Abstract
We have previously shown that erythroleukemia cells (K562) transfected with vascular adhesion molecule 1 (VCAM-1) are susceptible to human T-cell leukemia virus type 1 (HTLV-1)-induced syncytium formation. Since expression of VCAM-1 alone is not sufficient to render cells susceptible to HTLV-1 fusion, K562 cells appear to express a second molecule critical for HTLV-induced syncytium formation. By immunizing mice with K562 cells, we have isolated four monoclonal antibodies (MAbs), K5.M1, K5.M2, K5.M3, and K5.M4, that inhibit HTLV-induced syncytium formation between infected MT2 cells and susceptible K562/VCAM1 cells. These MAbs recognize distinct proteins on the surface of cells as determined by cell phenotyping, immunoprecipitation, and Western blot analysis. Since three of the proteins recognized by the MAbs appear to be GPI linked, we isolated lipid rafts and determined by immunoblot analysis that all four MAbs recognize proteins that sort entirely or in large part to lipid rafts. Dispersion of lipid rafts on the cells by cholesterol depletion with β-cyclodextrin resulted in inhibition of syncytium formation, and this effect was not seen when the β-cyclodextrin was preloaded with cholesterol before treating the cells. The results of these studies suggest that lipid rafts may play an important role in HTLV-1 syncytium formation.
Human T-cell leukemia virus type 1 (HTLV-1) is the causative agent of adult T-cell leukemia and tropical spastic paraparesis/HTLV-associated myelopathy (4, 45). Infection is spread mainly through direct contact between infected and uninfected cells, and infection by cell-free HTLV-1 is very inefficient (30). The envelope glycoprotein of HTLV-1 consists of the surface protein gp46 and the transmembrane protein gp21. Like the envelope glycoprotein gp120 of human immunodeficiency virus (HIV), gp46 is thought to be the virus's attachment protein (31, 47). The receptor(s) for this retrovirus has not yet been identified definitively but is theorized to be widely expressed, since many cell lines from various human and nonhuman sources, including mouse, rat, monkey, and dog, are susceptible to infection (44). Interestingly, despite the wide tropism of HTLV-1 in vitro, the virus shows a tropism for T cells in vivo (47). Despite the failure thus far to identify one protein as the receptor for this virus, various proteins have been reported to be implicated in syncytium formation by the virus, including vascular adhesion molecule 1 (VCAM-1) (23), heat shock cognate protein 70 (37), membrane glycoprotein C33 (11), CD2 (9, 12), HLA A2 (7), and interleukin-2 receptor (27). In a previous report, we showed that monoclonal antibodies (MAbs) to proteins highly expressed on the surface of HTLV-1-infected cells, such as major histocompatibility complex class II (MHC-2), could inhibit HTLV-1-induced syncytium formation while leaving HIV-1-induced syncytium formation unchanged (19). This suggested that the receptor that engages gp46 is, like gp46 itself, small and compact in relation to the proteins that surround it and thus cannot easily penetrate MAbs bound to proteins surrounding gp46. The gene encoding the receptor for HTLV-1 has been mapped to the long arm of chromosome 17 in studies employing mouse-human hybridomas (13, 43).
In previous studies we demonstrated that transfection of the erythroleukemia cell line K562 with the adhesion molecule VCAM-1 conferred sensitivity to HTLV-1-induced syncytium formation (23). Since VCAM-1 does not appear to directly bind gp46, our results suggest that K562 cells express a second molecule needed for HTLV-1 infection. In an attempt to identify this molecule, we have generated a panel of MAbs against K562 and screened them for inhibition of HTLV-1 syncytium formation. We have identified four MAbs that inhibit syncytium formation between the chronically infected MT2 cell line and K562 cells transfected with VCAM-1.
Characterization of these new MAbs showed that they do not recognize VCAM-1 but are specific for four distinct proteins expressed at various levels on many cell types. Further characterization showed that all four antibodies recognize proteins that are found mainly, if not solely, in specialized membrane domains known as lipid rafts. Lipid rafts are distinct regions of the membrane that are rich in sphingolipids and cholesterol. They are sites enriched in the expression of many glycosyl-phosphatidylinositol (GPI)-anchored proteins, as well as src family kinases, protein kinase C, heterotrimeric G proteins, actin and actin binding proteins, and caveolin (1, 6, 8, 41). Lipids in lipid rafts are much more tightly packed, and as a result, these domains are in a more ordered state compared to the surrounding membrane resulting in resistance to nonionic detergent treatment at low temperature (40).
We treated K562/VCAM1 and MT2 cells with β-cyclodextrin, which extracts cholesterol from the plasma membrane (26) and thereby partially disperses lipid rafts (25), and found that syncytium formation no longer occurred, implying that HTLV-1-induced cell fusion requires intact lipid rafts. Our results demonstrate for the first time that lipid rafts may play an important role in HTLV-1 biology and further indicate that the receptor for HTLV-1 or other molecules required for fusion may be localized in these membrane microdomains.
MATERIALS AND METHODS
Cells.
The K562, MT2, and MJG11 cell lines were obtained from the American Type Culture Collection (ATCC) (Manassas, Va.) and the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. The LD− and tLD− cell lines were kindly provided by Robert Brodsky (Johns Hopkins University Oncology Department). CEM × 174 cells were obtained from Janice Clements (Johns Hopkins University). All of the cell lines above were maintained in cRPMI (RPMI-1640 supplemented with 10 mM HEPES, 2 mM l-glutamine, and 10% fetal bovine serum). Construction of the pCEP4VCAM-1 expression vector and establishment of the K562/VCAM1 stable transfectant line were described previously (23). HOS and HeLa cell lines were obtained from ATCC and were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 10 mM HEPES, and 2 mM l-glutamine. Peripheral blood mononuclear cells were purified from buffy coats from healthy donors by Ficoll-Hypaque density centrifugation exactly as previously described (15).
Antibodies.
New MAbs against K562 cells were generated as previously described (20, 21). Briefly, female BALB/c mice were injected intraperitoneally (i.p.) three times, biweekly, with 107 K562 cells that had been washed three times with phosphate-buffered saline (PBS). Two weeks after the last i.p. injection, the mice were injected intravenously (i.v.) with 5 × 106 K562 cells. Four days after the i.v. injection, spleens were removed and fusions were performed as previously described (20, 22), using P3 × 653.Ag8 (ATCC) myeloma cells as fusion partners. Hybridoma culture supernatants were screened for inhibition of syncytium formation between MT2 and K562/VCAM1 cells, as previously described (19). Hybridomas from wells showing inhibition of fusion were subcloned by limiting dilution on BALB/c splenocyte feeder cells (19). The antibodies selected for further characterization were designated K5.M1, K5.M2, K5.M3, and K5.M4. MAb K5.M1 was determined to be of the immunoglobulin G2a,κ (IgG2a,κ) isotype, and the others were determined to be of the IgG1,κ isotype (MonoAb ID kit; Zymed). MAb against HTLV-1 gp46 was purchased from Cellular Products (Buffalo, N.Y.), and those against CD59 and CD71 (transferrin receptor) were purchased from PharMingen (San Diego, Calif.). Hybridoma VIII6G10 secreting MAb against VCAM-1 was obtained from ATCC. Fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse (FITC-RAM) and horseradish peroxidase-conjugated goat anti-mouse IgG (HRP-GAM IgG) were purchased from Jackson Immunoresearch (Avondale, Pa.). Other MAbs used in this study were produced in our laboratory as previously reported: MHM.5 (anti-MHC-1) (20) and MT.M5 (anti-ICAM-1) (19). All antibodies were used in the form of hybridoma culture supernatants (10 to 30 μg of IgG/ml) or purified IgG in the appropriate buffer or medium at a concentration of 20 μg/ml.
Syncytium formation assay.
