Abstract
Background
Breast carcinoma has become the leading fatal disease among women. The location of prohibitin in the chromosome is close to the breast cancer susceptibility gene 1 (BRCA1). Accumulated research reported that prohibitin could interact with a variety of transcription factors and cell cycle-regulating proteins.
Objective
This present study aims to comprehensively explore and reveal the biological functions of prohibitin on breast cancer via The Cancer Genome Atlas (TCGA) and validation experiment in vitro.
Methods
Exploring the expression level of prohibitin across 27 tumors based on the TGGA database by bioinformatic methods and its relationship with tumor immune infiltration. Furthermore, we thus analyzed the biological roles of prohibitin on human breast cancer cell line MCF-7 with pEGFP-prohibitin overexpression plasmid by western blotting and transwell-assay.
Results
Firstly, we found prohibitin is overexpressed in most tumors based on The Cancer Genome Atlas database, and the negative relationships between prohibitin and tumors infiltrating lymphocytes including B lymphocyte, CD4 T lymphocyte, CD8 T lymphocyte, Neutrophil, Macrophage and Dendritic, and its significant correlation with the prognosis of human cancer. In vitro, expression not only inhibited cell viability and invasive abilities but also increased the apoptosis percentage of cells with a decreased percentage of the S phase and an increased G2 phase. The reduction of Bcl-2 was observed when prohibitin was upregulated, although the expression of E2F-1 did not change.
Conclusion
Although prohibitin is over-expressed in various cancer types, it functions as an important tumor suppressor that may suppress breast cancer cell proliferation and the invasive ability of MCF-7 by influencing its DNA synthesis and promoting cell apoptosis. All these may be likely associated with P53, erbB-2, and Bcl-2.
Keywords: Breast cancer, prohibitin, bioinformatics, proliferation, invasion, pan-cancer analysis, TCGA
1. INTRODUCTION
Breast carcinoma has become the leading fatal disease and health problem among women [1-7]. Although many measures and scientific technologies have been applied to screening, diagnosis, therapy, and survival for breast cancer [8-11], they remain unavailable and insufficient for women due to the financial burden over the world [2, 11-13]. Breast cancer has a well-known complicated etiology and genetic characteristics [7, 14], which leads to classification diversity, clinical manifestations complexity, therapeutic disparities, and therapeutic specificity [15]. The morbidity and mortality rates were different among racial and ethnic groups [3]. For example, white women had the highest breast cancer incidence rates among women aged 40 years and older, but African American women gently increased with the poorest breast cancer survival of any racial/ethnic group [2] from 2006 to 2010. Lacking awareness of early detection, effective individualized therapy, or adequate financial support, the control of breast cancer is not still satisfied [16-18]. Fortunately, with timely screening and intervention of early breast cancer [6, 20], clinical use of tumor biomarkers [19-21], and continuous study of molecular mechanisms [22], the mortality rate of breast cancer has decreased gently [6]. Personalized treatment has gradually penetrated the consciousness of clinical researchers, which has brought good news to the survival rate and quality of life for breast cancer patients [3, 6, 23], especially in elderly patients. Various guidelines also suggest that precision treatment and improving quality of life should be given priority [6, 22]. In China, morbidity rates of breast cancer were becoming younger year by year [24] which same in other region of the world such as America [25]. Young women are more likely to develop more aggressive subtypes of breast cancer and have a unique biological feature, its survival rates in young women remain lower than those in older women which refers to more likely psychosocial concerns [25-28]. Therefore, it is urgently necessary to improve the diagnosis and treatments for different age stages, Complex molecular mechanisms, and highly specific biomarkers are eagerly called for individualized treatment of breast cancer. Bioinformatics analysis of high-throughput multiomics data including TCGA would be helpful to comprehensively identify the characteristics and roles of tumor-related genes, to explore their important biological roles in the progression of breast cancer [29].
Prohibitin (PHB/phb) is a highly conserved nuclear matrix protein with multiple functions, including proliferation, apoptosis, transcription, posttranslational modification, signal transduction, and energy metabolism [30-32], and it consists of two subtypes phb1 and phb2 in the mitochondria which acting as a molecular chaperone to maintain mitochondrial homeostasis [31, 32]. It is found to be associated with breast cancer [33, 34] in addition to liver cancer [35, 36], lung cancer, cervical cancer, and ovarian cancer [37] extensively. PHB is located extensively in the cell, such as the cytomembrane, cell unclear and mitochondria inner membrane, etc., and the location decides its function [30]. What’s more, PHB can shuttle between the nucleus and organelles under several stimulates [38, 39]. Thus, PHB may play diverse functions depending on transport. In general, nucleus PHB acts as a transcriptional regulation factor, membrane PHB plays signal transduction, and cytoplasm PHB participates in metabolism to keep a steady state [30]. Accumulated research reported PHB could participate in kinds of signal paths and interact with a variety of transcription factors and cell cycle regulating proteins [32, 40, 41]. However, the explicit mechanism has not been illuminated. The location of PHB in the chromosome (17q21) is close to breast cancer susceptibility gene 1 (BRCA1) which links it to breast cancer easily [35]. There is evidence indicating the mutations of BRCA1 were the key issue for breast cancer through bioinformatic methods [29, 42]. Many scholars have focused on signaling pathways and molecular mechanisms between PHB and breast cancer [43-46]. It is interesting that the mechanism of PHB, the same with other cancers, was not entirely consistent in breast cancer, more efforts are looking forward to elucidating it urgently. Our earlier study found an increased expression of PHB during the progression of normal breast tissue, hyperplasia tissue, atypical hyperplasia tissue, and breast cancer tissue [47], coincident with parts of research. Therefore, it has reason to believe PHB could serve as an essential factor in tumor development and a promising therapeutic target, including breast cancer [41, 43, 48]. However, the roles of PHB in cancer progression remain some controversial reports in the literature research. The present study aims to provide comprehensive and novel insights into the biological functions of PHB in the development, treatment, and prognosis of human pan-cancer, especially breast cancer, exploring the possible underlying molecular mechanisms associated with PHB in breast cancer.
2. MATERIALS AND METHODS
2.1. Bioinformatic Analysis of PHB Gene Expression and Functions
We analyzed the mRNA expression level of PHB in different tumor types based on the TGGA Database using an R package combined with standardized TPM expression data. Secondly, we performed a Protein-Protein Interaction (PPI) network in the STRING database (https://string-db.org/), a common online approach known to predict protein-protein interactions, to identify PHB-binding proteins and understand the possible underlying molecular mechanisms.
Furthermore, TIMER Database (cistrome.shinyapps.io/t) was used to perform correlation analysis between the expression levels of the PHB gene and tumor purity, as well as levels of tumor-infiltrating immune cells, including B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells. Meanwhile, we downloaded the unified and standardized pan-cancer dataset: TCGA TARGET GTEx (PANCAN, N=19131, G=60499), extracted the expression level of PHB gene in each sample and extracted the gene expression profile of each tumor, and then calculated the StromalScore, ImmuneScore, and ESTIMATEScore of each tumor using R software package ESTIMATE algorithm. The correlation coefficient of genes and immune infiltration scores in each tumor was obtained by the Spearman test.
