (namely, RAB36, TPM1, A2M, NGFR, COL11A2, and TXNIP) exhibited changes in protein abundance (p < 0.01 with fold changes >1.5), with RAB36 predicted to be involved in vesicle trafficking. Consistent with immunoblotting results, GCase abundance was unaffected by ONECUT2 activation (Supplementary Fig. 7). Intriguingly, MITF protein abundance was decreased after CRISPRa of ONECUT2 (log2FC = −0.74, p = 0.002).
Fig. 7. Identification of downstream mediators of ONECUT2’s effect on GCase activity.
a Transcriptomic profiling of selected DEGs following after CRISPRa of ONECUT2 in LN-229L444P cells (N = 3 replicates). Enzymatic activity in cell lysates following activation of top 30 DEGs in LN-229WT (b) or LN-229L444P (c) (N = 7 to 10). d Lysosomal enzymatic activity assessed with LysoFQ-GBA1 following CRISPRa of PLEKHG4 or PLEKHG4B in LN-229WT and LN-229L444P cells (N = 3). Median values of the NT controls set as 100%. Solid lines: medians and quartiles. Unpaired t-test.
In summary, screening the top 30 DEGs identified upon CRISPRa of ONECUT2 for their effect on GCase activity suggests that the GEFs PLEKHG4 and PLEKHG4B mediate, at least partially, the effect of ONECUT2 activation on GCase activity. Further, consistent with the hypothesis that ONECUT2 alters the trafficking of GCase to the lysosome, the impact on GCase was more pronounced when lysosomal enzymatic activity was assessed in live cells compared to total GCase activity in cell lysates (Fig. 7b–d).
Discussion
Taking advantage of CRISPR technology, we employed an unbiased approach to systematically define transcription factors modifying GCase activity in the presence of the pathogenic GBA1 L444P variant. Currently available CRISPRa libraries, unlike T.gonfio, are produced in a pooled format which limits their utility to assays reliant on selectable phenotypes like cell death, cell survival, or expression of cell-surface markers, and renders them unsuitable for complex biochemical screens as the one presented here. Therefore, we screened all human transcription factors employing a subset of the genome-wide T.gonfio arrayed CRISPRa library21.
In principle, overexpression can be achieved by transducing the genes of interest under transcriptional control of strong heterologous promoters. However, CRISPRa offers several advantages over traditional overexpression strategies in that it allows for the upregulation of TFs within their native context, thus preserving the cellular post-transcriptional processing. We had considered assaying GCase upon CRISPR-mediated arrayed gene ablation instead of activation. However, disrupting genes one by one is ineffective in identifying redundant pathways, as the ablation of single genes may not suffice to induce a phenotype. Moreover, ablating genes not endogenously expressed within the cell line under study cannot induce a phenotype. Lastly, unlike CRISPR ablation, CRISPRa enables studying the effect of essential genes on pathways of interest without causing cell death, thereby allowing interrogation of a larger number of genes.
Since transcriptional regulation likely alters GCase protein abundance, we screened for enzymatic activity in cell lysates. While lysate-based assays yield robust and reproducible results, they lack topological resolution since they report on total cellular GCase activity. Hence, these assays may overestimate residual enzymatic activity, especially with certain GBA1 mutations like the L444P variant, as the misfolded enzyme may be retained in the endoplasmic reticulum before being targeted to the proteasome. Moreover, changes in the lysosomal microenvironment may go undetected as lysosomes get disrupted (Supplementary Fig. 5c). Hence, TFs identified in the primary screen were validated in live cells using the cell-active substrate LysoFQ-GBA1 to measure the activity of the physiologically relevant lysosomal GCase fraction32. With this approach, we validated MITF, TFEC, USF2, and ONECUT2 as regulators of lysosomal GCase activity.
Identification of MITF and TFEC, members of the MiT/TFE family which includes TFEB and TFE3, is not unexpected and lends confidence to our findings. Conversely, USF2 and ONECUT2 have not been previously linked to regulating human GCase activity. Except for ONECUT2, these are all members of the basic helix-loop-helix leucine zipper (bHLH-LZ) family of TFs, which recognize DNA sequences known as enhancer (E)-boxes comprised of a CANNTG sequence. The CLEAR element harbors an E-box16, and according to the JASPAR database of TF binding profiles MITF, TFEC, and USF2 are all predicted to bind to the CLEAR motif45. In line with this prediction, CRISPRa of MITF and TFEC and ablation for USF2 changed GBA1 transcript levels as assessed by RT-qPCR or bulk RNASeq. Lending support to a direct transcriptional regulation via promoter binding, CRISPRa of MITF increased luminescence intensity in a GBA1 promoter luciferase assay28 (Supplementary Fig. 3d). The absence of a change in luminescence intensity upon CRISPRa of TFEC and USF2 could be related to the limitations of this assay, which lacks the endogenous genomic context. Additionally, small effects might go undetected due to high variance related to variable transfection efficiency.
Transcriptomic profiling revealed differential expression of lysosomal genes other than GBA1 upon MITF or TFEC activation and USF2 ablation. CRISPRa of MITF also increased lysosomal cathepsin B activity. Besides regulating GBA1 and CTSB transcript levels, transcriptomic profiling suggested that further factors, such as the regulation of lysosomal pH, might contribute to the increase in lysosomal hydrolase activities upon CRISPRa of MITF. However, this effect would have been missed in our primary screen employing a lysate-based assay, as lysosomes are disrupted, and the artificial substrate is provided in an acidic buffer. Hence, additional TFs affecting lysosomal pH or other components of the lysosomal microenvironment may exist beyond MITF.
Co-activation of MITF and TFEC led to a more pronounced increase in GBA1 RNA, GCase protein, and activity levels as compared to their individual activation, suggesting that heterodimer formation of MITF/TFEC results in higher affinity binding to the GBA1 promoter and/or more pronounced activation of GBA1 expression compared to their binding as homodimers. It remains to be seen in which cell type heterodimer formation is functionally relevant. However, simultaneously activating several genes is valuable for assessing the additive or synergistic effects of genes predicted to regulate the same pathways. We identified USF2 as a bidirectional regulator of GCase activity in both LN-229L444P and LN-229WT cells. The upstream factor USF2 and its homolog USF1 are evolutionarily conserved, ubiquitously expressed, and bind to DNA as homo- or heterodimers. USFs are implicated in regulating various cellular processes including proliferation, immune response, lipid metabolism, and histone acetylation46, and USFs have been proposed as regulators of lysosomal function in murine neurons41. As members of the bHLH-LZ family, USFs may compete with the MiT/TFE family members for binding to the CLEAR motifs in the promoters of lysosomal genes, thereby antagonizing their effects. A similar mechanism has been proposed for MYC, another member of the bHLH-LZ family41,47.
CRISPRa of all other identified TFs did not alter GBA1 transcript levels, and no predicted binding sites were found in the GBA1 promoter according to the JASPAR database. These intriguing results suggest that GCase is regulated through post-transcriptional mechanisms, some of which can be targeted by the TFs identified by our screen. Notably, the zinc finger proteins ZNF852, ZNF865, and SCRT2 exhibited increased GCase protein levels, despite no change in GBA1 RNA levels and lysosomal GCase activity measured with LysoFQ-GBA1. This could indicate alterations in GCase protein stability, as the mutant enzyme might partially evade premature proteasomal degradation but fail to enter the lysosome due to a trafficking defect.
Apart from MITF, none of the candidates we identified overlapped with transcription factors previously predicted or identified to modulate GBA1 expression17,48. This discrepancy might result from differences between the cell lines studied or variations in assay conditions, such as nutrient availability.