Syncytium formation assays were carried out as previously reported (23). All cells were washed with PBS and resuspended in phenol red-free cRPMI at a density of 2 × 106/ml. To test the MAbs' ability to inhibit syncytium formation, 50 μl of K562/VCAM1 cells was added to duplicate wells of a flat-bottom 96-well plate and mixed with cRPMI or MAb (20 μg of IgG/ml or neat hybridoma culture supernatants). After incubation for 1 h at room temperature, 50 μl of MT2 cells or, where indicated, MJG11 cells was added. For all other syncytium formation assays, 50 μl of K562/VCAM1 cells was incubated with 100 μl of MT2 cells. The contents of the wells were mixed, and the plates were incubated overnight at 37°C, 5% CO2. Syncytia were quantitated by counting syncytia (more than three or four cell diameters) in several high-power fields (HPF) (19, 23).
Flow cytometry.
Flow cytometry analysis was performed as described previously (15). Cells were washed and resuspended at 2 × 106/ml in PBS containing 5% normal rabbit serum. One hundred microliters of cells was incubated with 100 μl of neat hybridoma supernatant or 10 μg of purified IgG/ml for 45 min on ice. Cells were washed once with cold PBS and incubated with 100 μl of PBS-normal rabbit serum containing 10 μg of Fc-specific FITC-RAM/ml. After 45 min on ice, cells were washed once in cold PBS and resuspended in 500 μl of 2% paraformaldehyde in PBS. Flow cytometry analysis was carried out on an EPICS Profile II analyzer.
PI-PLC treatment.
CEM × 174 cells were washed once in PBS and resuspended to a density of 2 × 106/ml in complete medium containing 200 mU of phosphatidylinositol-specific phospholipase C (PI-PLC) before incubating 1 h at room temperature (RT). Flow cytometry analysis was conducted as described above.
Vectorial iodination and immunoprecipitation.
Vectorial iodination of cells and immunoprecipitations were performed as described previously (15, 20). Aliquots of 5 × 107 cells were washed three times with PBS and resuspended in 0.5 ml of PBS. 125I (NaI, 1 mCi; Amersham, Piscataway, N.J.) was added to the cells in PBS, followed by the addition of 50 μl each of lactoperoxidase (1 U), glucose oxidase (1 U), and 1% dextrose in PBS. After 15 min at RT, 25 μl of 1% dextrose was added, followed by an additional 15 min at RT before the cells were washed twice with 10 ml of PBS. The cell pellets were resuspended in 1 ml of 50 mM Tris (pH 7.5)–5 mM EDTA–150 mM NaCl (TEN) containing 1% NP-40 and protease inhibitor cocktail (2-μg/ml concentrations each of leupeptin, soybean trypsin inhibitor, antipain, aprotinin, and chymostatin) and 1 mM phenylmethylsulfonyl fluoride. After a 45-min incubation on ice, the lysate was transferred to an Eppendorf tube and spun for 10 min at 4°C, 13,800 × g. The supernatant was removed and further clarified by centrifugation at 100,000 × g for 45 min. The supernatant was incubated with 20 μg of aggregated human IgG 2 h on ice before adding 100 μl of 10% formalin-fixed Staphylococcus aureus Cowan strain cells and incubating 30 min on ice. After removing the S. aureus Cowan strain cells by centrifugation, 100 μl of the supernatant was mixed with 100 μl of MAb (neat hybridoma culture supernatant or purified IgG at 20 μg/ml) and incubated overnight at 4°C. Ten micrograms of rabbit anti-mouse Ig (Jackson Immunoresearch) was added followed by incubation for 1 h on ice before adding 50 μl of S. aureus Cowan strain cells. After 20 min on ice, the samples were washed twice with 2 M KCl in TEN–0.5% NP-40 and once with TEN–0.5% NP-40. The pelleted S. aureus Cowan strain cells were resuspended in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) reducing or nonreducing buffers and boiled for 3 to 4 min. After removing the S. aureus Cowan strain cells by centrifugation, samples were analyzed by SDS-PAGE on 10% bisacrylamide gels. Precipitated proteins were visualized by autoradiography.
Immunoblot assays.
Immunoblot assays were performed as previously described (32). Briefly, 5 × 107 cells were washed with PBS and resuspended in 500 μl of 50 mM Tris, pH 7.5, 25 mM KCl, 5 mM MgCl2, and 1 mM EDTA (TKM) with 1% Triton X-100 (TKMT) containing protease inhibitor cocktail and were incubated for 30 min on ice. Lysates were clarified by centrifugation for 10 min at 4°C, 8,160 × g. For isolation of the lipid rafts, 500 μl of the clarified lysates was mixed with 500 μl of 80% sucrose in TKM buffer. These samples were then overlaid with 6 ml of 38% sucrose in TKM buffer, which was in turn overlaid with 4 ml of 5% sucrose in TKM buffer. The samples were centrifuged at 200,000 × g for 18 h at 4°C. One-milliliter fractions were collected, diluted 1:10 with TKM, and blotted onto nitrocellulose. The nitrocellulose membranes were blocked for 2 h with Blotto (5% milk powder in PBS and 0.05% Tween 20 [PBST]). The primary antibodies (100 μl of neat hybridoma culture supernatant or 10 μg of IgG/ml) in Blotto were incubated with the membranes for 1 h at RT. The membranes were washed three times, 10 min each time, with PBST. The secondary antibody, HRP-GAM IgG, was diluted to 1:6,250 in Blotto and incubated with the membranes for 1 h at RT. The membranes were then washed five times as described above before visualizing bound antibodies with ECL reagent and Hyperfilm ECL.
Western blotting.
K562 cells were lysed in TKMT containing protease inhibitor cocktail for 1 h at 37°C. Protein samples (75 μg per lane) were run on a 4 to 20% acrylamide SDS-PAGE gradient gel (Bio-Rad, Hercules, Calif.) and transferred onto nitrocellulose. The nitrocellulose was blocked by a 1-h incubation in Blotto. The individual lanes were cut and each was incubated in 0.5% milk powder in PBST with the appropriate MAb for 1 h, followed by three 10-min washes in PBST. The blots were then incubated for 1 h with GAM-HRP in 0.5% milk powder, followed by five 10-min washes in PBST. The bound MAbs were visualized with ECL reagent and Hyperfilm ECL.
β-Cyclodextrin treatment of cells.
Cells were washed with PBS and resuspended at 2 × 106 cells/ml in cRPMI containing 20 mM 2-hydroxylpropyl-β-cyclodextrin (BCD) (CTD Technologies, Gainesville, Fla.) and incubated for 1 h at 37°C. Cells were washed with PBS and resuspended at 2 × 106 cells/ml in phenol red-free cRPMI and used in syncytium formation assays. Cell viability was measured by trypan blue exclusion.
Cholesterol-preloaded BCD.
BCD saturated with bound cholesterol was prepared as follows. Thirty milligrams (77 μmol) of cholesterol was added to 5 ml of 20 mM (100 μmol) BCD in cRPMI and vortexed. The mixture was incubated for 30 min in a 37°C water bath and vortexed. The undissolved cholesterol was removed by centrifugation and filtration of the supernatant through a 0.22-μm-pore-size filter. Cells were treated with the cholesterol-preloaded BCD as described above for control BCD.
BCD effects on VCAM-1 binding.
K562/VCAM1 cells were washed and pretreated with BCD or media as described above. Aliquots of 105 cells were incubated with an equal number of Jurkat T cells for 1 h at 37°C. Conjugate formation between the two cell types was assessed visually on an inverted microscope as previously described (23).
RESULTS
New MAbs inhibit HTLV-1 syncytium formation.
