Abstract
The small-ruminant lentiviruses ovine maedi-visna virus (MVV) and caprine arthritis-encephalitis virus (CAEV) cause encephalitis, progressive pneumonia, arthritis, and mastitis in sheep and goats. Icelandic MVV strains, which are lytic in tissue culture, have a wide species distribution of functional receptors, which includes human cells. In contrast, functional receptors for the nonlytic CAEV CO are absent from human cells. To determine if the wide species distribution of functional receptors is a common property of MVV strains or related to cytopathic phenotype, we tested the infectivity of viruses pseudotyped with the envelope glycoproteins of MVV K1514, CAEV CO, and lytic and nonlytic North American MVV strains to cells of different species. Replication-defective CAEV proviral constructs lacking the env, tat, and vif genes and carrying the neomycin phosphotransferase gene in the vif-tat region were developed for the infectivity assays. Cotransfection of human 293T cells with these proviral constructs and plasmids expressing CAEV, MVV, or vesicular stomatitis virus envelope glycoproteins produced infectious pseudotyped virus which induced resistance of infected cells to G418. Using these pseudotypes, we confirmed the wide species distribution of Icelandic MVV receptors and the narrow host range of CAEV. However, functional receptors for the two North American MVV strains tested, unlike the Icelandic MVV and similar to CAEV, were limited to cells of ruminant species, regardless of cytopathic phenotype. The results indicate a differential receptor recognition by MVV strains which is unrelated to cytopathic phenotype.
The ovine maedi-visna virus (MVV) and caprine arthritis-encephalitis virus (CAEV) are related but genetically distinct lentiviruses that cause encephalitis, chronic synovitis, progressive interstitial pneumonia, and mastitis in sheep and goats (27). Like other lentiviruses, CAEV and MVV infect cells of the monocyte/macrophage lineage and dendritic cells (26, 29), but the receptor used by these viruses and whether receptor utilization is a determinant of pathogenesis are unknown.
The small-ruminant lentivirus env genes have been segregated by phylogenetic analyses into at least four different clades (42). The Icelandic, South African, and British isolates of MVV form clade I, whereas the prototypic North American and French CAEV isolates form clade V. Other clades are composed of European (clade IV) and North American (clade II) strains of MVV. Ovine and caprine lentiviruses also differ in their biological properties, especially cytopathic phenotype (28), which may reflect their env sequence diversity. The Icelandic MVV strains induce syncytia and lysis of infected tissue culture monolayers and are classified as lytic. In contrast, CAEV strains induce syncytia with persistent infection of tissue culture monolayers and are classified as persistent or nonlytic.
Prototypic CAEV and MVV strains also differ in their in vitro species tropism. The Icelandic MVV strain K1514 has been shown to replicate in bovine, swine, and human cells in vitro (9, 19, 38). In addition, MVV K1514 virions or recombinant envelope glycoproteins can induce fusion of sheep, goat, human, monkey, mouse, and horse cells (1, 8, 43), indicating that Icelandic MVV can use receptors from a wide range of species for entry and/or cell-to-cell fusion. Similarly, the envelope glycoproteins of the British MVV strain EV-1 induce syncytia, and this strain enters cells from many different species (18), indicating that wide species tropism of MVV is not limited to the Icelandic strains. In contrast, human cells are refractory to infection by CAEV CO, and this restriction occurs at the receptor level (24), indicating a differential species tropism between ovine and caprine lentiviruses. In this regard, one study suggested that CAEV CO and MVV K1514 have different binding sites on the surface of susceptible sheep and goat cells, which may indicate differential receptor usage of these two viruses (12). In the same study, a nonlytic small-ruminant lentivirus (strain S93) isolated from a North American sheep (12, 13) utilized the same binding site as CAEV, suggesting a differential receptor usage between lytic and nonlytic small-ruminant lentiviruses. Here, we determined whether species distribution of receptors allowing entry of MVV and CAEV strains is related to cytopathic phenotype. Four small-ruminant lentivirus strains were selected for this study: CAEV CO, MVV K1514, and two independent North American MVV strains, the lytic strain MVV 85/34 (16, 17) and the nonlytic MVV S93 (13). In order to eliminate the possibility of differential postentry blocks to infection, we developed replication-deficient CAEV proviral clones which can be pseudotyped with the envelope glycoprotein of any small-ruminant lentivirus, allowing direct determination and comparison of species tropism for different strains. We show that North American MVV strains have a narrow species distribution of receptors, similar to CAEV, which is independent of their cytopathic phenotype.
MATERIALS AND METHODS
Cell lines and viruses.
The U87-MG (HTB-14), HeLa-S3 (CCL-2.2), Vero (CCL-81), NIH 3T3 (CRL-1658), MDBK (CCL-22), and CHO-K1 (CCL-61) cell lines were from the American Type Culture Collection. The 293T cell line was obtained from Richard Sutton. Goat synovial membrane (GSM) cells were derived as previously described (14). EK cells were expanded from equine kidney biopsies (20). All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mM glutamine and 10% fetal bovine serum (FBS). Culture medium for CHO-K1 cells was also supplemented with 40 mM l-proline. MVV S93 and K1514 were obtained from Opendra Narayan and grown in GSM cells in DMEM supplemented with 2% FBS.
Construction of Env-expressing plasmids.
Plasmid pMEVSV-G, expressing the vesicular stomatitis virus (VSV) G glycoprotein, was obtained from Richard Sutton. Plasmid pCMVCO2, expressing the CAEV CO env gene, was obtained by subcloning the 4.1-kbp SmaI-BglII fragment of plasmid pTATENV (3) between the HindIII and BamHI sites of pCR3 (Invitrogen). The 3.2-kbp EcoRI-NotI fragment from plasmid pLG1.35, containing the MVV 85/34 env gene (25), was subcloned into pLGCMV, a low-copy-number expression vector with the pSC101 origin of replication and the expression cassette of pCR3 (I. Hötzel, unpublished data) to create plasmid pCMV34. The MVV strain K1514 and S93 env genes were obtained by PCR amplification from genomic DNA of infected GSM cells. The MVV S93 env gene was amplified with primers 34ENVF (5′-GGGGAATTCAGGATGGCAAGCAAAAACAGACCGAG-3′; the restriction site used for cloning is underlined, and the initiation codon is bold) and VISNAENVRX (5′-GGGCTCGAGTTACATTTGCTGTGTCATCCTGACTA-3′) using 100 ng of genomic DNA from MVV S93-infected GSM cells as the template. The env gene of MVV K1514 was amplified with primers VISNAENVFE (5′-GGGGAATTCAGGATGGCCAGCAAAGAAAAGTAAGCC-3′) and VISNAENVRX using 100 ng of genomic DNA from MVV K1514-infected GSM cells as the template. All PCR amplifications were done with an annealing temperature of 50°C for 35 cycles. The amplification products were digested with EcoRI and XhoI, gel purified, and cloned between the EcoRI and XhoI sites of pLGCMV to yield plasmids pCMV93 and pCMV1514.
