Summary
Background
We apply endoscopic ultrasound-guided fine needle aspiration biopsy to cytopathologically diagnose and sample nucleic acids from primary tumours regardless of the disease stage.
Methods
397 patients with proven pancreatic adenocarcinoma were included and followed up in a multicentre prospective study. DNA and mRNA were extracted from materials of primary tumours obtained by endoscopic ultrasound-guided fine needle aspiration biopsy and analysed using targeted deep sequencing and RNAseq respectively.
Findings
The variant allele frequency of the KRAS mutation was used to evaluate the tumour cellularity, ranging from 15 to 20% in all cells, regardless of the tumour stage. The molecular profile of metastatic primary tumours significantly differed from other types of tumours, more frequently having TP53 mutations (p = 0.0002), less frequently having RNF43 mutations, and possessing more basal-like mRNA component (p = 0.001). Molecular markers associated with improved overall survival were: mutations in homologous recombination deficiency genes in patients who received first-line platinum-based chemotherapy (p = 0.025) and wild-type TP53 gene in patients with locally advanced tumours who received radio-chemotherapy (p = 0.01). The GemPred transcriptomic profile was associated with a significantly better overall survival in patients with locally advanced or metastatic pancreatic cancer who received a gemcitabine-based first-line treatment (p = 0.019).
Interpretation
The combination of genomic and transcriptomic analyses of primary pancreatic tumours enables us to distinguish metastatic tumours from other tumour types. Our molecular strategy may assist in predicting overall survival outcomes for platinum or gemcitabine-based chemotherapies, as well as radio-chemotherapy.
Funding
Institut National Du Cancer (BCB INCa_7294), CHU of Toulouse, Inserm and Ligue Nationale Contre le Cancer (CIT program).
Keywords: Pancreatic cancer, RNA sequencing, Targeted DNA deep sequencing, Translational medicine, Predictive medicine
Research in context.
Evidence before this study
Pancreatic ductal adenocarcinoma remains one of the deadliest types of cancer. The sole curative treatment is a surgical resection, but this option is applicable in less than 15% of cases. The remaining 85% of cases are treated in a palliative way using supportive care or chemotherapy (non-resectable locally advanced and metastatic tumours). As the use of neoadjuvant therapies, platinum-based chemotherapies, and targeted treatments continues to rise, a thorough comprehension of the molecular landscape of tumours is increasingly essential for precise pancreatic cancer treatment. Molecular profiles of pancreatic ductal adenocarcinoma have been investigated both at genomic and transcriptomic levels. Two main transcriptomic profiles have been described: “classical” and “basal-like”, the latter of which is associated with a worse prognosis. A predictive transcriptomic profile has been described for gemcitabine adjuvant treatment response. So far, all these studies have been mainly conducted on resected primary tumour samples that do not represent the entire breadth of pancreatic cancer. Endoscopic ultrasound-guided fine needle aspiration biopsy allows the investigation of primary pancreatic cancer tumours regardless of the disease stage. Therefore, it allowed us to compare the molecular profiles of metastatic, locally advanced, and resected primary pancreatic adenocarcinoma at the genomic and transcriptomic levels.
Added value of this study
We demonstrate the feasibility and the robustness of genomic and transcriptomic analyses from endoscopic ultrasound-guided fine needle biopsy materials in a real-life, prospective, multicentre study. We thus developed a robust bioinformatic approach to help navigate tumour molecular heterogeneity and contaminants. In this way, we confirm that the transcriptomic profile of metastatic (basal-like) primary tumours is different from other (classical) primary tumours. At a genomic level: homologous recombination mutated genes are predictive of responses to platinum-based chemotherapy, while wild-type TP53 gene predicts responsiveness to radio-chemotherapy. At a transcriptomic level: we confirm that transcriptomic profiles are predictive of response to gemcitabine treatment in locally advanced and metastatic tumours.