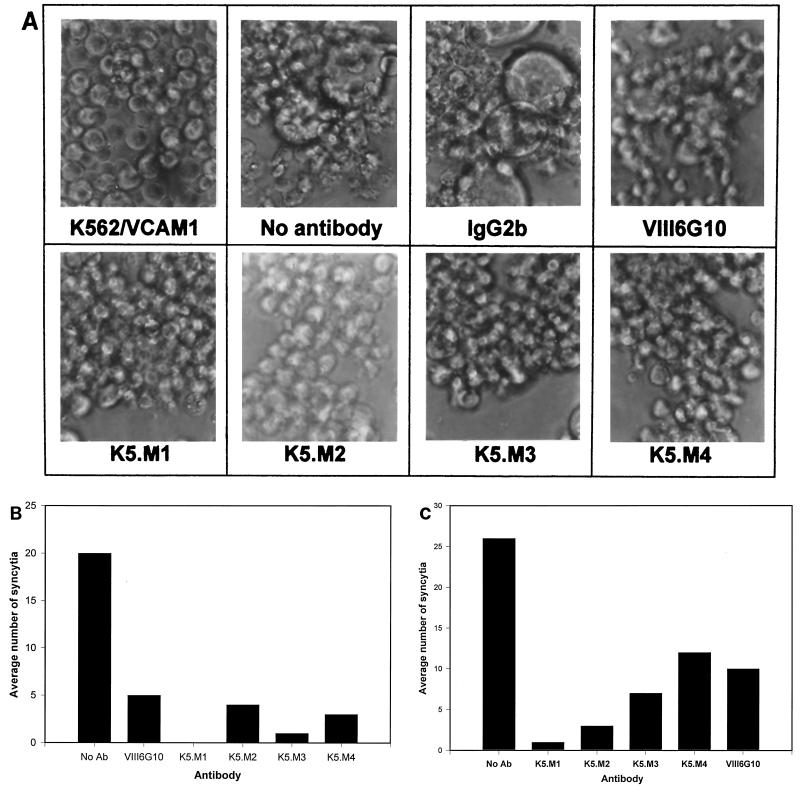
The new panel of MAbs against K562 cells was tested for inhibition of HTLV-1 syncytium formation as described in previous studies (19). K562/VCAM1 cells form gross syncytia when cocultured with chronically infected MT2 cells (Fig. 1A and B). When K562/VCAM1 cells were preincubated with the new MAbs, syncytia failed to form, whereas a control MAb, mouse myeloma IgG2b, had no effect. This experiment was repeated several times with similar results. The MAbs also blocked fusion between another HTLV-infected cell line, MJG11, and K562/VCAM1 cells (Fig. 1C). The data indicated that the new MAbs recognize structures that are potentially involved in HTLV-1-induced cell-cell fusion.
FIG. 1.
New MAbs against K562 cells block HTLV-1 syncytium formation. (A and B) MAbs produced against K562 cells (K5.M series) were tested for inhibition of HTLV-1-induced syncytium formation between K562/VCAM cells and MT2 cells, as described in Materials and Methods. Myeloma IgG2b and anti-VCAM-1 (VIII6G10) antibodies were used as negative and positive controls, respectively. (B) Data shown are mean numbers of syncytia per HPF. Syncytia in six HPFs were counted. (C) MAbs were tested for inhibition of HTLV-1-induced syncytium formation between K562/VCAM cells and MJG11 cells.
Characterization of new MAbs.
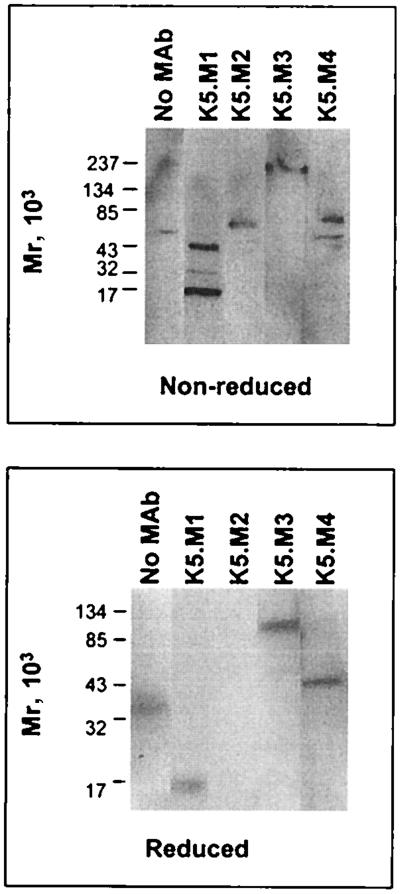
In order to begin characterizing the structures recognized by the new MAbs, Western blot studies were performed. To increase the solubility of all proteins, K562 cells were lysed with Triton X-100 for 1 h (in the presense of protease inhibitors) at 37°C instead of lysis at low temperature. Western blot analysis of this lysate under nonreducing conditions with the new MAbs showed proteins recognized by all four of the MAbs (Fig. 2). MAb K5.M1 recognized proteins with approximate molecular masses of 48, 24, and 16 kDa. MAb K5.M2 recognized a protein with a mass of approximately 76 kDa under nonreducing conditions. A protein with a mass of approximately 210 kDa was recognized by MAb K5.M3. MAb K5.M4 identified two proteins with molecular masses of 79 and 48 kDa (Fig. 2, top). Under reducing conditions, the pattern of protein bands recognized by the MAbs was different (Fig. 2, bottom). MAb K5.M1 recognized only one band, with a mass of approximately 15 kDa, while no bands were identified by MAb K5.M2. The latter probably reflects loss of a conformational epitope upon reduction of disulfide bridges in the protein. MAb K5.M3 recognized a 100-kDa protein under reducing conditions. Under reducing conditions, K5.M4 recognized only one band, with a molecular mass of approximately 51 kDa. There was a band in the negative control (no primary antibody) lane under both reducing and nonreducing conditions; however, it did not appear to correspond to any of the bands recognized by the MAbs. The Western blot analysis indicated that the four new MAbs that block HTLV-1 syncytium formation recognize four distinct proteins.
FIG. 2.
Western blotting of K562 lysate with K5.M MAbs. K562 cells were lysed in TKMT at 37°C for 1 h. Clarified lysate was run on a 4 to 20% gradient acrylamide SDS-PAGE gel under nonreducing and reducing conditions and subjected to Western blot analysis as described in Materials and Methods with the MAbs indicated.
Immunoprecipitation studies were also performed using these MAbs. K562 cells were vectorially labeled with 125I, lysed, and subjected to immunoprecipitation with the new MAbs. The precipitated proteins were analyzed by SDS-PAGE under reducing and nonreducing conditions (data not shown). Under both conditions, MAb K5.M1 pulled down a protein with a molecular mass of approximately 12 kDa, close to the 15- to 16-kDa value determined by Western blot analysis. MAb K5.M3 precipitated a single protein band with a mass of approximately 210 kDa under nonreducing conditions and a single band with a mass of 116 kDa under reducing conditions. No protein bands were seen after precipitation with either the K5.M2 or K5.M4 MAbs. The epitopes for these antibodies may have been destroyed under the conditions used for labeling the cells. These data are consistent with the Western blot results and support the suggestion that the four MAbs recognize distinct proteins.
Cell phenotyping with MAbs.
Since the MAbs were generated against erythroleukemia cells, we wished to determine if they also recognized antigens expressed on other cell types. We conducted flow cytometry analysis on a variety of cell lines and primary mononuclear cells. As seen in Table 1, the MAbs bound to most of the cells tested and the expression of the proteins recognized varied from cell type to cell type, consistent with recognition of distinct proteins by the MAbs. Interestingly, three of the MAbs (K5.M2, K5.M3, and K5.M4) also reacted with HTLV-1-infected MT2 cells. Since the parent cell type used to generate the MT2 cells was not available, we could not determine whether expression of the proteins recognized by the MAbs was affected by HTLV-1 expression.
TABLE 1.