Screening of plasmids expressing biologically active Env.
Clones pCMVCO2, pCMV34, and four independent pCMV93 and pCMV1514 clones were tested for biological activity in a syncytial assay with GSM cells. Plasmid DNA from clones containing the inserts were prepared using a Qiagen Midiprep kit, and 3 μg of DNA from each clone was used to transfect GSM cells in six-well plates using 25 μl of Lipofectamine reagent (Gibco-BRL). Transfected GSM cells were fixed with methanol 36 h posttransfection, stained with Giemsa, and observed for syncytium formation. The pCMV93 and pCMV1514 clones inducing syncytia in GSM cells were sequenced by the dideoxy termination method using an Applied Biosystems ABI 377 sequencer.
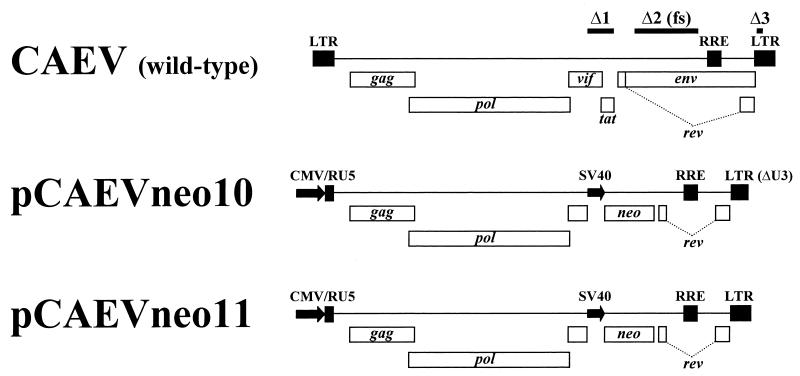
Construction of pCAEVneo10 and pCAEVneo11.
Two replication-deficient CAEV proviral clones carrying the neomycin phosphotransferase (neo) gene under the control of the simian virus 40 (SV40) early promoter in a CAEV CO background were created to produce pseudotyped viruses. The 7-kbp HindIII-BamHI fragment of the large 9.4-kbp CAEV CO plasmid (30) was subcloned into plasmid pCR3 to create pCR3CO5. Plasmid pCO5 was created by inserting a cytomegalovirus (CMV) promoter upstream from the 5′ HindIII site of the insert of pBSCO5, with the CMV TATA box placed in an identical position, relative to the RNA initiation site, as the wild-type CAEV long terminal repeat (LTR) TATA box. To eliminate the SV40-neo cassette of pCO5, the 8.8-kbp BamHI-PvuI fragment of this plasmid was ligated to the 1.7-kbp BamHI-PvuI fragment of pBluescript II SK(−) (Stratagene) to yield plasmid pBSCO5. Thus, the pBSCO5 plasmid contains the 5′ half of the CAEV proviral genome driven by a CMV promoter. Two similar plasmids containing the 3′ end of the CAEV genome, pCO3 and pCO4, were then created. The 195-bp HindIII-ApaI fragment of pCO5 containing the R/U5 regions of the LTR was inserted between the HindIII and ApaI sites of pCMVCO2 to create pCO3ΔHindIII. The 0.4-kbp HindIII insert of the small CAEV CO plasmid (17) was inserted into the HindIII site of pCO3ΔHindIII in the sense orientation to create pCO3. A U3 fragment with a 127-bp deletion from nucleotide 8951 to nucleotide 9077, which includes the CAEV TATA box, was created by recombinant PCR and inserted in the HindIII site of pCO3ΔHindIII in the sense orientation to produce pCO4. A 1.6-kbp fragment with the Rev response element (RRE), the third exon of rev, and the 3′ LTR was amplified by PCR and inserted between the HincII and the 3′ end of pBSCO5 to create pCAEVΔenv11. This plasmid includes the whole CAEV proviral sequence with a 1,330-bp deletion in env. An SV40-neo cassette obtained by PCR was inserted between the unique SbfI and SmaI sites of pCAEVΔenv11 in the vif-tat region to yield plasmid pCAEVneo11. pCAEV neo10 was then obtained by replacing the 3′ end of pCAEVneo11 with the equivalent fragment of pCO4 containing the deleted 3′ U3 region. All cloning steps involving PCR were checked by sequencing the inserts to confirm the absence of PCR misincorporations. All clones were stably propagated in Escherichia coli JM109 at 37°C except pCAEVΔenv11, pCO3ΔHindIII, pCO3, and pCO4, which were moderately unstable and allowed growth of small colonies only. Further details of plasmid construction and primers used are available on request.
Production of pseudotyped viruses.
CAEVneo pseudotyped with the various envelopes was produced by cotransfecting 106 293T cells in 6-cm plates with 6 μg of pCAEVneo10 or pCAEVneo11 plasmid and 6 μg of each Env-expressing clone or pCR3 without insert by the calcium phosphate coprecipitation procedure using a ProFection mammalian transfection kit (Promega). Culture medium was replaced the day after transfection with 4 ml of DMEM–10% FBS. Supernatants were collected 40 h posttransfection, cleared from cell debris by centrifugation at 3,000 × g for 20 min at 4°C, and used immediately.
Titration of pseudotyped viruses.
Supernatants with pseudotyped CAEVneo were titrated by a modification of a previously described procedure to titrate retrovirus vectors carrying the neo gene (21). On day 1, cells were plated in six-well plates (2.5 × 105 cells/well) and incubated overnight. On day 2, cells were infected with pseudotyped viruses diluted in 1 ml of DMEM–10% FBS without Polybrene. Cells were incubated with virus for 2 h at 37°C with occasional gentle agitation of plates, and 2 ml of DMEM–10% FBS was added to each well. On day 3, cells were detached from plates with trypsin and plated on 6-cm plates at 1:2 to 1:500 dilutions. Cells were incubated in growth medium with G418 (USB). The concentration of active G418 used was 1 mg/ml for HeLa, CHO-K1, and MDBK cells, 750 μg/ml for GSM, EK, NIH 3T3, and Vero cells, and 400 μg/ml for the U87-MG cells. Cells were allowed to grow for an additional 8 to 10 days until cells in control plates were dead, they were stained with a 2% crystal violet solution in 20% ethanol, and the colonies were counted. Titers are expressed as CFU per milliliter after dividing the number of colonies by the virus volume used and multiplying by the cell dilution factor. Titers of viruses for which no colonies were observed in plates with the lowest virus and cell dilutions are expressed as less than the minimum detectable titer for each case. Titrations were repeated at least twice, with titers differing less than threefold between experiments.