Implications of all the available evidence
This molecular strategy targeted at primary pancreatic tumours has the potential to improve patient management, regardless of tumour status, using methodologies that are clinically compliant to foster precision medicine strategies to help combat this disease.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) remains one of the deadliest types of cancer and several studies predict that it will be the second most prevalent cause of death by cancer within the next 15 years.1 The sole curative treatment is a surgical resection, but this option is applicable in less than 15% of cases. The remaining 85% of cases (i.e., 30% of patients with non-resectable locally advanced PDAC or 55% of patients with metastatic PDAC) are treated in a palliative way using supportive care or chemotherapy that do not significantly improve patients’ overall likelihood of survival.2,3 As the use of neoadjuvant therapies, platinum-based chemotherapies, and targeted treatments with PARP, NTRK, or KRAS-inhibitors continues to rise,2,4, 5, 6, 7, 8, 9 a thorough comprehension of the molecular landscape of tumours is increasingly essential for precise PDAC treatment.
In this context, a number of recent studies have investigated the molecular profiles of PDAC at the genomic and transcriptomic levels, to help improve patient management. Several prognostic and predictive molecular signatures were established and validated in various independent cohorts of patients.10, 11, 12, 13, 14, 15, 16, 17 Still, these studies are largely focused on resected tumours that correspond to only a fraction of pancreatic tumours, meaning that their translational value remains limited.
EUS-guided fine needle aspiration biopsy (EUS-FNAB) is currently the most effective procedure for cytopathological diagnosis of PDAC primary tumours, and is the most promising approach to address all PDAC tumour types. Still, EUS-FNAB is inconclusive in up to 20% of cases.18,19 We developed a molecular approach based on the KRAS mutation detection that is routinely performed on residual material left following core biopsy, showing clinical value.18,20, 21, 22 We and others extended this strategy to next-generation sequencing and transcriptomic analyses of EUS-FNAB material, improving the positive and differential diagnosis of PDAC as distinguished from pancreatitis.18,20,23 Thus, EUS-FNAB is now more and more considered as a robust and reliable method for the molecular characterisation of PDAC; however, its application in routine practice remains to be fully demonstrated and accepted.
Therefore, we set up a large, prospective, real-life, multicentre cohort in France that includes patients with PDAC regardless of their disease stage.24 Tumour material was obtained following EUS-FNAB. The aim of this study was to determine whether clinically relevant molecular information could be recovered from these samples to improve the management of patients with PDAC. We found that high-throughput genomic and transcriptomic analyses of primary pancreatic tumours is feasible, with the identification of diagnostic and prognostic biomarkers, paving the way towards molecularly based precision medicine strategies using clinically compliant methodologies.
Methods
Patients with PDAC and clinical data
The prospective BACAP cohort (Biological and clinical database for pancreatic adenocarcinoma, as described elsewhere)24,25 comprises patients with PDAC from 14 French centres coordinated by the Toulouse Hospital. Patients with proven PDAC (cytology and/or pathology) are included prior to receiving any treatment, regardless of their tumour status (resectable, borderline, locally advanced, or metastatic). The following are collected for all patients: baseline clinical, radiological and biological data; blood, tissue samples and corresponding nucleic acid; initial treatment and full clinical, radiological follow-up and subsequent treatment until death. Sex of the patients was obtained from clinical health records. The database is managed by the Montpellier Cancer Institute Data Centre using the Clinsight software.24
Ethics
The BACAP cohort was approved by: i) the national French committee for data processing related to health research (September 2013, Folder 13.490); ii) the National French Data Protection Authority (March 2014, Authorisation N°913,462); and iii) the ethics committee (CPP, March 2014). The biological collection was approved by the French Ministry of Research (#DC-2013-1974). The protocol was declared to ClinicalTrials.gov (NCT02818829). Each patient gave written informed consent for investigations, molecular analysis, and collection of the data.