HTLV-1 syncytium-inhibiting MAbs recognize proteins on various cell lines
| Cell or MAb | % Positive cellsa
|
|||||
|---|---|---|---|---|---|---|
| K562/VCAM1 | MT2 | HOS | PBMCs | 293 | CEM × 174 | |
| IgG2b | 1.1 (10.5) | 3.7 (19.8) | 2 (5.0) | 0.8 (0.2) | 0.3 (2.8) | 0.5 (3.9) |
| K5.M1 | 99.6 (98.7) | 0.4 (4.3) | 5.8 (7.0) | 50 (1.6) | 1.7 (4.2) | 99.8 (48.3) |
| K5.M2 | 98.2 (139.2) | 93.6 (209.8) | 100 (253.5) | 91.7 (13.1) | 99.9 (100.1) | 100 (120) |
| K5.M3 | 99.9 (217.1) | 99.5 (548.6) | 100 (111.9) | 64.2 (3.4) | 96.3 (38.5) | 99.9 (237.7) |
| K5.M4 | 99.9 (71.1) | 99.3 (311.3) | 100 (226.8) | 86.3 (8.1) | 99.7 (66.2) | 99.9 (54.2) |
| MT.M5 | 99.5 (49.7) | 98.7 (671.0) | 100 (271.5) | 65.4 (3.2) | 5 (4.9) | 100 (219.7) |
| VIII6G10 | 53.8 (54.6) | 72.3 (97.2) | 83.6 (17.3) | 31.8 (0.7) | 1.6 (3.4) | 85.7 (14.4) |
| MHM.5 | 5.5 (11.4) | 81.3 (89.2) | 100 (390.7) | 98.3 (92.4) | 22.4 (7.9) | 100 (826.1) |
MAbs were tested for binding to the indicated cell lines as described in Materials and Methods. Data shown are the percentages of positive cells (mean channel fluorescence in parentheses), with MAb IgG2b mouse myeloma used as negative control. MT.M5, VIII6G10, and MHM.5 recognize ICAM-1, VCAM-1, and MHC-1, respectively. PBMCs, peripheral blood mononuclear cells.
Cell phenotyping after treatment with PI-PLC.
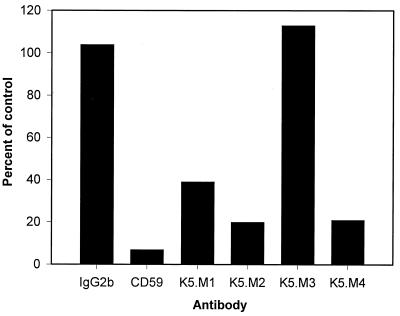
To further characterize the proteins recognized by the MAbs, we attempted to determine whether the proteins were bound to the cell membrane by GPI anchors. We treated K562 cells with PI-PLC, which releases GPI-anchored proteins from the cell surface, and determined expression of protein antigens by flow cytometry. As a positive control for GPI-anchored proteins, we used CD59. None of the proteins, including the positive control, changed expression levels after PI-PLC treatment of K562 cells (data not shown). We repeated the experiment with another cell line, CEM × 174, that expressed all four of the proteins recognized by the new MAbs at high levels, as seen in Table 1. Two of the four MAbs, K5.M2 and K5.M4, showed a large decrease in mean channel fluorescence, similar to that seen with CD59 (Fig. 3). This result suggested that the proteins recognized by these two MAbs are also GPI anchored. Expression of protein recognized by K5.M1 was also decreased by more than 50% after PI-PLC treatment of the cells. The smaller reduction of the K5.M1 protein expression may reflect two forms of the small protein recognized by K5.M1, one that is transmembrane and thus insensitive to PI-PLC treatment, and another that is GPI anchored. Alternatively, the K5.M1 protein may be less sensitive to PI-PLC than other GPI-anchored surface proteins.
FIG. 3.
PI-PLC treatment changes the ability of K5.M MAbs to bind cells. CEM × 174 cells were treated with PI-PLC and analyzed by flow cytometry as described in Materials and Methods. Data presented are the mean channel fluorescences of the PI-PLC-treated cells as a percent of that of the untreated control cells. Myeloma IgG2b was used to determine nonspecific binding. Anti-CD59 MAb was used as a positive control for release of GPI-anchored proteins by PI-PLC.
Phenotyping of LD− and tLD− cells.
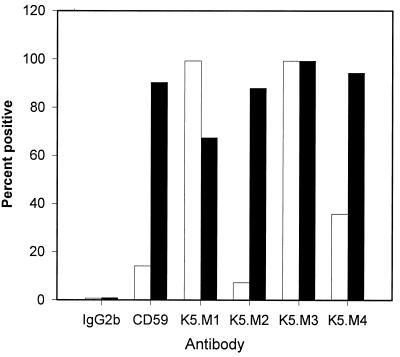
We confirmed the results of the PI-PLC studies using cells from a patient with paroxysmal nocturnal hemoglobinuria (PNH). PNH patients lack the PIG-A gene, which is required for synthesis of GPI anchors. The antibodies were tested for binding to an Epstein-Barr virus-transformed PNH cell line, LD−, and the same cell line with GPI anchor synthesis restored by transfection with the PIG-A gene (tLD−) (3). As seen in Fig. 4, K5.M2 and K5.M4 behaved like the MAb against the GPI-anchored protein CD59, binding poorly to LD− cells but very well to tLD− cells, confirming that these two MAbs recognize GPI-anchored proteins. K5.M1 and K5.M3 MAbs bound to both LD− and tLD− cells, indicating that these two MAbs recognize proteins that are not GPI anchored. In fact, the percent positive cells increased for MAb K5.M1, suggesting that the decreased number of GPI-anchored proteins on LD− cells may have increased the accessibility of the K5.M1 epitope. These results suggest that there are two forms of the K5.M1 protein, a PI-PLC-sensitive (GPI-anchored) form on CEM × 174 cells and a non-GPI-anchored form on LD− cells.
FIG. 4.
Flow cytometry analysis of MAb binding to GPI anchor-negative and GPI anchor-positive cells. MAbs were assayed for binding to LD− and tLD− cells by flow cytometry, as described in Materials and Methods. Data presented are the percent of positive cells. The open bars represent LD− cells (GPI anchor negative), and the shaded bars represent tLD− cells (GPI anchor positive).
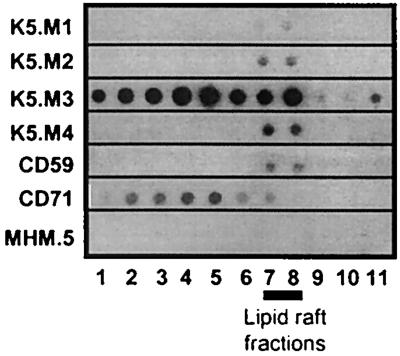
Immunoblot analysis of lipid raft proteins.
GPI-anchored proteins are found abundantly in lipid rafts—highly ordered, detergent-insoluble domains of the cell membrane, rich in cholesterol and sphingolipids (41). Lipid rafts are more buoyant than detergent-soluble membrane fractions and therefore can be separated by equilibrium centrifugation on sucrose gradients. We lysed K562 cells and isolated lipid rafts on sucrose gradients as previously described (26). The buoyant lipid rafts migrate upwards during centrifugation and accumulate at the interface between the dense 38% sucrose and the light 5% sucrose (fractions 6 to 9, as determined with anti-CD59 MAb). Solubilized proteins remain at the bottom of the gradient. The fractions were blotted onto nitrocellulose and immunoblot analysis was performed as described in Materials and Methods. As seen in Fig. 5, all four of the proteins recognized by the new MAbs associated with the lipid rafts, even those that were not GPI anchored. As expected, the GPI-anchored proteins were found primarily in the rafts. The proteins recognized by K5.M1, K5.M2, and K5.M4 were entirely in the rafts and that recognized by K5.M3 was found in both the soluble and raft fractions. The sorting of the proteins recognized by K5.M2 and K5.M4 to detergent-insoluble membrane domains in K562 cells may explain why they were undetectable by the immunoprecipitation protocol used; they would likely have been pelleted out of the lysate as part of detergent-insoluble material.