2.2. Survival Prognosis Analysis
PHB was evaluated for their prognostic value in breast cancer and other tumors using Kaplan–Meier Plotter (http://kmplot.com/analysis/), an online resource for survival analysis. Subsequently, the Kaplan–Meier method was used to analyze the overall survival (OS) of breast cancer patients, Kidney renal clear cell carcinoma, and so on based on the classification of patients into high and low groups according to their mRNA expression level. It was considered statistically significant using the criterion of log-rank P < 0.05.
2.3. Transient Transfection of MCF-7 and MTT Assay for Cell Viability
Human breast cancer cell MCF-7 was kindly provided by the Obstetrics and Gynecology Laboratory, First Affiliated Hospital of Xi’an Jiaotong University. It was cultured in Dulbecco's modified Eagle (DMEM) high glucose medium (HyClone, Logan, USA) containing 10% fetal bovine serum (Beyotime, Wuhan, China), 0.1% penicillin, and 0.1% streptomycin at 37℃ in 5% CO2 in the study. Enhance green fluorescence protein (pEGFP) (GenerayBiotech, Shanghai, China) was the vector of the recombinant plasmid. Transfection efficiency was confirmed by western blotting and real-time polymerase chain reaction (RT-PCR). LipofectamineTM2000 reagent (Invitrogen, Carlsbad, USA) acted as the overexpressed group (OE group), transfection with an empty pEGFP vector was the negative control (NC group), only MCF-7 cells group was the mock group (MOCK group). Primers for PHB (Ref Seq: NM_003634) were designed using primer 3.0 and synthesized by Sangon Biotechnology Company (Shanghai, China). The primer sequence is shown as follows: Forward 5’-GGCTGAGCAACAGAAAAAGG-3’, and Reverse3’-TTGTAGTGGATGGACGGTCG-5’. The total ribose nucleic acid (RNA) of each group (OE, NC, and MOCK group) was extracted with Trizol reagent (Invitrogen, Waltham, USA) and quantified with Nanodrop2000 (Thermo Fisher Scientific, Waltham, USA). SYBR Green (Takara, Dalian, China) was utilized to mark real-time polymerase chain reaction (RT-PCR). Genomic deoxyribonucleic acid (DNA) was eliminated with a gDNA eraser kit reaction before conducting RT-PCR on a PCR Detection System (Thermo Fisher Scientific, Waltham, USA) with program firstly 95℃ for 30 seconds, secondly 40 cycles at 95℃ for 5 seconds and 55℃ for 30 seconds, thirdly 95℃ for 15 seconds and 60℃ for 30 seconds, at last 95℃ for 15 seconds. Fold change difference in messenger ribose nucleic acid (mRNA) expression of PHB was calculated using the 2-△△Ct method which β-actin acted as an internal control.
Cell viability was detected with 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-Tetrazoliumbromide (MTT). Cells were cultivated in triplicates of 96-well plates and transfected with 0.2 ug plasmid per well for the over-expressed group and negative control. After transfected for 24 hours, 48 hours, and 72 hours, respectively, the culture medium was gently removed and 20 ul 0.01 mol/L (5 mg/l) MTT was added to stain live cells for 4 hours at 37ºC in 5% CO2, the same with the MOCK group. Then, 100 ul dimethyl sulfoxide (DMSO) was added to each well and absorbance was measured by a full wavelength enzyme marker (TECAN, MäNeeded, Switzerland) at 490 nanometers. Cell growth inhibition rate was estimated with the equation 1- average OD of experiment group/OD of control.
2.4. Transwell Matrigel Invasion Assay
Firstly, matrigel (Sigma, San Francisco, USA) was previously diluted with DMEM medium (1:8) and 100ul suspension was added into the transwell chamber after 24 hours of transfection in different groups. After 48 hours of transfection, cells were collected, and the cell density was adjusted to 25×104 cells/ml. Then, two hundred microliters of the cell solution were lightly dropped into each chamber and further incubated for 16 hours. Alcohol fixation and crystal violet staining were carried out finally. Cells penetrating through matrigel membranes of 10 fields were counted in each group randomly.
2.5. Cell Cycle and Apoptosis Analysis by Flow Cytometry
After transfection for 48 hours, cells were collected by trypsinization (no EDTA) and fixed with pre-cooled 75% ethanol overnight at 4℃, the same with the negative group and mock group. PropidiumIodide (PI) (Thermo Fisher Scientific, Waltham, USA) was added to stain the DNA for 30 minutes at room temperature in the dark. Flow Cytometer (Becton Dickinson, Franklin Lake, America) was used to detect DNA-PI fluorescence intensity at 530 and 488 nanometers, respectively, and Mod Fit LT was used to analyze the cell cycle.
We examined the apoptosis percentage of MCF-7 using flow cytometry with Annexin V-fluorescein isothiocyanate (FITC)/PI detection kit (KeyGENBioTECH, Nanjing, China). Cells were cultured in 6-well plates and transfected at 80% confluences. After 48 hours, cells were stained with Annexin V and analyzed by flow cytometry (cell Quest V.4.0) at 570 nanometers. The apoptosis rate was calculated with ModFit LT software.
2.6. Protein-protein Interactions Prediction and Checking for Tumor-Associated Proteins
STRING website, an online approach known to predict protein-protein interactions (PPI), showed PHB, P53, and E2F1 exhibited potential interactions. For western blotting, the total protein of each group was extracted from transfected cells after incubation for 72 hours with Radio-Immunoprecipitation Assay Buffer (RIPA) (HEART, Xi’an, China). All the lysates were added to a 5×loading buffer and stored at minus 80 ºC. Protein concentration was measured by BCA kit. Twenty micrograms of protein samples were isolated by Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE). The proteins were transferred onto polyvinylidene difluoride (PVDF) membrane and blocked in Tris-buffered saline and Tween-20 with 5% nonfat dry milk at room temperature for 2 hours. The membranes were incubated with primary antibodies (all primary antibodies: anti-prohibitin, anti-E2F1, anti-Bcl2, anti-βactin, and anti-erbB2 antibody from Abcam, Cambridge, UK) at 4 ℃ overnight. The membranes were washed with Tris-buffered saline (TBS) 3 times and incubated with HRP-linked secondary antibodies (Pierce Biotechnology, Rockford, USA) for 2 hours at room temperature. Meanwhile, the tumor-associated proteins, including P53, Bcl-2, E2F-1, and erbB-2 were detected. The signal was captured with Chemic Doc-ItTM UVP Imaging System Quantity One image processing software.
2.7. Statistical Analysis
SPSS Statistics (version 19.0 for Windows, IBM, Armonk, USA) was used for statistical analysis. Data are shown as mean ± standard deviation of 3 independent experiments. Comparisons among three groups were analyzed using one-way ANOVA and analysis of two groups was performed using unpaired two-tailed Student’s t-test. Graphs were generated by GraphPad Prism 5.0 software. A p value of < 0.05 was considered statistically significant.