The ONECUT family members (ONECUT1–3) participate in the development of the liver, pancreas, and retina but are not associated with lysosomal function49,50. Although ONECUT2 binds to the MITF promoter in melanocytes51, GBA1 RNA and GCase protein levels remained unchanged upon CRISPRa of ONECUT2, suggesting that it does not regulate GBA1 transcription. In addition to decreasing GCase activity, CRISPRa of ONECUT2 decreased lysosomal CatB activity. Transcriptomic profiling also did not show a significant decrease in CTSB expression, suggesting a broader regulatory effect on lysosomal function. Supporting this idea, several genes associated with vesicle trafficking were among the top DEGs (Fig. 7a). Thus, ONECUT2 may regulate GCase activity by influencing its trafficking. Despite small effect sizes and the likelihood of trafficking defects being overlooked in a lysate-based assay, screening the top 30 DEGs identified PLEKHG4 and PLEKHG4B as possible downstream mediators. Indeed, CRISPRa of PLEKHG4 and PLEKHG4B demonstrated a stronger effect on lysosomal GCase activity (assessed with LysoFQ-GBA1) than on total cellular GCase activity (assessed with 4MU-Glc) in both LN-229WT and LN-229L444P cells. Since PLEKHG4 and PLEKHG4B are guanine nucleotide exchange factors (GEFs) regulating cytoskeleton dynamics at the Golgi apparatus, their overexpression may enhance GCase trafficking and lysosomal GCase activity43,44.
Limitations of our study should also be acknowledged. The LN-229 cancer cell line was primarily chosen for practical reasons. LN-229 cells are polyploid and exhibit genomic instability due to a TP53 mutation, which may affect the generalizability of our results. While overexpression of MITF, TFEC, and USF2 had the same effect on lysosomal GCase activity in iPSC-derived forebrain neurons carrying the highly prevalent N370S and L444P GBA1 variants as we observed in LN-229L444P, ONECUT2 overexpression did not alter GCase activity in neurons. Possible reasons might be related to the experimental design. Overexpression vectors, unlike CRISPRa, only increase the expression of specific isoforms that may not be relevant to the cell type being tested. Alternatively, the ONECUT2 effectors might be cell-type specific and absent from glutamatergic neurons, or the phenotype might be mutation-specific. As the tested GD-derived neurons harboring the GBA1L444P/P415R mutation exhibit low baseline GCase activity, any further decrease induced by ONECUT2 might be below the detection limit of our assay.
It remains to be seen if these effects are of physiological relevance, as both endogenous MITF and TFEC levels are predicted to be low in neurons. Given the emerging role of neuroinflammation in the pathogenesis of neuronopathic GD and PD, testing these candidates in microglia will be of interest52,53.
We chose the GBA1L444P mutation based on biological and technical considerations. The L444P mutation results in premature proteasomal degradation of GCase and causes GD with neurological involvement (type II/III) in homozygous individuals and is associated with a high PD risk in heterozygous carriers. Therefore, it would be desirable to identify pathways that modulate ER-associated degradation and enhance GCase stability and activity. Importantly, a two- to three-fold increase in lysosomal GCase activity in homozygous GBA1L444P/ L444P was sufficient to reverse the toxic accumulation of glucosylsphingosine. This suggests that even small increases in GCase activity (to around 10% of WT levels) can partially reverse some of the intracellular metabolic effects associated with GD, and might therefore be clinically meaningful37,54. From a practical viewpoint, the GBA1L444P locus is highly amenable to base editing with no bystander edits. Finally, the drastically reduced enzymatic activity in LN-229L444P cells provided a large dynamic range for identifying upregulators of GCase activity, which we considered more clinically relevant than downregulators.
Considering the extensive premature proteasomal degradation associated with this mutation, it is remarkable that the twofold increases in GBA1 transcript levels, as measured in RT-qPCR upon CRISPRa of MITF and TFEC, translated into a comparable increase in lysosomal GCase activity. Possible explanations include the saturation of the ubiquitin-proteasome system in the mutant cell line or the influence of MITF and TFEC on factors beyond GCase abundance contributing to increased lysosomal enzymatic activity, such as chaperone levels, the unfolded protein response48, trafficking, or the intralysosomal milieu. Moreover, the >300 distinct GBA1 mutations that impair its enzymatic activity do so through mechanisms extending beyond proteasomal degradation, including disrupted interaction with the activating protein saponin C as seen in the case of the N370S mutation. Thus, the effect of the identified TFs on GCase activity may vary depending on the specific mutation and, quite possibly, cell type55.
Curiously, TFEB and TFE3, two TFs known to regulate the expression of numerous lysosomal genes including GBA139, were not among our top candidates. Constitutive activation of the mTORC1 pathway in glioblastoma cells may explain their absence, as phosphorylation by mTORC1 prevents their nuclear translocation56. In contrast, TFEC and MITF-M (the isoform corresponding to CRISPRa of MITF TSS2) lack a mTORC1 phosphorylation site and may therefore be able to enter the nucleus under such conditions57. Moreover, functional redundancy of the MiT/TFE family members contributes to their cell-type-dependent regulation of metabolic pathways38.
To address the functional effects of altering the expression of the TFs we identify as regulators of lysosomal GCase activity, TFs could be tested for their effect on -synuclein aggregation, glucosyl sphingosine levels, or endoplasmic reticulum stress, which have all been associated with PD risk in the context of GBA1 mutations2.
In conclusion, by conducting an arrayed CRISPRa screen in cell lysates, we identified four TFs that modulate lysosomal GCase activity when assessed with the cell-active substrate LysoFQ-GBA1 within live cells. These results point to an intricate network of TFs beyond TFEB and TFE3 that regulates lysosomal GCase activity, with mechanisms beyond direct transcriptional regulation of GBA1 being at play. Identifying MITF, TFEC, USF2, and ONECUT2 as regulators of GCase activity illuminates an underappreciated interplay between transcriptional regulation of lysosomal function and PD risk. While the pharmacological modulation of TFs remains challenging58, discovering an ONECUT2-mediated non-transcriptional mechanism of GCase regulation may yield targets more amenable to pharmacotherapy.
Materials and methods
Generation of the LN-229L444P cell line
Cells were cultured in DMEM (Gibco) supplemented with 10% Fetal Bovine Serum (FBS). All experiments were performed in cells under passage 40. The GBA1 L444P mutation was introduced into the LN-229 human glioblastoma cells (CRL-2611, ATCC).
gRNA was ordered as forward (5′- CACCGTCCAGGTCGTTCTTCTGAC -3′) and reverse (5′ - AAACGTCAGAAGAACGACCTGGAC -3′) primers from Microsynth (Balgach, Switzerland).
gRNA cloning
The lentiGuide-Puro plasmid (Addgene #52963) was digested with Esp3I (BsmBI) (ER0451, Thermo Scientific) in Tango Buffer containing DTT. The reaction mix was incubated overnight at 37 °C. On the next day, 0.5 µL of calf intestinal alkaline phosphatase (CIP) (M0290, New England Biolabs (NEB)) was added and the reaction was incubated for 1 h at 37 °C. The digested plasmid was gel purified (NucleoSpin Gel and PCR Clean-up, 740609.50, Macherey-Nagel). To generate the gRNA, the forward and reverse primers were phosphorylated and annealed with T4-PNK (M0201S, NEB). The phosphorylation/annealing reaction was incubated for 30 min at 37 °C followed by 5 min denaturation step at 95 °C before ramping down the reaction to 25 °C at 5 °C/min. The annealed oligonucleotides were diluted at 1:200 in sterile water. Ligation of digested plasmid and annealed oligonucleotides was performed with T4 DNA Ligase (M1801, Promega). The reaction was incubated for 3 h at room temperature.
Transformation into NEB® Stable Competent E. coli (C3040H) was performed as follows: 5 µL of ligation reaction were mixed with 50 µL of E. coli. The mix was kept on ice for 30 min before performing a heat shock (30 s at 42 °C). After another 5 min on ice, 300 µL of S.O.C. medium (15544034, ThermoFisher) was added and E. coli were shaken at 37 °C for 1 h before plating them on an ampicillin agarose plate (100 μg/mL) and incubating them overnight at 37 °C. Single colonies were picked the next day and grown in LB medium containing ampicillin (100 μg/mL) for one night at 37 °C in a shaker. Plasmid DNA was extracted (QIAprep Spin Miniprep Kit, 27106×4 Qiagen) and sent for Sanger sequencing (Microsynth). 5′ sequencing primer: EF-1a-F (TCAAGCCTCAGACAGTGGTTC); 3′ sequencing primer: WPRE-R (CATAGCGTAAAAGGAGCAACA). A negative control ligation and transformation (digested vector with water in place of oligonucleotides) was run in parallel.
