Abstract
Nucleocytoplasmic transport of viral ribonucleoproteins (vRNPs) is an essential aspect of the replication cycle for influenza A, B, and C viruses. These viruses replicate and transcribe their genomes in the nuclei of infected cells. During the late stages of infection, vRNPs must be exported from the nucleus to the cytoplasm prior to transport to viral assembly sites on the cellular plasma membrane. Previously, we demonstrated that the influenza A virus nuclear export protein (NEP, formerly referred to as the NS2 protein) mediates the export of vRNPs. In this report, we suggest that for influenza B and C viruses the nuclear export function is also performed by the orthologous NEP proteins (formerly referred to as the NS2 protein). The influenza virus B and C NEP proteins interact in the yeast two-hybrid assay with a subset of nucleoporins and with the Crm1 nuclear export factor and can functionally replace the effector domain from the human immunodeficiency virus type 1 Rev protein. We established a plasmid transfection system for the generation of virus-like particles (VLPs) in which a functional viral RNA-like chloramphenicol acetyltransferase (CAT) gene is delivered to a new cell. VLPs generated in the absence of the influenza B virus NEP protein were unable to transfer the viral RNA-like CAT gene to a new cell. From these data, we suggest that the nuclear export of the influenza B and C vRNPs are mediated through interaction between NEP proteins and the cellular nucleocytoplasmic export machinery.
Influenza A, B, and C viruses are human pathogens of the Orthomyxoviridae family. These negative-sense RNA viruses replicate and transcribe their genomes in the nuclei of infected cells. The genomes of influenza A and B viruses are composed of eight segments, while influenza C virus genomes have seven segments (46, 48). These RNA segments are encapsidated by the nucleoprotein (NP) and are associated with the viral polymerase (the three P proteins), which together are termed the viral ribonucleoprotein (vRNP) complex (4, 23). After the initial binding, penetration, and uncoating of the viral particle, the vRNPs are released into the cytoplasm of the infected cell. Influenza A vRNP transport into the nucleus is mediated by soluble cellular nuclear import factors karyopherin α, karyopherin β, Ran, and p10 by a direct interaction between the viral NP protein and karyopherin α (42, 43, 58). Genomic vRNPs are amplified within the nucleus and then must exit the nucleus to accumulate with other viral proteins at the plasma membrane, where packaging and assembly of viral particles occur.
The majority of cellular and viral RNA export from the nucleus is thought to be protein mediated. The export of human immunodeficiency virus type 1 (HIV-1) unspliced RNA, for example, is mediated by the virally encoded export protein, Rev. The Rev protein interacts with both a cis-acting sequence present on the viral RNA (the Rev-responsive element, or RRE) and with the karyopherin β family member, Crm1 (reviewed in reference 12).
It was originally suggested by competitive inhibition of RNA transport in Xenopus oocytes and genetic analyses or RNA transport in Saccharomyces cerevisiae that there are several distinct pathways for the export of specific classes of RNA (for recent reviews see references 35 and 52). Crm1 is thought to specifically mediate the transport of export factors that contain the Rev class of nuclear export sequences (NES) and are rich in bulky hydrophobic amino acids, such as leucine and methionine (16, 19). In addition to HIV-1 Rev-bound RNA, cellular U snRNA and 5S RNAs also exit the nucleus in a Crm1-dependent manner, whereas mRNA export, for example, is thought to be Crm1 independent. Furthermore, Crm1-mediated export requires the GTP-bound form of Ran (2, 27). Export of leucine-rich export factors (and their RNA cargo) occurs upon formation of a trimolecular complex between the NES motif, Crm1, and Ran-GTP. The specific steps following formation of this complex leading to active transport through the nuclear pore are poorly understood.
We and others, using several distinct experimental approaches, have shown that the influenza A virus NEP (nuclear export protein) is required for proper nuclear egress of vRNPs (20, 37, 38, 44). Originally named the NS2 (for nonstructural 2) protein, the influenza A viral NEP has since been found to be associated with purified viral particles and is, therefore, by definition a structural protein (47, 60). Furthermore, the function of nuclear export can now be assigned to this influenza A viral protein. We therefore proposed that the influenza A virus NS2 protein be renamed NEP.
Influenza B and C virus genomic RNAs are also amplified within the nucleus and must also be transported to the cytoplasm prior to assembly into progeny viral particles at the cellular plasma membrane. The influenza B and C viruses share a common replication strategy with influenza A virus and have several functionally homologous proteins. However, several of the viral proteins possess different activities. For example, the glycoprotein of influenza C virus has an esterase activity (4, 24, 32, 53) not found with the influenza A and B viruses. The genomic organizations of influenza A, B, and C viruses have several differences from each other (4). For example, the neuraminidase (NA) gene of influenza B virus codes for two open reading frames (49) while those of influenza A viruses code for only one open reading frame and influenza C viruses lack an NA gene. Furthermore, an amino acid comparison of the second open reading frame (ORF) of the influenza A, B, and C viral NS genes shows limited sequence identity (data not shown). In this report, we have functionally characterized the second ORFs (NS2 proteins) of the influenza B and C viral NS genes (1, 7, 8, 33, 34) and found that they demonstrate properties indicating their role as nuclear export factors. We therefore propose that the NS2 proteins from influenza B and C viruses also be renamed NEP.
Through a yeast two-hybrid assay we have determined that the NEP proteins from influenza B and C viruses are able to interact with nucleoporins and with the Crm1 nuclear receptor. Second, we show that when fused to a Rev mutant which contains a functional RRE-binding domain but which lacks an NES (amino acids [aa] 1 to 69 of the wild-type Rev protein) (44), influenza B and C viral NEP proteins are able to promote the export of an RRE-containing reporter. Third, in a newly established virus-like particle (VLP) assay for influenza B virus, the NEP protein was shown to be essential to transfer a viral RNA-like chloramphenicol acetyltransferase (CAT) gene to a new cell. We propose that in a process analogous to that of influenza A virus, the influenza B and C viral NEP proteins facilitate nuclear export of the vRNPs by bridging the interaction between vRNP complexes and the cellular Crm1 export pathway.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney (MDCK) and 293T (generous gift of Y. Kawaoka, University of Wisconsin) cells were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco Life Technologies, Grand Island, N.Y.) supplemented with 10% fetal calf serum. Influenza virus strains B/Yamagata/73, B/Lee/40, and C/California/78 were used in this study. Virus was propagated in MDCK cells at 35 or 33°C for 72 h. Infections of MDCK cells were performed in DMEM supplemented with 0.1% bovine albumin (BA) and 1-μg/ml concentrations of trypsin 1:250 (Difco Laboratories, Detroit, Mich.)