FIG. 5.
Immunoblot of K562 cell fractions shows that proteins recognized by K5.M MAbs are in lipid rafts. Nonionic detergent lysates were prepared from K562 cells and subjected to equilibrium centrifugation on sucrose gradients, and fractions were collected as described in Materials and Methods. The fractions were subjected to immunoblot analysis with the MAbs indicated. Lipid raft and soluble fractions were identified by anti-CD59 and anti-CD71 MAbs, respectively. Anti-MHC-1 MAb (MHM.5) served as a negative control.
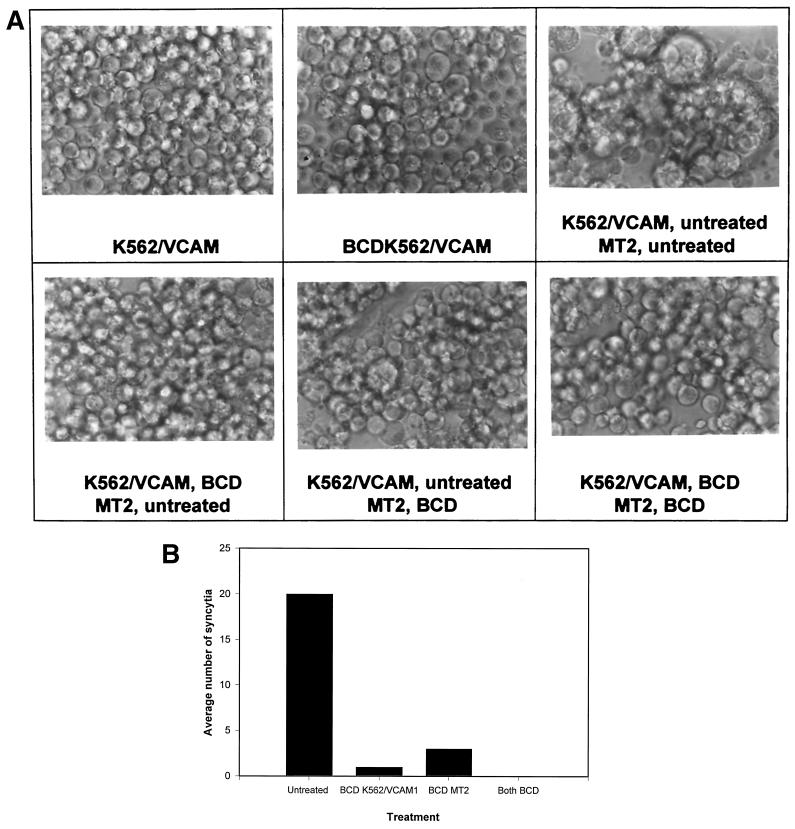
β-Cyclodextrin treatment blocks syncytium formation.
Since the new MAbs were found to recognize proteins that were in lipid rafts, we wanted to determine if these specialized membrane domains were critically involved in HTLV-1-induced syncytium formation. Cholesterol has been shown to be required for the formation and integrity of lipid rafts (5, 16, 28). BCD effectively depletes cholesterol from cell membranes and, in doing so, disperses lipid rafts (25, 39). If lipid rafts are critically important for HTLV-1 fusion, their dispersion should result in inhibition of HTLV-1 syncytium formation. Both K562/VCAM1 cells and MT2 cells were treated with 20 mM BCD in culture medium and then cocultured in different combinations to determine the contribution of the lipid rafts on each cell type to virus-induced cell fusion (Fig. 6). Cell viability after treatment was 100% that of control cells, as assayed by trypan blue staining. When both cell types were treated with BCD, there were none to very few syncytia compared to untreated controls. When the K562/VCAM1 cells were treated and cocultured with untreated MT2 cells, syncytium formation was also blocked. When the MT2 cells were treated and mixed with untreated K562/VCAM1 cells, syncytium formation was only partially inhibited. These data suggest that intact lipid rafts are required for HTLV-1-induced fusion and therefore are important for cell-to-cell transmission of the virus. Moreover, it seems that intact lipid rafts in the target membrane are more critical than lipid rafts in the infected cell membrane for infection to occur. To determine if treatment with BCD affected surface expression of cellular proteins, we treated the cells with BCD as we had for the syncytium formation assay and analyzed them by flow cytometry. When analyzing K562/VCAM1 cells, we saw a slight increase in surface expression of proteins recognized by the four K5.M MAbs and a slight decrease in VCAM-1 and ICAM-1 expression (Table 2). Similar results were found for MT2 cells except that expression of VCAM-1 and ICAM-1 was essentially the same or slightly higher (Table 2). The increase in the surface expression of proteins could be the result of the cells attempting to restore the cholesterol content of their plasma membranes after BCD treatment. These proteins could be components of cholesterol-rich vesicles being shuttled to the surface after cholesterol depletion. The decrease in VCAM-1 expression was very small and could not account for the complete inhibition in fusion that is seen in Fig. 6. Thus, we concluded that BCD did not block syncytium formation by changing surface expression of proteins.
FIG. 6.
BCD treatment blocks HTLV-1-induced syncytium formation. K562/VCAM and MT2 cells were pretreated with 20 mM BCD and incubated as described in Materials and Methods. The cells were then cocultured in syncytium formation assays in the combinations shown. Photomicroscopy (A) and quantitation of syncytia (B) were performed at 16 h after mixing the cells. (B) Data shown are mean numbers of syncytia per HPF. Syncytia in three HPFs were counted.
TABLE 2.
BCD treatment does not drastically alter cell surface expression of proteins
| Cell or MAb | % Positive cellsa
|
|||
|---|---|---|---|---|
| K562/VCAM1 | BCD K562/VCAM1 | MT2 | BCD MT2 | |
| IgG2b | 5.5 (7.5) | 4 (6.4) | 4.2 (10.3) | 2.3 (7.7) |
| K5.M1 | 88.4 (40.2) | 9.7 (45.1) | 0.5 (8.4) | 1.3 (7.7) |
| K5.M2 | 96.4 (53.6) | 98.9 (70.4) | 100 (391.6) | 100 (475.3) |
| K5.M3 | 99.7 (533.4) | 99.9 (618.5) | 100 (415.8) | 100 (596.0) |
| K5.M4 | 99.6 (164.3) | 99.7 (235.4) | 100 (510.2) | 100 (540.6) |
| CD59 | 99.2 (194.0) | 99.5 (216.3) | 86.8 (31.1) | 81.3 (29.2) |
| VIII6G10 | 59.9 (24.5) | 46.5 (17.0) | 99.2 (108.7) | 99.6 (125.0) |
| MT.M5 | 99.9 (342.4) | 100 (294.6) | 100 (965.9) | 100 (970.0) |
Cells were treated with media with or without BCD and then were tested for binding the indicated MAbs as described in Materials and Methods. Data shown are percentages of positive cells (mean channel fluorescence in parentheses) with MAb IgG2b mouse myeloma used as a negative control. VIII6G10 and MT.M5 recognize VCAM-1 and ICAM-1, respectively.
