Abstract
Dimethylsulfoniopropionate (DMSP) is a marine organosulfur compound with important roles in stress protection, marine biogeochemical cycling, chemical signalling and atmospheric chemistry. Diverse marine microorganisms catabolize DMSP via DMSP lyases to generate the climate‐cooling gas and info‐chemical dimethyl sulphide. Abundant marine heterotrophs of the Roseobacter group (MRG) are well known for their ability to catabolize DMSP via diverse DMSP lyases. Here, a new DMSP lyase DddU within the MRG strain Amylibacter cionae H‐12 and other related bacteria was identified. DddU is a cupin superfamily DMSP lyase like DddL, DddQ, DddW, DddK and DddY, but shares <15% amino acid sequence identity with these enzymes. Moreover, DddU proteins forms a distinct clade from these other cupin‐containing DMSP lyases. Structural prediction and mutational analyses suggested that a conserved tyrosine residue is the key catalytic amino acid residue in DddU. Bioinformatic analysis indicated that the dddU gene, mainly from Alphaproteobacteria, is widely distributed in the Atlantic, Pacific, Indian and polar oceans. For reference, dddU is less abundant than dddP, dddQ and dddK, but much more frequent than dddW, dddY and dddL in marine environments. This study broadens our knowledge on the diversity of DMSP lyases, and enhances our understanding of marine DMSP biotransformation.
INTRODUCTION
The organosulfur compound dimethylsulfoniopropionate (DMSP) produced by marine phytoplankton, macroalgae, corals, angiosperms, animals and heterotrophic bacteria can reach several petagrams each year (Curson et al., 2017, 2018; Ksionzek et al., 2016; Otte, 2004; Raina et al., 2013; Stefels, 2000; Zhang et al., 2019). DMSP not only participates in the nutrient cycling to provide an important source of organic carbon and/or reduced sulphur for microbial communities (Kiene et al., 2000; Yoch, 2002), but also possesses important physiological functions that include, but are not limited to, acting as an osmolyte, cryoprotectant, predator deterrent, antioxidant, chemoattractant and protectant against high hydrostatic pressure (Cosquer et al., 1999; Karsten et al., 1996; Seymour et al., 2010; Sunda et al., 2002; Wolfe et al., 1997; Zheng et al., 2020). After DMSP is released into marine environment from the producer organisms during exudation, senescence, viral lysis, or grazer attack (Bratbak et al., 1995; Hill et al., 1998; Stefels & Boekel, 1993; Wolfe et al., 1997), it is degraded largely by bacteria (Yoch, 2002; Zhang et al., 2019).
To date, three DMSP metabolic pathways have been reported, the demethylation pathway, the oxidation pathway and the lysis pathway. The demethylation pathway is initiated by the DMSP demethylase enzyme DmdA, in which DMSP is finally catabolized to acetaldehyde and volatile methanethiol (MeSH) (Howard et al., 2006; Reisch et al., 2011). It is estimated that 50% to 90% of DMSP is catabolized through this pathway (Kiene et al., 2000). In the oxidation pathway, DMSP is first oxidized to dimethylsulfoxonium propionate (DMSOP) that can then be catabolized to dimethylsulfoxide (DMSO) and acrylate via unidentified DMSOP lyase enzymes (Thume et al., 2018). In the lysis pathway, DMSP is cleaved by diverse enzymes, including a DMSP CoA‐transferase DddD (Todd et al., 2007), a DMSP‐CoA ligase DddX (Li et al., 2021), and seven DMSP lyases (or DMSP dethiomethylases) (Alcolombri et al., 2015; Curson et al., 2008; Curson, Sullivan, et al., 2011; Sun et al., 2016; Todd et al., 2009, 2011, 2012), to generate the climate‐active volatile dimethyl sulphide (DMS) and 3‐hydroxypropionate‐CoA (3‐HP‐CoA) (for DddD catalysis), acryloyl‐CoA (for DddX catalysis) or acrylate (for DMSP lyases catalyses). DMS, which is generated with DMSP as its main precursor (de Souza & Yoch, 1995), is an important nutrient and an info‐chemical in signalling pathways for diverse organisms (Nevitt, 2011; Shemi et al., 2021; Teng, Wang, et al., 2021). DMS represents the largest biogenic sulphur source entering the atmosphere (Andreae, 1990), and its oxidation products aid cloud formation and may influence weather and climate (Vallina & Simo, 2007). Acrylate, another product in the DMSP lysis pathway, can not only be utilized as a carbon source by some microorganisms (Curson, Todd, et al., 2011), but also possesses important physiological functions, for example, it protects marine bacteria from grazing by ciliate predators (Teng, Wang, et al., 2021). In marine bacteria, acrylate can be metabolized through the AcuN‐AcuK pathway (Todd et al., 2010) and/or PrpE‐AcuI pathway (Reisch et al., 2013).
Among the seven reported DMSP lyases, Alma1 is the only known algal DMSP lyase (Alcolombri et al., 2015), while the other six DMSP lyases (DddP, DddL, DddQ, DddW, DddY and DddK) were all identified from bacteria. Alma1 is a tetrameric, redox‐sensitive enzyme of the aspartate racemase superfamily, yet its structure and catalytic mechanism is not elucidated (Alcolombri et al., 2015). DddP, existing in the marine Roseobacter group (MRG) (Simon et al., 2017), the SAR116 clade, Gammaproteobacteria and some ascomycete fungi (Curson, Todd, et al., 2011), is a member of the M24 metallopeptidase family (Todd et al., 2009), and the most abundant bacterial DMSP lyase in marine metagenomes database (Teng, Qin, et al., 2021). The crystal structure and the catalytic mechanism of DddP have been revealed (Hehemann et al., 2014; Wang et al., 2015). DMSP lyases DddL, DddQ, DddW, DddY and DddK all belong to the cupin superfamily, which comprises a functionally highly diverse group of proteins (Dunwell et al., 2004). DMSP lyases of the cupin superfamily share two conserved cupin motifs, and require divalent metal ions as cofactors for their enzymatic activities (Brummett et al., 2015; Li et al., 2014, 2017; Peng et al., 2019). Among them, DddL is membrane‐associated (Teng, Wang, et al., 2021), DddY is periplasmic (Curson, Sullivan, et al., 2011), while the others are cytoplasmic. The biochemical properties, catalytic mechanism and ecological distribution for DddQ, DddY and DddK have been reported (Li et al., 2014, 2017; Peng et al., 2019). The biochemical properties of DddW have also been studied (Brummett et al., 2015). The dddL, dddQ and dddW genes mainly occur in MRG, while dddK is only found in SAR11 bacteria, and dddY is sporadically distributed in Betaproteobacteria, Gammaproteobacteria, Deltaproteobacteria and Epsilonproteobacteria (Curson, Todd, et al., 2011). Recently, researchers found that many marine microorganisms that liberate DMS from DMSP do not contain known DMSP lyase genes in their genomes, suggesting that there are still more unrecognized DMSP lyases in the natural environments yet to be identified (Liu et al., 2018; Zhang et al., 2019).