Derivation of GSM cells stably carrying CAEVneo proviruses and semiquantitative RT-PCR assays.
GSM cells in 25-cm2 flasks (6 × 105 cells per flask) were infected with CAEVneo10 or CAEVneo11 virus pseudotyped with the vesicular stomatitis virus (VSV) G glycoprotein, obtained from transfected 293T cells, at a multiplicity of infection of 0.25 CFU per cell. Infected cells were selected with G418 as described above and passaged under selective conditions until all the cells in the control plate were dead. Infected and uninfected GSM cells were plated on 75-cm2 flasks (2 × 106 cells per flask), the culture medium was changed daily, and cells were used to isolate total RNA 4 days postplating. Total RNA was extracted from infected and uninfected GSM cells using the Trizol reagent (Gibco-BRL) as recommended by the manufacturer. RNA (1 μg in a 20-μl reaction) was reverse transcribed with a 1st Strand cDNA synthesis kit with avian myeloblastosis virus reverse transcriptase (RT) (Roche) using oligo(dT)15 as a primer. Five microliters of cDNA was amplified with 100 ng of primers P1 (5′-CTTCGGGGACGCCTGAAGGAGTAA-3′) and P3 (5′-GGCACGGCTCCAAGCTTTCTGTAC-3′) or P2 (5′-TTCGCAGCGCATCGCCTTCTATCG-3′) and P3 in a 50-μl reaction. In addition, amplifications were also done with primers P1, P2, and P3 (80 ng each) in the same 50-μl reaction. All amplifications were done with an annealing temperature of 51°C for 30 s, extension at 72°C for 2 min, and denaturation at 94°C for 30 s for 35 cycles using Platinum Taq DNA polymerase (Gibco-BRL). Ethidium bromide-stained agarose gels with the amplification products were scanned and analyzed by one-dimensional densitometry using a ChemiImager 4000 (Alpha Innotech).
Nucleotide sequence accession numbers.
The MVV S93 and K1514 env sequences described here are available from GenBank with accession numbers AF338226 and AF338227, respectively.
RESULTS
Characterization of plasmids expressing functional envelope glycoproteins of small-ruminant lentivirus strains.
The env gene of the nonlytic small-ruminant lentivirus strains CAEV CO and MVV S93 and the lytic MVV strains K1514 and 85/34 were cloned in the mammalian expression vectors pCR3 and pLGCMV. GSM cells transfected with plasmids pCMVCO2, pCMV34, pCMV93, and pCMV1514, encoding the env genes of CAEV CO and MVV strains 85/34, S93, and K1514, respectively, but not those transfected with pCR3 formed large syncytia (not shown), indicating that these clones express biologically active envelope glycoproteins.
The MVV S93 env gene had not been sequenced. Therefore, we sequenced the insert of plasmid pCMV93 to determine its similarity to the env genes of other MVV and CAEV strains. Blast searches indicated that the sequence of MVV S93 env was most closely related to the env gene of MVV 85/34 (Fig. 1). However, amino acid sequence identity between MVV 85/34 and S93 was only 86.8% in the full-length Env precursor and 87.8% in the SU region. The MVV S93 env had a 3-bp insertion in the leader region of the Env precursor relative to MVV 85/34 env. Amino acid sequence identity between the SU of MVV S93 and CAEV CO or MVV K1514 was 75 and 73.2%, respectively, similar to the sequence identity between the SU of these strains and MVV 85/34 (40). Thus, the MVV S93 env gene is closely related to the env genes of clade II North American MVV strains.
FIG. 1.
Alignment of predicted amino acid sequence of MVV 85/34 (GenBank accession number U64439) and S93 (AF338226) Env precursors. Only the residues of S93 Env not conserved with 85/34 are shown. Dots represent amino acid identity with MVV 85/34 Env, and dash represents a gap introduced into the 85/34 sequence for optimal alignment. The amino acid residue positions are indicated on the right of the alignment. The boundaries of the leader peptide, SU, and TM subunits of the Env precursor are indicated above the alignment. The putative cleavage site between the leader peptide and SU is in the position homologous to the chemically defined amino terminus of CAEV 63 SU (15).
The insert of plasmid pCMV1514 was also sequenced to confirm that it was indeed derived from MVV K1514. The env gene in this plasmid was most closely related to the env gene from a defective MVV K1514 provirus (35), although with a few differences, including an insertion of a serine residue at position 581 in the principal neutralization domain (32). The insertion and all nonsynonymous point mutations (S495L, I534T, Q818R, and H900R) except one in the leader peptide of Env (V32A) were consistently found in the sequences of other clones derived from the same or independent PCR amplifications, indicating that these variations were present in our MVV K1514 stock and were not due to PCR errors. Thus, the env gene of pCMV1514 is closely related to previously described MVV K1514 env sequences.
Construction and characterization of CAEV proviral clones encoding replication-deficient viruses carrying the neomycin phosphotransferase gene.
Plasmids pCAEVneo10 and pCAEVneo11, carrying modified CAEV CO proviral sequences, were produced for the infectivity assays. Both clones have a 1,330-bp out-of-frame deletion (Δ2) in env and a 525-bp deletion (Δ1) in the vif-tat region between the unique SbfI and SmaI restriction sites, where the 1,161-bp SV40-neo cassette was inserted (Fig. 2). These clones lack tat and encode a truncated and defective Vif. Thus, viruses produced by pCAEVneo10 and pCAEVneo11 are expected to be replication deficient in GSM cells. pCAEVneo10 has a 3′ LTR with a 127-bp deleted sequence (Δ3) which includes the TATA box and some promoter-enhancer elements of the LTR (Fig. 2) and was intended to function as a self-inactivating virus to minimize the production of replication-competent virus by recombination events between the proviral and envelope plasmids. However, the 3′ LTR of pCAEVneo10 retains 131 bp of the U3 region, including one of the 70-bp repeat units containing AP1 and GAS enhancer elements (10, 11, 39). In contrast, pCAEVneo11 has a full-length 3′ LTR (Fig. 2) and should produce virus which is transcriptionally competent in permissive cells. Expression of proviruses in either plasmid is driven by a CMV early promoter replacing the U3 region in the 5′ LTR, which obviates the possible need for Tat trans-activation of the LTR (31) in the producer 293T cells.