EUS-FNAB procedure and nucleic acid extraction
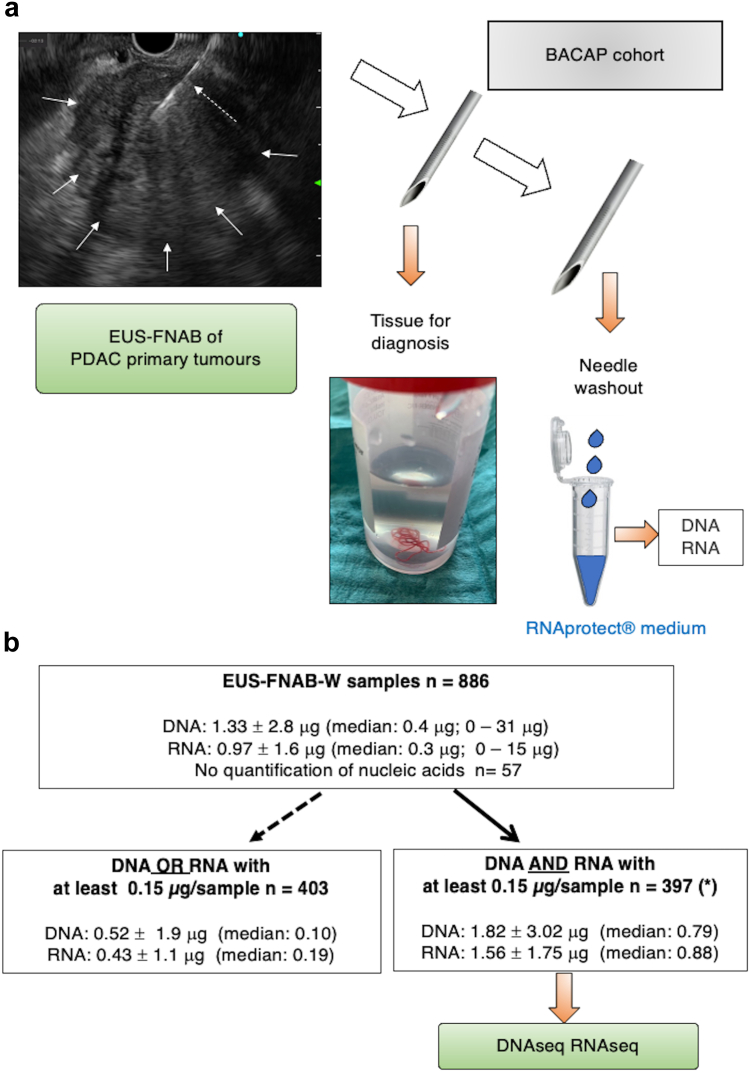
EUS-FNAB from primary tumours was performed prior to any treatment, using 19 to 22 G FNA needles as previously described.20,22 The material for cytopathology was instantly fixed in formalin. The remaining material was obtained by flushing the FNA needle with air via 10 mL syringe (the so called EUS-FNAB wash – EUS-FNAB-W), before immediately storing in 500 μl of RNAprotect® cell reagent (Qiagen, Les Ulis, France). This procedure was previously validated in the coordinating centre.22,23 EUS-FNAB-W were centrifuged at 8000 rpm. Cell pellets were immediately frozen at −80 °C until nucleic acid extraction. Fig. 1a depicts the sampling procedure of both cytopathology and material for nucleic acid extraction. This procedure was standardised across all centres.
Fig. 1.
Procedure of nucleic acid sampling during endoscopic ultrasound-guided fine needle aspiration biopsy of pancreatic adenocarcinoma primary tumour and subsequent quantification. a: Schematic representation of the BACAP protocol for RNA and DNA sequencing from EUS-FNAB-W (endoscopic ultrasound-guided fine needle aspiration biopsy wash) materials. The tissue is reserved for anatomopathological diagnostics. Thereafter, the needle is washed (by flushing with air) and kept in RNAprotect Cell Reagent® before pellet isolation and storage at −80 °C until subsequent extraction of both DNA and total RNA and then DNA and RNA sequencing, respectively. In the upper left part: EUS view of a PDAC tumour (white arrows) and the needle within the tumour (dashed white arrow). b: Detail of distribution of EUS-FNAB-W samples and quantification of nucleic acids: for each sample, DNA and total RNA were dissolved in 40 and 20 μL, respectively. Total quantification is given for each sub-group (mean ± SD; median and extremes in parenthesis). Among the 886 samples from the BACAP cohort, nucleic acids (DNA and RNA) were not detectable in 57 samples. From the remaining 829 samples, we selected 426 EUS-FNAB-W samples upon a threshold of both DNA and RNA of at least 0.15 μg per sample. Final analyses were performed on 397 samples as 29 samples were in duplicate. For the 403 remaining samples, either DNA or RNA did not reach a threshold of 0.15 μg.