For base editing, LN-229 (passage 9) were transfected in 24-well plates. The adenine base editor ABE8e (Addgene #138489)23 and the lentiGuide-Puro vector encoding gRNAL444P were co-transfected using Lipofectamine 3000® (Thermo Fisher) in a ratio of 3:1 (i.e., 375 ng and 125 ng). Selection with Puromycin (Gibco) 1 µg/µL was started after 24 h. Medium was changed daily. On day 5, cells from a single well were dissociated with Trypsin and diluted in 1000 µL medium. 2–50 µL of the cell suspension was transferred to 10 cm dishes to obtain single colonies. Medium containing Puromycin was changed twice weekly and single colonies were picked manually after 2–3 weeks when reaching an adequate size and expanded. Genomic DNA was extracted (DNeasy Blood & Tissue Kit, Qiagen), amplified with primers L444P_fwd (5′- CCT GAA CCC CGA AGG AGG AC -3′) and L444P_rev (5′- ACT TCC CAG ACC TCA CCA TTG -3′) and sent for Sanger sequencing (Microsynth) with primers L444P_fwd2 (5′- CCA ATT GGG TGC GTA ACT TTG TC - 3′) and L444P_rev specific for GBA1 (i.e., not amplifying GBAP1).
To ensure that all the alleles of the polyploid LN-229 clones were edited, Illumina® next-generation sequencing (NGS) was performed. Genomic DNA extracted from base-edited clones showing the desired point mutation with Sanger sequencing underwent PCR with primers L444P_fwd and L444P_rev to ensure that only GBA1 and not GBAP1 was amplified. The amplicon was used as a template for the second round of PCR with primers introducing adapters for Illumina sequencing with NGS_L444P_fwd (5′ CTT TCC CTA CAC GAC GCT CTT CCG ATC TCC ATC TGT TCC CAC ATT CAG C – 3′) and NGS_L444P_rev(5′- GGA GTT CAG ACG TGT GCT CTT CCG ATC TAC TTC CCA GAC CTC ACC ATT G -3′). Q5 High-Fidelity DNA Polymerase (M0491S, NEB) was used with 200 ng of template DNA in a total reaction volume of 20 µL. PCR products were purified using AMPure XP beads (Beckman Coulter) and subsequently amplified for eight cycles using barcoding primers. Approximately equal amounts of PCR products from each sample were pooled, gel purified (QIAquick Gel Extraction Kit, Qiagen), and quantified using a Qubit 3.0 fluorometer and the dsDNA HS Assay Kit (Thermo Fisher). Paired-end sequencing of purified libraries was performed on an Illumina® MiSeq.
Sequencing reads were demultiplexed using MiSeq Reporter (Illumina). Amplicon sequences were aligned to their reference sequences using CRISPResso2 and the tool was used to generate Fig. 1b59 (Amplicon: CCATCTGTTCCCACATTCAGCAAGTTCATTCCTGAGGGCTCCCAGAGAGTGGGGCTGGTTGCCAGTCAGAAGAACGACCTGGACGCAGTGGCACTGATGCATCCCGATGGCTCTGCTGTTGTGGTCGTGCTAAACCGGTGAGGGCAATGGTGAGGTCTGGGAAGT; sgRNA: GTCCAGGTCGTTCTTCTGAC).
Generation of LN-229ΔGBA1 cells
LN-229 cells were co-transfected with the plasmid encoding the sgRNAs as outlined below and hCas9 (Addgene #41815) using Lipofectamine 3000 in a ratio of 3:1. Single colonies were selected as described above. Knockout was confirmed by performing Sanger sequencing, immunoblotting, and a lysate-based enzyme activity assay. The sequence of the sgRNAs to target the catalytic domain of GBA1 was: The sgRNA1 (sgRNA1 sequence) oligonucleotide sequence is: 5′- ACAGAAGTTCCAGAAAGTGA-3′; sgRNA2 (sgRNA2 sequence, N20sg2); 5′-GAGAGCAGCAGCATCTGTCA-3′; sgRNA3 5′- AGTGATGGAGCAGATACTCA -3′; sgRNA4 5′- GATGGAGCAGATACTCAAGG -3′.
Lentiviral packaging
HEK293T cells were grown to 80–90% confluency in DMEM + 10% FBS on poly-D-lysine coated 24-well plates and transfected with the three different plasmids (Transfer plasmid, psPAX2 (#12260), VSV-G (#8454); ratios: 5:3:2) with Lipofectamine 3000 for lentivirus production. After 6 h or overnight incubation, the medium was changed to virus harvesting medium (DMEM + 10% FBS + 1% BSA (Cytiva HyClone)). The supernatant containing the lentiviral particles was harvested 48–72 h after the change to virus harvesting medium. Suspended cells or cellular debris were pelleted with centrifugation at 200 × g for 5 min. Clear supernatant was titrated by FACS and stored at −80 °C in single-use aliquots.
For the titration of the lentiviral particles, HEK293T cells were grown in 24-well plates and infected with 4 mL (V) of the above-mentioned viral supernatant. Cells of two representative wells were counted at the time of infection (N). 72 h after infection, the cells were harvested and analyzed by flow cytometry to determine the fraction of infected cells (BFP positive) and determine the viral titer (T) according to the following formula: T = (p * N)/V. Flow cytometry analysis was performed with a BD Canto II or LSRFortessaTM Cell Analyzer at the core facility center of the University of Zurich. Data were analyzed with FlowJo.
Generation of cell lines stably expressing dCas9-VPR or Cas9
Lentiviruses of plasmids pXPR_120 (Addgene #96917) and lentiCas9-Blast (Addgene #52962) were produced. Cells in 24-well plates at 70% confluency were infected and put under Blasticidin selection (15 µg/mL) 24 h post-infection. Blasticidin-containing medium was changed every 48 h and cells were gradually expanded when confluent. Successful expression of the Cas9 construct was determined by means of RT-qPCR and Western Blot of two target genes.
Proteasome inhibitor treatment
For immunoblotting, 250,000 cells per well were seeded in 6-well plates 24 h before adding MG-132 (C2211, Sigma-Aldrich) at a concentration of 0.375 µM dissolved in DMSO. 0.375 µM MG-132 containing medium was replaced after 24 h. Control cells were treated with an equal volume of DMSO. Cells were lysed 48 h after starting the treatment and immunoblotting for GCase was performed as outlined above. To assess enzymatic activity, 100,000 cells grown in 96-well plates were treated for 24 h with either DMSO or 0.5 µM MG-132 in full medium. On the day of the assay, cells were washed twice in Phenol-red free medium before Hoechst 20 mM (dilution of 1:7,500) was added to three wells per condition, whereas the remaining cells were kept in full medium. Cells were incubated for 20 min at 37 °C before imaging of the nuclei with a Pico ImageXpress (Molecular Devices) was performed to assess cell number, as the treatment might result in toxicity/cell death. Thereafter, the enzyme activity assay was assessed in cell lysates as described above. Fluorescence intensities were normalized by the number of nuclei.
Lysate-based enzyme activity assay in 96 well plates
The protocol was adapted from a previously published protocol60. Medium was removed by tilting the plates. Cells were washed with 200 µL PBS once before 50 µL of ice-cold McIlvaine buffer (0.1 M citrate and 0.2 M Na2PO4 buffer at pH 5.2 containing 0.4% Triton-X100 and Protease inhibitor) were added per well. Plates were shaken for 1 h at 4 °C at 350 rpm. 50 µL of 4MU-Glc (at a final concentration of 5 mM) in McIlvaine buffer containing 5 mM sodium taurocholate and 1% DMSO was added. Plates were incubated for 1 h at 37 °C before 100 µL of a stop solution (0.3 M Glycine, 0.2 M NaOH solution; pH 10.8) was added.
The plate was centrifuged at 1000 × g for 10 min before fluorescence intensity was read at a VersaMax Microplate Reader (Molecular Devices) or a GloMax® Discover (Promega) (Excitation: 365 nm, Emission: 440 nm).