FIG. 2.
Structure of pCAEVneo10 and pCAEVneo11 proviral constructs compared to wild-type CAEV provirus. Positions of viral and neo genes are shown with boxes. Bars labeled Δ1, Δ2, and Δ3 indicate the deletions in the vif-tat and env genes and the 3′ U3 region, respectively, present in the pCAEVneo constructs. Deletion Δ2 also introduced a frameshift (fs) mutation in env. Bent dotted lines indicate the intron separating rev exons 2 and 3. The human CMV and SV40 early promoters are shown as arrows. R/U5, R and U5 regions of the 5′ LTR.
As neither pCAEVneo clone encodes a functional envelope glycoprotein, infectious particles should only be produced if Env is provided in trans. To test whether the CAEV proviral clones produce infectious virus, 293T cells were cotransfected with pCAEVneo10 or pCAEVneo11 and plasmid pMEVSV-G, expressing the pantropic VSV-G glycoprotein. Clarified supernatants from transfected cells were used to infect GSM cells which were then grown in culture medium with G418. After an initial phase of moderate cell death, GSM cells infected with CAEVneo10 or CAEVneo11 pseudotyped with VSV-G [CAEVneo10(VSV) and CAEVneo11(VSV), respectively] grew under selective conditions. In contrast, all the cells incubated with CAEVneo10 and CAEVneo11 without envelope glycoproteins died after a few days under the same conditions. Thus, both pCAEVneo10 and pCAEVneo11 were able to produce infectious virus in 293T cells when trans-complemented with an envelope glycoprotein.
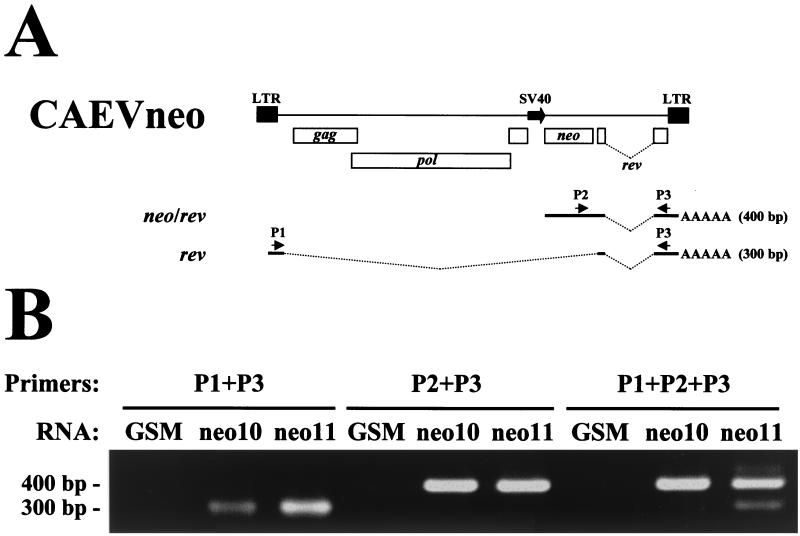
The self-inactivating phenotype of CAEVneo10 was tested by measuring the level of LTR-derived transcripts in infected cells by a semiquantitative RT-PCR assay using the spliced SV40-derived neo/rev transcript as an internal standard. The forward primers used were P1, which hybridizes to the 5′ leader sequence, and P2, which hybridizes to the neo sequence (Fig. 3A). Reverse primer P3, which hybridizes to the third exon of rev and the predicted second exon of neo/rev, was used in reactions with either forward primer. Reactions with forward primers P1 and P2 are predicted to yield products of 300 and 400 bp, respectively. Semiquantitative RT-PCR was performed with all three primers simultaneously, with a limiting concentration of reverse primer P3 relative to the forward primers to stop the reaction when either of the forward primers was exhausted. Semiquantitative reactions were done using oligo(dT)-primed cDNAs from total RNA isolated from uninfected GSM cells and GSM cells infected with CAEVneo10(VSV) or CAEVneo11(VSV) selected with G418. As expected, the neo/rev single-spliced 400-bp RT-PCR product was detected in reactions with CAEVneo10- and CAEVneo11-infected GSM cell cDNA (Fig. 3B), confirming the infectivity of CAEVneo to GSM cells. The 300-bp RT-PCR product derived from the double-spliced rev transcript was also detected in reactions using cDNA from GSM cells infected with CAEVneo10 or CAEVneo11 (Fig. 3B). No amplification products were detected in reactions with cDNA from uninfected GSM cells (Fig. 3B). HaeIII digestion of PCR products yielded restriction fragments of the expected size, confirming the specificity of the products (not shown). The 300- and 400-bp products were detected in the semiquantitative RT-PCR with cDNA from CAEVneo11-infected GSM cells at a ratio of 1:3, as determined by one-dimensional densitometry of scanned gels. However, while the signal of the 400-bp product was very strong in the semiquantitative RT-PCR using cDNA from CAEVneo10-infected GSM cells, the signal of the 300-bp fragment was very weak. In PCRs with cDNA derived from CAEVneo10-infected cells, the ratio of the concentration of these products was only 1:42. Using the SV40-derived 400-bp product as an internal standard, the level of LTR-derived transcripts was determined to be 14 times lower in CAEVneo10-infected cells than in CAEVneo11-infected cells. The results indicate that the LTR of CAEVneo10 proviruses was still able to direct RNA synthesis in permissive cells, although at a very low level, probably due to the presence of AP-1 or other enhancer elements in the nondeleted regions of U3 of pCAEVneo10.
FIG. 3.
Semiquantitative RT-PCR assay of double-spliced rev transcripts in GSM cells infected with CAEVneo10 or CAEVneo11. (A) The CAEVneo10 and CAEVneo11 integrated proviral structure is shown to indicate the relative positions of primers P1, P2, and P3. Thick lines indicate the rev and predicted neo/rev exons. Bent dotted lines indicate introns. The expected sizes of amplicons obtained by RT-PCR for each transcript are shown on the right. (B) Ethidium bromide-stained agarose gel with RT-PCR products. The size of the amplified fragments is indicated on the left. Primers for PCR are indicated above the gel. The RNA used for RT-PCR is indicated above each lane. GSM, uninfected GSM cell RNA; neo10, CAEVneo10-infected GSM cell RNA; neo11, CAEVneo11-infected GSM RNA.