DNA/RNA extraction and storage
Extraction of nucleic acids was centralised in a single platform for molecular biology at the University Institute Oncopole of Toulouse. DNA and total RNA were extracted using AllPrep DNA/RNA/miRNA Universal kit (Qiagen), according the manufacturer’s protocol. Quantification of DNA and RNA was performed using Qubit® dsDNA and RNA Kits, respectively (Thermofisher Scientific, Saint-Herblain, France). DNA and RNA were stored at −80 °C until further analysis.
Targeted deep sequencing
Library preparation, exome capture, sequencing and data analysis were performed by IntegraGen SA (Evry, France) using a panel of 74 genes using Twist Bioscience’s instructions and protocols without modification, except for library preparation, which was performed with the NEBNext® Ultra II kit (New England Biolabs®, Evry, France) using 150 ng of input. DNA samples were sequenced with an Illumina NovaSeq 6000 apparatus as paired-end 100 reads. Sequence reads were mapped to the human genome build (hg38) using BWA (Burrows-Wheeler Aligner software). Duplicate reads and variants (SNV and indels) were removed using GATK’s (Genome Analysis ToolKit) Haplotype Caller GVCF tool (GATK3.8.1). The normal panels of the Broad Institute and of the constitutional samples sequenced at Integragen were used in somatic variant analysis. Ensembl’s VEP (Variant Effect Predictor, release VEP95.1) was used to annotate variants as well as clinicopathological significance from ClinVar (https://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg38&g=clinvar) and predicted pathogenicity using DANN (Deep learning approach for ANNotating the pathogenicity of genetic variants), FATHMM (http://fathmm.biocompute.org.uk) MutationTaster, SIFT (https://sift.bii.a-star.edu.sg) and Polyphen (Polymorphism Phenotyping). A homologous recombination deficiency (HRD) was concluded if a somatic non-synonymous mutation in one of the homologous recombination genes (BRCA2, BRCA1, PALB2, BARD1, BLM, BRIP1, CDK12, CHEK1, CHEK2, FANCC, FANCD2, FANCE, FANCF, FANCI, FANCL, FANCM, MRE11, NBN, RAD51, RAD51B, RAD51C, RAD51D, RAD52, RAD54L, RPA1) was found. To evaluate the tumour cellularity of EUS-FNAB-W samples, the variant allele frequency of the KRAS oncogene hotspots mutation was chosen and compared to the one obtained from resected tumours for which the DNA was extracted from a punch sampled26 in a tissue area of high cellularity, as defined by anatomopathological analysis.
Transcriptomic analysis
150 ng of total RNA was used to generate RNA-seq sequencing libraries using Lexogen Quantseq 3’ FWD kit and sequenced by the Institut du Cerveau (Paris, France) iGenseq platform on a NovaSeq 6000 apparatus aiming for a minimum of 10 million reads. Raw RNAseq reads were mapped to the human genome (Ensembl GRCH38) and to Ensembl’s reference transcriptome using STAR (RNA-seq aligner). Gene counts were obtained using FeatureCount, normalised by a UpperQuartile procedure, and logged on a base 2.
Marker-based deconvolution
The marker-based deconvolution (mDeconv) approach aims to identify the prototypic gene expression profiles of pre-specified cell populations and their proportions in a given set of bulk transcriptomic profiles simultaneously, using only a set of non-overlapping marker genes. This is achieved using a constrained non-negative matrix factorisation (NMF), with a number of components equal to the number of provided cell types plus one for the remaining variance and an additional constraint ensuring that marker genes have their maximum weight in the cell type components they are assigned to. Allowing any marker gene to be expressed in any cell type makes marker selection less stringent. Moreover, learning the gene weights in a specific dataset allows us to capture the inherent domain-specific variance. The implementation of this semi-supervised NMF is available at https://github.com/GeNeHetX/semiSupNMF. This unsupervised approach was evaluated: i) on 400 pseudo-bulks from single-cell RNA-seq27 to re-identify initial mixture proportions; ii) on the TCGA data28; and iii) on a series of 50 resected tumours29 to identify the proportion of tumour cells in correlation with the unmatched exome-based estimation of cellularity and pan-cytokeratin staining, respectively.