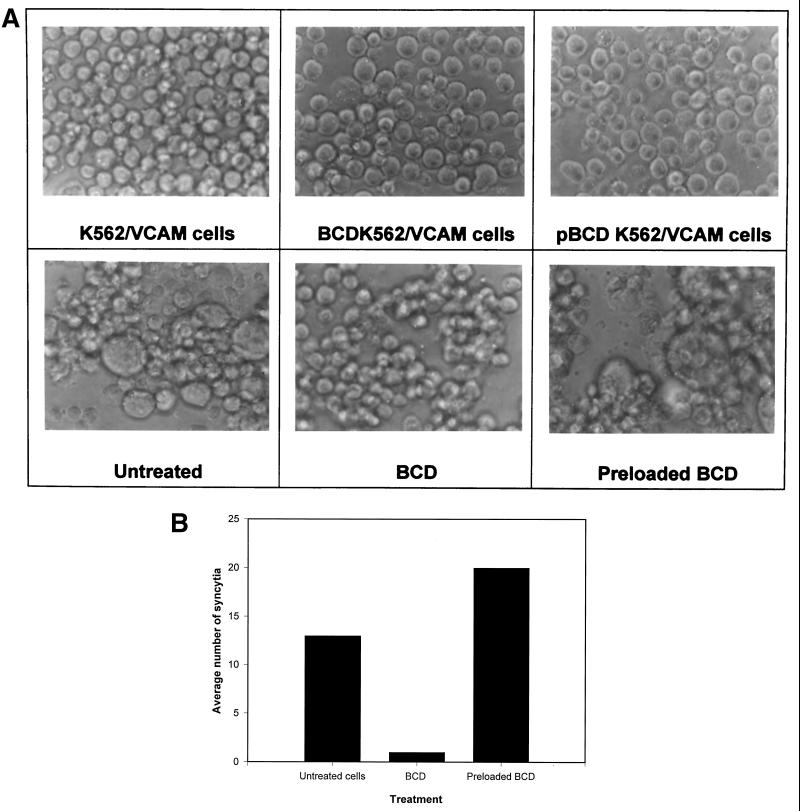
Preloading BCD with cholesterol prevents inhibition of fusion.
To be certain that the effect of the BCD on HTLV-1 syncytium formation was due to its depletion of cholesterol from cell membranes, we preloaded the BCD with cholesterol and incubated the cells in media containing the cholesterol-preloaded BCD or BCD alone before testing them in syncytium formation assays as described above. Syncytium formation was not blocked in cells treated with cholesterol-loaded BCD, confirming that depletion of cholesterol by BCD was required for inhibition of syncytium formation (Fig. 7).
FIG. 7.
BCD preloaded with cholesterol does not inhibit HTLV-1-induced syncytium formation. BCD was preloaded with cholesterol and used along with BCD alone to treat cells as described in Materials and Methods. Treated and untreated cells were then mixed, and syncytium formation was allowed to occur over 16 h. Photomicroscopy (A) and quantitation of syncytia (B) were then performed at 16 h as described previously. Preloaded BCD and pBCD refer to cholesterol-saturated BCD.
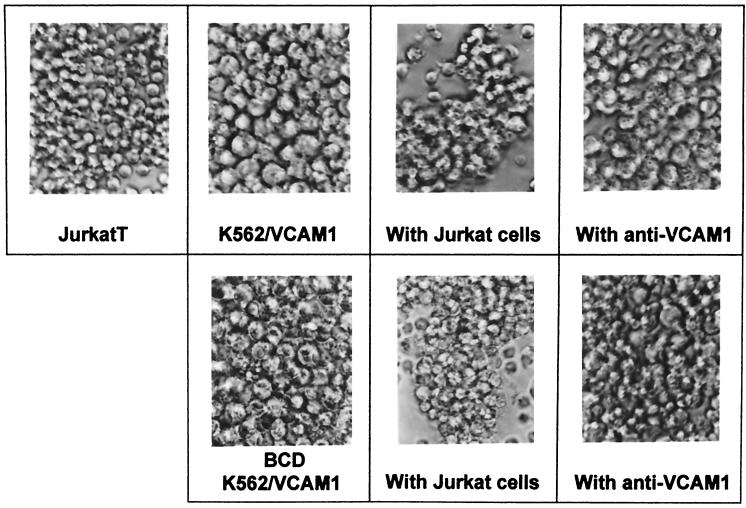
BCD treatment does not alter VCAM-1 binding.
Since BCD does not drastically alter VCAM-1 surface expression as seen by flow cytometry (Table 2), we examined whether the mode by which BCD inhibited syncytium formation was by preventing interactions between VCAM-1 and its major integrin receptor, VLA-4. K562/VCAM1 cells were pretreated with BCD as described above and incubated with Jurkat T cells, which express VLA-4. As is shown in Fig. 8, Jurkat T cells formed conjugates with K562/VCAM1 cells, regardless of pretreatment conditions, suggesting that the function of VCAM-1 was unchanged. In addition, the antibody to VCAM-1, VIII6G10, inhibited the clustering of the two cell types, showing that the clustering is VCAM-1 dependent and unaltered in the BCD-treated cells.
FIG. 8.
BCD does not alter VCAM-1 binding. K562/VCAM cells were pretreated with BCD, followed by preincubation with anti-VCAM-1 MAb VIII6G10 or media alone. They were then incubated with Jurkat T cells for 1 h at 37°C as described in Materials and Methods. Conjugate formation was visualized by photomicroscopy 1 h after mixing the cells. Top row, left to right: untreated Jurkat cells and untreated K562/VCAM1 cells alone, mixed with Jurkat cells, and mixed with Jurkat cells in the presence of anti-VCAM1 MAb (with anti-VCAM1). Bottom row, left to right: BCD-treated K562/VCAM1 cells alone, mixed with Jurkat cells (middle panel), and mixed with Jurkat cells in the presence of anti-VCAM1 MAb (right panel).
DISCUSSION
In a previous study, we showed that the adhesion molecule VCAM-1 could confer to resistant K562 cells sensitivity to HTLV-1 syncytium formation (21). Because not all VCAM-1-positive cells are sensitive to HTLV-1 fusion, we concluded that K562 cells must express a second molecule required for HTLV-1 infection. We have produced a panel of four new MAbs against K562 cells that block HTLV-1 syncytium formation. Characterization of these MAbs has revealed that they recognize four distinct molecules, three of which localize predominantly to lipid rafts and one of which is found at high levels in all cellular fractions, including the lipid rafts. Furthermore, dispersion of rafts by cholesterol depletion blocks HTLV-1 syncytium formation, strongly indicating that lipid rafts are critical for HTLV-1 infection.
It is unlikely that all four molecules recognized by the MAbs are true receptors for the virus. While it is possible that these four proteins may share the same polypeptide motif that interacts with gp46, this seems improbable since there has yet to be a virus identified whose binding site resides on a polypeptide sequence in four distinct proteins. It is possible that these molecules may share a common posttranslational modification, such as a carbohydrate moiety, that interacts with the virus. For example, the receptor for influenza viruses, sialic acid, can be borne by many distinct proteins (18, 35, 46). Alternatively, these four proteins recognized by the MAbs may associate with the true receptor and inhibition of syncytium formation by the MAbs could result from steric effects (19) or conformation changes in the associated receptor.
One characteristic these four molecules do have in common is their localization to lipid rafts. Lipid rafts are microdomains of the cell membrane rich in cholesterol, GPI-anchored proteins, and sphingolipids (24, 28, 41). Cholesterol appears to be required for their formation, and the resulting membrane domains are in a liquid-ordered phase, whereas the surrounding membrane regions take on a more fluid liquid-crystalline or gel phase state (6). Rafts have been shown to contain a multitude of signaling proteins, implying that they serve as sites of integration for multiple signaling pathways (41). In the formation of these membrane domains, certain integral membrane proteins with bulky cytoplasmic tails, such as the transmembrane phosphatase CD45 (34), are excluded. This could result in areas on the cytoplasmic membrane leaflet which facilitate interactions between cytoplasmic proteins and small membrane-bound signaling molecules. Since excluded molecules like CD45 are very tall and heavily negatively charged (2), lipid rafts may represent sites at which gp46, relatively small and compact, can gain easy access to its receptor.