In this study, Amylibacter cionae H‐12, a member of the MRG (Wang et al., 2017), was found to exhibit DMSP‐dependent DMS production (Ddd+). However, strain H‐12 lacked known DMSP lyase genes in its genome sequence, indicating that this strain likely utilizes a novel enzyme to generate DMS from DMSP. Using genetic and biochemical approaches on strain H‐12, a new cupin superfamily DMSP lyase, termed DddU, was discovered and characterized. Structural prediction and mutational work were done to investigate the DddU DMSP lysis mechanism and its key catalytic amino acid residues. Bioinformatic analysis was also conducted to study the environmental importance of DddU in comparison to the other known DMSP lyase genes. The results presented here broaden our knowledge on the diversity of DMSP lyases, and enhance our understanding of DMSP catabolism in marine environments.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
Amylibacter cionae H‐12, the ΔdddU mutant strain and the complemented ΔdddU strain (ΔdddU/pHGPtac‐dddU, ΔdddU/pHGPtac) were cultured in the marine broth 2216 medium at 25°C, and kanamycin (Km) with a final concentration of 50 μg/mL was added into culture of complemented strain. The E. coli strains WM3064, DH5α and BL21 (DE3) were cultured in the Lysogeny Broth (LB) medium at 37°C, and diaminopimelic acid (DAP) with a final concentration of 0.3 mM was added into culture of E. coli WM3064. Strains were listed in Table S5.
Quantification of DMS by gas chromatography
Cells cultured in the marine broth 2216 medium to the late exponential phase were harvested, washed with sterilized artificial seawater three times and diluted to the OD600 ≈ 0.03. Then the cells were incubated in the minimal medium supplied with 1 mM DMSP in gas‐tight sealing bottles at 25°C for 3 h. The cultures were assayed for DMS production on gas chromatography (GC) (GC‐2030, Shimadzu, Japan) equipped with a flame photometric detector according to the method described by Zhang (Zhang et al., 2014). The culture medium without bacteria and culture medium without DMSP were set as controls. A DMS standard was used as a positive control. A six‐point calibration curve of DMS standards was used to quantify DMS production (Curson et al., 2017). Bacterial cells were lysed by ultrasonication, and proteins content in the cell extracts were measured by Pierce BCA Protein Assay Kit (Thermo, USA). DMS production is expressed as nmol DMS min−1 mg protein−1.
Growth assay with DMSP as the sole carbon source
Cells were grown in the marine broth 2216 medium at 180 rpm and 25°C to the late exponential phase (OD600 ≈ 0.8), and washed three times with sterilized artificial seawater. Then, 1% (v/v) cells were inoculated into the minimal medium with sodium pyruvate, DMSP or acrylate (5 mM) as the sole carbon source. Strains were cultured in the dark at 25°C and 180 rpm. The bacteria growths were measured by detecting the OD600 of the cultures using a spectrophotometer V‐550 (Jasco Corporation, Japan).
Transcriptome sequencing of strain H‐12
The strain H‐12 was cultured in the marine broth 2216 medium at 180 rpm and 25°C to the late exponential phase (OD600 ≈ 0.8). Subsequently, the cells were washed with sterilized artificial seawater three times and incubated in sterilized artificial seawater for 2 h. Then, cells were inoculated into the minimal medium with DMSP (5 mM) or with carbon mixture (1.11 g/L glucose, 1.01 g/L fructose, 1.38 g/L disodium succinate, 1.25 g/L sodium pyruvate, 1 mL/L glycerol, 1.39 g/L sodium acetate) as the carbon source for 4 h at 180 rpm and 25°C. Cells cultured in minimal medium with carbon mixture were set as control groups and with DMSP were set as experimental groups. Total RNA was extracted using a RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's protocol. After validating the quality, RNA samples were sent to Novogene Biotechnology Co., Ltd (China) for transcriptome sequencing and subsequent bioinformatic analyses.
Genetic manipulations of Amylibacter cionae H‐12
Genomic DNA from strain H‐12 was extracted using a bacterial genomic DNA isolation kit (BioTeke Corporation, China) according to the manufacturer's instructions. For in‐frame deletion of the target gene dddU, the upstream 771‐bp and downstream 816‐bp flanking regions of dddU were amplified by PCR with the primer pairs dddU‐5′O/dddU‐5′I and dddU‐3′I/dddU‐3′O respectively, and cloned into pHGM01. The resulting plasmid was transformed into E. coli WM3064 and then mobilized into wild‐type strain H‐12 by conjugation as previously described (Fu et al., 2015). The dddU deletion mutant was obtained through homologous recombination.
For complementation of the ΔdddU mutant, the dddU gene was amplified using the primers set dddU‐pHGPtac‐F/dddU‐pHGPtac‐R, and then inserted into pHGPtac. The recombinant vector or empty vector was transformed into E. coli WM3064, and then mobilized into the ΔdddU mutant by conjugation, as detailed above. Colony PCR was used to confirm the presence of the transferred plasmid.
All PCR‐amplified sequences were verified by DNA sequencing. The primers used were listed in Table S6.
Gene synthesis, point mutation, protein expression and purification
Full‐length dddU gene, codon optimized for expression in Escherichia coli, was chemically synthesized and then subcloned into the pET‐22b (Novagen, USA) by BGI gene Co., Ltd (China). The plasmid was also used as the template for site‐directed mutagenesis. Point mutations in dddU were introduced using the PCR‐based method and were verified by DNA sequencing. The primers used were listed in Table S6. The DddU protein and its enzyme variants were overexpressed in E. coli BL21 (DE3). The cells were cultured in the LB medium with 100 μg/mL ampicillin (Am) at 37°C to an OD600 of 0.8 ~ 1.0 and then induced by 0.5 mM isopropyl‐β‐D‐thiogalactopyranoside (IPTG) at 18°C for 16 h. Briefly, cells were collected and resuspended in the lysis buffer (50 mM Tris–HCl, 100 mM NaCl, 0.5% glycerol, pH 8.0), and then lysed by cryogenic pressure crusher. The lysates were purified by affinity chromatography on a Ni2+‐nitrilotriacetic acid (NTA) column (GE healthcare, USA), and subsequently fractionated by gel filtration on a Superdex‐G75 column (GE healthcare, USA). The purities of these recombinant proteins were analysed by SDS‐PAGE, and protein concentrations were determined by the BCA assays.
Enzyme assay and characterization
The enzymatic activity of DddU was measured by detecting the production of acrylate as previously described (Wang et al., 2015). DddU (at a final concentration of 0.1 μM) and DMSP (at a final concentration of 5 mM) were mixed with reaction buffer containing 100 mM Tris–HCl (pH 7.0) in a total volume of 200 μL. After the mixture was incubated at 40°C for 10 min, the reaction was stopped by adding perchloric acid. The amount of acrylate in the reaction mixture was detected by high‐performance liquid chromatography (HPLC) on a Sunfire C18 column (Waters, Ireland). The reaction mixture without DddU enzymes was set as the control group.