Eukaryotic expression constructs.
Influenza B virus cDNAs were cloned into the vector pCAGGS containing a multiple cloning site (kindly provided by Y. Kawaoka) using standard techniques (41). The PB1, PB2, PA, M1, and BM2 ORFs were amplified from purified influenza B/Yamagata/73 virus RNA by reverse transcription-PCR (RT-PCR) and inserted between the EcoRI and XhoI restriction sites of pCAGGS to generate the plasmids pCAGGS-B/Yamagata/73/PB1, pCAGGS-B/Yamagata/73/PB2, pCAGGS-B/Yamagata/73/PA, pCAGGS-B/Yamagata/73/M1, and pCAGGS-B/Yamagata/73/BM2. The B/Lee/40 NP cDNA was cloned into the KpnI and XhoI sites of pCAGGS. The B/Lee/40 HA cDNA was subcloned from the construct pT3-BHALEE (3) into the EcoRI and XhoI sites of this vector. pCAGGS-B/Yamagata/73/NANB (encoding both NA and NB ORFs) was generated by subcloning the full-length cDNA of B/Yamagata NA from pT3-BNAYA#6 between EcoRI and XhoI sites. To generate pCAGGS-B/Lee/40/NEP, the BNEP cDNA was subcloned from pEG202-BNS2 into EcoRI and XhoI sites (see below). For pCAGGS-B/delNES/NEP, specific primers were designed to PCR amplify aa 20 to 121 of the ORF and to include a MET initiation codon at position 19. The purified DNA fragment was cloned between the EcoRI and XhoI sites of the vector pCAGGS. The clone pCAGGS-A21/B/NEP was constructed by subcloning the EcoRI and SacI fragments from pCAGGS-A/NEP into the EcoRI and SacI sites of the clone pCAGGS-B/Lee/40/NEP. pPOLI-B/Lee/40/NSCAT was constructed by PCR using oligonucleotide primers containing the 5′ and 3′ noncoding ends corresponding to the sequence of segment 8 from the influenza B/Lee/40 virus and a portion of the CAT ORF derived from pSV2-CAT (3). The purified PCR fragment was then cloned between the SapI sites of the vector pPOLI-version II (15).
For the yeast two-hybrid assay, the NEP (NS2) ORFs from influenza B/Lee/40 and C/California/78 viruses were cloned by standard RT-PCR cloning techniques between the EcoRI and XhoI sites of the vector pEG202 (bait plasmid containing the LexA DNA binding domain). These constructions resulted in the generation of the constructs pEG-202-BNEP and pEG-202-CNEP.
To generate the constructs pRev*-BNEP and pRev*-CNEP, the ORFs of pEG-202-BNEP and pEG-202-CNEP were amplified by PCR and inserted into the pRev* vector (44). This results in expression of a translational fusion of the Rev* protein and either the influenza B or C virus NEP proteins. Site-directed and truncation mutant plasmid derivatives were constructed by standard PCR mutagenesis techniques. All constructs were confirmed by DNA sequencing.
VLP assay.
293T cells were cotransfected with 11 plasmids, including 10 protein expression plasmids and pPOLI-B/Lee/40/NSCAT (Table 1, WT) to generate VLPs. DNA mixtures were adjusted to 250 μl with OPTI-MEM (Gibco-BRL) containing 12 ml (1 mg/ml) of Lipofectamine 2000 reagent (Gibco-BRL) and 106 cells in suspension (5). In some assays, the plasmid pCAGGS-B/Lee/40/NEP was omitted (Table 1, −NEP). Transfection medium was removed 6 h later and replaced with 2 ml of DMEM supplemented with 0.1% BA (ICN Biomedicals, Aurora, Ohio) and 1-μg/ml concentrations of trypsin 1:250. Seventy-two hours posttransfection cells and media were harvested and separated by low-speed centrifugation (5,000 × g for 5 min). Cells were assayed for CAT activity using previously described assay conditions (44). Supernatants were further clarified by high-speed centrifugation (14,000 × g) for 5 min. Clarified supernatant was used to infect confluent 35-mm-diameter dishes of MDCK cells, which were superinfected with influenza B/Yamagata/73 virus at a multiplicity of infection of 1 or mock superinfected with phosphate-buffered saline (PBS). After 1 h of viral adsorption at room temperature cells were washed five times with PBS supplemented with 0.1% BA. Two milliliters of DMEM containing 0.1% BA and 1 μg of trypsin 1:250 per ml was added to the cells, which were incubated in a humidified incubator at 35°C for 12 h. Cells and media were harvested as described above. Cell extracts were assayed for CAT activity, and supernatants were assayed for hemagglutination (HA) titer to confirm consistent infection efficiencies.
TABLE 1.
Constructs transfected to generate VLPsa
| Plasmid | Amt (μg) of construct used
|
|||
|---|---|---|---|---|
| WT | A21/B/NEP | delNES/NEP | −NEP | |
| pCAGGS-B/Yamagata/73/PB1 | 1 | 1 | 1 | 1 |
| pCAGGS-B/Yamagata/73/PB2 | 1 | 1 | 1 | 1 |
| pCAGGS-B/Yamagata/73/PA | 1 | 1 | 1 | 1 |
| pCAGGS-B/Lee/40/HA | 1 | 1 | 1 | 1 |
| pCAGGS-B/Lee/73/NP | 1 | 1 | 1 | 1 |
| pCAGGS-B/Yamagata/73/NANB | 1 | 1 | 1 | 1 |
| pCAGGS-B/Yamagata/73/M1 | 1 | 1 | 1 | 1 |
| pCAGGS-B/Yamagata/73/BM2 | 1 | 1 | 1 | 1 |
| pCAGGS-B/Yamagata/73/NS1 | 1 | 1 | 1 | 1 |
| pCAGGS-B/Lee/40/NEP | 1 | 0 | 0 | 0 |
| pCAGGS-A21/B/NEP | 0 | 1 | 0 | 0 |
| pCAGGS-B/delNES/NEP | 0 | 0 | 1 | 0 |
| pPOLI-B/Lee/NSCAT | 1 | 1 | 1 | 1 |
Eukaryotic expression plasmids were constructed for the influenza B virus gene products and used to generate VLPs after transfection into 293T cells. WT, the plasmids corresponding to the complete set of plasmids representing the entire set of influenza B virus gene products, including a viral RNA-like CAT gene. A21/B/NEP replaces the wild-type NEP with the construct pCAGGS-A21/B/NEP. In the column labeled delNES/NEP, wild-type NEP is replaced with a mutant version of NEP lacking the first 20 aa. −NEP, the complete set without the plasmid encoding the NEP protein from influenza B virus. In all cases 1 μg per plasmid was transfected for a total of 10 or 11 μg (−NEP or others, respectively). Supernatants of transfected cells showed HA titers of log 25 in repeated experiments.