In a previous report, we showed that MAbs to proteins highly expressed on the surface of HTLV-1-infected cells, such as MHC-2, could inhibit HTLV-1-induced syncytium formation while leaving HIV-1-induced syncytium formation unchanged (19). This suggested that the receptor that engages gp46 is, like gp46 itself, small and compact in relation to the proteins that surround it and cannot easily penetrate MAbs bound to proteins surrounding gp46. The levels of the proteins on K562/VCAM1 cells recognized by the K5.M MAbs do not approach those of antigens on MT2 cells bound by MAbs used in the previous study. MAb K5.M3 is the only one whose level of binding approaches that of antibodies that block HTLV-1 fusion by protein crowding, and that was true of its binding to MT2 cells but not to K562/VCAM1 cells. However, a similar phenomenon may explain the results obtained with the MAbs characterized in this report. It is possible that the receptor for HTLV-1 may reside in lipid rafts and that MAbs bound to other proteins in lipid rafts are able to mask the receptor by localized protein crowding in such a way that gp46 cannot bind to it and promote membrane fusion. This could happen even if the overall expression of the proteins recognized by the MAbs was modest or low.
The many reports of HTLV-1 infection of a variety of cell types strongly imply that the HTLV-1 receptor is widely expressed. Lipid rafts and their caveolin-containing counterparts, caveolae, are ubiquitous, being found on all cell types (24, 28). Localization of the HTLV-1 receptor to rafts may provide insights into many unanswered questions. Caveolae form invaginated flasks of plasma membrane with an opening of ∼70 nm. If the receptor is in this “flask,” it may be inaccessible to free virus particles, which have a diameter of approximately 100 nm. The ineffectiveness of HTLV-1 free virus infection would be explained by the “protected” nature of the receptor. However, an infected cell expressing gp46 along with various cell adhesion molecules, through cell-cell interactions and the appropriate signaling pathways, could induce changes in the membrane structure of the uninfected cell so as to undo the invaginated flask morphology of caveolae, making the receptor accessible to gp46 at the cell surface. In the case of T lymphocytes, which do not express caveolin or have caveolae, adhesion molecule mediated cell-cell interactions could result in coalescence of lipid rafts (48), thereby facilitating gp46-receptor binding. A “virological synapse” could result, in which gp46 has access to its receptor while the two membranes are held together by surrounding adhesion molecules (VLA-4, α4β7, LFA-1) binding to their ligands (VCAM-1, ICAM-1), in a process analogous to that shown to occur when T-cell receptors engage their peptide-MHC complexes in the formation of an “immunological synapse” between T cells and antigen-presenting cells (14). This model of sequestration of the HTLV-1 receptors to lipid rafts and/or caveolae could explain why HTLV-1 seems to be transmitted much more effectively in a cell-to-cell manner than does free virus.
Lipid rafts have been implicated in the infection pathways of other pathogens. We have shown that HIV-1 budding occurs primarily at lipid rafts (32). Evidence has also been published that influenza and measles viruses bud from lipid rafts (29, 38). Other examples are Semliki Forest virus and Sindbis virus, both of which seem to require cholesterol and/or sphingolipids for fusion and entry (33, 42). These viruses may therefore preferentially enter at lipid rafts and caveolae. In addition, Sendai virus and rotaviruses appear to use gangliosides as receptors (10, 36), and these structures are highly enriched in lipid rafts (17). We have previously demonstrated that lipid rafts may be required for HIV-1 entry as well (28a; J. E. K. Hildreth, D. Nguyen, Z. Liao, and R. Hampton, J. Assoc. Acad. Minority Phys., abstract, 10:106.a).
In this paper, we show for the first time that lipid rafts may play a role in HTLV-1-induced syncytium formation and infection. These specialized regions of the cell membrane may represent sites at which the HTLV-1 receptor is expressed, or they may provide the needed lipid composition for virus-induced membrane fusion. For the reasons given above, lipid rafts may provide viruses the most accessible sites for entry and, through as-yet-unidentified signaling mechanisms, allow viruses to prime cells for postentry replication steps. Further studies on the role of lipid rafts in HTLV-1 binding and entry may provide important new insights in the biology of this virus.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant no. AI31806 and NIH grant no. 2T32-GM07445.
REFERENCES
- 1.Arni S, Ilangumaran S, van Echten-Deckert G, Sandoff K, Poincelet M, Briol A, Rungger-Brandle E, Hoessli D C. Differential regulation of Src-family protein tyrosine kinases in GPI domains of T lymphocyte plasma membranes. Biochem Biophys Res Commun. 1996;225:801–807. doi: 10.1006/bbrc.1996.1254. [DOI] [PubMed] [Google Scholar]
- 2.Barclay A N, Beyers A D, Birkeland M L, Brown M H, Davis S J, Somoza C, Williams A F. The leukocyte antigen facts book. New York, N.Y: Academic Press; 1993. [Google Scholar]
- 3.Bessler M, Mason P J, Hillmen P, Miyata T, Yamada N, Takeda J, Luzzatto L, Kinoshita T. Paroxysmal nocturnal haemoglobinuria (PNH) is caused by somatic mutations in the PIG-A gene. EMBO J. 1994;13:110–117. doi: 10.1002/j.1460-2075.1994.tb06240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cann A J, Chen I S Y. Human T-cell leukemia virus types I and II. In: Fields B N, Knipe D M, Howley P N, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1849–1880. [Google Scholar]
- 5.Cereneus D P, Ueffing E, Posthuma G, Strous G J, van der Ende A. Detergent insolubility of alkaline phophatase during biosynthetic transport and endocytosis. Role of cholesterol. J Biol Chem. 1993;268:3155. [PubMed] [Google Scholar]
- 6.Cinek T, Horejsi V. The nature of large noncovalent complexes containing glycosyl-phosphatidylinositol-anchored membrane glycoproteins and protein tyrosine kinases. J Immunol. 1992;149:2262–2270. [PubMed] [Google Scholar]
- 7.Clarke M F, Gelmann E P, Reitz M S J. Homology of human T-cell leukaemia virus envelope gene with class I HLA gene. Nature. 1983;305:60–62. doi: 10.1038/305060a0. [DOI] [PubMed] [Google Scholar]
- 8.Collette Y H, Benziane A, Ramos-Morales F, Benarous R, Harris M, Olive D. Physical and functional interaction of Nef with Lck. J Biol Chem. 1996;271:6333–6341. doi: 10.1074/jbc.271.11.6333. [DOI] [PubMed] [Google Scholar]
- 9.Dodon M D, Bernard A, Gazzalo L. Peripheral T-lymphocyte activation by human T-cell leukemia virus type I interferes with the CD2 but not with the CD3/TCR pathway. J Virol. 1989;63:5413–5419. doi: 10.1128/jvi.63.12.5413-5419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epand R M, Nir S, Parolin M, Flanagan T D. The role of ganglioside GD1a as a receptor for Sendai virus. Biochemistry. 1995;34:1084–1089. doi: 10.1021/bi00003a045. [DOI] [PubMed] [Google Scholar]
- 11.Fukudome K, Furuse M, Imai T, Nishimura M, Takagi S, Himuna Y, Yoshie O. Identification of membrane antigen C33 recognized by monoclonal antibodies inhibitory to human T-cell leukemia virus type 1 (HTLV-1)-induced syncytium formation: altered glycosylation of C33 antigen in HTLV-1-positive cells. J Virol. 1992;66:1394–1401. doi: 10.1128/jvi.66.3.1394-1401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavalchin J, Fan N, Lane M J, Papsidero L, Poiesz B J. Identification of a putative cellular receptor for HTLV-I by a monoclonal antibody, Mab 34–23. Virology. 1993;194:1–9. doi: 10.1006/viro.1993.1228. [DOI] [PubMed] [Google Scholar]