To determine the optimal temperature for PiDddU, reaction mixtures were incubated at 0, 10, 20, 30, 40, 50 or 60°C for 10 min. The optimum pH for PiDddU was examined at 40°C (the optimal temperature for PiDddU enzymatic activity) using Britton‐Robinson buffer at pH values of 4 to 10. The Britton–Robinson buffer is a mixture of 0.04 M H3BO3, 0.04 M H3PO4 and 0.04 M CH3COOH (Barek et al., 1999). The kinetic parameters of PiDddU were determined by adding PiDddU (0.1 μM) into the reaction systems containing different concentrations of DMSP (1, 2, 3, 5, 10, 20, 25, 60 and 80 mM) under conditions of pH 7.0 at 40°C for 10 min, and then the non‐linear analysis was performed based on the initial rates determined with different DMSP concentrations. The enzymatic activities of the PiDddU enzyme variants were also examined under the optimum pH and temperature.
Circular dichroism spectroscopy assays
Circular dichroism (CD) spectra for wild‐type PiDddU and its enzyme variants were carried out in a 0.1 cm‐path length cell on a JASCO J‐1500 Spectrometer (Japan) at 25°C. All proteins were adjusted to a final concentration of 20 μM in 10 mM Tris–HCl (pH 8.0) and 100 mM NaCl. CD Spectra were recorded from 250 to 200 nm at a scan speed of 200 nm min−1.
Bioinformatics
The Basic Local Alignment Search Tool (BLAST) was used to perform similarity searches and searched for sequences through the National Center for Biotechnology Information (NCBI) BLAST webpage interface (http://www.ncbi.nlm.nih.gov/BLAST/). DddU of Amylibacter cionae H‐12 was used as the query sequence to search for homologues in the NCBI Reference Sequence Database using BLastP with the percentage identity >70%, and a total of 40 hits was retrieved. Among them, 28 sequences in genome‐sequenced bacteria were selected for further phylogenetic analysis. Trees were constructed using the algorithm in the MEGA 7.0 software package. The distribution analyses of DddU homologues according to the method described by Teng (Teng, Qin, et al., 2021). Briefly, hidden Markov models (HMM) were created using protein sequences that are biochemically characterized. DddU homologues from metagenomes/metatranscriptomes were obtained using hmmsearch (http://hmmer.org). The cut‐off value used was <e‐30. The DddU homologues sequences retrieved from our bioinformatics pipeline were further scrutinized for the presence of conserved key residues involved in substrate binding or catalysis. The distribution of dddU homologues was analysed in metagenomic data from 60 polar seawater samples (NCBI BioProject accession no. PRJNA588686) and 174 non‐polar Tara Ocean samples (fraction size, 0.22–3 μm; http:// www.pangaea.de/). The amino acid sequences of 10 conserved bacterial marker genes (Sunagawa et al., 2013) were retrieved from the NCBI database, and the average abundance of these marker genes was used to normalize the abundance of the dddU gene as described previously (Curson et al., 2017), that is, the relative abundance of dddU in each site is calculated by the ratio of dddU abundance to the average abundance of 10 selected bacterial marker genes. The distribution of dddU transcripts was analysed in Tara Oceans Microbiome Reference Gene Catalogue v2+ meta T Arctic Inside (prokaryotes) dataset from Tara Ocean samples (https://tara-oceans.mio.osupytheas.fr/ocean-gene-atlas/) and used the percent of mapped reads to express the relative abundance. The geographical distributions of dddU and its transcripts were constructed by Ocean Data View.
RESULTS AND DISCUSSION
Discovery of a novel DMSP lyase in a marine Roseobacter strain
Amylibacter cionae H‐12 was isolated from a sea squirt Ciona savignyi collected from Jiaozhou Bay, China (Wang et al., 2017). Through routine screening of available marine bacteria by GC analysis, the strain H‐12 was found to be able to catabolize DMSP and produce DMS (485 ± 62 nmol DMS min−1 mg protein−1) in the minimal medium supplemented with DMSP (Figure 1A), although it was unable to utilize DMSP as a sole carbon source (Figure S1).
FIGURE 1.
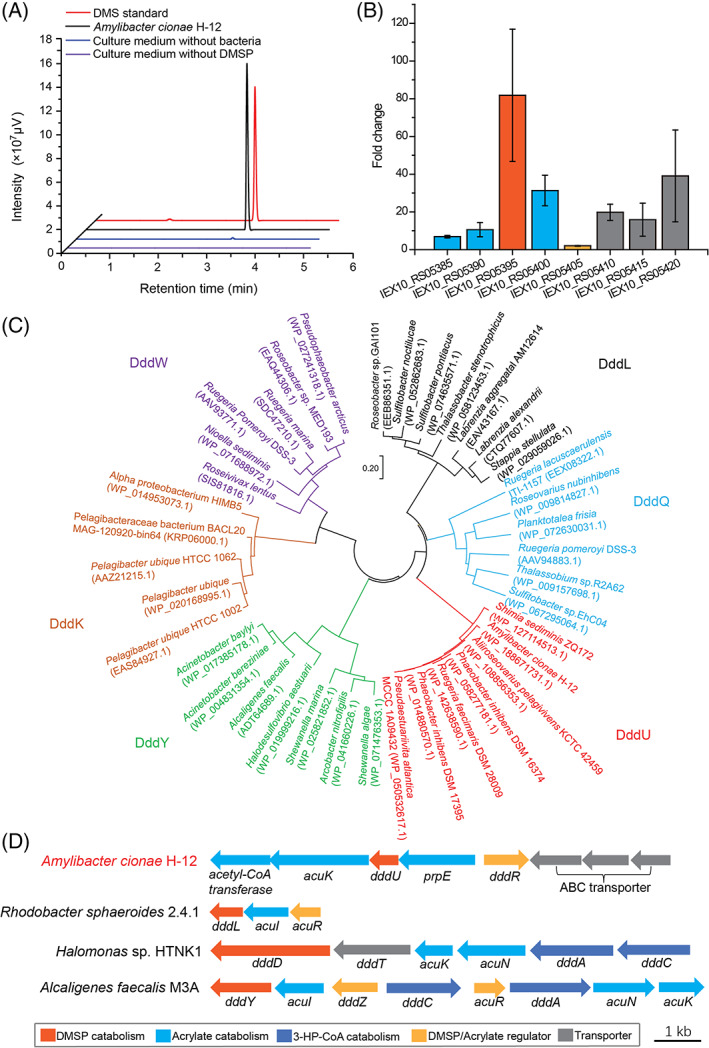
Analysis of Amylibacter cionae H‐12 dimethylsulfoniopropionate (DMSP) lyase activity and its candidate DMSP lyase. (A) Gas chromatography detection of DMSP‐dependent dimethyl sulphide (DMS) production by strain H‐12. The culture medium without bacteria and the medium without DMSP were used as the controls. The DMS standard was used as a positive control. (B) Transcriptomic analysis of putative DMSP/acrylate‐catabolizing genes from H‐12. The fold changes were calculated by dividing the gene transcripts in the presence of 5 mM DMSP by those in the absence of DMSP. The error bar represents standard deviation of triplicate experiments. (C) Neighbour‐joining phylogenetic tree of DddU and representative proteins of the five other reported cupin‐containing DMSP lyases. Phylogenetic analysis was performed using MEGA version 7.0 (Tamura et al., 2013). (D) Genetic organization of the putative DMSP‐catabolizing gene cluster in strain H‐12. DMSP catabolic genes and their regulators from Rhodobacter sphaeroides 2.4.1, Halomonas sp. HTNK1 and Alcaligenes faecalis M3A were shown (Curson, Sullivan, et al., 2011).