Infectivity of pseudotyped CAEVneo10 and CAEVneo11 to cell lines of different species.
It has been shown that CAEV pseudotyped with the VSV-G glycoprotein can infect human cells and that there is no postentry block to infection in human cells (24). We extended these results to cells from other species. Both CAEVneo10 and CAEVneo11 were titrated to determine whether the deletion in the 3′ LTR of pCAEVneo10 has any effect on virus production or infectivity. CAEVneo10 and CAEVneo11 pseudotypes were produced in 293T cells and used to infect cell lines from goat (GSM), human (HeLa and U87-MG), African green monkey (Vero), bovine (MDBK), equine (equine kidney [EK]), Chinese hamster (CHO-K1), and NIH Swiss mouse (NIH 3T3) origin.
Infection with CAEVneo10(VSV) and CAEVneo11(VSV) but not with CAEVneo10 or CAEVneo11 produced without envelope glycoproteins was confirmed in all cell lines tested, with similar titers for CAEVneo10 and CAEVneo11 pseudotypes in all cell types (Table 1 and not shown), indicating that the 127-bp deletion in the U3 region of pCAEVneo10 did not affect virus production or infectivity. Titers of pseudotyped viruses varied between cells of different species. The goat, bovine, and monkey cell lines were the most permissive to entry. The human cell lines HeLa and U87-MG were about 10 times less permissive to entry than monkey cells. For the U87-MG cell line, this is at least partially due to the lower plating efficiency of this cell line at the low cell densities required for efficient G418 selection because many cells that survived the initial drug selection failed to form visible colonies. Cells of rodent origin were up to 25 times less permissive to infection than cells of ruminant or primate origin. EK cells were 8 to 500 times less permissive to entry by VSV-G-pseudotyped virus than other cell types. The results indicate that cells of many species are permissive to postentry steps of infection by CAEV.
TABLE 1.
Host range of CAEVneo10 pseudotyped with the envelope glycoproteins of CAEV and MVV strains
| Target cell | Titer (CFU/ml) of CAEVneo10 pseudotyped with envelope glycoproteina:
|
|||||
|---|---|---|---|---|---|---|
| Noneb | CAEV CO | MVV S93 | MVV 85/34 | MVV K1514 | VSV-G | |
| GSM | <2 | 8.8 × 104 | 5.3 × 104 | 2.1 × 105 | 9.5 × 104 | 1.1 × 105 |
| MDBK | <20 | 1.1 × 104 | 6.0 × 103 | 8.1 × 104 | 3.1 × 104 | 5.6 × 105 |
| Vero | <20 | <10 | <10 | <10 | 3.7 × 104 | 7.3 × 105 |
| HeLa | <20 | <4 | <4 | 4 | 2.1 × 103 | 7.3 × 104 |
| U87-MG | <20 | <4 | <4 | <4 | 8.0 × 103 | 2.6 × 104 |
| EK | <4 | 4 | <2 | <2 | 3.6 × 102 | 1.3 × 103 |
| NIH 3T3 | <20 | <10 | <10 | <10 | <10 | 6.7 × 104 |
| CHO-K1 | <20 | <10 | <10 | 10 | 10 | 3.4 × 104 |
Cells were infected with 0.1 ml of virus pseudotyped with the VSV-G glycoprotein or 0.5 ml of virus without envelope and passaged 24 h postinfection undiluted or at dilutions of 1:2 to 1:500 in selective medium as indicated in Materials and Methods. Representative results from at least two titration experiments are shown. Titers of each pseudotype in independent titrations varied by less than threefold.
Control virus without envelope glycoprotein, produced by cotransfection of 293T cells with pCAEVneo10 and control plasmid pCR3.
We tested whether the same cell lines could be infected by virus pseudotyped with the envelope glycoproteins of small-ruminant lentiviruses. For these experiments we used only CAEVneo10, as the 14-fold-lower transcriptional activity of its LTR should minimize secondary infection events by env+ replication-competent recombinant viruses that could arise in the transfected producer cells.
Results of a representative titration experiment are shown in Table 1. The CAEVneo10(K1514) pseudotype infected the human HeLa and U87-MG cells and monkey Vero cells as well as goat, bovine, and equine cells, confirming the broad species distribution of host cell receptors for the MVV K1514 strain. As expected, GSM cells were the most permissive to entry by CAEVneo10(K1514). The titer of CAEVneo(K1514) in EK cells was much lower than in other permissive cell types. However, as the titer of CAEVneo10(VSV) in EK cells was also very low, it appears that the MVV K1514 envelope can use the equine receptor for entry with relative efficiency. The Chinese hamster cell line CHO-K1 was resistant to infection by CAEVneo10(K1514). However, in contrast to previous results showing the induction of syncytia in mouse cells by MVV K1514 Env (57), titers of CAEVneo10(K1514) in mouse NIH 3T3 cells were below detection level (<10 CFU/ml), indicating that these cells do not express functional MVV K1514 receptors allowing virus entry.
CAEVneo10(CO) was able to efficiently infect not only goat cells but also the bovine kidney cell line MDBK, although with lower efficiency, indicating that the CAEV receptor is expressed not only in goat and sheep cells (5) but also in bovine cells (Table 1). Human cells (HeLa and U87-MG) that were susceptible to CAEVneo10(K1514) were resistant to CAEVneo10(CO), confirming previous results showing the absence of functional CAEV receptor in human cells. In addition, monkey, equine, mouse, and Chinese hamster cells were also resistant to CAEVneo10(CO). In the case of equine cells, the low infection observed in the experiment shown in Table 1 is probably due to background infection or a rare spontaneous cell mutant, as EK cells were refractory (<4 CFU/ml) to CAEVneo10(CO) entry in other experiments. These results confirm and extend previous reports demonstrating the narrow species distribution of CAEV CO receptors.
Results with CAEVneo10(S93) and CAEVneo10(85/34) were similar to those obtained with CAEVneo10(CO) (Table 1). CAEVneo10(S93) and CAEVneo10(85/34) infected GSM and MDBK cells with titers ranging from 6 × 103 to 2.1 × 105 CFU/ml. However, infectivity of CAEVneo10(S93) and CAEVneo10(85/34) to the human, monkey, and horse cells susceptible to CAEVneo10(K1514) was limited to below or near background levels. The mouse and Chinese hamster cells resistant to CAEVneo(K1514) were also resistant to CAEVneo10 pseudotyped with the env of MVV 85/34 or S93. These results indicate that North American MVV strains can enter cells of a narrow range of species, more similar to the limited tropism of CAEV than to the wide tropism of the Icelandic MVV K1514.