3. RESULTS & DISCUSSION
3.1. Analysis of PHB Expression Level and Its Correlation with Tumor Immune Microenvironment
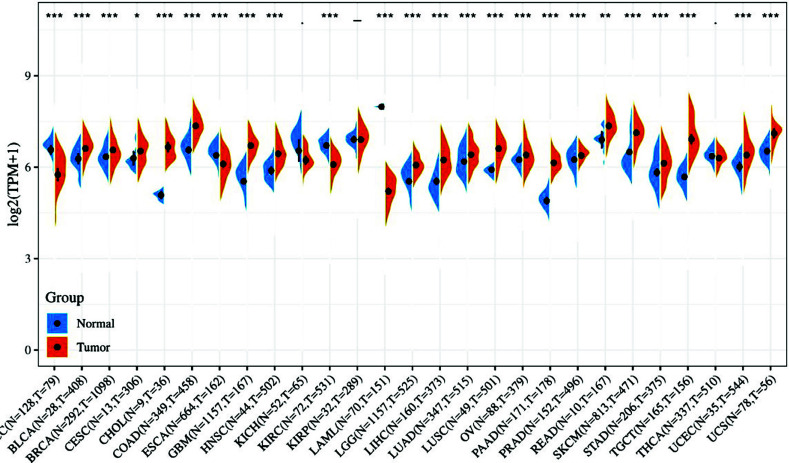
The pan-cancer analysis showed that PHB was over-expressed in most tumor types relative to normal tissues or paracological tissue based on the TCGA database, including in breast cancer (BRCA), as shown in Fig. (1) (p < 0.05). However, only three cancer types showed the opposite level of downregulation, including adrenal carcinoma (ACC), kidney renal clear cell carcinoma (KIRC), and acute myeloid leukemia (LAML). This result was consistent with most studies and our earlier research in breast cancer [47].
Fig. (1).
Expression of PHB in a multitude of tumors. The differences in expression levels of PHB mRNA in 27 kinds of tumors and normal tissues from TCGA and GTEx database.
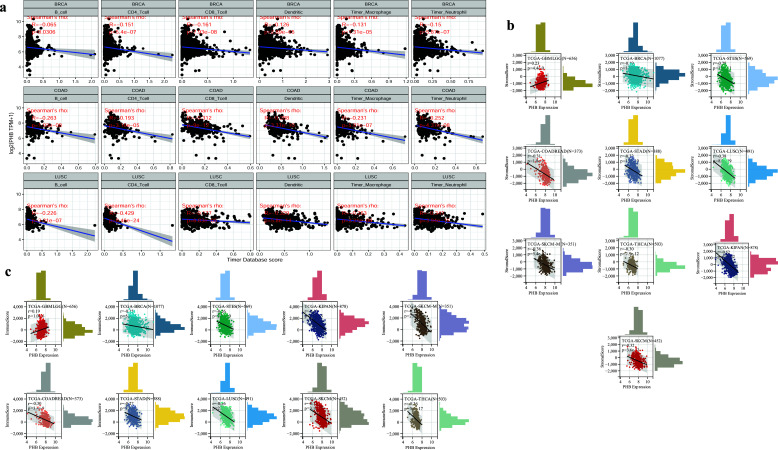
Meanwhile, the negative correlation between tumor-infiltrating immune cells and PHB expression was evident (Fig. 2a) in most tumor types using the Timer database, and breast cancer was located top 3 behind Chronic Obstructive Air Way Disease (COAD) and Lung Squamous cell Carcinoma (LUSC) as showed in Fig. (2a). Pan-cancer analysis including 33 kinds of tumors revealed the significantly negative correlations between PHB expression and ImmuneScore and StromalScore in most tumors (Fig. 2b and 2c), but among them, only the positive correlations in a few tumors including glioma (N = 656, R = 0.19, P= 1.0e-6 with ImmuneScore; R = 0.23, P = 4.4e-9 with StromalScore), uveal Melanoma (N = 79, R = 0.26, P = 0.02 with ImmuneScore; R = 0.24, P = 0.03 with StromalScore), acute Lymphoblastic Leukemia (N= 86, R = 0.23, P = 0.03 with ImmuneScore) (Fig. 3).
Fig. (2).
Correlation between expression of PHB and tumors immune infiltration. (a) Most tumor's immune infiltrating lymphocytes including B lymphocyte, CD4 T lymphocyte, CD8 T lymphocyte, Neutrophil, Macrophage, and Dendritic were all negatively correlated with expression of PHB. Breast cancer is one of the top 3 behind Chronic Obstructive Air Way Disease (COAD) and Lung Squamous Cell Carcinoma (LUSC) among 33 kinds of tumor samples. (b) Correlation between expression of PHB and Stromal score. (c) Correlation between expression of PHB and immune. The negative correlation between the expression of PHB and ImmuneScore (R= -0.116, p < 0.05) and StromalScore (R= -0.195, p < 0.05) of 33 kinds of tumor samples in the Timer database was obvious.
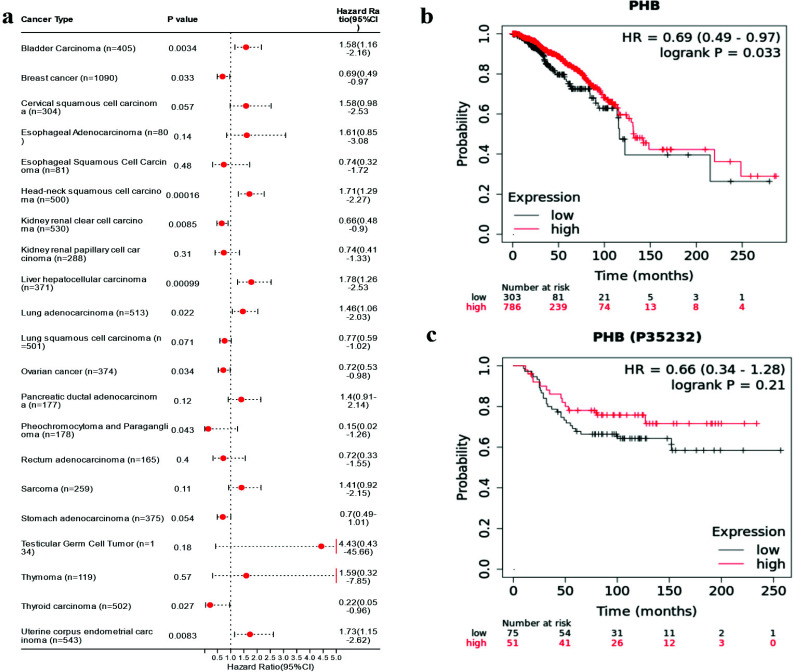
Fig. (3).
Survival prognosis analysis and Prognostic significance of PHB for pan-cancer. (a) Pan-cancer analysis revealed the prognosis correlation between the PHB gene and cancer patient OS. (b) The prognosis correlation between PHB gene and breast cancer. (c) The prognosis correlation between PHB protein and breast cancer.
3.2. Survival Prognosis Analysis and Prognostic Significance of PHB for Pan-cancer
We observed PHB gene was not only significantly over-expressed in most tumors compared with normal tissue, but also showed significant prognostic value by pan-cancer analysis. We noted that PHB mRNA should have a significant protective effect on the prognosis of breast cancer but with the lack of significant evidence that its protein form showed prognostic value on breast cancer. Therefore, we would further explore the biological effect of PHB on breast cancer cells through the overexpression plasmid recombinant plasmid pEGFP-PHB.