To identify the DMSP lyase enzymes in strain H‐12, the sequenced genome (GenBank accession number: GCA_014643735.1) was searched for gene products homologous to DddD, DddP, DddQ, DddX, DddY, DddW, DddL, DddK and Alma1. However, no proteins homologous to these enzymes with amino acid sequence identity >25% were found (Table S1), implying that H‐12 may possess novel DMSP lyase enzyme(s). The transcriptomes of strain H‐12 in the presence and absence of 5 mM DMSP were then sequenced. Transcriptomic analysis showed that the transcripts of eight genes that compose a gene cluster were highly upregulated (Figure 1B; Table S2) when strain H‐12 was induced by DMSP.
In the gene cluster, IEX10_RS05385 was annotated as an acetyl‐CoA C‐acyltransferase. Although no significant amino acid sequence identity was found between IEX10_RS05385 and AcuN (Todd et al., 2010), they both belong to the CoA‐transferase family III. IEX10_RS05390 was annotated as an enoyl‐CoA hydratase, sharing 32% amino acid sequence identity with AcuK in Halomonas sp. HTNK1 (Todd et al., 2010). IEX10_RS05395 was annotated as a DMSP lyase family protein, however, the amino acid sequence of this protein possesses <15% identity to any reported DMSP lyases. IEX10_RS05400 was annotated as a fatty acid‐CoA ligase, and shared 25% sequence identity with PrpE in Ruegeria pomeroyi DSS‐3 (Reisch et al., 2013). AcuN, AcuK and PrpE were reported to participate in acrylate catabolism (Reisch et al., 2013; Todd et al., 2010). IEX10_RS05405 was annotated as a LysR family transcriptional regulator, sharing 30% sequence identity with DddR in Pseudomonas sp. J465 (Curson et al., 2010). IEX10_RS05410, IEX10_RS05415 and IEX10_RS05420 constituted an ABC (ATP‐binding cassette)‐type transporter, which may play an important role in importing DMSP for strain H‐12 (Li et al., 2023). The results above suggested that the gene cluster IEX10_RS05385‐IEX10_RS05420 in strain H‐12 is highly possible to be involved in DMSP catabolism, and IEX10_RS05395 may encode a new DMSP lyase, which we term as DddU hereafter. Sequence analysis showed that the DddU protein contained two conserved cupin motifs (Dunwell et al., 2004), and thus belongs to the cupin superfamily. Neighbour‐joining phylogenetic analysis revealed that DddU was most closely related to DddQ but it clearly formed a separate clade to DddQ and the other known cupin‐containing DMSP lyases (Figure 1C), suggesting the divergent evolution of DddU from other cupin DMSP lyases. Among reported DMSP lyase genes, dddL, dddD and dddY were found in the ddd‐acu cluster (Figure 1D). The gene dddU also locates in the ddd‐acu cluster. However, the pattern of the DMSP‐catabolizing cluster in strain H‐12 is different from the patterns of those reported DMSP‐catabolizing gene clusters (Figure 1D).
Functional verification of DddU
To confirm that dddU encoded a DMSP lyase in strain H‐12, a ΔdddU mutant strain was constructed in which the majority of this gene was deleted by homologous recombination (Figure S2A). The H‐12 ΔdddU mutant strain was shown produce ~2‐fold less DMS from DMSP compared wild type H‐12 and this reduction in DMSP lyase activity was fully complemented back to wild type levels by cloned dddU on pHGPtac‐dddU (Figure 2A; Figure S2B). These data are consistent with dddU encoding an enzyme with in vivo DMSP lyase activity. Given that the ΔdddU mutation did not completely abolish the DMSP lyase activity of strain H‐12, this strain likely contains other unidentified DMSP lyase(s). The phenomenon of multiple DMSP lyases co‐occurring in the same host is very common, particularly in MRG bacteria (Teng, Qin, et al., 2021). For example, Ruegeria pomeroyi DSS‐3 possesses DddP, DddW and DddQ, and Roseovarius nubinhibens ISM contains DddP and DddQ. Why a bacterium possesses multiple DMSP lyases is unknown, but may be linked to potentially different roles for DMSP catabolism in an organism, for example a DMSP lyase for growth on DMSP and one for signalling processes that may be differentially regulated.
FIGURE 2.
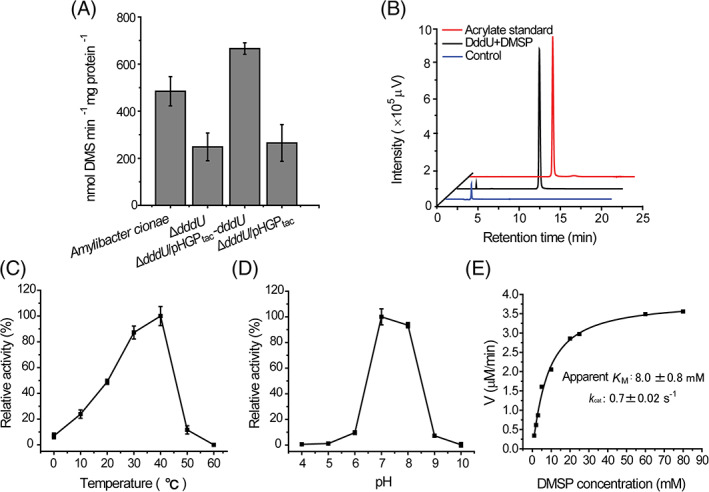
DddU is a dimethylsulfoniopropionate (DMSP) lyase. (A) The DMSP‐dependent dimethyl sulphide production from the wild‐type strain Amylibacter cionae, the ΔdddU mutant, the complemented mutant ΔdddU/pHGPtac‐dddU, and the mutant containing an empty vector ΔdddU/pHGPtac. The error bar represents standard deviation of triplicate experiments. (B) High‐performance liquid chromatography detection of the acrylate produced from the cleavage of DMSP by recombinant DddU. The reaction system without DddU was used as the control. The acrylate standard was used as a positive control. (C) Effect of temperature on the enzymatic activity of PiDddU. The error bar represents standard deviation of triplicate experiments. (D) Effect of pH on the enzymatic activity of PiDddU. The error bar represents standard deviation of triplicate experiments. (E) Nonlinear fit curves for DMSP cleavage by PiDddU. The kinetic parameters were determined under pH 7.0 at 40°C.
Expression and characterization of DddU
To demonstrate in vitro DMSP lyase activity for H‐12 DddU, full‐length dddU, codon optimized for expression in Escherichia coli, was chemically synthesized, overexpressed in E. coli BL21 (DE3) cells, and the recombinant DddU was purified.