Dose-response curves to assess the specificity of the lysate-based assay
4000 LN-229WT cells were seeded in 96-well plates (ViewPlate-96 Black #6005182, PerkinElmer). The next day, cells were washed in PBS and lysed in 50 mL McIlvaine buffer containing EDTA-free cOmplete Mini Protease Inhibitors (Roche). Cells were shaken at 4 °C and 450 rpm for 10 min followed by one hour of incubation at 4 °C. Serial dilutions of inhibitors (CBE [575, 383, 256, 170, 114, 76, 50, 33.7 mM final conc.], AT3375 [400, 200, 100, 50, 25, 12.5, 6.25, 3.125 mM final conc.], and miglustat [200, 100, 50, 25, 12.5, 6.25, 3.125, 1.56 mM final conc.]) or an equal volume of DMSO (serving as the negative control) were prepared in an empty 96-well plate in McIlvaine buffer. Cell lysates (three wells per condition) and 4MUG in assay buffer were added (final conc. of 4MUG 2.5 mM) with a multichannel pipette and incubated for one hour at 37 °C before the addition of stop solution. Following a final centrifugation step (300 g for 10 min), plates were read at a VersaMax Microplate Reader (Molecular Devices) (Excitation wavelength: 365 nm, Emission wavelength: 440 nm). AT3375 was kindly provided by Amicus Therapeutics®.
Workflow of primary arrayed CRISPR screen
The generation of the arrayed CRISPR activation library is outlined in ref. 21
The oligonucleotide sequence targeting the TSS of GBA1 was: sgRNA1 (sgRNA1 sequence) 5′- cATGTATGGGTGACAACTTT -3′; sgRNA2 (sgRNA2 sequence); 5′- TTGCCCTATAGAGGTGTGTG -3′; sgRNA3 5′-TATAATCTGTAACAGATGAG-3′; sgRNA4 5′- GGCACAAGAGGGTGGGACAC -3′.
The oligonucleotide sequence of the NT ctrl was: sgRNA1 (sgRNA1 sequence) 5′- GTGTCGTGATGCGTAGACGG -3′; sgRNA2 (sgRNA2 sequence); 5′- GTCATCAAGGAGCATTCCGT -3′; sgRNA3 5′- GGACCCTGACATGTATGTAG -3′; sgRNA4 5′- GCACTTAGCAGTTTGCAATG -3′.
LN-229L444P cells stably expressing dCas9-VPR were cultured in T75 tissue culture flasks (TPP) in DMEM (Gibco) supplemented with 10% FBS (Takara) and 15 mg/mL Blasticidin (Gibco). In preparation for the primary screen, cells were expanded, washed with PBS (Gibco), harvested using Trypsin-EDTA 0.025% (Gibco), resuspended in DMEM supplemented as outlined above, pooled, and counted using a TC20 (BioRad) Cell Counter with trypan blue (Gibco). For the primary screen, all LN-229L444P cells were at the same passage number.
2000 LN-229L444P dCas9-VPR cells were dispensed in 50 µL/well of medium using a multi-drop dispenser (MultiFlo FX, Biotek) in 384-well plates (#781091, Greiner). Plates were centrifuged at 10 × g for 30 s (5804 R, Eppendorf) and incubated overnight in a rotating tower incubator (StoreX STX, LiCONiC). Cells were transduced after 18–20 h using a handheld electronic multichannel pipetting system (ViaFlo, Integra) with an MOI of four with lentiviruses encoding the qgRNA plasmids targeting each human TF61. Outermost wells were spared to avoid edge effects related to evaporation. 14 wells/plate were transduced with lentiviruses encoding negative and positive controls (i.e., 14 NT ctrls and 14 GBA1 TSS1-targeting 4sgRNA). Each 4sgRNA plasmid targeting a TF was transduced in triplicates on three separate plates in the same well position. Plates were incubated in a rotating tower incubator for five days. Subsequently, one replicate plate was used to determine cell viability by applying CellTiter-Glo® (Promega) following the instructions of the manufacturer. All solutions were dispensed with a multi-drop dispenser. Briefly, medium was removed by inverting the plates and replaced with 25 µL of fresh medium and 25 µL of CellTiter-Glo® solution, both solutions were at RT. Plates were incubated on a plate shaker (ThermoMixer Comfort, Eppendorf) for two min (400 rpm shaking) at RT. After further 10 min of incubation at RT, luminescence was determined with a fluorescence plate reader (EnVision, Perkin Elmer).
GCase activity was assessed on the two remaining plates. Medium was removed by inverting the plates, and cells were lysed in 10 μL lysis buffer (McIlvaine buffer with 0.4% Triton-X), supplemented with EDTA-free cOmplete protease inhibitor. Following lysis, assay plates were incubated on a plate shaker for 10 min at 4 °C (400 rpm shaking) prior to centrifugation at 200 × g for 1 min and incubation at 4 °C for 1 h. Following incubation, plates were centrifuged under the same conditions mentioned above and 10 μL of assay buffer containing 4MUG was added. After centrifugation, plates were incubated for 1.5 h at 37 °C in a rotating tower incubator. After centrifugation, 20 μL of stop solution was added to each well. Following a final centrifugation step (300 × g for 10 min), plates were read at a VersaMax Microplate Reader (Molecular Devices) (Excitation: 365 nm, Emission: 440 nm).
Reproduction of results with a higher number of replicates
The same protocol as for the primary screening was carried out. The only difference was that lentiviruses were pipetted manually. To test for bidirectional regulation (CRISPRko of candidates), 500 cells/well stably expressing Cas9 were seeded in 384-well plates on day 0 and lysis was performed on day seven post-transduction.
RNA extraction and real-time quantitative PCR
Total RNA was isolated using the High Pure RNA Isolation Kit (Roche). 800–1000 ng of RNA were reversed transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen). Real-time quantitative PCR was performed with SYBR green (Roche) according to the manual with the primer sets for each gene listed in Table 1. ACTB was used as the housekeeping controls. A ViiA7 Real-Time PCR system (Applied Biosystems) was used for fluorescence detection. Primer sequences are provided in Table 1.
Table 1.
List of primers for RT-qPCR
| Gene | FWD Primer Sequence 5′-3′ | REV Primer Sequence 5′-3′ |
|---|---|---|
| ACTB | CAT GTA CGT TGC TAT CCA GGC | CTC CTT AAT GTC ACG CAC GAT |
| TFEC | CTA GAA ATT CAG GCT CGT AC | AGC ACC TAA ATC AAC CGT GC |
| MITF (total) | CGA GCG TCC TGT ATG CAG AT | AAG CAG GAT CCA TCA AGC CC |
| M-MITF | AGA GGG AGG GAT AGT CTA CCG | GGT GGG TCT GCA CCT GAT AG |
| USF2 | GCT TCA GAC AGG AAC ACA GA | CTG GGC TCT TCT CCT CTC AT |
| ONECUT2 | CAG CGT GCA AAC GCA AAG AG | TCT TCT GGG AAT TGT TCC TG |
| GBA1 | ATT GGG TGC GTA ACT TTG TC | TCC AGG TCG TTC TTC TGA CT |
| GBAP1 | GGA CCG ACT GGA ACC CAT | TCC AGG TCG TTC TTC TGA CTG |
Immunoblotting and deglycosylation
On day zero, 40,000 cells were seeded in 6-well plates. On day one, cells were infected with lentiviruses (MOI of 4). Medium was changed after 24 h. Three days (CRISPRa) or four days (CRISPRko) post-transduction.
On day five (CRISPRa) or seven (CRISPRko) post-transduction, cells were lysed in RIPA buffer supplemented with Protease Inhibitor. Samples were centrifuged at 17,000 × g for 10 min at 4 °C and the supernatant was subjected to further downstream analyses. BCA assay (Pierce, Thermo Fisher) was used to determine the total protein concentration of each sample. For immunoblotting, samples were diluted in RIPA buffer to obtain the same total protein concentration. Deglycosylation was performed with either PNGase F or EndoH (both from NEB), following the protocol of the manufacturer.