3.3. Efficiency of pEGFP-PHB Plasmid Transfection, Cell Proliferation, and Invasive Ability of PHB
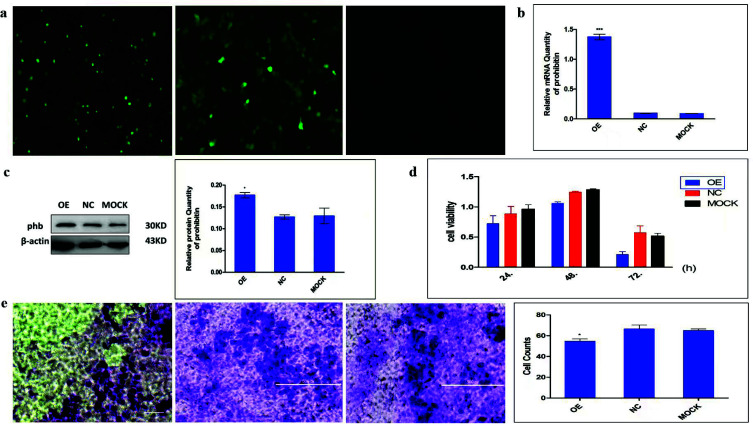
Plasmid pEGFP-PHB was transiently transfected into MCF-7 breast cancer cells successfully and the transfection efficiency was measured by fluorescence microscopy, RT-PCR, and western blotting. As shown in Fig. (4), the fluorescence intensity of overexpressed-PHB cells and negative control was significantly higher than the MOCK control group, which indicates the model of over-expressed PHB was successfully constructed. Expressions of PHB at mRNA and protein levels were predominantly higher in the transfection group compared with the other two groups (Fig. 4b and 4c). Collectively, these results demonstrated that pEGFP-PHB was successfully transfected into the MCF-7 breast cancer cell, as well as increasing PHB expression.
Fig. (4).
Transient Transfection of MCF-7, MTT Assay for Cell viability, and Invasion Assay of PHB. (a) Green fluorescence protein intensity among groups (OE, NC, and MOCK group ordinally). Cells were imaged with a fluorescence microscope (200×). Relative mRNA expression of PHB. GAPDH was used as an internal reference gene. (b) Representative western blot image of PHB and β-actin gene and densitometric analysis. (c) Cell viability was measured by MTT assay at 24, 48, or 72 hours following transfection of MCF-7 with pEGFP-PHB plasmid. (d) Representative transmembrane cell image of three groups after staining with crystal violet, and its statistical analysis of the total number of transmembrane cells in the three groups from 10 random fields. (e) the invasive ability of MCF-7 cells was measured by Transwell Ma trigel assay. *p < 0.001 or p < 0.05 versus NC control; Abbreviations: GAPDH=Glyceraldehyde 3-phosphate Dehydrogenase; OE=Overexpressed Group; NC=Negative Control Group.
To investigate the effect of PHB in tumor growth in vitro, an MTT assay was used to analyze cell viability at first. As shown in Fig. (4), all three groups showed a decreased proliferation in a time-dependent manner, and the overexpressed group showed more suppressive compared with the control cells (p < 0.05). The inhibition rates of proliferation were 20.98%, 14.93%, and 62.94% at 24, 48 and 72 hours, respectively in Fig. (4d). It was interesting that the inhibition rate was the most significant after 72 hours of transfection. Next, the effect of PHB on the invasive ability of MCF-7 cells was measured by transwell matrigel assay. As illustrated in Fig. (4e), the number of cells crossing the matrigel transwell membrane in the over-expressed group was 54.5 ± 7.4 compared with the negative control group (66 ± 12.6) and mock group (64 ± 7.4). These data suggested that PHB could decrease cell proliferation and invasive abilities in vitro.
3.4. PHB Influences Cell Cycle and Promotes Apoptosis Analysis
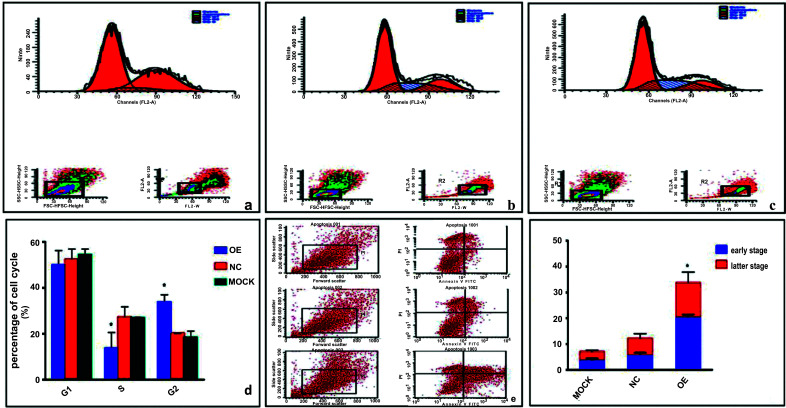
Cell proliferation and apoptosis in MCF-7 cells were tested by flow cytometry. The results showed that the cell percentage of the S phase in the overexpressed group, negative control, and mock group was 14.30 ± 6.97, 27.07 ± 4.44, and 26.54 ± 0.87, respectively. The cell percentages of the G2 phase were 34.76 ± 3.51, 21.57 ± 2.32, and 19.04 ± 2.83 in the over-expressed group, negative control, and mock group, respectively. The percentage of cells in the S phase was decreased, while those in the G2 phase were increased compared to the control groups in Fig. (5a-5d). No significant difference was observed in the G1 phase among the three groups (p > 0.05). The results indicated that PHB changed the cell cycle distribution, arrested DNA synthesis in the S phase, and induced cell cycle S/G2 arrest. Thereby inhibiting cell proliferation in ER-positive breast cancer cells MCF-7. Moreover, cellular apoptosis was higher in the over-expressed group vs. controls both in the early and late stages of apoptosis in Fig. (5e-5f). The apoptosis rate in the over-expressed group was 33.67 ± 8.68% compared with the negative control group (12.13 ± 3.76%) and the mock group (6.99 ± 2.33%) (p < 0.05). Collectively, these data further suggested that PHB inhibited breast cancer cell proliferation and promoted cell apoptosis.
Fig. (5).
Effect of PHB on proliferation and apoptosis in MCF-7 cells. (a-d) The distribution of cell cycle among the three groups by flow cytometry. Cells were collected and washed with phosphate-buffered saline fixed with alcohol, and stained with DNA with propidiumiodide. (e and f) Effect of PHB on apoptosis. After transfection with the plasmid for 48 h, cells were collected and stained with annexin. Propidium Iodide and fluorescein isothiocyanate (FITC) were used to stain phosphatidylserine in the early apoptosis stage and DNA in the later apoptosis, respectively. Analysis of variance was used for comparison. OE = overexpressed group; NC = negative control group, *p < 0.05 vs. controls.