DISCUSSION
Here we examined the ability of envelope glycoproteins of North American strains of MVV with different cytopathic phenotypes to mediate entry into cells of different species. A pseudotype system based on a selectable, replication-defective CAEV was developed for the infectivity assays. This system allowed unambiguous determination of virus entry, which is independent of blocks in postentry steps of replication due to differences in the gag and pol genes encoding the internal structural proteins in different small-ruminant lentiviruses. Using these pseudotypes, we showed that the functional receptor(s) for two North American MVV strains was limited to ruminant species, although we cannot eliminate the possibility of a low level of infection of EK cells due to the low postentry permissiveness of this cell type. The system was sensitive enough to allow detection of infection of nonruminant cells, as demonstrated by the ability of CAEVneo10(K1514) to infect all primate cell lines tested. Therefore, the lack of infectivity of CAEVneo10(S93) and CAEVneo10(85/34) to nonruminant cells was not due to low sensitivity of the assay. Furthermore, CAEVneo10(85/34) titers in GSM and MDBK cells were consistently higher than those of CAEVneo10(K1514), indicating that the difference in host range of the two pseudotypes is not due to a less efficient incorporation of MVV 85/34 Env into virions or a generally lower infectivity of the MVV 85/34 pseudotypes. Similar results were also obtained with a CAEV pseudotype system encoding the puromycin resistance gene (Hötzel, unpublished).
The pattern of receptor distribution for MVV 85/34 and S93 is more similar to the narrow species distribution of the CAEV receptor than to wide receptor distribution for Icelandic and British MVV strains. Differential species tropism of MVV strains may be related to previous results showing that virus binding sites in goat and sheep cells appear to differ between the nonlytic MVV S93 and lytic MVV K1514 (12). The different pattern of species tropism in MVV strains would support the interpretation that North American MVV and CAEV strains use a different receptor(s) than MVV K1514. We are currently determining whether small-ruminant lentivirus strains use the same or different receptors for entry into sheep and goat cells.
The MVV strains used in this study allowed determination of the relationship between cytopathic phenotype and host range of small-ruminant lentiviruses. Human immunodeficiency virus type 1 (HIV-1) strains vary in their cytopathic potential, and this variation is related to coreceptor usage (6, 33, 36). However, as both lytic and nonlytic North American MVV strains have a narrow receptor distribution, the possible differential receptor usage between these strains and the Icelandic MVV K1514 seems to be unrelated to cytopathic phenotype, as previously suggested (12). Interestingly, pseudotype titers appeared to correlate with the cytopathic phenotype of the strain from which env was derived. MVV 85/34 is the most fusogenic and lytic of the MVV strains used here, and its envelope consistently yielded higher titers of pseudotyped virus in GSM and MDBK cells than the envelopes of other strains, including CAEV CO. Conversely, MVV S93 env, which induces very few syncytia in infected GSM cells, consistently yielded the lowest pseudotype titers. The reason for the difference in titers is not known, but it could be due to better surface expression and/or incorporation into virions of envelope glycoproteins from the lytic strains, or it could be related to the efficiency with which different envelopes bind their receptors or trigger membrane fusion.
Some discrepancies between our results and previously published work were noticed. First, syncytium formation does not appear to reliably indicate the presence of functional receptors for small-ruminant lentivirus strains. For instance, 293T cells transfected with the MVV K1514 env plasmid formed large syncytia upon cocultivation with Vero or GSM cells but not with susceptible HeLa cells (not shown), similar to previous results with an MVV K1514-related env plasmid (43). This indicates that, similar to other retroviruses (34), receptor density or additional factors are involved in syncytium formation by small-ruminant lentivirus envelope glycoproteins and that failure to induce fusion by Env does not necessarily indicate absence of functional receptors. Therefore, infectivity assays appear to be more reliable than syncytium assays to determine the presence of functional receptors for small-ruminant lentiviruses. Another significant discrepancy between our results and published work is the species distribution of MVV K1514 receptors. Mouse cells have been shown to form syncytia when expressing MVV EV-1 Env or cocultured with cells expressing MVV K1514 (LV1–1 variant) Env (18, 43). In addition, MVV EV-1 can enter mouse cells, indicating that mouse cells can express functional MVV receptors (18). Furthermore, hamster-mouse somatic cell hybrids have been used to map the location of the gene encoding the MVV EV-1 receptor to mouse chromosome 2 and/or 4 (18). Although the host range of the CAEVneo10(K1514) pseudotype described here was in general agreement with previously published results, we were unable to infect mouse NIH 3T3 cells with CAEVneo10(K1514) or even to observe any significant extent of syncytia in cocultures of NIH 3T3 and 293T cells expressing the MVV K1514 envelope (not shown). Considering that mouse NIH 3T3 cells were about 2 to 10 times less susceptible to CAEVneo10(VSV) than cell lines from other species, their susceptibility to CAEVneo10(K1514) was at least 100- to 1,000-fold lower than that of ruminant or primate cell lines. Although this discrepancy could be due to differences in the systems used to determine virus tropism, it is more likely explained by genetic differences between our envelope clone and other MVV K1514 or EV-1 env clones, genetic differences in the MVV receptor between mouse strains or cell lines, or differential expression of this receptor in different mouse cell lines. Determining the susceptibility of mouse cells to MVV as a function of virus strain and cell types will be necessary to establish the usefulness of mouse cells for receptor-cloning experiments.
The techniques used for cloning of retroviral receptor genes have been greatly improved in recent years. These cDNA-cloning protocols require a selectable recombinant retrovirus pseudotyped with the envelope for which a receptor is being sought. Currently available vectors based on either murine leukemia virus or HIV-1 do not incorporate and are not easily adaptable to use with envelopes from small-ruminant lentiviruses, probably due to interference with Env incorporation by the long internal domain of lentiviral TM (43). The vectors developed here should allow the production of selectable viruses pseudotyped with MVV and CAEV envelope glycoproteins necessary for receptor-cloning experiments. In addition, this and similar CAEV pseudotype systems could also be used to map the location of the CAEV and MVV receptor genes in human (for MVV K1514) and bovine or sheep (for all MVV and CAEV strains) chromosomes, as none of the strains is able to infect Chinese hamster cells, which are commonly used for the development of whole-genome radiation hybrid-cell panels (7, 41). It should be noted that Syrian hamster cells have been shown to be susceptible to MVV K796 (4). In addition, the Chinese hamster CHO-K1 cells are resistant to retroviruses due to hyperglycosylation of receptors and production of inhibitory factors rather than lack of functional receptors (22). Thus, other hamster cell lines or CHO-K1 sublines may be susceptible to MVV K1514 infection.