Rev nuclear export assay.
Duplicate 35-mm-diameter dishes of 293T cells were transfected with the reporter plasmid pDM128 and either pRev*-BNEP or pRev*-CNEP (26, 30, 36, 44) using the liposomal reagent DOTAP (Roche Molecular Biochemicals, Indianapolis, Ind.) as previously described. The plasmid pDM128 was a generous gift of Tristram G. Parslow (University of California, San Francisco). Transfection efficiencies were normalized using the β-galactosidase-expressing reporter plasmid pCH110 (Pharmacia, Piscataway, N.J.) according to the manufacturer's instructions. Cells were collected 48 h posttransfection and assayed for CAT activity by previously described methods (44).
Yeast two-hybrid assay.
S. cerevisiae (MATa trp1 ura3 his3 LEU::pLEX-Aop6-Leu2), pEG202, and pSH18-34 were kindly provided by R. Brent (The Molecular Sciences Institute) and have been previously described (22, 43, 59). Yeast two-hybrid constructs encoding the yeast nucleoporins yRip1, yNup100, yNup1, and yCrm1 in the vector pJG4-5 (prey plasmid containing an acidic activation domain) were generously provided by M. Rosbash (Brandeis University). pVP16/RAB was provided by B. R. Cullen (Duke University). In addition, the ORFs from influenza B and C virus NEP proteins were expressed as fusion proteins with the LexA protein from the vector pEG-202 (bait plasmid; see above). β-Galactosidase expression in yeast cells transformed with various combinations of bait and prey plasmids was analyzed as described previously (43).
RESULTS
Influenza B and C virus NEP proteins interact with nucleoporins and with Crm1.
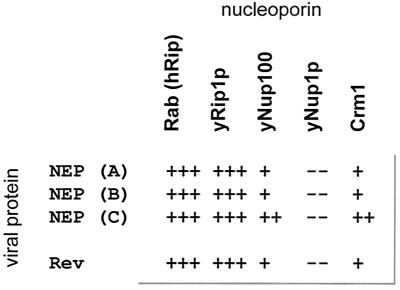
For both HIV-1 Rev and influenza A virus NEP, a positive correlation has previously been demonstrated for the ability to bind particular nucleoporins and Crm1 in yeast and/or mammalian two-hybrid systems and function as a nuclear export chaperon (14, 17, 18, 29, 37, 40, 44, 50, 51). The NEP proteins from influenza B and C viruses were also tested for the ability to interact with cellular nucleoporins and the Crm1 nuclear export receptor in the yeast two-hybrid assay. Three distinct types of nucleoporins were tested: Rab/hRip1, which contains an XXFG type of repeat; yNUP100, which contains a GLFG type of FG repeat; and yNUP1p, which has an FXFG type of FG repeat. The NEP proteins from influenza B and C viruses demonstrated a positive interaction with Rab/hRIP1, yRip1, and yNup100 but did not interact with yNup1p (Fig. 1). We were able to confirm a positive interaction between the influenza A viral NEP and Crm1 and were also able to identify an interaction between the influenza B and C viral NEP proteins and this cellular protein. These results are consistent with results obtained in other studies for both the influenza A virus NEP and the HIV-1 Rev protein. (Fig. 1 and reference 37).
FIG. 1.
Yeast two-hybrid analysis of NEP interactions. Influenza A, B, and C virus NEP proteins were fused to a LexA binding domain and used as bait to test for interaction with nucleoporins and Crm1, which were fused to an acidic activation domain (for details see Materials and Methods). A β-galactosidase gene containing upstream LexA binding sites was used as a reporter. The number of plus signs indicates the relative strength of blue color on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside indicator plates. The scoring system is 1 for the weakest relative strength to 3 for the strongest relative strength. Rev (wild-type) and influenza A virus NEP protein results are included for comparison (6, 14, 37, 40, 44, 51)
Influenza B and C virus NEP proteins can functionally replace the Rev effector domain.
The interaction of the influenza B and C NEP proteins with nucleoporins and Crm1 suggested a role for these proteins in nucleocytoplasmic trafficking. We therefore took advantage of a Rev-based nuclear export assay to test the ability of these proteins to mediate the transport of CAT mRNA transcribed from the reporter plasmid pDM128 (26, 30, 36, 44). The reporter plasmid pDM128 expresses a CAT messenger mRNA, in which the ORF is within an intron containing an RRE. CAT expression requires the function of Rev to promote the nuclear export of unspliced mRNA.
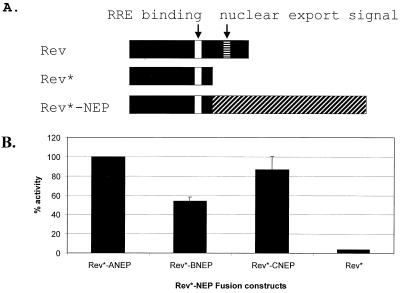
Rev* lacks a functional NES (Fig. 2) and cannot promote the nuclear export of the unspliced CAT mRNA. Without a functional NES (i.e., Rev* [see Fig. 2]), the Rev* protein can interact with the reporter RNA via its RRE but not with the cellular export machinery. In this case, unspliced transcripts are retained in the nucleus and CAT activity is reduced. However, if a protein containing a functional NES (such as the influenza A viral NEP) is fused to Rev*, the chimeric protein is then able to interact with both the reporter RRE and with the nuclear export machinery and CAT activity can be detected (44). Using this system we show that the NEP proteins from influenza B and C viruses substitute for the Rev effector domain (Fig. 2B). When fused to Rev*, full-length influenza B and C virus NEP proteins had 53 and 87% activity of the Rev*-ANEP fusion protein, respectively.
FIG. 2.