HPLC analysis showed that DddU catalysed the release of acrylate from DMSP (Figure 2B; Table S3), confirming that DddU was a functional DMSP lyase in vitro. However, the enzymatic activity of DddU from H‐12 is relatively weak (0.15 μM acrylate min−1 μM protein−1; Table S3). A DddU homologue from Phaeobacter inhibens P66 (PiDddU), sharing 93% sequence identity with H‐12 DddU, exhibited much higher activity towards DMSP (5.03 μM acrylate min−1 μM protein−1; Table S3), and was selected for further characterization. The optimal temperature for PiDddU enzymatic activity was 40°C (Figure 2C), and the optimal pH was 7.0 (Figure 2D). This optimal temperature for PiDddU was relatively high compared to that in most marine environments. However, PiDddU maintained ~50% of its highest enzymatic activity at 20°C and ~85% at 30°C (Figure 2C), indicating that PiDddU is a viable enzyme in physiological environments.
The recombinant PiDddU exhibited a K M value of 8.0 mM for DMSP at pH 7.0 and 40°C (Figure 2E), which is lower than that of the reported DMSP lyases DddP, DddQ, DddW and Alma1, but higher than that of DddY and DddK (Table 1). These relatively high K M values at the millimolar level are common in DMSP lyases and may help bacteria maintain intracellular DMSP at in vivo levels to perform physiological functions, for example, in stress protection. The k cat of PiDddU for DMSP was 0.7 s− 1 (Figure 2E), lower than that of most other reported DMSP lyases (Table 1).
TABLE 1.
Kinetic parameters of different dimethylsulfoniopropionate (DMSP) lyases towards DMSP.
| Protein | Organism | K M (mM) | k cat (s−1) | k cat/K M (M−1 s−1) | References |
|---|---|---|---|---|---|
| PiDddU | Phaeobacter inhibens P66 | 8.0 ± 0.8 | 0.7 ± 0.02 | 87.8 | This study |
| DddY | Acinetobacter bereziniae | 5.0 ± 0.6 | 8.3 ± 0.5 × 103 | 1.7 × 106 | Li et al. (2017) |
| DddP | Roseovarius nubinhibens ISM | 13.8 ± 5.5 | 0.3 ± 0.1 | 18.7 | Kirkwood et al. (2010) |
| Phaeobacter inhibens DSM 17395 | 34.8 ± 4.7 | 3.9 ± 0.18 | 1.1 × 102 | Burkhardt et al. (2017) | |
| DddQ | Ruegeria lacuscaerulensis ITI_1157 | 21.5 ± 6.8 | 1.0 ± 0.3 | 46.5 | Li et al. (2014) |
| Ruegeria pomeroyi DSS‐3 | 28.6 ± 3.3 | 0.9 ± 0.04 | 32 | Burkhardt et al. (2017) | |
| DddW | Ruegeria pomeroyi DSS‐3 | 8.7 ± 0.7 | 18.3 | 2.1 × 103 | Brummett et al. (2015) |
| Ruegeria pomeroyi DSS‐3 | 12.8 ± 0.8 | 16.8 ± 0.4 | 1.3 × 103 | Burkhardt et al. (2017) | |
| DddK | Pelagibacter ubique HTCC 1062 | 3.7 ± 0.6 | 0.9 ± 0.1 | 2.4 × 102 | Peng et al. (2019) |
| Alma1 | Emiliania huxleyi | 9.0 ± 0.9 | 7.0 ± 0.3 × 102 | 7.8 × 104 | Alcolombri et al. (2015) |
Key residues involved in DddU catalysis
To further investigate the catalytic mechanism of PiDddU DMSP lysis, we tried to crystallize PiDddU, but unfortunately all attempts failed. Instead, the structure of PiDddU was predicted using AlphaFold2 (Jumper et al., 2021). The PiDddU structure was predicted to contain a β‐barrel fold surrounded by several α‐helices (Figure 3A). Despite the low amino acid sequence identity between PiDddU and DddQ (~13%), their structures were similar (Figure 3B), with a root mean square deviation of 1.8 Å over 104 Cα atoms between these two structures. Moreover, structural alignment of PiDddU and DddQ indicated that residues involved in metal ion binding (His146, Glu150, Tyr152 and His185 in PiDddU) and catalysis (Tyr152 in PiDddU) were perfectly superposed (Figure 3C), implying that PiDddU utilizes a similar catalytic mechanism to DddQ (Li et al., 2014), with Tyr152 being the catalytic residue to initiate the β‐elimination reaction. Substitution of these His146, Glu150, Tyr152 and His185 residues to alanine almost abolished the enzymatic activity of PiDddU (Figure 3D), supporting the important roles of these residues. Furthermore, CD spectroscopy analysis showed that the secondary structures of the enzyme variants exhibited little deviation from that of wild‐type (WT) PiDddU (Figure 3E), indicating that the loss of DMSP lyase activity was due to amino acid replacement rather than any major structural change.
FIGURE 3.
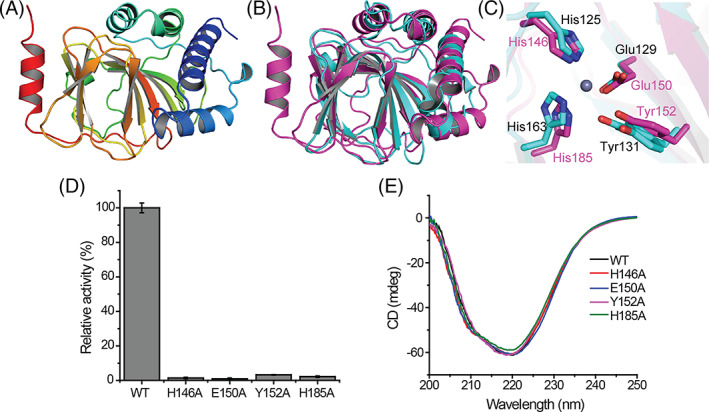
Structural and mutational analyses of PiDddU. (A) The predicted structure of PiDddU. (B) Alignment of the PiDddU (purple) and DddQ (cyan, PDB ID: 4LA2) protein structures. (C) Key residues predicted to be involved in PiDddU dimethylsulfoniopropionate (DMSP) cleavage. Residues of PiDddU are coloured in purple, and of DddQ in cyan. (D) DMSP lyase activity of PiDddU amino acid substitution mutants. The activity of native PiDddU was defined as 100%. The error bar represents standard deviation of triplicate experiments. (E) Circular dichroism (CD) spectra of native PiDddU and its enzyme variants. CD spectra of the proteins at a final concentration of approximately 20 μM were collected from 250 to 200 nm.
Distribution of DddU in bacteria
The distribution of dddU in sequenced genomes available on the NCBI Reference Sequence database was analysed to predict those with DMSP lyase activity. DddU proteins (with >70% amino acid sequence identity to H‐12 DddU) were predicted in many Alphaproteobacteria, most being MRG (Figure 4A). Multiple sequence alignment showed that the key amino acid residues His146, Glu 150, Tyr152 and His185 were highly conserved (Figure S3), suggesting that these candidates DddU were likely functional DMSP lyases. Indeed, several of these representative candidate DddU proteins were expressed and purified from E. coli BL21 (DE3) and all exhibited DMSP lyase activity (Figure 4B; Table S3), implying that bacteria expressing this enzyme would cleave DMSP.