Samples were boiled at 100 °C for 10 min after the addition of NuPAGE 4x LDS loading buffer (Thermo Fisher) to which 10% b-mercaptoethanol (Sigma) was added. Samples were then loaded onto a NuPAGE 4–12% Bis-Tris gradient gel (Invitrogen, Thermo Fisher) and blotted onto a nitrocellulose membrane using the iBlot2 dry transfer system (Invitrogen, Thermo Fisher). Membranes were blocked using 5% SureBlock (LubioScience) diluted in 1x PBS containing 0.1% Tween-20 (Sigma Aldrich) for 60 min. Membranes were then incubated with primary antibodies diluted in 1% SureBlock-PBST overnight at 4 °C under shaking conditions. For detection, anti-mouse HRP or anti-rabbit HRP (Bio-Rad) were diluted 1:5000 in 1% SureBlock-PBST. Imaging was performed with a VILBER Fusion Solo S after the addition of Immobilon Crescendo Western HRP Substrate (Milipore). Antibodies are listed in Table 2. Samples for individual blots were derived from the same experiment if not stated otherwise and were processed in parallel.
Table 2.
List of antibodies for immunoblotting
| Target | Manufacturer | Dilution |
|---|---|---|
| GCase | G4171 (Sigma-Aldrich), produced in rabbot | 1: 4000 |
| Vinculin | ab91459 (Abcam) produced in rabbit | 1:5000 |
| Beta-Actin (HRP-coupled) | PA1-183-HRP produced in mouse; Invitrogen | 1: 10,000 |
| USF2 | clone 5E9 H00007392-M01 (Abnova) produced in rabbit | 1: 1000 |
| MITF | Clone D5G7V #12590 (CellSignaling), produced in rabbit | 75.9 ug/mL at 1:1000 |
| ONECUT2 | 21916-1-AP (Proteintech), produced in rabbit | 0.6 mg/mL at a 1:750 dilution |
Live cell assays to determine GCase, NAGAL, and Cathepsin B activities in LN-229L444P
The protocols were adapted according to an established protocol obtained from the group of David Vocadlo, Simon Fraser University, Burnaby, British Columbia CA. LysoFQ-GBA1 and NAGAL-BABS were kindly provided by the group of David Vocadlo32,40. LN-229 cells were plated in 12-well plates (20,000 cells/well). Cells were seeded into 12-well plates. 24 h later, lentiviruses were dispensed manually at an MOI of ~4. Cells were collected and counted four days post-transduction and one day before imaging. The cell suspension was plated in 96- or 384-well plates for treatment and imaging (#4680 or 4681, Corning). Dispensing of cells (LN-229: 1800 cells/well in 36 μL for 384-well plates or 4,000 cells/well in 90 μL for 96-well plates), with three to six replicates per plate and condition. Plates were centrifuged (150 × g for 30 s) and placed in a humidified incubator. AT3375 was prepared and diluted (in DMEM) and added to the cell-containing plate 24 h before imaging (10 μM final conc.). On the day of imaging, the substrate solution was prepared in culture medium. For LysoFQ-GBA1, the substrate was added to the cell plate at a final conc of 5 μM final. After 1 h of incubation at 37 °C, cells were washed three times in Phenol-Red free MEM (Gibco) using a multichannel pipette before 40 or 90 μL imaging buffer was added (Phenol-red free MEM containing 10% FBS with 1:7,500 dilution of Hoechst and 10 μM AT3375). After another 20 min incubation at 37 °C, imaging of live cells was performed using an ImageXpress HT.ai high-content imager (Molecular Devices) connected to environmental control (37 °C, 5% CO2). Image acquisition was carried out using a 40x water immersion objective. For each well, 25 (for 384-well plates) to 36 (for 96-well plates) sites (regions of interest) were imaged using the DAPI and FITC channels. Before acquisition, the focus was adjusted for both the DAPI and FITC channels and set to adjust on plate and well bottom. Exposure times were set at 50 ms for the DAPI and 500 ms for the FITC channel. To assess NAGAL activity, 250 cells/well were seeded in 96-well plates (#4680, Corning) in 90 μL and infected on the following day at an MOI of 4 (assuming a doubling time of 24 h). 24 h before imaging (4 days post-transduction), control wells were treated with the NAGAL-specific inhibitor DGJNAc (10 μM final conc.). On the day of imaging, cells were treated with NAGAL-BABS at a final concentration of the substrate of 10 μM for 3 h. DGJNAc was added to the imaging medium at a final concentration of 10 μM as a negative control. To assess Cathepsin B activity, the Cathepsin B Assay Kit (Magic Red; Abcam) was used following the protocol of the manufacturer.
Image analysis
Following acquisition, images were analyzed using the MetaXpress software suite (Molecular Devices). Data were analyzed using the Multiwavelength Cell Scoring module. Nuclei were detected using the DAPI channel and analyzed using a 5/50 μm constraint (min/max width) and a 500 gray-level minimal intensity above background. BODIPY-FL fluorescence was analyzed from the FITC channel image using a 1/7 μm constraint (min/max width) and various minimal intensities above background (200–1000 gray-level threshold), with the background threshold being set so that there were a few positive pixels in the negative control wells (AT3375- or DGJNAc-treated cells). For each region of interest in each well, this analysis module returned the mean integrated intensity for the FITC or TRITC channel and the number of cells. After combining sites for each well, the integrated intensity normalized by the number of cells for each well was generated and transformed to percentage of activity of the NT ctrl treated cells.
Luciferase GBA1 promoter assay
A previously published GBA1 promoter construct was kindly provided by Dr. Letizia Straniero, Humanitas University Campus in Milan, Italy28. 5000 LN-229L444P dCas9-VPR cells were seeded in white 96-well culture plates in 100 µL (Perkin Elmer). 24 h later, lentiviruses containing 4sgRNA to activate expression of the candidate TFs or the NT ctrl were transduced at an MOI of 4. The following day, 97.5 ng of the GBA1-pGL2 plasmid and 2.5 ng of the pRL-TK plasmid (ratio of 1:25) (Thermo Fisher Scientific) were co-transfected in quadruplicates for each condition using Lipofectamine 3000 (Thermo Fisher Scientific). Medium was changed 6 h post-transfection. 48 h post-transfection, the Dual-Glo Luciferase Reagent (Promega) was added to each well and the luminescence of the firefly luciferase was measured using a GloMax plate reader (Promega) after 20 min incubation at RT. Subsequently, Dual-Glo Stop & Glo Reagent (1:100) was added to each well and the luminescence of the renilla luciferase was measured after 20 min incubation at RT. For data analysis, the Firefly luciferase signal was divided by the Renilla luciferase signal and normalized to the mean of the NT ctrl values.
Bulk RNA sequencing in LN229L444P
High Pure RNA Isolation Kit (Roche) was used for RNA extraction according to the manufacturer’s protocol. Libraries were prepped with the Illumina TruSeq stranded mRNA protocol (Illumina, San Diego, CA, USA), and quality control (QC) was assessed on the Agilent 4200 TapeStation System (Agilent Technologies, Santa Clara, CA, USA). Subsequently, libraries were pooled equimolecular and sequenced on the Illumina NovaSeq6000 platform with single‐end 100 bp reads. Sequencing depth was around 20 million reads per sample. Experiments were run in biological triplicates. Data analysis was performed as previously described62.