Influenza virus Rev*-NEP fusions can promote the nuclear export of CAT mRNA. (A) Full-length HIV-1 Rev (aa 1 to 116) contains both an RRE and a nuclear export signal (aa 75 to 83) and is therefore able to promote the nuclear export of pDM128 reporter mRNA. In Rev* (aa 1 to 69), the NES has been removed (44). Rev*, therefore, cannot interact with the cellular export machinery to promote the export of unspliced CAT mRNA containing an RRE. Fusion of the influenza A, B, and C virus NEP proteins (not drawn to scale) to Rev* results in the nuclear export of pDM128 mRNA, indicating the presence of a functional NES within the NEP. Rev*-ANEP contains 190 aa. Rev*-BNEP contains 191 aa. Rev*-CNEP contains 252 aa. (B) Thirty-five-millimeter-diameter dishes of 293T cells were transfected with pDM128 reporter (2 μg) and the indicated Rev* fusion plasmid (3 μg). Cell lysates were harvested 48 h posttransfection, and CAT assays were performed as described in Materials and Methods. CAT values are the mean percentages of duplicate transfections and are normalized to levels induced by Rev*-ANEP.
Determination of the regions within B and C viral NEPs which confer nuclear export activity.
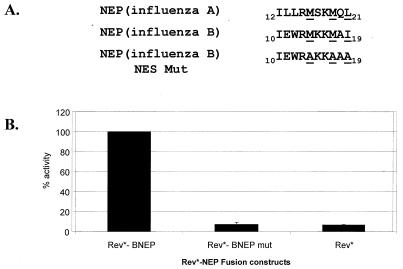
To further characterize the nuclear export activity of the influenza B and C virus NEP proteins, attempts to define the NES signals were made. Although the influenza A and B virus NEP proteins share limited overall amino acid sequence identity (less than 25% identity using the CLUSTAL program [data not shown; CLUSTAL analysis was provided by the Institute of Computational Biomedicine, Mount Sinai School of Medicine of New York University]), a 10-aa peptide located near the N terminus of the influenza B virus NEP is 50% identical (5 out of 10 aa) to the NES of influenza A virus NEP (37, 44). We hypothesized that this region may represent the NES for the influenza B virus NEP protein. In order to test this possibility, bulky hydrophobic residues representing a potential Rev-like export sequence within this region were mutated to alanine. The exact residues altered are depicted in Fig. 3A. The mutant and wild-type constructs were then tested in the Rev assay for nuclear export function. Mutation of the bulky hydrophobic residues to alanine resulted in greater than 90% reduction of CAT activity compared to that of wild type (Fig. 3B).
FIG. 3.
Mapping of the NES for the influenza B virus NEP protein. (A) The amino acid sequence of influenza A and B virus NEP proteins are compared, and a 10-aa stretch with homology to the influenza A virus NEP NES was observed. Key hydrophobic residues (underlined) within the influenza B virus NEP NES motif were mutated to alanine and tested in the pDM128 CAT reporter assay. (B) CAT values are the mean percentages of duplicate transfections and are normalized to levels induced by Rev*-BNEP.
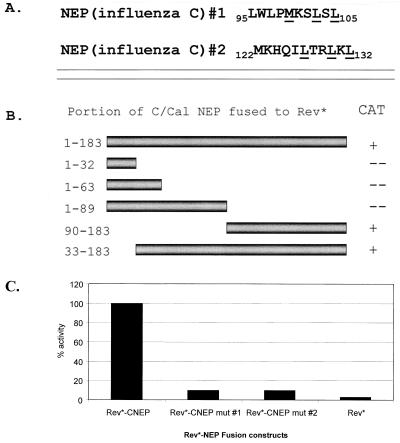
We were unable to identify peptides that were similar to the NES of influenza A virus NEP by examination of the amino terminus of the influenza C virus NEP protein sequence. Therefore a series of N-terminal and C-terminal deletion mutants were constructed and tested in the Rev assay to directly assay for such activity (Fig. 4B). The C-terminal 94 aa were sufficient to restore nuclear export activity to the Rev* mutant (Fig. 4B and 3C). Within these 94 aa, two potential NESs were identified by comparison of influenza C virus NEP amino acid sequences corresponding to a Rev-like NES (Fig. 4A). Again, key hydrophobic residues within the Rev-like NES were mutated to alanine and tested in the Rev assay. Mutation of either signal resulted in the inability of the Rev* fusion to catalyze the nuclear export of the CAT-containing mRNA. Both mutants had less than 10% CAT activity compared to that of the wild-type influenza C virus Rev*-NEP fusion (Fig. 4C).
FIG. 4.
Mapping of the influenza C virus NEP putative NES. (A) Within the C-terminal 94 aa, two Rev-like nuclear export signals were detected. Key hydrophobic amino acids (underlined) were mutated to alanine in each motif separately and tested in the CAT export assay. (B) A series of N- and C-terminal deletion mutants in the influenza C virus NEP ORF were made and fused to Rev*. The C-terminal 94 aa were found to be sufficient to promote the export of the CAT reporter gene. Plus signs indicate a positive CAT signal comparable to that of the full-length Rev*-CNEP fusion. A minus sign indicates CAT activity at or below background levels. (C) CAT values are the mean percentages of duplicate transfections and are normalized to levels induced by Rev*-CNEP.
NEP protein from influenza B virus is essential for passage of a viral RNA-like CAT gene in a VLP assay.
In order to test the significance of the nuclear export function of the influenza B virus NEP protein in formation of functional viral particles, we have established a VLP system for influenza B virus. Similar systems have previously been established for influenza A virus (31, 38) and Thogoto virus (54). Using this VLP system we tested whether the influenza B virus NEP protein is an essential protein to transfer a functional viral RNA-like CAT gene to a new cell. The ORFs of the eleven viral proteins coded for by the influenza B virus genome were cloned into the eukaryotic expression vector pCAGGS (41). An influenza viral RNA-like CAT gene construct was engineered containing the CAT ORF inserted between the 5′ and 3′ NC (noncoding) regions from an influenza B virus NS segment (3) flanked by a truncated human polymerase I promoter and a hepatitis delta virus ribozyme (15, 39, 45). When transfected into 293T cells, this construct produces a precise negative-sense RNA that will only be replicated and transcribed by the influenza B virus polymerase (3, 11, 28).
Cotransfection of the 10 plasmids expressing influenza B viral proteins along with a viral RNA-like CAT reporter construct into 293T cells resulted in the formation of VLPs, as determined by detection of HA activity in the supernatant 48 h posttransfection (Table 1). Both transfected cell supernatants (Table 1, WT and −NEP) were positive for HA (HA titer = 32), suggesting that influenza B virus VLPs were released into the transfected cell media and that the process of hemagglutinating particle formation is NEP protein independent. Transfected cell lysates were assayed for CAT activity (Fig. 5A). In independent experiments the absence of the influenza B virus NEP protein resulted in an approximately 1-log increase in detected CAT activity (Fig. 5A and 6).