FIGURE 4.
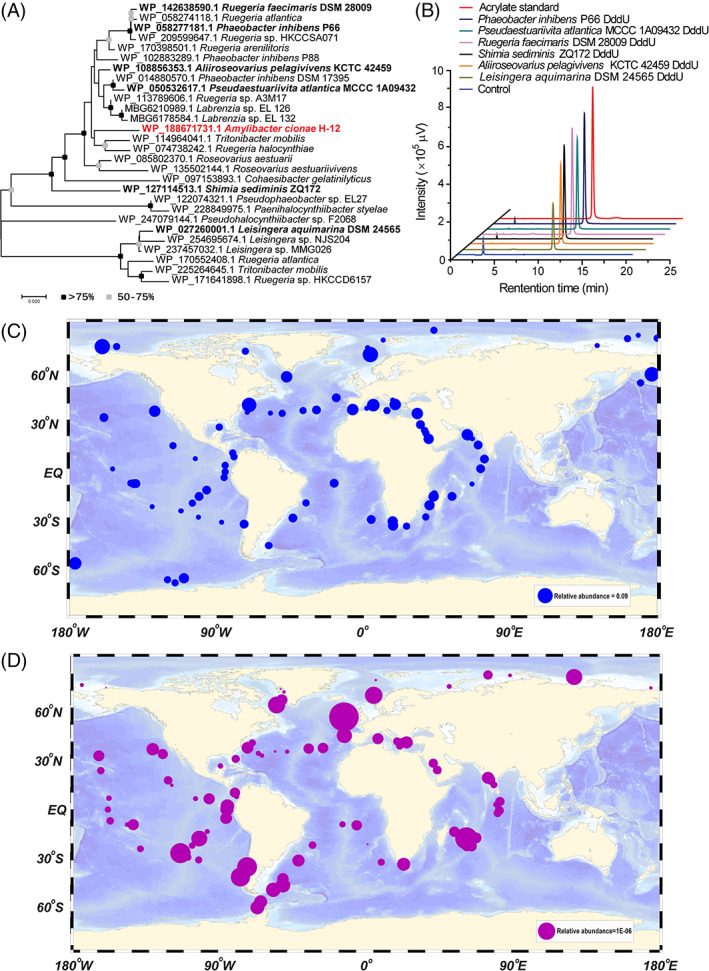
Distribution of DddU in bacteria. (A) The phylogenetic tree of DddU enzymes in Alphaproteobacteria. The black dots represented >75% of the bootstrap values and grey dots represented 50%–75%. Phylogenetic analysis was performed using MEGA version 7.0 (Tamura et al., 2013). Those DddU homologues that are functionally characterized are highlighted in bold. (B) Detection of the dimethylsulfoniopropionate cleavage activity of DddU homologues via high‐performance liquid chromatography. The reaction system without DddU enzymes was used as the control. The acrylate standard was used as a positive control. (C) The geographic distribution of dddU (blue symbols). The relative abundance of dddU at each station was represented by different sizes symbols. (D) The geographic distribution of dddU transcripts (purple symbols). The relative abundance of dddU transcripts at each station was represented by different sizes symbols.
To infer the environmental importance of dddU compared with other bacterial DMSP lyase genes, the relative abundance of these genes in metagenomic data from polar waters (60 samples) and non‐polar Tara Ocean (174 samples) was assayed. The relative abundance of DddU homologues is lower than DddP, DddQ and DddK but higher than DddW, DddY, and DddL (Table S4; Figure S4). In addition, dddU and its transcripts widely dispersed among the marine metagenomic and metatranscriptomic samples in the Atlantic, Pacific, Indian and polar oceans (Figure 4C,D), suggesting that dddU may play an important role in the global sulphur cycling.
CONCLUSION
DMSP is ubiquitous in marine environments, and its cleavage produces the climate‐active gas DMS. In this study, a new DMSP lyase DddU from an MRG strain Amylibacter cionae H‐12 was identified. DddU is a cupin superfamily DMSP lyase that is phylogenetically distinct to other reported cupin‐containing DMSP lyases. Despite the lack of sequence similarity, DddU likely possesses a very similar catalytic mechanism with DddQ. dddU exists in many Alphaproteobacteria, particularly MRG, and is widely distributed in the global oceans. This study broadens our knowledge on the diversity of DMSP lyases, and enhances our understanding of the DMSP biotransformation in marine environments.
AUTHOR CONTRIBUTIONS
Shu‐Yan Wang: Formal analysis (lead); investigation (lead); writing – original draft (lead). Nan Zhang: Formal analysis (equal); investigation (equal). Zhao‐Jie Teng: Data curation (lead); formal analysis (equal). Xiao‐Di Wang: Data curation (equal); formal analysis (equal). Jonathan D. Todd: Writing – review and editing (equal). Yu‐Zhong Zhang: Conceptualization (equal); funding acquisition (equal); supervision (supporting). Hai‐Yan Cao: Formal analysis (equal); writing – original draft (supporting); writing – review and editing (supporting). Chun‐Yang Li: Conceptualization (lead); funding acquisition (lead); supervision (lead); writing – review and editing (lead).
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
Supporting information
Data S1. Supporting Information
ACKNOWLEDGEMENTS
This work was supported by the National Key Research and Development Program of China (2022YFC2807500, 2021YFA0909600), the National Science Foundation of China (grants 92251303, 42276102, 42076229, 91851205, 31961133016, 42206156), the Fundamental Research Funds for the Central Universities (202172002, 202041011), the Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817), the Program of Shandong for Taishan Scholars (tspd20181203), the Biotechnology and Biological Sciences Research Council, UK, grant (BB/X005968) and Natural Environment Research Council, UK, Standard grants (NE/X000990, NE/V000756 and NE/S001352).
Wang, S.‐Y. , Zhang, N. , Teng, Z.‐J. , Wang, X.‐D. , Todd, J.D. , Zhang, Y.‐Z. et al. (2023) A new dimethylsulfoniopropionate lyase of the cupin superfamily in marine bacteria. Environmental Microbiology, 25(7), 1238–1249. Available from: 10.1111/1462-2920.16355
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in the figures, table and supporting information of this article.