Cloning of overexpression vectors
The LentiCas9-Blast vector (Addgene #52962) was taken as backbone and digested with high-fidelity BamHI (R3136, NEB) and XBaI (R0145S, NEB). Digestion was performed overnight at 37 °C. The digested plasmid was loaded onto a 0.5% agarose gel and the upper band (8.7 kB) was cut under UV light. Gel purification was performed (NucleoSpin Gel and PCR Clean-up, Macherey-Nagel). cDNA derived from LN229L444P was taken as a template to generate the inserts into the digested Lenticas9-Blast vector. PAGE-purified primers (synthesis scale 0.04 μmol) for Gibson assembly were ordered from Microsynth (Balgach, Switzerland). Two rounds of PCR using Phusion high-fidelity polymerase (F630S, ThermoScientific) were performed (first round with primers specific for the gene of interest and its reverse primer; second PCR taking the first PCR product as template was performed with the “outer_primer_Fwd_ChIP” primer (Table 2) for all constructs to ligate the digested backbone with the template DNA. Annealing temperature was 60 °C, extension time depended on the size of the PCR product (between 30 and 90 s; assuming a synthesis speed of 15–30 s/kB) with 36 repeat cycles. Correct sizes of the PCR products were checked on an agarose gel. If there were multiple bands, the PCR product at the correct height was cut and gel purified. For Gibson assembly, 5 µL Builder HiFi DNA Assembly Master Mix (NEB) was used in a final reaction volume of 10 µL. DNA molar ratio of vector: insert was 1:2. The reaction mix was incubated for 60 min at 50 °C. Transformation of 5 µL of the reaction mix into NEB® Stable Competent E. coli (C3040H) was performed as outlined above. Bacteria were plated on agarose plates containing ampicillin. Extracted plasmid DNA was sent for Sanger sequencing (Microsynth) using the blast-R primer (5′-gctctttcaatgagggtgga-3′). The constructs for GBA1, TFEC, and ONECUT2 were synthesized at TWIST bioscience (San Francisco, CA, USA). Primer sequences are provided in Table 3.
Table 3.
Primer sequences for generation of overexpression vectors
| outer_primer_Fwd_ChIP |
5′-GCC AGA ACA CAG GAC CGG TTG CCA CCA TGG ACT ACA AAG ACC ATG ACG GTG ATT ATA AAG ATC ATG AC-3' |
| USF2 isoform 2 | |
| USF2_inner_primer_Fwd_ChIP |
5′-CGG TGA TTA TAA AGA TCA TGA CAT CGA TTA CAA GGA TGA CGA TGA CAA GGA CAT GCT GGA CCC GGG TCT-3' |
| USF2_rev_ChIP |
5′-GAG AGA AGT TTG TTG CGC CGG ATC CCT GCC GGG TGC CCT CGC CCA CCA TCT-3' |
| M-MITF | |
| M_MITF_inner_primer_Fwd_ChiP |
5′-CGG TGA TTA TAA AGA TCA TGA CAT CGA TTA CAA GGA TGA CGA TGA CAA GCT GGA AAT GCT AGA ATA TAA TCA CTA TCA GGT GCA GAC CC-3' |
| M_MITF_Rev_ChIP |
5′-AGC AGA GAG AAG TTT GTT GCG CCG GAT CCA CAA GTG TGC TCC GTC TCT T-3' |
| empty vector with BSD resistance | |
| BSD for vector GB |
5′-GCC AGA ACA CAG GAC CGG TTC TAG AGC GCT GCC ACC ATG GCC AAG CCT TTG TCT CA-3' |
| BSD fwd 1st round PCR | 5′-ATG GCC AAG CCT TTG TCT CA-3' |
| BSD reverse for vector GB |
5′-TAC CGA TAA GCT TGA TAT CGA ATT CTT AGC CCT CCC ACA CAT AAC-3' |
Human induced pluripotent stem cell (iPSC) lines
Human iPSC lines PGPC17, derived from healthy individuals with whole-genome sequencing-based annotation, are generous gifts from Dr. James Ellis at the Hospital for Sick Children, Toronto, Canada63. Line UOXFi001-B was obtained from EBiSC and was derived from a patient with Parkinson’s Disease (PD) carrying the heterozygous mutation GBA1N370S/WT. The Gaucher Disease (GD) iPSC line C43-1260 (GBA1L444P/P415R)32 was generated from a GD patient’s skin fibroblast line (GM01260, Coriell) and characterized using a contract service provided by the Tissue and Disease Modeling Core (TDMC) at the BC Children’s Hospital Research Institute following their standard protocols. The genomic integrity of this line was confirmed by G-banded karyotyping performed at WiCell. The pluripotency of C43-1260 was tested by in vitro differentiation to the three germ layers using the STEMdiff™ Trilineage Differentiation Kit (STEMCELL Tech, Catalog # 05230). All iPSCs were cultured and expanded in mTeSR™ Plus medium (STEMCELL Tech, Catalog # 100-0276) according to the manual.
Generation of iPSC-derived neural progenitor cells (NPCs) and forebrain mature neurons
iPSC lines including PGPC17, UOXFi001-B, and C43-1260 were differentiated to NPCs using the STEMdiff™ Neural Induction Medium + SMADi (STEMCELL Tech, Catalog # 08581) following the monolayer-based differentiation protocol described in the technical manual. Generated NPCs were either used directly or cryopreserved in STEMdiff™ Neural Progenitor Freezing Medium (STEMCELL Tech, Catalog # 05838) and stored in a liquid nitrogen Dewar until use. NPCs were further differentiated to neuronal precursors using the STEMdiffTM Forebrain Neuron Differentiation Kit (STEMCELL Tech, Catalog # 08600) and then matured to forebrain neurons using the STEMdiffTM Forebrain Maturation Kit (STEMCELL Tech, Catalog # 08605) following the standard protocol outlined in the technical manual.
Live-cell GCase assay on iPSC-derived forebrain mature neurons transduced with lentiviral vectors
Neuronal precursors were seeded in 96-well High Content Imaging (HCI) plates (Corning, Catalog # 4680) precoated with poly-L-ornithine (PLO, 15 μg/mL) and laminin (5 μg/mL) at a seeding density of approximately 100,000 cells/cm2 and cultured using the Forebrain Maturation Kit as described above. Half-medium changes were performed every 2–3 days. Seven days post-seeding, neurons were infected with an appropriate amount of lentiviruses encoding different transcription factors or with lentiviral vectors encoding GBA1 isoform two or an empty vector serving as positive and negative controls, respectively. The following day, an 80% medium exchange was carried out to reduce toxicity related to lentiviral particles and to minimize the detachment of neurons, followed by an additional 80% medium exchange 48 hours later before returning to half medium exchanges every 2–3 days. Live-cell assays to assess GCase activity using the fluorescence-quenched probe LysoFQ-GBA132 were carried out on days 7 and 14 post-infection. The assay was initiated by replacing the culture medium with fresh medium containing 10 µM of LysoFQ-GBA1 and then incubating in the TC incubator for designated lengths of time (2 h for the GBA1 WT line PGPC1, 3 h for the PD-GBA1 line UOXFi001-B, and 4 h for the GD line C43-1260). The assay was stopped by two consecutive 85% medium exchanges with BrainPhys™ Imaging Optimized Medium (IOM) (STEMCELL Tech, Catalog # 05796) and one 85% medium exchange with IOM supplemented with NeuroCult™ SM1 Neuronal Supplement (SM1) (STEMCELL Tech, Catalog # 05711) containing 10 μM of AT3375. After 20 min of incubation at 37 °C, 10 μL of 11 x Hoechst (11 μg/mL) prepared in IOM + SM1 were added to each well (final conc 1 μg/mL). The plate was gently tapped for distribution, centrifuged at 300 × g for 30 s, and incubated for another 10 min. Images were then acquired using a high-content widefield microscope (ImageXpress XLS, Molecular Devices) with a 40x objective. Micrographs were analyzed using the in-built HCI analysis software MetaXpress (Molecular Devices) to yield the mean integrated intensity level of vesicle-like structures (representing lysosomal GCase activity) per cell in the FITC channel for each well. To assess neuron quality using immunofluorescence, forebrain neurons cultured in a 96-well HCI plate were fixed with 4% PFA in PBS for 20 min at RT and permeabilized using 0.1% Triton X-100 in PBS for 5 min on ice. After blocking with 5% BSA in PBS, neurons were incubated with primary anti-GFAP (Aves Lab, SKU: GFAP) or anti-MAP2 (Abcam, Catalog # Ab5392) antibodies at 4 °C overnight. On the following day, primary antibodies were washed away, and fixed neurons were stained with corresponding secondary fluorescent antibodies. Neurons were finally stained with Hoechst for 30 min and imaged using the ImageXpress XLS high-content widefield microscope (Molecular Devices).