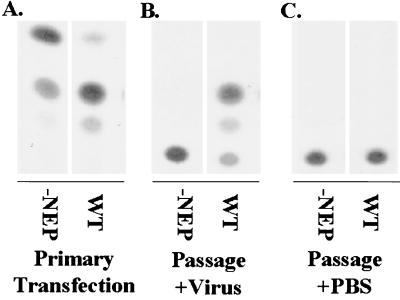
FIG. 5.
Effect of the NEP protein on the ability to package and passage a functional viral RNA-like CAT gene. 293T cells were transfected with either wild-type (WT) or NEP-lacking (−NEP) combinations of plasmids (Table 1). Forty-eight hours posttransfection clarified supernatants were used to infect MDCK cells. Cells were either superinfected with influenza B/Yamagata/73 virus at a multiplicity of 1 or mock infected with PBS. Twelve hours postinfection cells were harvested and assayed for CAT activity. The WT lane shows a positive signal for passage of the viral RNA-like CAT gene. The supernatant derived from the −NEP-transfected cells are not able to passage the viral RNA-like CAT gene. (A) CAT activity (103 dilution of cell extract) from the primary transfection; (B) CAT activity (undiluted cell extract) detected following infection with clarified supernatants shown in panel A that were superinfected with influenza B/Yamagata/73 virus; (C) CAT activity (undiluted cell extract) from MDCK cells infected with clarified supernatants shown in panel A or mock superinfected with PBS.
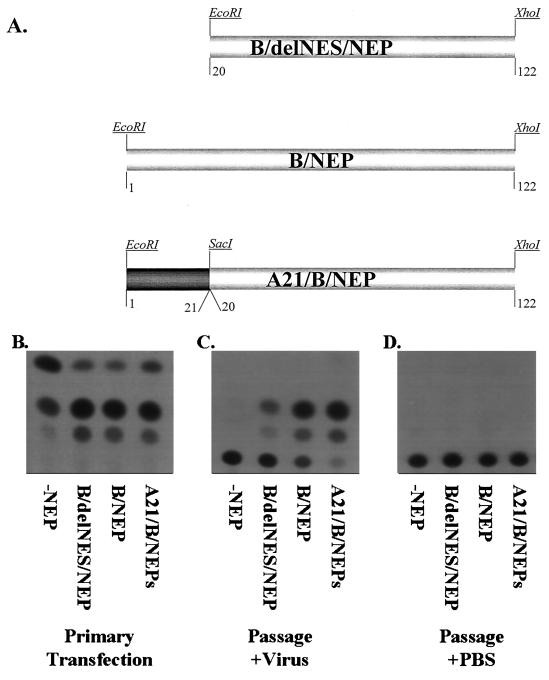
FIG. 6.
The NES contained in the influenza B virus NEP can be functionally replaced by the influenza A virus NEP NES. (A) Diagram of alterations of the influenza B virus NEP gene. B/delNES/NEP is the wild-type NEP with the first 20 aa removed. A MET codon has been added for proper protein translational initiation. B/NEP is the wild-type NEP gene from influenza B/Lee/40 virus. A21/B/NEP has the first 20 aa of the influenza B virus NEP replaced with the established NES contained within the first 21 aa of the influenza A virus NEP. Both ORFs were cloned into the vector pCAGGS. (B through D) 293T cells were transfected with each of the DNA combinations listed in Table 1. Forty-eight hours posttransfection supernatants were mixed with either 106 PFU of influenza B/Yamagata virus or an equivalent volume of PBS and then used to infect fresh MDCK cells. (B) Primary transfection refers to the CAT activity (103 dilution of cell extract) from the transfection of 293T cells. (C) Passage+Virus refers to the CAT activity (undiluted cell extract) detected following infection of MDCK cells with supernatants from the 293T transfection mixed with influenza B/Yamagata/73 virus. (D) Passage+PBS is the CAT activity from MDCK cells infected with supernatants from 293T transfected cells and mixed with PBS.
In passaging experiments, clarified transfected cell supernatants (Table 1, WT) were used to infect fresh MDCK cells (Fig. 5C). No CAT reporter gene activity was detectable. This is the expected result, because vRNPs transferred to the new cells are not expected to transcribe and replicate to detectable reporter gene levels in the absence of a complementing source of NP, PB1, PB2, and PA protein expression. However, when VLP-infected MDCK cells were superinfected with wild-type influenza B/Yamagata/73 virus, the viral RNA-like CAT gene could be amplified. The superinfecting virus serves as a de novo source of NP and polymerase to drive replication and transcription. From these influenza B virus VLP-infected MDCK cells, CAT reporter gene activity could be detected 12 h postinfection (Fig. 5B). This result suggests that the produced VLPs are competent in delivering a viral RNA-like gene into new cells. VLP-mediated passage of the viral RNA-like CAT gene was inhibited by the addition of neutralizing antibodies or by the omission of the HA and/or the NA expression plasmids (data not shown). When a preparation of VLPs generated in the absence of the NEP protein (Table 1, −NEP) was used to infect MDCK cells together with influenza B virus, no CAT activity could be detected (Table 1). Therefore, these VLPs, which are indistinguishable to wild-type VLPs in HA activity, proved to be functionally distinguishable in passaging experiments (Fig. 5B). These results are consistent with the formation of empty VLPs (without viral RNA) in the absence of NEP (Fig. 5B).
Influenza A virus NEP NES can substitute for the influenza B virus NEP NES in the VLP system.
To determine that the function of the proposed NES from influenza B virus NEP is that of a bona fide NES, we have replaced it with an established NES from influenza A virus. The first 20 aa from the influenza B virus NEP were deleted and a methionine start codon was added (Fig. 6A, B/delNES/NEP). In place of the first 20 aa of influenza B virus NEP, the first 21 aa from the influenza A virus NEP was added (Fig. 6A, A21/B/NEP). The VLP assay was used to test the ability of each of the constructs to deliver a functional viral RNA-like CAT gene to a new cell. DNA mixtures (Table 1, WT, −NEP, A21/B/NEP, or B/delNES/NEP) were transfected into 293T cells. Forty-eight hours posttransfection cell supernatants were tested for viral titers. All supernatants contain equal amounts of HA (HA titer = 32). The same clarified supernatants were used to infect fresh MDCK cells. Twelve hours postinfection no CAT activity could be detected (Fig. 6D). When the same supernatants were mixed with 106 PFU of influenza B virus, comparable CAT activity could be detected in the wild type and A21/B/NEP, low levels of activity could be detected in B/delNES/NEP, and no activity could be detected in −NEP (Fig. 6C).