Transcriptomic analysis in the prediction of response to gemcitabine treatment
To investigate the association between the improved mRNA signature and OS in patients who had been predicted to be sensitive and treated with gemcitabine-based treatment, we applied the Gem-predictive profile recently characterised and validated and known as GemPred.30,31
Statistics
Differential analysis of gene expression was performed using a non-parametric Mann–Whitney test. The association between the proportion of mutation and the stage was tested using Fisher’s exact test. Initial unsupervised discovery of sample composition in BACAP samples was performed using an independent component analysis on the transcriptome using 20 components. Components were manually annotated by projecting components in other datasets (normal and tumour samples from the TCGA and ICGC), by gene set enrichment analysis of each component gene weight and by projection in single-cell RNA-seq datasets.27,32 Spearman’s correlation was used to evaluate the association of two continuous variables. Survival analyses were performed only using data from patients with advanced diseases (locally advanced or metastatic), with no reported surgery and with a reported first-line treatment. Survival analyses were performed using a Cox proportional hazard regression model stratified by ECOG status and reporting the Log rank test.
Role of funders
The funders had no role in the study design, data collection, data analyses, interpretation, or writing of the report.
Results
Patients and samples
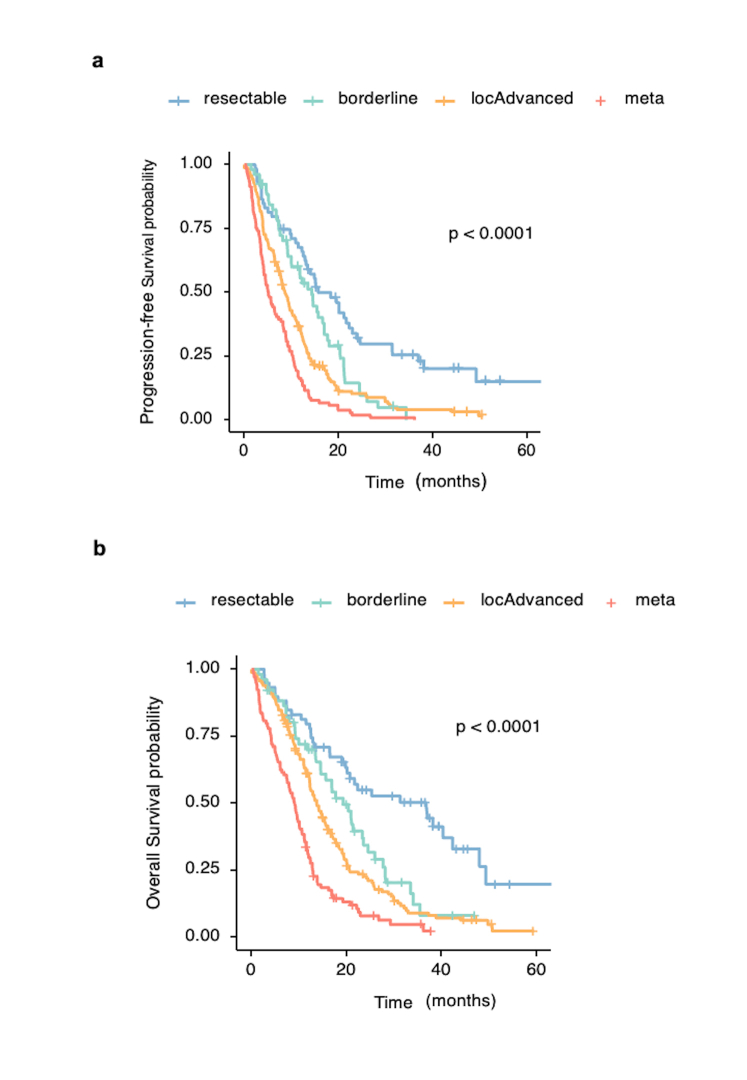
From the BACAP cohort, a total of 886 EUS-FNAB-W samples were extracted using the same nucleic acid extraction method. For 57 samples (6.4%), nucleic acids (DNA and RNA) were not detectable. From the remaining 829 samples, we selected 426 EUS-FNAB-W samples of proven primary adenocarcinoma tumours corresponding to 397 different patients, as 29 samples were in duplicate. These samples were selected upon a threshold of both DNA and RNA of at least 0.15 μg per sample according to the sequencing facility recommendations. For the remaining 403 samples, DNA or RNA did not reach a threshold of 0.15 μg (distribution and quantifications are shown in Fig. 1b). The demographic and main clinical characteristics of these patients are detailed in Table 1 (main demographic data separating female and male are given in the Supplementary File 2). The survival curves of patients depending