3.5. PHB Influences Expression of Multiple Tumors Associated Proteins
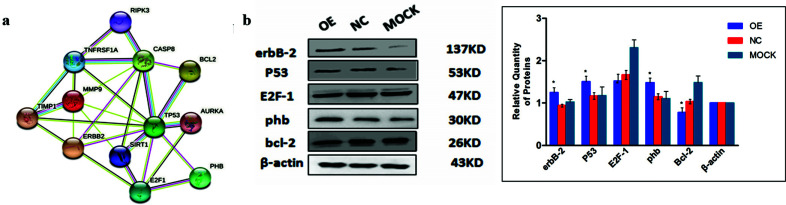
We explored related molecular mechanisms of PHB regulating cell growth and apoptosis as the results showed above that PHB inhibits the MCF-7 cell growth. STRING website, an online approach known to predict protein-protein interactions (PPI), showed PHB, P53, and E2F1 exhibited the potential interactions (Fig. 6a). Several critical indicators, including erbB-2, tumor gene P53, transcription factor E2F-1, and cell cycle protein Bcl-2, were measured by western blotting. As shown in Fig. (6b), erbB-2, which is closely related to breast cancer diagnosis and treatment, and P53, a tumor suppressor protein, were increased when PHB overexpressed. In contrast, PHB decreased the expression level of cell cycle protein Bcl-2, an anti-apoptotic protein. However, the transcription factor E2F-1 remained unchanged. These results suggested that PHB modulates tumor cell growth and apoptosis by regulating the expression of proteins that are associated with tumor development (Fig. 6c).
Fig. (6).
Effect of PHB on tumor-associated proteins. (a) PHB-binding proteins and PPI network. (b) Representative western blot image. KD = kilodalton; OE = over expressed group; NC= negative control, β-actin was used as a loading control. (c) Statistic analysis of tumor correlative protein levels. OE = overexpressed group; NC = negative control group, *p < 0.05 vs. negative control.
CONCLUSION
The morbidity and mortality rates of breast carcinoma rank high for women all over the world [26, 49]. It is not linked only to its unique biological characteristics, but also to close living environment [10, 50]. It is exciting that current scientific screening techniques and molecular diagnosis research have greatly reduced the mortality rate of early breast cancer [51, 52]. In Combination with the heterogeneity and specificity of breast cancer, non-responsiveness or resistance to hormone-related drugs, and complexity of molecular mechanisms, comprehensive consideration in the process of the screening, diagnosis, and treatment of breast cancer is needed, such as age, hormone level, drug resistance, races, and economic level [14, 28, 49, 53-56]. More, current treatments for breast cancer are expected to extend 5-year survival and improve life quality based on the control tumor development [6]. Biomarkers and precision therapy the promising prospects for breast cancer patients [57, 58].
PHB was one of the potential genes in tumorigenesis for breast cancer [59]. Many studies have shown that it is highly expressed in breast cancer, the same with our early study [47]. It is a highly homologous protein in a variety of cancer cells linked to cell proliferation, apoptosis [30], single transduction, transcription, energy metabolism, and immune regulation [58]. What’s more, PHB was reported to express differently under the inflammatory stimulation of cancers and diseases [41, 59-66]. PHB was shown to be linked to kinds of signal paths such as Ras-Raf-MEK-ERK [23], P53 signal pathway [38], and Rb-E2Fs signal pathway [64], and so on [65, 66]. When it comes to PHB in breast cancer, mRNA encoded by 3 '-UTR of PHB was directly microinjected into the nucleus of MCF-7 breast cancer, and the cell cycle was blocked in the G1/S phase [67]. PHB at a physiological dose can not only interact with P53 and enhance its ability to bind to DNA promoters, inhibiting cell proliferation subsequent, but its overexpression also increases the transcriptional activity regulated by P53 and inhibits the transcriptional activity regulated by E2F1 [38]. PHB combines with Rb and E2Fs in the nucleus to form a complex triplet, which prevents the binding of E2Fs to the DNA promoter, thus inhibiting the transcriptional activity of E2F and preventing the cell from entering the S phase from G1 [64]. All these indicated that breast cancer could be linked with PHB through multipaths [68]. The correlation of PHB expression and 33 kinds of tumor samples was negative, with the top 3 based on bioinformatic analysis in this study. PHB was expected as a biomarker of the diagnosis, therapy, and prognosis of diseases, especially for breast cancer [5, 45, 54, 69, 70].
To further explore the function of PHB in breast cancer tumorigenesis following a previous study, we first explored bioinformation roles based on the Cancer Genome Atlas (TGGA) Database and conducted biological functions in vitro. Significant upregulation of PHB could be found in different tumors, compared to normal tissues, including breast cancer, lung adenocarcinoma, and non-small cell lung cancer. High PHB mRNA levels were associated with a favorable prognosis in breast cancer patients, but in fact, the potential effect of PHB on breast cancer needed further study. Notably, our study found that PHB showed a favorable or unfavorable prognostic effect in different tumors by pan-cancer analysis. Therefore, the distinct roles of PHB in breast cancer remain to be elucidated by a series of experiments to determine whether PHB acts as a tumor suppressor molecule in breast cancer.
Based on pan-cancer analysis using bioinformatics methods and our previous study [47], we explored the biological functions of PHB in MCF-7 human breast cancer cells by transfection with pEGFP-PHB plasmid. Our data indicate that higher expression of PHB inhibited cell proliferation, arrested cell cycle in the S/G2 phase, decreased cell viability and invasive ability of MCF-7, and accelerated cell apoptosis in vitro. Moreover, these changes were likely related to the expression of tumor-associated proteins such as P53, erbB-2, Bcl-2, etc. Previous studies have shown that PHB could individually or jointly act with oncogene P53 [38], positive transcription factor E2F-1, and Rb family members to inhibit cell proliferation. Meanwhile, regulatory factors such as BrgI, Brm, HDAC1, and HP1 are also involved in proliferation. Further studies discovered that the Ras-Raf-MEK-ERK signal pathway might affect proliferation [23]. In this study, prohibitin changed cell cycle distribution and arrested cells in the S/G2 phase. Moreover, cell viability was reduced compared with control cells. All these results demonstrate that PHB inhibited breast cancer cell DNA replication and affected cell cycle distribution, thereby restraining cell viability and cell proliferation. The data involved in cellular apoptosis showed that the percentage of cells was increased in the over-expressing prohibitin, no matter the early stages and the later stages of apoptosis. As some studies indicated that P53 could not only inhibit cell proliferation but also mediate cell apoptosis [30, 38], our study also supported this possibility, as we observed an increased expression of P53 in cells with overexpressed PHB. As apoptosis tightly depends on the balance of pro-apoptosis proteins (e.g., Bax, Bak, and Noxa proteins) and the opposite anti-apoptosis proteins (e.g., Bcl-2, Bcl-xl, and Bcl-w), any change in the expression of these proteins could induce or inhibit apoptosis [30]. In the present study, the reduction of Bcl-2 was more likely responsible for the induction of apoptosis by PHB. Consistently, cell invasive ability was decreased which may be associated with an increase in apoptosis of the over-expressing group. A human epidermal growth factor erbB-2 [17], also known as Her-2, triggers cell invasive ability, and a high level of Her-2 could promote cancer cell growth, invasive ability, and resistance to chemotherapy drugs in ER-positive cells. We found that Her-2 was increased, and the invasive ability was reduced which indicates PHB could regulate Her-2 through unclear downstream mechanisms, further studies are required to explore this possibility.
Finally, Tumor Microenvironment (TME) was essential in the development and progression of cancer [50, 57]. Many recent studies have demonstrated that tumor-infiltrating immunocytes (TICs) could harbor either tumor-promoting or tumor-suppressing activities due to the regulation of tumor progression, invasion, metastasis, and lymph node metastasis. Our study showed that the expression of PHB significantly correlated with the infiltration of six immune cell types, including B cells, CD4+T cells, and CD8+ T cells, suggesting that PHB could also indicate the immune status besides the disease prognosis.