DISCUSSION
We have determined that the NEP proteins from influenza B and C viruses possess characteristics indicative of nuclear export function. From the data presented in this report, we postulate that the viral NEP proteins serve as adapters between influenza vRNPs and the nuclear export receptor Crm1 to form a functional export complex. There are three lines of evidence to support this hypothesis. First, the NEP proteins from influenza B and C viruses interact in the yeast two-hybrid system with a discrete subset of nucleoporins and the nuclear receptor Crm1 (Fig. 1). Second, the influenza B and C virus NEP proteins can substitute for the Rev effector domain in a functional assay when fused to a Rev mutant lacking a functional NES (Fig. 1B, Rev*). Third, when the function of the influenza B virus NEP protein is assayed in the VLP assay, it is essential to produce infectious VLPs (Fig. 4B). These three sets of data suggest that the NEP proteins from influenza B and C viruses act in a manner similar to that of influenza A virus for the export of vRNPs from the nucleus to the cytoplasm.
Evidence that the NEP proteins from influenza B and C viruses are factors in promoting nuclear export is the ability of these proteins to functionally substitute for the effector domain from the Rev protein in the export of an RRE-containing CAT reporter gene (Fig. 2). Previously, this assay had been used to show that the NEP from influenza A virus could also functionally substitute for the Rev effector domain (44). CAT activity is dependent on the nuclear export of an unspliced CAT reporter transcript. The ability of full-length NEP proteins from influenza B and C viruses to substitute for the activity of the effector domain suggests that the NEP molecules contain an authentic NES (26, 36). The various CAT activities obtained in the Rev assay may reflect subtle differences in the strengths of the different wild-type NEP proteins to function as NEP proteins. Alternatively, these differences may be due to different protein stabilities or expression levels of the fusion proteins in transfected cells. When translated in vitro using a coupled transcription and translation system, wild-type and mutant influenza B and C virus NEP proteins appeared to be stably expressed (data not shown). We utilized the Rev nuclear export assay to further map the regions within the influenza B and C virus NEP proteins that can functionally complement the Rev effector domain. The influenza B virus NEP protein contains a functional NES with a sequence that is highly similar to the defined sequence for influenza A virus NEP protein. For the influenza C virus NEP protein, there appear to be two motifs important for nuclear export. Site-directed mutation of either motif resulted in less than 10% activity compared to that of wild-type influenza C virus NEP protein. This preliminary finding may suggest that the influenza C virus NEP protein contains a complex (bipartite) signal. However, at this point we are unable to exclude the possibility that the mutant proteins are misfolding or have altered stabilities. This preliminary result requires further investigation. For the correct sequence of the NS gene for influenza C virus see the report by Hongo et al. (25), later confirmed by Alamgir et al. (1). We have confirmed that there are four G residues at positions 698 to 701 of the NS gene from influenza C/California/78 virus (data not shown) and not three G residues, as an earlier report found (33). Consequently, the deduced number of amino acids for the influenza C virus NEP protein is changed from 116 to 182.
Yeast two-hybrid studies provided direct evidence of the ability of the influenza B and C virus NEP proteins to interact with different components of the cellular nuclear export machinery. The interaction of these two proteins with nucleoporins and the nuclear export receptor Crm1 paralleled the results seen with other viral NEP proteins, such as the HIV-1 Rev protein or the NEP protein from influenza A virus (37, 44). In a report by Neville et al., the authors found that binding of the HIV Rev to nucleoporins is bridged by Crm1 (40). In fact, the NEP proteins from influenza A, B, and C viruses and the HIV-1 Rev protein interact with a specific subset of nucleoporins and not with others. Interaction between the soluble nuclear export receptor Crm1 and the NEP proteins from influenza B and C viruses suggests that the influenza A, B, and C and HIV-1 vRNPs all utilize this specific cellular export pathway. It should be noted that the influenza B virus NEP NES mutant retained the ability to interact with Crm1 in the yeast two-hybrid assay (data not shown). Surprisingly, it was found that an export dead mutant of the influenza B virus NEP retained the ability to bind to Crm1. This result may suggest that a functional signal for nuclear export and a Crm1 recognition motif are separated. This was previously seen for an influenza A virus NEP NES mutant (37). However, neither of the putative influenza C virus NEP NES mutants was able to interact with Crm1 in this assay (data not shown). This may reflect altered stability, as suggested above, or limited sensitivity of the yeast two-hybrid assay to detect this specific interaction. The significance of these differences awaits a convenient genetic system with which to study influenza C virus replication.
The influenza A virus NEP protein is likely to function as an adapter molecule between the vRNP complex and the nuclear pore complex through the Crm1 interaction (37 and this report). However, further work is needed to clarify the specific steps leading to transport through the nuclear pore following formation of a functional vRNP/NEP/Crm1/Ran-GTP complex. In addition, specific interactions between the influenza A virus NEP protein and the vRNP complex are presumably bridged by the influenza A viral matrix (M1) protein, as this protein is known to interact with both the NEP protein and vRNPs (55). Furthermore, several studies have confirmed the requirement of this influenza A viral protein for vRNP export (9, 56, 57). Thus, we favor a model where the NEP protein mediates the export of vRNPs at late times of infection by binding to both M1-containing vRNPs and to Crm1. However, a direct interaction between the NEP protein and the vRNP complex cannot be ruled out. Interactions between the NEP and M1 proteins or between the NEP protein and vRNPs have not yet been analyzed for either influenza B or C virus.
Alternative models of vRNP export have been proposed. A recent report from Bui et al. suggests that the NEP from influenza A virus is not required for the nuclear export of vRNPs but rather that the viral M1 protein is sufficient (9). This conclusion is based on inhibitor studies in virus-infected cells using a broadly acting kinase inhibitor, H7. The authors demonstrate by immunofluorescense that in the presence of the H7 inhibitor, levels of the NEP and M1 proteins are reduced. Transfection of the M1 protein from an expression plasmid was able to complement the defect in nuclear export. Although this study suggests that the M1 protein may be an important player in the nuclear export of influenza A vRNPs, these experiments do not effectively eliminate NEP expression. Undetectable, catalytic amounts of the NEP protein may be present and sufficient for export of vRNPs. In contrast, Elton et al. have reported that the NP protein of influenza A virus interacts with Crm1 and that a vaccinia T7 virus-driven expression of the influenza A virus NP protein was retained in the nucleus in response to the Crm1 inhibitor leptomycin B, suggesting that NP is sufficient for vRNP export (13). The same group found that the subcellular localization of neither the NEP nor M1 proteins was sensitive to leptomycin B treatment. Neither group has studied the packaging of functional vRNPs into VLPs or recombinant viruses. Careful analyses of the requirements for vRNP movement through the pore are likely best accomplished with mutant viruses generated using reverse-genetics techniques. (15, 37, 38).