Attempts have been made to use small-ruminant lentiviruses to develop nonhuman lentivirus vectors. However, the titers obtained with vectors based on either CAEV (23) or MVV (2) have been much lower than the titers obtained with vectors based on other lentiviruses. The low titers of these vectors appear to be due to defects in vector RNA packaging (23) or reverse transcription and integration (2). Although these studies cannot be directly compared to ours, the recombinant CAEV constructs developed here appear to be much more efficient when pseudotyped with the VSV-G glycoprotein than these two previously described systems, approaching the titers obtained with similar HIV-1 constructs (37). The reason for the difference in titers between our constructs and similar MVV vectors (for example, vector VXCG) is not obvious, as the only apparent differences in the two systems besides the marker genes are the locations of the marker genes, in the vif-tat region in our CAEV constructs or in the env gene in VXCG, and the internal promoters driving their expression (SV40 or CMV early promoters). Thus, the constructs described here demonstrate that CAEV-based constructs can be developed as nonhuman lentivirus vectors.
ACKNOWLEDGMENTS
We thank Richard Sutton (Baylor College of Medicine, Houston, Tex.) for the 293T cell line and the pMEVSV-G plasmid, Travis McGuire (Washington State University, Pullman) for the EK cells, James C. DeMartini (Colorado State University, Ft. Collins) for plasmid pLG1.35, and Opendra Narayan (University of Kansas Medical Center, Kansas City) for MVV strains K1514 and S93. We also thank Kathy Pretty On Top for technical assistance.
This work was supported by NIH grants RO1 AR 43718 and R21 AI 42690 and by a grant from the Adler Endowment.
REFERENCES
- 1.August M J, Harter D H. Visna virus-induced fusion of continuous simian kidney cells. Arch Ges Virusforsch. 1974;44:92–101. doi: 10.1007/BF01250217. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz R D, Ilves H, Plavec I, Veres G. Gene transfer systems derived from visna virus: analysis of virus production and infectivity. Virology. 2001;279:116–129. doi: 10.1006/viro.2000.0659. [DOI] [PubMed] [Google Scholar]
- 3.Beyer J C, Chebloune Y, Mselli-Lakhal L, Hötzel I, Kumpula-McWhirter N, Cheevers W P. Immunization with plasmid DNA expressing the caprine arthritis-encephalitis virus envelope gene: quantitative and qualitative aspects of antibody response to viral surface glycoproteins. Vaccine. 2001;19:1643–1651. doi: 10.1016/s0264-410x(00)00418-7. [DOI] [PubMed] [Google Scholar]
- 4.Brown H R, Thormar H. Persistence of visna virus in murine and hamster cell cultures without the appearance of cell transformation. Microbios. 1975;13:51–60. [Google Scholar]
- 5.Chebloune Y, Sheffer D, Karr B M, Stephens E, Narayan O. Restrictive type of replication of ovine/caprine lentiviruses in ovine fibroblast cell cultures. Virology. 1996;222:21–30. doi: 10.1006/viro.1996.0394. [DOI] [PubMed] [Google Scholar]
- 6.Chesebro B, Wehrly K, Nishio J, Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox D R, Burmeister M, Price E R, Kim S, Myers R M. Radiation hybrid mapping: a somatic cell genetic method for constructing high-resolution maps of mammalian genomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 8.Harter D H, Choppin P W. Plaque assay of visna virus using a secondary cellular overlay as an indicator. Virology. 1967;31:176–178. doi: 10.1016/0042-6822(67)90025-6. [DOI] [PubMed] [Google Scholar]
- 9.Harter D H, Hsu K C, Rose H M. Multiplication of visna virus in bovine and porcine cell lines. Proc Soc Exp Biol (NY) 1968;129:295–300. doi: 10.3181/00379727-129-33306. [DOI] [PubMed] [Google Scholar]
- 10.Hess J L, Pyper J M, Clements J E. Nucleotide sequence and transcriptional activity of the caprine arthritis-encephalitis virus long terminal repeat. J Virol. 1986;60:385–393. doi: 10.1128/jvi.60.2.385-393.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess J L, Small J A, Clements J E. Sequences in the visna virus long terminal repeat that control transcriptional activity and respond to viral trans-activation: involvement of AP-1 sites in basal activity and trans-activation. J Virol. 1989;63:3001–3015. doi: 10.1128/jvi.63.7.3001-3015.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jolly P E, Narayan O. Evidence for interference, coinfections, and intertypic virus enhancement of infection by ovine-caprine lentiviruses. J Virol. 1989;63:4682–4688. doi: 10.1128/jvi.63.11.4682-4688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy-Stoskopf S, Zink C, Narayan O. Pathogenesis of ovine lentivirus-induced arthritis: phenotypic evaluation of T lymphocytes in synovial fluid, synovium, and peripheral circulation. Clin Immunol Immunopathol. 1989;52:323–330. doi: 10.1016/0090-1229(89)90183-9. [DOI] [PubMed] [Google Scholar]
- 14.Klevjer-Anderson P, Cheevers W P. Characterization of the infection of caprine synovial membrane cells by the retrovirus caprine arthritis-encephalitis virus. Virology. 1981;110:113–119. doi: 10.1016/0042-6822(81)90012-x. [DOI] [PubMed] [Google Scholar]
- 15.Knowles D P, Cheevers W P, McGuire T C, Brassfield A L, Harwood W G, Stem T A. Structure and genetic variability of envelope glycoproteins of two antigenic variants of caprine arthritis-encephalitis lentivirus. J Virol. 1991;65:5744–5750. doi: 10.1128/jvi.65.11.5744-5750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lairmore M D, Akita G Y, Russel H R, DeMartini J C. Replication and cytopathic effects of ovine lentivirus strains in alveolar macrophages correlate with in vivo pathogenicity. J Virol. 1987;61:4038–4042. doi: 10.1128/jvi.61.12.4038-4042.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lairmore M D, Rosadio R H, DeMartini J C. Ovine lentivirus lymphoid interstitial pneumonia: rapid induction in neonatal lambs. Am J Pathol. 1986;125:173–181. [PMC free article] [PubMed] [Google Scholar]