REFERENCES
- Alcolombri, U. , Ben‐Dor, S. , Feldmesser, E. , Levin, Y. , Tawfik, D.S. & Vardi, A. (2015) Identification of the algal dimethyl sulfide–releasing enzyme: a missing link in the marine sulfur cycle. Science, 348, 1466–1469. [DOI] [PubMed] [Google Scholar]
- Andreae, M.O. (1990) Ocean‐atmosphere interactions in the global biogeochemical sulfur cycle. Marine Chemistry, 30, 1–29. [Google Scholar]
- Barek, J. , Pumera, M. , Muck, A. , KaderÏaÂbkova, M. & Zima, J. (1999) Polarographic and voltammetric determination of selected nitrated polycyclic aromatic hydrocarbons. Analytica Chimica Acta, 393, 141–146. [Google Scholar]
- Bratbak, L.M. , Michaud, S. , Cantin, G. & Heldal, M. (1995) Viral activity in relation to Emiliania huxleyi blooms: a mechanism of DMSP release? Marine Ecology Progress Series, 128, 133–142. [Google Scholar]
- Brummett, A.E. , Schnicker, N.J. , Alexander, C. , Todd, J.D. , Mishtu, D. & Ashraf, A.S. (2015) Biochemical, kinetic, and spectroscopic characterization of Ruegeria pomeroyi DddW—a mononuclear iron‐dependent DMSP lyase. PLoS One, 10, e0127288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt, I. , Lauterbach, L. , Brock, N.L. & Dickschat, J.S. (2017) Chemical differentiation of three DMSP lyases from the marine Roseobacter group. Organic & Biomolecular Chemistry, 15, 4432–4439. [DOI] [PubMed] [Google Scholar]
- Cosquer, A. , Pichereau, V. , Pocard, J.A. , Minet, J. , Cormier, M. & Bernard, T. (1999) Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli . Applied and Environmental Microbiology, 65, 3304–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curson, A.R.J. , Liu, J. , Martínez, A.B. , Green, R.T. , Chan, Y. , Carrión, O. et al. (2017) Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nature Microbiology, 2, 17009. [DOI] [PubMed] [Google Scholar]
- Curson, A.R.J. , Rogers, R. , Todd, J.D. , Brearley, C.A. & Johnston, A.W.B. (2008) Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate‐changing gas dimethylsulfide in several marine alpha‐proteobacteria and Rhodobacter sphaeroides . Environmental Microbiology, 10, 757–767. [DOI] [PubMed] [Google Scholar]
- Curson, A.R.J. , Sullivan, M.J. , Todd, J.D. & Johnston, A.W.B. (2010) Identification of genes for dimethyl sulfide production in bacteria in the gut of Atlantic Herring (Clupea harengus). The ISME Journal, 4, 144–146. [DOI] [PubMed] [Google Scholar]
- Curson, A.R.J. , Sullivan, M.J. , Todd, J.D. & Johnston, A.W.B. (2011) DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. The ISME Journal, 5, 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curson, A.R.J. , Todd, J.D. , Sullivan, M.J. & Johnston, A.W.B. (2011) Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nature Reviews. Microbiology, 9, 849–859. [DOI] [PubMed] [Google Scholar]
- Curson, A.R.J. , Williams, B.T. , Pinchbeck, B.J. , Sims, L.P. , Martínez, A.B. , Rivera, P.P.L. et al. (2018) DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nature Microbiology, 3, 430–439. [DOI] [PubMed] [Google Scholar]
- de Souza, M.P. & Yoch, D.C. (1995) Comparative physiology of dimethyl sulfide production by dimethylsulfoniopropionate lyase in Pseudomonas doudoroffii and Alcaligenes sp. strain M3A. Applied and Environmental Microbiology, 61, 3986–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell, J.M. , Purvis, A. & Khuri, S. (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry, 65, 7–17. [DOI] [PubMed] [Google Scholar]
- Fu, H.H. , Jin, M. , Wan, F. & Gao, H.C. (2015) Shewanella oneidensis cytochrome c maturation component CcmI is essential for heme attachment at the non‐canonical motif of nitrite reductase NrfA. Molecular Microbiology, 95, 410–425. [DOI] [PubMed] [Google Scholar]
- Hehemann, J.H. , Law, A. , Redecke, L. & Boraston, A.B. (2014) The structure of RdDddP from Roseobacter denitrificans reveals that DMSP lyases in the DddP‐family are metalloenzymes. PLoS One, 9, e103128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, R.W. , White, B.A. , Cottrell, M.T. & Dacey, J.W.H. (1998) Virus‐mediated total release of dimethylsulfoniopropionate from marine phytoplankton: a potential climate process. Aquatic Microbial Ecology, 14, 1–6. [Google Scholar]
- Howard, E. , Henriksen, J. , Buchan, A. , Reisch, C. , Buergmann, H. , Welsh, R. et al. (2006) Bacterial taxa that limit sulfur flux from the ocean. Science, 314, 649–652. [DOI] [PubMed] [Google Scholar]
- Jumper, J. , Evans, R. , Pritzel, A. , Green, T. & Hassabis, D. (2021) Highly accurate protein structure prediction with AlphaFold. Nature, 596, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten, U. , Kueck, K. , Vogt, C. & Kirst, G.O. (1996) Dimethylsulfoniopropionate production in phototrophic organisms and its physiological functions as a cryoprotectant. Springer, 304, 143–153. [Google Scholar]
- Kiene, R.P. , Linn, L.J. & Bruton, J.A. (2000) New and important roles for DMSP in marine microbial communities. Journal of Sea Research, 43, 209–224. [Google Scholar]
- Kirkwood, M., Le Brun, N.E., Todd, J.D., & Johnston, A.W.B. (2010) The dddP gene of Roseovarius nubinhibens encodes a novel lyase that cleaves dimethylsulfoniopropionate into acrylate plus dimethyl sulfide. Microbiology, 156,1900–1906. [DOI] [PubMed] [Google Scholar]
- Ksionzek, K.B. , Lechtenfeld, O.J. , McCallister, S.L. , Schmitt‐Kopplin, P. , Geuer, J.K. , Geibert, W. et al. (2016) Dissolved organic sulfur in the ocean: biogeochemistry of a petagram inventory. Science, 354, 456–459. [DOI] [PubMed] [Google Scholar]
- Li, C.Y. , Mausz, M.A. , Murphy, A. , Zhang, N. , Chen, X.L. , Wang, S.Y. et al. (2023) Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria. The ISME Journal. Available from: 10.1038/s41396-023-01375-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.Y. , Wang, X.J. , Chen, X.L. , Sheng, Q. , Zhang, S. , Wang, P. et al. (2021) A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl‐CoA. eLife, 10, e64045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.Y. , Wei, T.D. , Zhang, S.H. , Chen, X.L. , Gao, X. , Wang, P. et al. (2014) Molecular insight into bacterial cleavage of oceanic dimethylsulfoniopropionate into dimethyl sulfide. Proceedings of the National Academy of Sciences of the United States of America, 111, 1026–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.Y. , Zhang, D. , Chen, X.L. , Wang, P. , Shi, W.L. , Li, P.Y. et al. (2017) Mechanistic insights into dimethylsulfoniopropionate lyase DddY, a new member of the cupin superfamily. Journal of Molecular Biology, 429, 3850–3862. [DOI] [PubMed] [Google Scholar]
- Liu, J.L. , Liu, J. , Zhang, S.H. , Liang, J.C. , Lin, H.Y. , Song, D.L. et al. (2018) Novel insights into bacterial dimethylsulfoniopropionate catabolism in the East China Sea. Frontiers in Microbiology, 9, 3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt, G.A. (2011) The neuroecology of dimethyl sulfide: a global‐climate regulator turned marine infochemical. Integrative and Comparative Biology, 51, 819–825. [DOI] [PubMed] [Google Scholar]