Results using biochemical, VLP, or recombinant virus systems from different groups have found that the NEP protein from influenza A virus is essential for the packaging and passaging of functional vRNPs (21, 31, 37, 38, 44). To further analyze the importance of the influenza B virus NEP protein in viral replication, the functionality of this viral protein was tested in an influenza B virus-based VLP assay. When the NEP protein expression plasmid was omitted from the transfection mix it was still possible to generate VLPs as measured by HA titer. However, there was a profound functional difference between VLPs generated in the presence or absence of NEP expression. While the former were fully competent for vRNP delivery into new cells, the latter were unable to perform this activity. These differences most likely reflect a retention of the RNA-like CAT gene in the nucleus of cells when NEP expression is omitted, therefore resulting in the generation of empty VLPs. When the proposed NES of influenza B virus NEP was removed, the ability to produce a functional VLP was substantially inhibited. The low levels of delivery of the viral RNA-like CAT gene to new cells when the wild-type NEP was replaced by the B/delNES/NEP construct may reflect low levels of binding to nuclear export factors. Furthermore, the NES from the influenza A virus NEP was able to substitute for the proposed NES of influenza B virus NEP.
These data are consistent with the findings from two other groups using VLP-based experiments from influenza A virus (21, 31, 38). Mena et al. (31) found that the generation of infectious VLPs was dependent on NEP expression. However, another group (20) reported the presence of viral RNA-like structures using RT-PCR assays. Nevertheless, these authors could not rule out the possibility that this finding is an artifact of their vaccinia virus-based expression system (20). Neumann et al. confirmed that the influenza A virus NEP protein and the corresponding NES signal are essential for the export of vRNP complexes. Importantly, the authors found that recombinant viruses containing an altered NEP protein were not viable and attributed the defect in viral replication to the failure of the vRNP to be properly exported from the nucleus, as demonstrated by nuclear retention of the viral NP protein in cells infected with NEP-defective viruses. These data are in complete agreement with previous findings (44) and furthermore appear to be consistent with the conclusions in this report.
Recently, Bullido et al. suggested that the influenza A virus NEP downregulates RNA synthesis in a model template RNA replication/transcription system (10). Although such an activity may point towards a pleiotrophic effect of the influenza virus NEP (NS2) proteins, this finding may also be compatible with the nuclear export function of the NEP (NS2) proteins (37, 44). The transport of RNP complexes out of the nucleus would reduce the amount of available template for viral RNA transcription/replication.
In summary, the NEP proteins from influenza B and C viruses show characteristics similar to those of the NEP protein from influenza A virus. The NEP proteins interact with components of the nuclear pore complex and with Crm1. Each of these viral proteins can functionally substitute for the activity of the Rev effector domain in a Rev-based export assay. Finally, the NEP protein from influenza B virus is essential for the formation of functional VLPs. Taken together we suggest that the vRNPs from influenza B and C viruses are exported from the nucleus in an NEP-dependent manner.
ACKNOWLEDGMENTS
J. Paragas and J. Talon contributed equally to this work.
We thank Bryan R. Cullen and Michael Rosbash for kindly providing yeast expression plasmids and Tristram G. Parslow for providing pDM128. We also thank Roger Brent for providing the yeast two-hybrid system and Yoshihiro Kawaoka for providing cells and plasmids. In addition, we thank Mirella Salvatore for helpful discussions.
This work was supported by grants from the National Institutes of Health to P.P. and A.G.-S.
REFERENCES
- 1.Alamgir A S, Matsuzaki Y, Hongo S, Tsuchiya E, Sugawara K, Muraki Y, Nakamura K. Phylogenetic analysis of influenza C virus nonstructural (NS) protein genes and identification of the NS2 protein. J Gen Virol. 2000;81:1933–1940. doi: 10.1099/0022-1317-81-8-1933. [DOI] [PubMed] [Google Scholar]
- 2.Askjaer P, Bachi A, Wilm M, Bischoff F R, Weeks D L, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj I W, Fornerod M. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barclay W S, Palese P. Influenza B viruses with site-specific mutations introduced into the HA gene. J Virol. 1995;69:1275–1279. doi: 10.1128/jvi.69.2.1275-1279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler C, Palese P. Influenza viruses. In: Creighton T, editor. Encyclopedia of molecular medicine. New York, N.Y: John Wiley and Sons; 2001. [Google Scholar]
- 5.Basler C F, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk H D, Garcia-Sastre A, Palese P. The ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 7.Briedis D J, Lamb R A. Influenza B virus genome: sequences and structural organization of RNA segment 8 and the mRNAs coding for the NS1 and NS2 proteins. J Virol. 1982;42:186–193. doi: 10.1128/jvi.42.1.186-193.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briedis D J, Lamb R A, Choppin P W. Influenza B virus RNA segment 8 codes for two nonstructural proteins. Virology. 1981;112:417–425. doi: 10.1016/0042-6822(81)90289-0. [DOI] [PubMed] [Google Scholar]
- 9.Bui M, Wills E G, Helenius A, Whittaker G R. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J Virol. 2000;74:1781–1786. doi: 10.1128/jvi.74.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullido R, Gomez-Puertas P, Saiz M J, Portela A. Influenza A virus NEP (NS2 protein) downregulates RNA synthesis of model template RNAs. J Virol. 2001;75:4912–4917. doi: 10.1128/JVI.75.10.4912-4917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crescenzo-Chaigne B, Naffakh N, van der Werf S. Comparative analysis of the ability of the polymerase complexes of influenza viruses type A, B and C to assemble into functional RNPs that allow expression and replication of heterotypic model RNA templates in vivo. Virology. 1999;265:342–353. doi: 10.1006/viro.1999.0059. [DOI] [PubMed] [Google Scholar]
- 12.Cullen B R. Nuclear RNA export pathways. Mol Cell Biol. 2000;20:4181–4187. doi: 10.1128/mcb.20.12.4181-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elton D, Simpson-Holley M, Archer K, Medcalf L, Hallam R, McCauley J, Digard P. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J Virol. 2001;75:408–419. doi: 10.1128/JVI.75.1.408-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 15.Fodor E, Devenish L, Engelhardt O G, Palese P, Brownlee G G, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 17.Fritz C C, Green M R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- 18.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 20.Gomez-Puertas P, Albo C, Perez-Pastrana E, Vivo A, Portela A. Influenza virus matrix protein is the major driving force in virus budding. J Virol. 2000;74:11538–11547. doi: 10.1128/jvi.74.24.11538-11547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Puertas P, Mena I, Castillo M, Vivo A, Perez-Pastrana E, Portela A. Efficient formation of influenza virus-like particles: dependence on the expression levels of viral proteins. J Gen Virol. 1999;80:1635–1645. doi: 10.1099/0022-1317-80-7-1635. [DOI] [PubMed] [Google Scholar]