- 18.Lyall J W, Solansky N, Tiley L S. Restricted species tropism of maedi-visna virus strain EV-1 is not due to limited receptor distribution. J Gen Virol. 2000;81:2919–2927. doi: 10.1099/0022-1317-81-12-2919. [DOI] [PubMed] [Google Scholar]
- 19.MacIntyre E H, Wintersgill C J, Thormar H. Morphological transformation of human astrocytes by visna virus with complete virus production. Nat New Biol. 1972;237:111–113. doi: 10.1038/newbio237111a0. [DOI] [PubMed] [Google Scholar]
- 20.McGuire T C, Tumas D B, Byrne K M, Hines M T, Leib S R, Brassfield A L, O'Rourke K I, Perryman L E. Major histocompatibility complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J Virol. 1994;68:1459–1467. doi: 10.1128/jvi.68.3.1459-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller A D, Garcia J V, Von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller D G, Miller A D. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol. 1992;66:78–84. doi: 10.1128/jvi.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mselli-Lakhal L, Favier C, Da Silva Teixeira M F, Chettab K, Legras C, Ronfort C, Verdier G, Mornex J F, Chebloune Y. Defective RNA packaging is responsible for low transduction efficiency of CAEV-based vectors. Arch Virol. 1998;143:681–695. doi: 10.1007/s007050050323. [DOI] [PubMed] [Google Scholar]
- 24.Mselli-Lakhal L, Favier C, Leung K, Guiguen F, Grezel D, Miossec P, Mornex J, Narayan O, Quérat G, Chebloune Y. Lack of functional receptors is the only barrier that prevents caprine arthritis-encephalitis virus from infecting human cells. J Virol. 2000;74:8343–8348. doi: 10.1128/jvi.74.18.8343-8348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mwaengo D M, Grant R F, DeMartini J C, Carlson J O. Envelope glycoprotein nucleotide sequence and genetic characterization of North American ovine lentiviruses. Virology. 1997;238:135–144. doi: 10.1006/viro.1997.8813. [DOI] [PubMed] [Google Scholar]
- 26.Narayan O, Kennedy-Stoskopf S, Sheffer D, Griffin D E, Clements J E. Activation of caprine arthritis-encephalitis virus expression during maturation of monocytes to macrophages. Infect Immun. 1983;41:67–73. doi: 10.1128/iai.41.1.67-73.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayan O, Zink M, Gorrel M, Crane S, Huso D, Jolly P, Saltarelli M, Adams R R, Clements J. The lentiviruses of sheep and goats. In: Levy J A, editor. The retroviridae. New York, N.Y: Plenum Press; 1993. pp. 229–256. [Google Scholar]
- 28.Quérat G, Barban V, Sauze N, Filippi P, Vigne R, Russo P, Vitu C. Highly lytic and persistent lentiviruses naturally present in sheep with progressive pneumonia are genetically distinct. J Virol. 1984;52:672–679. doi: 10.1128/jvi.52.2.672-679.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan S, Tiley L, McConnell I, Blacklaws B. Infection of dendritic cells by the maedi-visna lentivirus. J Virol. 2000;74:10096–10103. doi: 10.1128/jvi.74.21.10096-10103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saltarelli M, Quérat G, Konings D A M, Vigne R, Clements J E. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology. 1990;179:347–364. doi: 10.1016/0042-6822(90)90303-9. [DOI] [PubMed] [Google Scholar]
- 31.Saltarelli M J, Schoborg R, Gdovin S L, Clements J E. The CAEV tat gene trans-activates the viral LTR and is necessary for efficient viral replication. Virology. 1993;197:35–44. doi: 10.1006/viro.1993.1564. [DOI] [PubMed] [Google Scholar]
- 32.Skraban R, Matthíasdóttir S, Torsteinsdóttir S, Agnarsdóttir G, Gudmundsson B, Georgsson G, Meloen R H, Andrésson O S, Staskus K A, Thormar H, Andrésdottir V. Naturally occurring mutations within 39 amino acids in the envelope glycoprotein of maedi-visna virus alter the neutralization phenotype. J Virol. 1999;73:8064–8072. doi: 10.1128/jvi.73.10.8064-8072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth R J, Yi Y, Singh A, Collman R G. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sommerfelt M A, Weiss R A. Receptor interference groups of 20 retroviruses plating on human cells. Virology. 1990;176:58–69. doi: 10.1016/0042-6822(90)90230-o. [DOI] [PubMed] [Google Scholar]
- 35.Sonigo P, Alizon M, Staskus K, Klatzmann D, Cole S, Danos O, Retzel E, Tiollais P, Haase A, Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985;42:369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- 36.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutton R E, Littman D R. Broad host range of human T-cell leukemia virus type 1 demonstrated with an improved pseudotyping system. J Virol. 1996;70:7322–7326. doi: 10.1128/jvi.70.10.7322-7326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thormar H, Sigurdardóttir B. Growth of visna virus in primary tissue cultures from various animal species. Acta Microbiol Pathol Scand. 1962;55:180–186. doi: 10.1111/j.1699-0463.1962.tb04115.x. [DOI] [PubMed] [Google Scholar]
- 39.Tong-Starksen S E, Sepp T, Pagtakhan S. Activation of caprine arthritis-encephalitis virus long terminal repeat by gamma interferon. J Virol. 1996;70:595–599. doi: 10.1128/jvi.70.1.595-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valas S, Benoit C, Guionaud C, Perrin G, Mamoun R Z. North American and French caprine arthritis-encephalitis viruses emerge from ovine maedi-visna viruses. Virology. 1997;237:307–318. doi: 10.1006/viro.1997.8800. [DOI] [PubMed] [Google Scholar]
- 41.Womack J E, Johnson J S, Owens E K, Rexroad III C E, Schläpfer J, Yang Y-P. A whole-genome radiation hybrid panel for bovine gene mapping. Mamm Genome. 1997;8:854–856. doi: 10.1007/s003359900593. [DOI] [PubMed] [Google Scholar]
- 42.Zanoni R G. Phylogenetic analysis of small-ruminant lentiviruses. J Gen Virol. 1998;79:1951–1961. doi: 10.1099/0022-1317-79-8-1951. [DOI] [PubMed] [Google Scholar]
- 43.Zeilfelder U, Bosch V. Properties of wild-type, C-terminally truncated, and chimeric maedi-visna virus glycoprotein and putative pseudotyping of retroviral particles. J Virol. 2001;75:548–555. doi: 10.1128/JVI.75.1.548-555.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]