Proteomics analysis
LN-229L444P dCas9-VPR cells transduced with qgRNAs targeting ONECUT2 or the non-targeting control at an MOI of 4 were prepared in four biological replicates (separate wells of a 6-well plate), washed twice in PBS, harvested by scraping of cells in PBS, and snap-frozen in liquid nitrogen. Native proteins were extracted by resuspending the pellets in a buffer (100 mM HEPES, 150 mM KCl, 1 mM MgCl2, pH 7.4), and lysed with 10 cycles of 10 dounces each using a pellet pestle on ice. Protein concentrations were measured with bicinchoninic acid assay and adjusted. Samples were boiled for 5 min at 99 °C and cooled down on ice. The samples were diluted in a 10% sodium deoxycholate to a final DOC concentration of 5%, reduced with tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in a final concentration of 5 mM at 37 °C for 40 min, and alkylated using a final concentration of 40 mM iodoacetamide (IAA) for 30 min at room temperature in the dark. The samples were then diluted to 1% DOC using 10 mM ammonium bicarbonate. Proteins were digested overnight at 37 °C with LysC (Wako Chemicals) and trypsin (Promega) in a 1/100 enzyme-to-substrate ratio, under constant shaking. Formic acid (FA) (Carl Roth GmbH) was added at a final concentration of 2% to stop digestion and precipitate DOC. DOC was removed by three centrifugation cycles, and subsequently, the supernatant was desalted on Sep-Pak tC18 cartridges (Waters). Samples were eluted with 50% acetonitrile and 0.1% formic acid. LC-MS/MS experiments were performed on an Orbitrap Eclipse mass spectrometer (Thermo Fisher Scientific). Data-independent acquisition (DIA, ref) scans were performed in 41 variable-width isolation windows. MS data were searched with Spectronaut (Biognosis AG). Statistical analysis was performed in R.
Ref: Mol Cell Proteomics 11, O111 016717. 10.1074/mcp.O111.016717.
Supplementary information
Acknowledgements
We thank Prof. em. Konrad Sandhoff for his valuable input and explanations. We thank Buket Züllig for her assistance in cell culture, immunoblotting, and RT-qPCR. We thank Lennart Opitz and Dr. Timothy Sykes and the Functional Genomics Center Zurich for their help with the RNA-Sequencing data analysis. We thank Joana Delgado Martinez and the Centre for Microscopy and Image Analysis of the University of Zurich for her help with and access to the microscopes. We thank Amicus Therapeutics for providing AT3375. Figuress. 1a, 2a, 3a/e/i, Supplementary Figs. 3b and 5c were created with BioRender.com. A.A. is supported by institutional core funding by the University of Zurich and the University Hospital of Zurich, and Parkinson Schweiz. He received a Distinguished Scientist Award from the NOMIS Foundation Switzerland. This study was supported by the Michael J. Fox Foundation (grant ID MJFF-022156; MJFF-024255) and the Swiss National Science Foundation SNSF, the GELU Foundation, the swissuniversities CRISPR4ALL grant, the Alzheimer’ Association grant ID 23AACSF-1026662 (JS), the Sanfilippo Children’s Foundation Australia (JS), the Fondation Sanfilippo Suisse Switzerland (JS), and the Natural Sciences and Engineering Research Council of Canada Discovery. The funders played no role in the study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author contributions
Aguzzi, Adriano: proposed the project, supervised its execution, secured funding, helped writing and correcting the manuscript. Armani, Andrea: supervision, experimental design, writing of manuscript. Böck, Desirée: supervision of adenine base editing; next-generation sequencing. Chen, Xi: assistance with cell culture work; experimental design and optimization of live cell assays; writing of manuscript. Deen, Matthew: design and synthesis of LysoFQ6-GBA1; writing of manuscript. Dhingra, Ashutosh: supervision, experimental design and generation of lentiviruses encoding the qgRNA plasmids targeting the human TFs used for the primary screen. Frick, Lukas: design of CRISPR library; supervision and experimental design, data analysis and visualization; writing of manuscript. Gilormini, Pierre-André: supervision, experimental design of lysate-based and live cell enzyme activity assays. Ging, Kathi: generated cellular model system, designed and performed, or contributed to all experiments; writing of manuscript. Marques, Ana: experimental design; assistance in cell culture, live cell assay, immunoblotting, and RT-qPCR; writing of manuscript. Pisano, Claudio: experimental design; assistance in cell culture, lysate-based assay, immunoblotting, and RT-qPCR Schlachetzki, Johannes: supervision, experimental design, writing of manuscript. Sellitto, Stefano: experimental design; assistance in cell culture; writing of manuscript. Serdiuk, Tetiana: experimental design, mass spectrometry, data analysis. Trevisan, Chiara: assistance in primary CRISPR activation screen; assistance in cell culture; writing of manuscript. Yin, Jiang-An: design and production of CRISPR library; supervision and experimental design; writing of manuscript. Zhu, Yanping: supervision and experimental design. Heutink, Peter; Vocadlo, David; Glass, Christopher K: supervision of parts of the project, secured funding, manuscript writing. All authors read and approved the final manuscript.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Statistical analysis and code availability
Screening data were analyzed using an in-house developed, open-source, R-based HTS analysis pipeline developed by Dr. Lukas Frick. The underlying code for this study is available on GitHub and can be accessed via github.com/Ginka21/CRISPRa_TF. Initially, Z’-Factor and strictly standardized mean difference (SSMD) scores36,64 were calculated to assess the discrimination between the positive and NT controls. Subsequently, heat maps of individual plates were generated to examine gradients or dispensing errors. Additionally, fluorescence values were plotted to check for row and/or column effects. Correlation between duplicates was assessed by calculating Pearson’s correlation coefficient. Candidate genes were selected with the following cut-off criteria: Fold change in fluorescence intensity >1.25 as compared to NT controls and a p (t-test) of <0.05. Volcano plots and dual flashlight plots were generated for data visualization. Unless stated otherwise, unpaired t-tests were performed in R to assess statistical significance. Supplementary Fig. 4 was generated in Prism GraphPad 8.0.0.
Competing interests
D.J.V. is cofounder of and holds equity in the company Alectos Therapeutics. D.J.V. serves as CSO and Chair of the Scientific Advisory Board (SAB) of Alectos Therapeutics. All other authors declare they have no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiang-An Yin, Email: jiang-an.yin@usz.ch.
David J. Vocadlo, Email: dvocadlo@sfu.ca
Adriano Aguzzi, Email: adriano.aguzzi@uzh.ch.
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-024-00819-7.
References
- 1.Vazquez-Velez, G. E. & Zoghbi, H. Y. Parkinson’s disease genetics and pathophysiology. Annu. Rev. Neurosci.44, 87–108 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Morris, H. R., Spillantini, M. G., Sue, C. M. & Williams-Gray, C. H. The pathogenesis of Parkinson’s disease. Lancet403, 293–304 (2024). [DOI] [PubMed] [Google Scholar]
- 3.Hannun, Y. A. & Obeid, L. M. Author correction: sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol.19, 673 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Sidransky, E. & Lopez, G. The link between the GBA gene and parkinsonism. Lancet Neurol.11, 986–998 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidransky, E. et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med.361, 1651–1661 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidransky, E., Samaddar, T. & Tayebi, N. Mutations in GBA are associated with familial Parkinson disease susceptibility and age at onset. Neurology73, 1424–1425 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Billingsley, K. J., Bandres-Ciga, S., Saez-Atienzar, S. & Singleton, A. B. Genetic risk factors in Parkinson’s disease. Cell Tissue Res.373, 9–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blauwendraat, C. et al. Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia. Brain143, 234–248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Do, J., McKinney, C., Sharma, P. & Sidransky, E. Glucocerebrosidase and its relevance to Parkinson disease. Mol. Neurodegener.14, 36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan-Or, Z. et al. The Alzheimer disease BIN1 locus as a modifier of GBA-associated Parkinson disease. J. Neurol.262, 2443–2447 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Jinn, S. et al. TMEM175 deficiency impairs lysosomal and mitochondrial function and increases alpha-synuclein aggregation. Proc. Natl Acad. Sci. USA114, 2389–2394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krohn, L. et al. Genetic, structural, and functional evidence link TMEM175 to synucleinopathies. Ann. Neurol.87, 139–153 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Manolio, T. A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med.363, 166–176 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Lambert, S. A. et al. The human transcription factors. Cell175, 598–599 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Settembre, C. & Perera, R. M. Lysosomes as coordinators of cellular catabolism, metabolic signalling and organ physiology. Nat. Rev. Mol. Cell Biol.10.1038/s41580-023-00676-x (2023). [DOI] [PubMed]
- 16.Sardiello, M. et al. A gene network regulating lysosomal biogenesis and function. Science325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Blech-Hermoni, Y. N. et al. In silico and functional studies of the regulation of the glucocerebrosidase gene. Mol. Genet Metab.99, 275–282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kampmann, M. A CRISPR approach to neurodegenerative diseases. Trends Mol. Med.23, 483–485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampmann, M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol.16, 465–480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron104, 239–255.e212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin, J.-A. et al. Robust and versatile arrayed libraries for human genome-wide CRISPR activation, deletion and silencing. bioRxiv, 2022.2005.2025.493370. 10.1101/2022.05.25.493370 (2023).