Under the guidance of bioinformatic analysis for pan-cancer, we have successfully constructed the model of higher PHB breast cancer cell with transient transfection, and explored the biological functions that inhibited cell proliferation, arrested cell cycle in the S/G2 phase, decreased cell viability, and invasive ability of MCF-7, and accelerated cell apoptosis in vitro, this study indicate PHB regulates breast cancer cell survival and predicts biomarker function. However, there are many drawbacks in the present study. In addition, to improve this experiment, we intend to further study the expression level of PHB in the tissue and serum samples from breast cancer patients or use gene sequencing technology to validate the biomarker role and clinical value of PHB.
ACKNOWLEDGEMENTS
We are pleased to benefit from the funding of the International Collaborative Program of Shaanxi province, and we are grateful to all the staff of the Laboratory Animal Center of the medical school, Xi’an Jiaotong University for providing help and an experiment site.
LIST OF ABBREVIATIONS
- ACC
Adrenal Carcinoma
- BRCA
Breast Cancer
- BRCA1
Breast Cancer Susceptibility Gene 1
- COAD
Chronic Obstructive Air Way Disease
- DMEM
Dulbecco's Modified Eagle
- DNA
Deoxyribonucleic Acid
- KIRC
Kidney Renal Clear Cell Carcinoma
- LAML
Acute Myeloid Leukemia
- NC group
Negative Control
- OE group
Overexpressed Group
- OS
Overall Survival
- PHB/phb
Prohibitin
- PI
PropidiumIodide
- PPI
Protein-protein Interaction
- PVDF
Polyvinylidene Difluoride
- RIPA
Radio-Immunoprecipitation Assay Buffer
- RNA
Ribose Nucleic Acid
- RT-PCR
Real-time Polymerase Chain Reaction
- TBS
Tris-buffered Saline
- TCGA
The Cancer Genome Atlas
- TICs
Tumor-infiltrating Immunocytes
- TME
Tumor Microenvironment
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
We declare that all data and materials are valid and available.
FUNDING
We are pleased to benefit from the financial support of the International Collaborative Program of Shaanxi province (No. 2012KW-35) and the natural science basic research program of Shaanxi province (2020JZ-40) greatly.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Harbeck N., Gnant M. Breast cancer. Lancet. 2017;389(10074):1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C., Siegel R., Bandi P., Jemal A. Breast cancer statistics, 2011. CA Cancer J. Clin. 2011;61(6):408–418. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 3.Fahad Ullah M. Breast cancer: Current perspectives on the disease status. Adv. Exp. Med. Biol. 2019;1152:51–64. doi: 10.1007/978-3-030-20301-6_4. [DOI] [PubMed] [Google Scholar]
- 4.Libson S., Lippman M. A review of clinical aspects of breast cancer. Int. Rev. Psychiatry. 2014;26(1):4–15. doi: 10.3109/09540261.2013.852971. [DOI] [PubMed] [Google Scholar]
- 5.Wang W., Xu L., Yang Y., Dong L., Zhao B., Lu J., Zhang T., Zhao Y. A novel prognostic marker and immunogenic membrane antigen: prohibitin (PHB) in pancreatic cancer. Clin. Transl. Gastroenterol. 2018;9(9):e178. doi: 10.1038/s41424-018-0044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wörmann B. Breast cancer: Basics, screening, diagnostics and treatment. Med. Monatsschr. Pharm. 2017;40(2):55–64. [PubMed] [Google Scholar]
- 7.Kawiak A. Molecular research and treatment of breast cancer. Int. J. Mol. Sci. 2022;23(17):9617. doi: 10.3390/ijms23179617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barzaman K., Karami J., Zarei Z., Hosseinzadeh A., Kazemi M.H., Moradi-Kalbolandi S., Safari E., Farahmand L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020;84:106535. doi: 10.1016/j.intimp.2020.106535. [DOI] [PubMed] [Google Scholar]
- 9.Maughan K.L., Lutterbie M.A., Ham P.S. Treatment of breast cancer. Am. Fam. Physician. 2010;81(11):1339–1346. [PubMed] [Google Scholar]
- 10.Khokhar A. Breast cancer in India: where do we stand and where do we go? Asian Pac. J. Cancer Prev. 2012;13(10):4861–4866. doi: 10.7314/APJCP.2012.13.10.4861. [DOI] [PubMed] [Google Scholar]
- 11. Seminars in oncology nursing. Elsevier; 2015. Genetic tests to identify risk for breast cancer. In: Lynch, J.A.; Venne, V.; Berse, B., Eds.; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francken A.B., Schouten P.C., Bleiker E.M.A., Linn S.C., Rutgers E.J.T. Breast cancer in women at high risk: The role of rapid genetic testing for BRCA1 and -2 mutations and the consequences for treatment strategies. Breast. 2013;22(5):561–568. doi: 10.1016/j.breast.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 13.Merino Bonilla J.A., Torres Tabanera M., Ros Mendoza L.H. Breast cancer in the 21st century: from early detection to new therapies. Radiologia (Madr.) 2017;59(5):368–379. doi: 10.1016/j.rx.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Criscitiello C., Corti C. Breast cancer genetics: Diagnostics and treatment. Genes (Basel) 2022;13(9):1593. doi: 10.3390/genes13091593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braden A., Stankowski R., Engel J., Onitilo A. Breast cancer biomarkers: risk assessment, diagnosis, prognosis, prediction of treatment efficacy and toxicity, and recurrence. Curr. Pharm. Des. 2014;20(30):4879–4898. doi: 10.2174/1381612819666131125145517. [DOI] [PubMed] [Google Scholar]
- 16.Majeed W., Aslam B., Javed I., Khaliq T., Muhammad F., Ali A., Raza A. Breast cancer: Major risk factors and recent developments in treatment. Asian Pac. J. Cancer Prev. 2014;15(8):3353–3358. doi: 10.7314/APJCP.2014.15.8.3353. [DOI] [PubMed] [Google Scholar]
- 17.Bécourt S., Espié M. Reference Module in Biomedical Sciences. 2018. Hormonal treatment of breast cancer. [DOI] [Google Scholar]
- 18.Newman L.A. Breast cancer in African-American women. Oncologist. 2005;10(1):1–14. doi: 10.1634/theoncologist.10-1-1. [DOI] [PubMed] [Google Scholar]
- 19.Adrada B.E., Candelaria R., Rauch G.M. MRI for the staging and evaluation of response to therapy in breast cancer. Top. Magn. Reson. Imaging. 2017;26(5):211–218. doi: 10.1097/RMR.0000000000000147. [DOI] [PubMed] [Google Scholar]
- 20.Rooney M.M., Miller K.N., Plichta J.K. Genetics of breast cancer. Surg. Clin. North Am. 2023;103(1):35–47. doi: 10.1016/j.suc.2022.08.016. [DOI] [PubMed] [Google Scholar]
- 21.Noor F., Noor A., Ishaq A.R., Farzeen I., Saleem M.H., Ghaffar K., Aslam M.F., Aslam S., Chen J.T. Recent Advances in diagnostic and therapeutic approaches for breast cancer: A comprehensive review. Curr. Pharm. Des. 2021;27(20):2344–2365. doi: 10.2174/18734286MTE06NzEAx. [DOI] [PubMed] [Google Scholar]