- Otte, M.L. (2004) Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. Journal of Experimental Botany, 55, 1919–1925. [DOI] [PubMed] [Google Scholar]
- Peng, M. , Chen, X.L. , Zhang, D. , Wang, X.J. , Wang, N. , Wang, P. et al. (2019) Structure‐function analysis indicates that an active‐site water molecule participates in dimethylsulfoniopropionate cleavage by DddK. Applied and Environmental Microbiology, 85, e03127‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina, J.B. , Tapiolas, D.M. , Foret, S. , Lutz, A. , Abrego, D. , Ceh, J. et al. (2013) DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature, 502, 677–680. [DOI] [PubMed] [Google Scholar]
- Reisch, C.R. , Crabb, W.M. , Gifford, S.M. , Teng, Q. , Stoudemayer, M.J. , Moran, M.A. et al. (2013) Metabolism of dimethylsulphoniopropionate by Ruegeria pomeroyi DSS‐3. Molecular Microbiology, 89, 774–791. [DOI] [PubMed] [Google Scholar]
- Reisch, C.R. , Stoudemayer, M.J. , Varaljay, V.A. , Amster, I.J. , Moran, M.A. & Whitman, W.B. (2011) Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature, 473, 208–211. [DOI] [PubMed] [Google Scholar]
- Seymour, J.R. , Simo, R. , Ahmed, T. & Stacker, R. (2010) Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science, 329, 342–345. [DOI] [PubMed] [Google Scholar]
- Shemi, A. , Alcolombri, U. , Schatz, D. , Farstey, V. , Vincent, F. , Rotkopf, R. et al. (2021) Dimethyl sulfide mediates microbial predator–prey interactions between zooplankton and algae in the ocean. Nature Microbiology, 6, 1357–1366. [DOI] [PubMed] [Google Scholar]
- Simon, M. , Scheuner, C. , Meier‐Kolthoff, J.P. , Brinkhoff, T. , Wagner‐Döbler, I. , Ulbrich, M. et al. (2017) Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non‐marine habitats. The ISME Journal, 11, 1483–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefels, J. (2000) Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. Journal of Sea Research, 43, 183–197. [Google Scholar]
- Stefels, J. & Boekel, W.V. (1993) Production of DMS from dissolved DMSP in axenic cultures of the marine‐phytoplankton species Phaeocystis sp. Marine Ecology Progress Series, 97, 11–18. [Google Scholar]
- Sun, J. , Todd, J.D. , Thrash, J.C. , Qian, Y. , Qian, M.C. , Temperton, B. et al. (2016) The abundant marine bacterium Pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol. Nature Microbiology, 1, 16065. [DOI] [PubMed] [Google Scholar]
- Sunagawa, S. , Mende, D.R. , Zeller, G. , Izquierdo‐Carrasco, F. , Berger, S.A. , Kultima, J.R. et al. (2013) Metagenomic species profiling using universal phylogenetic marker genes. Nature Methods, 10, 1196–1199. [DOI] [PubMed] [Google Scholar]
- Sunda, W. , Kieber, D.J. , Kiene, R.P. & Huntsman, S. (2002) An antioxidant function for DMSP and DMS in marine algae. Nature, 418, 317–320. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. & Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, Z.J. , Qin, Q.L. , Zhang, W.P. , Li, J. , Fu, H.H. , Wang, P. et al. (2021) Biogeographic traits of dimethyl sulfide and dimethylsulfoniopropionate cycling in polar oceans. Microbiome, 9, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, Z.J. , Wang, P. , Chen, X.L. , Guillonneau, R. , Li, C.Y. , Zou, S.B. et al. (2021) Acrylate protects a marine bacterium from grazing by a ciliate predator. Nature Microbiology, 6, 1351–1356. [DOI] [PubMed] [Google Scholar]
- Thume, K. , Gebser, B. , Chen, L. , Meyer, N. , Kieber, D.J. & Pohnert, G. (2018) The metabolite dimethylsulfoxonium propionate extends the marine organosulfur cycle. Nature, 563, 412–415. [DOI] [PubMed] [Google Scholar]
- Todd, J.D. , Curson, A.R.J. , Dupont, C.L. , Nicholson, P. & Johnston, A.W.B. (2009) The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environmental Microbiology, 11, 1376–1385. [DOI] [PubMed] [Google Scholar]
- Todd, J.D. , Curson, A.R.J. , Kirkwood, M. , Sullivan, M.J. , Green, R.T. & Johnston, A.W.B. (2011) DddQ, a novel, cupin‐containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria: Cupin‐containing DMSP lyase. Environmental Microbiology, 13, 427–438. [DOI] [PubMed] [Google Scholar]
- Todd, J.D. , Curson, A.R.J. , Nikolaidou‐Katsaraidou, N. , Brearley, C.A. , Watmough, N.J. , Chan, Y. et al. (2010) Molecular dissection of bacterial acrylate catabolism ‐ unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environmental Microbiology, 12, 327–343. [DOI] [PubMed] [Google Scholar]
- Todd, J.D. , Kirkwood, M. , Newton‐Payne, S. & Johnston, A.W.B. (2012) DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS‐3. The ISME Journal, 6, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, J.D. , Rogers, R. , Li, Y.G. , Wexler, M. , Bond, P.L. , Sun, L. et al. (2007) Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science, 315, 666–669. [DOI] [PubMed] [Google Scholar]
- Vallina, S.M. & Simo, R. (2007) Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science, 315, 506–508. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Wei, Y. , Qiu, C. & Li, W. (2017) Amylibacter cionae sp. nov., isolated from the sea squirt Ciona savignyi . International Journal of Systematic and Evolutionary Microbiology, 67, 3462–3466. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Chen, X.L. , Li, C.Y. , Gao, X. , Zhu, D.Y. , Xie, B.B. et al. (2015) Structural and molecular basis for the novel catalytic mechanism and evolution of DddP, an abundant peptidase‐like bacterial dimethylsulfoniopropionate lyase: a new enzyme from an old fold. Molecular Microbiology, 98, 289–301. [DOI] [PubMed] [Google Scholar]
- Wolfe, G.V. , Steinke, M. & Kirst, G.O. (1997) Grazing‐activated chemical defence in a unicellular marine alga. Nature, 387, 894–897. [Google Scholar]
- Yoch, D.C. (2002) Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Applied and Environmental Microbiology, 68, 5804–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.H. , Yang, G.P. , Zhang, H.H. & Yang, J. (2014) Spatial variation of biogenic sulfur in the south Yellow Sea and the East China Sea during summer and its contribution to atmospheric sulfate aerosol. Science of the Total Environment, 488–489, 157–167. [DOI] [PubMed] [Google Scholar]
- Zhang, X.H. , Liu, J. , Liu, J. , Yang, G. , Xue, C.X. , Curson, A. et al. (2019) Biogenic production of DMSP and its degradation to DMS‐their roles in the global sulfur cycle. Science China. Life Sciences, 62, 1296–1319. [DOI] [PubMed] [Google Scholar]
- Zheng, Y. , Wang, J. , Zhou, S. , Zhang, Y. , Liu, J. , Xue, C.‐X. et al. (2020) Bacteria are important dimethylsulfoniopropionate producers in marine aphotic and high‐pressure environments. Nature Communications, 11, 4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the figures, table and supporting information of this article.