- 22.Iorio, F. et al. A landscape of pharmacogenomic interactions in cancer. Cell166, 740–754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol.38, 883–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendikov-Bar, I., Ron, I., Filocamo, M. & Horowitz, M. Characterization of the ERAD process of the L444P mutant glucocerebrosidase variant. Blood Cells Mol. Dis.46, 4–10 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Bendikov-Bar, I. & Horowitz, M. Gaucher disease paradigm: from ERAD to comorbidity. Hum. Mutat.33, 1398–1407 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Lu, J. et al. Histone deacetylase inhibitors prevent the degradation and restore the activity of glucocerebrosidase in Gaucher disease. Proc. Natl Acad. Sci. USA108, 21200–21205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goker-Alpan, O. et al. Divergent phenotypes in Gaucher disease implicate the role of modifiers. J. Med. Genet42, e37 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straniero, L. et al. The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p. Sci. Rep.7, 12702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson, B. A., Hassan, S., Garcia, E. J., Tayebi, N. & Sidransky, E. Exploring genetic modifiers of Gaucher disease: the next horizon. Hum. Mutat.39, 1739–1751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ysselstein, D. et al. Evaluation of strategies for measuring lysosomal glucocerebrosidase activity. Mov. Disord.36, 2719–2730 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deen, M. C. et al. Selective fluorogenic beta-glucocerebrosidase substrates for convenient analysis of enzyme activity in cell and tissue homogenates. ACS Chem. Biol.15, 824–829 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Deen, M. C. et al. A versatile fluorescence-quenched substrate for quantitative measurement of glucocerebrosidase activity within live cells. Proc. Natl Acad. Sci. USA119, e2200553119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breiden, B. & Sandhoff, K. Lysosomal glycosphingolipid storage diseases. Annu Rev. Biochem.88, 461–485 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Kuo, C. L. et al. In vivo inactivation of glycosidases by conduritol B epoxide and cyclophellitol as revealed by activity-based protein profiling. FEBS J.286, 584–600 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sultana, S. et al. Characterization of the zebrafish homolog of beta-glucosidase 2: a target of the drug miglustat. Zebrafish13, 177–187 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Zhang, J. H., Chung, T. D. & Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Ron, I. & Horowitz, M. ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum. Mol. Genet.14, 2387–2398 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Pastore, N. et al. TFE3 regulates whole-body energy metabolism in cooperation with TFEB. EMBO Mol. Med.9, 605–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raben, N. & Puertollano, R. TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu Rev. Cell Dev. Biol.32, 255–278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cecioni, S. et al. Quantifying lysosomal glycosidase activity within cells using bis-acetal substrates. Nat. Chem. Biol.18, 332–341 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Yamanaka, T. et al. Genome-wide analyses in neuronal cells reveal that upstream transcription factors regulate lysosomal gene expression. FEBS J.283, 1077–1087 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Vu, H. N., Dilshat, R., Fock, V. & Steingrimsson, E. User guide to MiT-TFE isoforms and post-translational modifications. Pigment Cell Melanoma Res.34, 13–27 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Gupta, M. et al. Plekhg4 is a novel Dbl family guanine nucleotide exchange factor protein for rho family GTPases. J. Biol. Chem.288, 14522–14530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan, Q. et al. Pleckstrin homology and RhoGEF domain containing G4 (PLEKHG4) leads to the activation of RhoGTPases promoting the malignant phenotypes of thyroid cancer. Apoptosis28, 1315–1331 (2023). [DOI] [PubMed] [Google Scholar]
- 45.Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res.48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corre, S. & Galibert, M. D. USF as a key regulatory element of gene expression. Med. Sci.22, 62–67 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Annunziata, I. et al. MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat. Nat. Commun.10, 3623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braunstein, H. et al. UPR activation and CHOP mediated induction of GBA1 transcription in Gaucher disease. Blood Cells Mol. Dis.68, 21–29 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Sapkota, D. et al. Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development. Proc. Natl Acad. Sci. USA111, E4086–E4095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanhorenbeeck, V. et al. Role of the Onecut transcription factors in pancreas morphogenesis and in pancreatic and enteric endocrine differentiation. Dev. Biol.305, 685–694 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Jacquemin, P. et al. The transcription factor onecut-2 controls the microphthalmia-associated transcription factor gene. Biochem. Biophys. Res. Commun.285, 1200–1205 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Boddupalli, C. S. et al. Neuroinflammation in neuronopathic Gaucher disease: Role of microglia and NK cells, biomarkers, and response to substrate reduction therapy. Elife11, e79830 (2022). [DOI] [PMC free article] [PubMed]
- 53.Brunialti, E. et al. Inhibition of microglial beta-glucocerebrosidase hampers the microglia-mediated antioxidant and protective response in neurons. J. Neuroinflamm.18, 220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, D. et al. Development of quantitative high-throughput screening assays to identify, validate, and optimize small-molecule stabilizers of misfolded β-glucocerebrosidase with therapeutic potential for Gaucher disease and Parkinson’s disease. bioRxiv, 2024.2003.2022.586364. 10.1101/2024.03.22.586364 (2024).
- 55.Tamargo, R. J., Velayati, A., Goldin, E. & Sidransky, E. The role of saposin C in Gaucher disease. Mol. Genet. Metab.106, 257–263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mecca, C., Giambanco, I., Donato, R. & Arcuri, C. Targeting mTOR in glioblastoma: rationale and preclinical/clinical evidence. Dis. Markers2018, 9230479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goding, C. R. & Arnheiter, H. MITF-the first 25 years. Genes Dev.33, 983–1007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henley, M. J. & Koehler, A. N. Advances in targeting ‘undruggable’ transcription factors with small molecules. Nat. Rev. Drug Discov.20, 669–688 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol.37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goddard-Borger, E. D. et al. Rapid assembly of a library of lipophilic iminosugars via the thiol-ene reaction yields promising pharmacological chaperones for the treatment of Gaucher disease. J. Med. Chem.55, 2737–2745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lambert, S. A. et al. The human transcription factors. Cell172, 650–665 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Hatakeyama, M. et al. SUSHI: an exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinforma.17, 228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hildebrandt, M. R. et al. Precision health resource of control iPSC lines for versatile multilineage differentiation. Stem Cell Rep.13, 1126–1141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, X. D. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J. Biomol. Screen16, 775–785 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.
Screening data were analyzed using an in-house developed, open-source, R-based HTS analysis pipeline developed by Dr. Lukas Frick. The underlying code for this study is available on GitHub and can be accessed via github.com/Ginka21/CRISPRa_TF. Initially, Z’-Factor and strictly standardized mean difference (SSMD) scores36,64 were calculated to assess the discrimination between the positive and NT controls. Subsequently, heat maps of individual plates were generated to examine gradients or dispensing errors. Additionally, fluorescence values were plotted to check for row and/or column effects. Correlation between duplicates was assessed by calculating Pearson’s correlation coefficient. Candidate genes were selected with the following cut-off criteria: Fold change in fluorescence intensity >1.25 as compared to NT controls and a p (t-test) of <0.05. Volcano plots and dual flashlight plots were generated for data visualization. Unless stated otherwise, unpaired t-tests were performed in R to assess statistical significance. Supplementary Fig. 4 was generated in Prism GraphPad 8.0.0.