- 22.Sachdev J.C., Sandoval A.C., Jahanzeb M. Update on precision medicine in breast cancer. Cancer Treat. Res. 2019;178:45–80. doi: 10.1007/978-3-030-16391-4_2. [DOI] [PubMed] [Google Scholar]
- 23.Rajalingam K., Wunder C., Brinkmann V., Churin Y., Hekman M., Sievers C., Rapp U.R., Rudel T. Prohibitin is required for Ras-induced Raf–MEK–ERK activation and epithelial cell migration. Nat. Cell Biol. 2005;7(8):837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 24.Fan L., Strasser-Weippl K., Li J.J., St Louis J., Finkelstein D.M., Yu K.D., Chen W.Q., Shao Z.M., Goss P.E. Breast cancer in China. Lancet Oncol. 2014;15(7):e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 25.Ferzoco R.M., Ruddy K.J. Unique aspects of caring for young breast cancer patients. Curr. Oncol. Rep. 2015;17(2):1. doi: 10.1007/s11912-014-0425-x. [DOI] [PubMed] [Google Scholar]
- 26.Menen R.S., Hunt K.K. Considerations for the treatment of young patients with breast cancer. Breast J. 2016;22(6):667–672. doi: 10.1111/tbj.12644. [DOI] [PubMed] [Google Scholar]
- 27.Freedman R.A., Partridge A.H. Management of breast cancer in very young women. Breast. 2013;22(Suppl. 2):S176–S179. doi: 10.1016/j.breast.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 28.Anastasiadi Z., Lianos G.D., Ignatiadou E., Harissis H.V., Mitsis M. Breast cancer in young women: An overview. Updates Surg. 2017;69(3):313–317. doi: 10.1007/s13304-017-0424-1. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Dong W., Zhang P., Zhang T., Ma L., Qu M., Ma X., Zhou X., He Q. Comprehensive analysis of regulatory factors and immune-associated patterns to decipher common and BRCA1/2 mutation-type-specific critical regulation in breast cancer. Front. Cell Dev. Biol. 2021;9:750897. doi: 10.3389/fcell.2021.750897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y.T., Chen P., Ouyang R.Y., Song L. Multifaceted role of prohibitin in cell survival and apoptosis. Apoptosis. 2015;20(9):1135–1149. doi: 10.1007/s10495-015-1143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artal-Sanz M., Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol. Metab. 2009;20(8):394–401. doi: 10.1016/j.tem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F.T., von Kleist-Retzow J.C., Waisman A., Westermann B., Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22(4):476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanz M.A., Tsang W.Y., Willems E.M., Grivell L.A., Lemire B.D., van der Spek H., Nijtmans L.G.J. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem. 2003;278(34):32091–32099. doi: 10.1074/jbc.M304877200. [DOI] [PubMed] [Google Scholar]
- 34.Webster L.R., Provan P.J., Graham D.J., Byth K., Walker R.L., Davis S., Salisbury E.L., Morey A.L., Ward R.L., Hawkins N.J., Clarke C.L., Meltzer P.S., Balleine R.L. Prohibitin expression is associated with high grade breast cancer but is not a driver of amplification at 17q21.33. Pathology. 2013;45(7):629–636. doi: 10.1097/PAT.0000000000000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan W., Yang H., Liu T., Wang J., Li T.W.H., Mavila N., Tang Y., Yang J., Peng H., Tu J., Annamalai A., Noureddin M., Krishnan A., Gores G.J., Martínez-Chantar M.L., Mato J.M., Lu S.C. Prohibitin 1 suppresses liver cancer tumorigenesis in mice and human hepatocellular and cholangiocarcinoma cells. Hepatology. 2017;65(4):1249–1266. doi: 10.1002/hep.28964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koushyar S., Jiang W.G., Dart D.A. Unveiling the potential of prohibitin in cancer. Cancer Lett. 2015;369(2):316–322. doi: 10.1016/j.canlet.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Fusaro G., Dasgupta P., Rastogi S., Joshi B., Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J. Biol. Chem. 2003;278(48):47853–47861. doi: 10.1074/jbc.M305171200. [DOI] [PubMed] [Google Scholar]
- 38.Rastogi S., Joshi B., Fusaro G., Chellappan S. Camptothecin induces nuclear export of prohibitin preferentially in transformed cells through a CRM-1-dependent mechanism. J. Biol. Chem. 2006;281(5):2951–2959. doi: 10.1074/jbc.M508669200. [DOI] [PubMed] [Google Scholar]
- 39.Zi Xu Y.X., Ande S.R., Mishra S. Prohibitin: A new player in immunometabolism and in linking obesity and inflammation with cancer. Cancer Lett. 2018;415:208–216. doi: 10.1016/j.canlet.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimaru T., Ono M., Bando Y., Chen Y.A., Mizuguchi K., Shima H., Komatsu M., Imoto I., Izumi K., Honda J., Miyoshi Y., Sasa M., Katagiri T. A-kinase anchoring protein BIG3 coordinates oestrogen signalling in breast cancer cells. Nat. Commun. 2017;8(1):15427. doi: 10.1038/ncomms15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato T., Saito H., Swensen J., Olifant A., Wood C., Danner D., Sakamoto T., Takita K., Kasumi F., Miki Y. The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res. 1992;52(6):1643–1646. [PubMed] [Google Scholar]
- 42.Mishra S., Murphy L.C., Nyomba B.L.G., Murphy L.J. Prohibitin: A potential target for new therapeutics. Trends Mol. Med. 2005;11(4):192–197. doi: 10.1016/j.molmed.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Theiss A.L., Jenkins A.K., Okoro N.I., Klapproth J.M.A., Merlin D., Sitaraman S.V. Prohibitin inhibits tumor necrosis factor alpha-induced nuclear factor-kappa B nuclear translocation via the novel mechanism of decreasing importin alpha3 expression. Mol. Biol. Cell. 2009;20(20):4412–4423. doi: 10.1091/mbc.e09-05-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimaru T., Komatsu M., Miyoshi Y., Honda J., Sasa M., Katagiri T. Therapeutic advances in BIG 3‐ PHB 2 inhibition targeting the crosstalk between estrogen and growth factors in breast cancer. Cancer Sci. 2015;106(5):550–558. doi: 10.1111/cas.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takagi H., Moyama C., Taniguchi K., Ando K., Matsuda R., Ando S., Ii H., Kageyama S., Kawauchi A., Chouha N., Désaubry L., Nakata S. Fluorizoline blocks the interaction between prohibitin-2 and γ -glutamylcyclotransferase and induces p21 waf1/cip1 expression in MCF7 breast cancer cells. Mol. Pharmacol. 2022;101(2):78–86. doi: 10.1124/molpharm.121.000334. [DOI] [PubMed] [Google Scholar]
- 46.He Q., Zhang S.Q., Chu Y.L., Jia X.L., Zhao L.H., Wang X.L. Separation and identification of differentially expressed nuclear matrix proteins in breast carcinoma forming. Acta Oncol. 2010;49(1):76–84. doi: 10.3109/02841860903287213. [DOI] [PubMed] [Google Scholar]