- 13.Gavalchin J, Fan N, Waterbury P G, Corbett E, Faldasz B D, Peshick S M, Poiesz B J, Papsidero L, Lane M J. Regional localization of the putative cell surface receptor for HTLV-I to human chromosome 17q23.2–17q25.3. Virology. 1995;212:196–203. doi: 10.1006/viro.1995.1468. [DOI] [PubMed] [Google Scholar]
- 14.Grakoui A, Bromley S K, Sumen C, Davis M M, Shaw A S, Allen P M, Dustin M L. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 15.Guo M L, Hildreth J E K. HIV-induced loss of CD44 expression in monocytic cell lines. J Immunol. 1993;151:2225–2236. [PubMed] [Google Scholar]
- 16.Hanada K, Nishijima M, Akamatsu Y, Pagano R E. Both sphingolipids and cholesterol participate in the detergent insolubility of alkaline phosphatase, a glycosylphosphatidylinositol-anchored protein. J Biol Chem. 1995;270:6254–6260. doi: 10.1074/jbc.270.11.6254. [DOI] [PubMed] [Google Scholar]
- 17.Harder T, Scheiffle P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrier G, Klenk H D. The surface receptor is a major determinant of the cell tropism of influenza C virus. Virology. 1987;159:102–108. doi: 10.1016/0042-6822(87)90352-7. [DOI] [PubMed] [Google Scholar]
- 19.Hildreth J E K. Syncytium-inhibiting monoclonal antibodies produced against human T-cell lymphotropic virus type I-infected cells recognize class II major histocompatibility complex molecules and block by protein crowding. J Virol. 1998;72:9544–9552. doi: 10.1128/jvi.72.12.9544-9552.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hildreth J E K, August J T. The human lymphocyte function-associated (HFLA) antigen and a related macrophage differentiation antigen (Hmac-1): functional effects of subunit-specific monoclonal antibodies. J Immunol. 1985;134:3272–3280. [PubMed] [Google Scholar]
- 21.Hildreth J E K, Holt V, August J T, Pescovitz M D. Monoclonal antibodies against porcine LFA-1: species cross-reactivity and functional affects of β subunit-specific antibodies. Mol Immun. 1989;26:883–895. doi: 10.1016/0161-5890(89)90145-4. [DOI] [PubMed] [Google Scholar]
- 22.Hildreth J E K, Hyman J A. Production and characterization of monoclonal anti-CD18 anti-iodotype antibodies. Mol Immun. 1989;26:1155–1167. doi: 10.1016/0161-5890(89)90060-6. [DOI] [PubMed] [Google Scholar]
- 23.Hildreth J E K, Subramanium A, Hampton R A. Human T-cell lymphotropic virus type 1 (HTLV-1)-induced syncytium formation mediated by vascular cell adhesion molecule-1 (VCAM-1): evidence for involvement of cell adhesion molecules in HTLV-1 biology. J Virol. 1997;71:1173–1180. doi: 10.1128/jvi.71.2.1173-1180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper N M. Detergent-insoluble glycosphingolipid/cholesterol-rich membrane domains, lipid rafts and caveolae. Mol Membr Biol. 1999;16:145–156. doi: 10.1080/096876899294607. [DOI] [PubMed] [Google Scholar]
- 25.Ilangumaran S, Hoessli D C. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem J. 1998;335:433–440. doi: 10.1042/bj3350433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein U, Gimpl G, Fahrenholz F. Alteration of the myometrial plasma membrane cholesterol content with beta-cyclodextrin modulates the binding affinity of the oxytocin receptor. Biochemistry. 1995;34:13784–13793. doi: 10.1021/bi00042a009. [DOI] [PubMed] [Google Scholar]
- 27.Kohtz D S, Altman A, Kohtz J D, Puszkin S. Immunological and structural homology between human T-cell leukemia virus type I envelope glycoprotein and a region of human interleukin-2 implicated in binding the beta receptor. J Virol. 1988;62:659–662. doi: 10.1128/jvi.62.2.659-662.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurzchalia T V, Parton R G. Membrane microdomains and caveolae. Curr Opin Cell Biol. 1999;11:424–431. doi: 10.1016/s0955-0674(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 28a.Liao Z, Cimakasky L M, Hampton R, Nguyen D H, Hildreth J E K. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res Hum Retrovir. 2001;17:1009–1017. doi: 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]
- 29.Manie S N, Debreyne S, Vincent S, Gerlier D. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J Virol. 2000;74:305–311. doi: 10.1128/jvi.74.1.305-311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markham P D, Salahuddin S Z, Kalyanaraman V S, Popovic M, Sarin P, Gallo R C. Infection and transformation of fresh human umbilical cord blood cells by multiple sources of human T-cell leukemia-lymphoma virus (HTLV) Int J Cancer. 1983;31:413–420. doi: 10.1002/ijc.2910310404. [DOI] [PubMed] [Google Scholar]
- 31.Nagy K, Weiss R A, Clapham P, Cheinsong-Popov R. Biological properties of human T-cell leukemia virus envelope antigens. In: Gallo R C, Essex M, Guo M L, editors. Human T-cell leukemia viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 121–131. [Google Scholar]
- 32.Nguyen D H, Hildreth J E K. Evidence for selective budding of HIV-1 from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J Cell Biol. 1991;112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodgers W, Rose J K. Exclusion of CD45 inhibits activity of p56lck associated with glycolipid-enriched membrane domains. J Cell Biol. 1996;135:1515–1523. doi: 10.1083/jcb.135.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers G N, Paulson J C, Daniels R S, Skehel J J, Wilson I A, Wiley D C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 36.Rolsma M D, Kuhlenschmidt T B, Gelberg H B, Kuhlenschmidt M S. Structure and function of a ganglioside receptor for porcine rotavirus. J Virol. 1999;72:9079–9091. doi: 10.1128/jvi.72.11.9079-9091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sagara Y, Ishida C, Inoue Y, Shiraki H, Maeda Y. 71-Kilodalton heat shock protein acts as a cellular receptor for syncytium formation induced by human T-cell lymphotropic virus type 1. J Virol. 1998;72:535–541. doi: 10.1128/jvi.72.1.535-541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheiffle P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Virol. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 39.Scheiffle P, Roth M G, Simons K. Interaction of influenza virus hemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domains. EMBO J. 1997;16:5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroeder R, Sharmin J, Ahmed N, Zhu Y, London E, Brown D A. Cholesterol and sphingolipids enhance the Triton X-100 insolubility of GPI-anchored proteins by the formation of detergent-insoluble ordered membrane domains. J Biol Chem. 1998;273:1150–1157. doi: 10.1074/jbc.273.2.1150. [DOI] [PubMed] [Google Scholar]
- 41.Simons K, Ikonen E. Functional rafts in membranes. Nature. 1997;387:569–570. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 42.Smit J A, Bittman R, Wilschut J. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J Virol. 1999;73:8476–8484. doi: 10.1128/jvi.73.10.8476-8484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommerfelt M A, Williams B P, Clapham P R, Soloman E, Goodfellow P N, Weiss R A. Human T cell leukemia viruses use a receptor determined by human chromosome 17. Science. 1988;242:1557–1559. doi: 10.1126/science.3201246. [DOI] [PubMed] [Google Scholar]
- 44.Sutton R E, Littman D R. Broad host range of human T-cell leukemia virus type I demonstrated with improved pseudotyping system. J Virol. 1996;70:7322–7326. doi: 10.1128/jvi.70.10.7322-7326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchiyama T. Human T cell leukemia virus type I (HTLV-I) and human diseases. Annu Rev Immunol. 1997;15:15–37. doi: 10.1146/annurev.immunol.15.1.15. [DOI] [PubMed] [Google Scholar]
- 46.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 47.Weiss R A, Clapham P, Nagy K, Hoshino H. Envelope properties of human T-cell leukemia viruses. Curr Top Microbiol Immunol. 1985;115:235–246. doi: 10.1007/978-3-642-70113-9_15. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Trible R P, Samelson L E. LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 1998;9:239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]