- 22.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 23.Hayden F, Palese P. Influenza virus. In: Richman D D, editor. Clinical virology. New York, N.Y: Churchill Livingstone; 1997. [Google Scholar]
- 24.Herrler G, Durkop I, Becht H, Klenk H D. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J Gen Virol. 1988;69:839–846. doi: 10.1099/0022-1317-69-4-839. [DOI] [PubMed] [Google Scholar]
- 25.Hongo S, Kitame F, Sugawara K, Nishimura H, Nakamura K. Cloning and sequencing of influenza C/Yamagata/1/88 virus NS gene. Arch Virol. 1992;126:343–349. doi: 10.1007/BF01309708. [DOI] [PubMed] [Google Scholar]
- 26.Hope T J, Huang X J, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jambrina E, Barcena J, Uez O, Portela A. The three subunits of the polymerase and the nucleoprotein of influenza B virus are the minimum set of viral proteins required for expression of a model RNA template. Virology. 1997;235:209–217. doi: 10.1006/viro.1997.8682. [DOI] [PubMed] [Google Scholar]
- 29.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 30.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mena I, Vivo A, Perez E, Portela A. Rescue of a synthetic chloramphenicol acetyltransferase RNA into influenza virus-like particles obtained from recombinant plasmids. J Virol. 1996;70:5016–5024. doi: 10.1128/jvi.70.8.5016-5024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakada S, Creager R S, Krystal M, Aaronson R P, Palese P. Influenza C virus hemagglutinin: comparison with influenza A and B virus hemagglutinins. J Virol. 1984;50:118–124. doi: 10.1128/jvi.50.1.118-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakada S, Graves P N, Desselberger U, Creager R S, Krystal M, Palese P. Influenza C virus RNA 7 codes for a nonstructural protein. J Virol. 1985;56:221–226. doi: 10.1128/jvi.56.1.221-226.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakada S, Graves P N, Palese P. The influenza C virus NS gene: evidence for a spliced mRNA and a second NS gene product (NS2 protein) Virus Res. 1986;4:263–273. doi: 10.1016/0168-1702(86)90005-5. [DOI] [PubMed] [Google Scholar]
- 35.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 36.Neufeld K L, Nix D A, Bogerd H, Kang Y, Beckerle M C, Cullen B R, White R L. Adenomatous polyposis coli protein contains two nuclear export signals and shuttles between the nucleus and cytoplasm. Proc Natl Acad Sci USA. 2000;97:12085–12090. doi: 10.1073/pnas.220401797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neumann G, Hughes M T, Kawaoka Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 2000;19:6751–6758. doi: 10.1093/emboj/19.24.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann G, Watanabe T, Kawaoka Y. Plasmid-driven formation of influenza virus-like particles. J Virol. 2000;74:547–551. doi: 10.1128/jvi.74.1.547-551.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann G, Zobel A, Hobom G. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology. 1994;202:477–479. doi: 10.1006/viro.1994.1365. [DOI] [PubMed] [Google Scholar]
- 40.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 41.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 42.O'Neill R E, Jaskunas R, Blobel G, Palese P, Moroianu J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem. 1995;270:22701–22704. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill R E, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 44.O'Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pleschka S, Jaskunas R, Engelhardt O G, Zurcher T, Palese P, Garcia-Sastre A. A plasmid-based reverse genetics system for influenza A virus. J Virol. 1996;70:4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Racaniello V R, Palese P. Influenza B virus genome: assignment of viral polypeptides to RNA segments. J Virol. 1979;29:361–373. doi: 10.1128/jvi.29.1.361-373.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richardson J C, Akkina R K. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Arch Virol. 1991;116:69–80. doi: 10.1007/BF01319232. [DOI] [PubMed] [Google Scholar]
- 48.Ritchey M B, Palese P, Kilbourne E D. RNAs of influenza A, B, and C viruses. J Virol. 1976;18:738–744. doi: 10.1128/jvi.18.2.738-744.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw M W, Choppin P W, Lamb R A. A previously unrecognized influenza B virus glycoprotein from a bicistronic mRNA that also encodes the viral neuraminidase. Proc Natl Acad Sci USA. 1983;80:4879–4883. doi: 10.1073/pnas.80.16.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stutz F, Izaurralde E, Mattaj I W, Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 52.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 53.Vlasak R, Krystal M, Nacht M, Palese P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor-destroying (esterase) activities. Virology. 1987;160:419–425. doi: 10.1016/0042-6822(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 54.Wagner E, Engelhardt O G, Weber F, Haller O, Kochs G. Formation of virus-like particles from cloned cDNAs of thogoto virus. J Gen Virol. 2000;81:2849–2853. doi: 10.1099/0022-1317-81-12-2849. [DOI] [PubMed] [Google Scholar]
- 55.Ward A C, Castelli L A, Lucantoni A C, White J F, Azad A A, Macreadie I G. Expression and analysis of the NS2 protein of influenza A virus. Arch Virol. 1995;140:2067–2073. doi: 10.1007/BF01322693. [DOI] [PubMed] [Google Scholar]
- 56.Whittaker G, Bui M, Helenius A. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J Virol. 1996;70:2743–2756. doi: 10.1128/jvi.70.5.2743-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whittaker G, Kemler I, Helenius A. Hyperphosphorylation of mutant influenza virus matrix protein, M1, causes its retention in the nucleus. J Virol. 1995;69:439–445. doi: 10.1128/jvi.69.1.439-445.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whittaker G R, Kann M, Helenius A. Viral entry into the nucleus. Annu Rev Cell Dev Biol. 2000;16:627–651. doi: 10.1146/annurev.cellbio.16.1.627. [DOI] [PubMed] [Google Scholar]
- 59.Wolff T, O'Neill R E, Palese P. Interaction cloning of NS1-I, a human protein that binds to the nonstructural NS1 proteins of influenza A and B viruses. J Virol. 1996;70:5363–5372. doi: 10.1128/jvi.70.8.5363-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasuda J, Nakada S, Kato A, Toyoda T, Ishihama A. Molecular assembly of influenza virus: association of the NS2 protein with virion matrix. Virology. 1993;196:249–255. doi: 10.1006/viro.1993.1473. [DOI] [PubMed] [Google Scholar]