- 47.Kim N.H., Yoshimaru T., Chen Y.A., Matsuo T., Komatsu M., Miyoshi Y., Tanaka E., Sasa M., Mizuguchi K., Katagiri T. BIG3 inhibits the estrogen-dependent nuclear translocation of PHB2 via multiple karyopherin-alpha proteins in breast cancer cells. PLoS One. 2015;10(6):e0127707. doi: 10.1371/journal.pone.0127707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hwang K.T. Clinical databases for breast cancer research. Adv. Exp. Med. Biol. 2021;1187:493–509. doi: 10.1007/978-981-32-9620-6_26. [DOI] [PubMed] [Google Scholar]
- 49.Roulot A., Héquet D., Guinebretière J.M., Vincent-Salomon A., Lerebours F., Dubot C., Rouzier R. Tumoral heterogeneity of breast cancer. Ann. Biol. Clin. (Paris) 2016;74(6):653–660. doi: 10.1684/abc.2016.1192. [DOI] [PubMed] [Google Scholar]
- 50.Printz C. Breast cancer screening for women in their 40s reduces mortality. Cancer. 2021;127(4):497. doi: 10.1002/cncr.33439. [DOI] [PubMed] [Google Scholar]
- 51.Kolak A., Kamińska M., Sygit K., Budny A., Surdyka D., Kukiełka-Budny B., Burdan F. Primary and secondary prevention of breast cancer. Ann. Agric. Environ. Med. 2017;24(4):549–553. doi: 10.26444/aaem/75943. [DOI] [PubMed] [Google Scholar]
- 52.Li Z., Wei H., Li S., Wu P., Mao X. The role of progesterone receptors in breast cancer. Drug Des. Devel. Ther. 2022;16:305–314. doi: 10.2147/DDDT.S336643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu P., Xu Y., Zhang W., Li Y., Tang L., Chen W., Xu J., Sun Q., Guan X. Prohibitin promotes androgen receptor activation in ER-positive breast cancer. Cell Cycle. 2017;16(8):776–784. doi: 10.1080/15384101.2017.1295193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chigira T., Nagatoishi S., Tsumoto K. Differential binding of prohibitin-2 to estrogen receptor α and to drug-resistant ERα mutants. Biochem. Biophys. Res. Commun. 2015;463(4):726–731. doi: 10.1016/j.bbrc.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Kahlert S., Nuedling S., van Eickels M., Vetter H., Meyer R., Grohé C. Estrogen receptor alpha rapidly activates the IGF-1 receptor pathway. J. Biol. Chem. 2000;275(24):18447–18453. doi: 10.1074/jbc.M910345199. [DOI] [PubMed] [Google Scholar]
- 56.Rios A.C., van Rheenen J., Scheele C.L.G.J. Multidimensional imaging of breast cancer. Cold Spring Harb. Perspect. Med. 2023;13(5):a041330. doi: 10.1101/cshperspect.a041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X. Molecular classification of breast cancer: Relevance and challenges. Arch. Pathol. Lab. Med. 2023;147(1):46–51. doi: 10.5858/arpa.2022-0070-RA. [DOI] [PubMed] [Google Scholar]
- 58.Nuell M.J., Stewart D.A., Walker L., Friedman V., Wood C.M., Owens G.A., Smith J.R., Schneider E.L., Dell’ Orco R., Lumpkin C.K. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol. Cell. Biol. 1991;11(3):1372–1381. doi: 10.1128/mcb.11.3.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Too I.H.K., Bonne I., Tan E.L., Chu J.J.H., Alonso S. Prohibitin plays a critical role in Enterovirus 71 neuropathogenesis. PLoS Pathog. 2018;14(1):e1006778. doi: 10.1371/journal.ppat.1006778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tortelli Junior T.C., de Godoy L.M.F., de Souza G.A., Bonatto D., Otake A.H., de Freitas Saito R., Rosa J.C., Greene L.J., Chammas R. Accumulation of prohibitin is a common cellular response to different stressing stimuli and protects melanoma cells from ER stress and chemotherapy-induced cell death. Oncotarget. 2017;8(26):43114–43129. doi: 10.18632/oncotarget.17810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kahl A., Anderson C.J., Qian L., Voss H., Manfredi G., Iadecola C., Zhou P. Neuronal expression of the mitochondrial protein prohibitin confers profound neuroprotection in a mouse model of focal cerebral ischemia. J. Cereb. Blood Flow Metab. 2018;38(6):1010–1020. doi: 10.1177/0271678X17720371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woodlock T.J., Bethlendy G., Segel G.B. Prohibitin expression is increased in phorbol ester-treated chronic leukemic B-lymphocytes. Blood Cells Mol. Dis. 2001;27(1):27–34. doi: 10.1006/bcmd.2000.0348. [DOI] [PubMed] [Google Scholar]
- 63.Raut G.K., Chakrabarti M., Pamarthy D., Bhadra M.P. Glucose starvation-induced oxidative stress causes mitochondrial dysfunction and apoptosis via Prohibitin 1 upregulation in human breast cancer cells. Free Radic. Biol. Med. 2019;145:428–441. doi: 10.1016/j.freeradbiomed.2019.09.020. [DOI] [PubMed] [Google Scholar]
- 64.Wang S., Fusaro G., Padmanabhan J., Chellappan S.P. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21(55):8388–8396. doi: 10.1038/sj.onc.1205944. [DOI] [PubMed] [Google Scholar]
- 65.Fu P., Yang Z., Bach L.A. Prohibitin-2 binding modulates insulin-like growth factor-binding protein-6 (IGFBP-6)-induced rhabdomyosarcoma cell migration. J. Biol. Chem. 2013;288(41):29890–29900. doi: 10.1074/jbc.M113.510826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bai Y., Ludescher M., Poschmann G., Stühler K., Wyrich M., Oles J., Franken A., Rivandi M., Abramova A., Reinhardt F., Ruckhäberle E., Niederacher D., Fehm T., Cahill M.A., Stamm N., Neubauer H. PGRMC1 promotes progestin-dependent proliferation of breast cancer cells by binding prohibitins resulting in activation of ERα signaling. Cancers (Basel) 2021;13(22):5635. doi: 10.3390/cancers13225635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang K., Long B., Zhou L.Y., Liu F., Zhou Q.Y., Liu C.Y., Fan Y.Y., Li P.F. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014;5(1):3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 68.Huang X., Liu J., Ma Q. Prohibitin participates in the HIRA complex to promote cell metastasis in breast cancer cell lines. FEBS Open Bio. 2020;10(10):2182–2190. doi: 10.1002/2211-5463.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong N., Cui Y., Zhou X., Li T., Han J. Identification of prohibitin 1 as a potential prognostic biomarker in human pancreatic carcinoma using modified aqueous two-phase partition system combined with 2D-MALDI-TOF-TOF-MS/MS. Tumour Biol. 2015;36(2):1221–1231. doi: 10.1007/s13277-014-2742-y. [DOI] [PubMed] [Google Scholar]
- 70.Satheesh Kumar M.K., Nair S., Mony U., Kalingavarman S., Venkat R., Sivanarayanan T.B., Unni A.K.K., Rajeshkannan R., Anandakuttan A., Radhakrishnan S., Menon K.N. Significance of elevated Prohibitin 1 levels in Multiple Sclerosis patients lymphocytes towards the assessment of subclinical disease activity and its role in the central nervous system pathology of disease. Int. J. Biol. Macromol. 2018;110:573–581. doi: 10.1016/j.ijbiomac.2017.12.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We declare that all data and materials are valid and available.