on the disease stage are shown in Supplementary Fig. S1. The patient’s profiles and clinical features agree with the expected clinical and evolutive profile of patients with PDAC. The ranking of overall survival (OS) and progression-free survival (PFS) values was as follows: patients with resectable > borderline > locally advanced > metastatic PDAC (median overall survivals of 36.8, 19.2, 13.7 and 9.1 months respectively; p < 0.0001 between each classification) (Log rank test).
Table 1.
Demographic characteristics of the patients with pancreatic cancer included in the DNA and RNA sequencing analysis from endoscopic ultrasound-guided fine needle aspiration biopsy washing of material from primary tumours.
| Characteristics | Resectable N = 61a | Borderline N = 55a | Loc. Advanced N = 170a | Meta N = 106a | Overall N = 392b | p-value |
|---|---|---|---|---|---|---|
| Age | 71 (64, 75) | 69 (60, 77) | 70 (63, 76) | 68 (61, 76) | 70 (62, 76) | 0.5c |
| Sex | 0.4d | |||||
| F | 26 (43%) | 19 (35%) | 81 (48%) | 46 (43%) | 172 (44%) | |
| M | 35 (57%) | 36 (65%) | 89 (52%) | 60 (57%) | 220 (56%) | |
| Size of primary tumour (mm) | 29 (23, 34) | 35 (25, 40) | 40 (30, 47) | 36 (30, 48) | 35 (30, 44) | <0.001c |
| Unknown | 4 | 2 | 9 | 4 | 19 | |
| ECOG2 | 0.042d | |||||
| PS0 | 27 (52%) | 31 (65%) | 59 (40%) | 36 (37%) | 153 (44%) | |
| PS1 | 18 (35%) | 13 (27%) | 70 (48%) | 49 (51%) | 150 (44%) | |
| PS2–S3 | 7 (13%) | 4 (8.3%) | 18 (12%) | 12 (12%) | 41 (12%) | |
| Unknown | 9 | 7 | 23 | 9 | 4 | |
| Treatments | ||||||
| Resected | 40 (66%) | 22 (40%) | 6 (3.5%) | 0 (0%) | 68 (17%) | <0.001d |
| Radiotherapy | 11 (18%) | 12 (22%) | 39 (23%) | 2 (1.9%) | 64 (16%) | <0.001d |
| Chemotherapy | 49 (80%) | 37 (67%) | 141 (83%) | 81 (76%) | 308 (79%) | 0.090c |
| Measured survival | 59 (97%) | 52 (95%) | 164 (96%) | 104 (98%) | 379 (97%) | 0.6e |
Loc. Advanced: locally advanced tumour; Meta.: Metastatic tumour; ECOG2: Eastern Cooperative Oncology Group General Status score; PS: clinical performance status. Hepatic, peritoneal and pulmonary metastasis were present in 70, 15.5 and 14.5% of patients, respectively. 30% of patients had at least two metastatic sites.
Median (IQR); N (%).
Given that there were 5 missing stages, the data pertain to 392 patients (from a total of 397 patients).
Kruskal-Wallis rank sum test.
Pearson’s Chi-square test.
Fisher’s exact test.
Use of the mutational landscape for evaluating tumour cellularity of EUS-FNAB-W samples
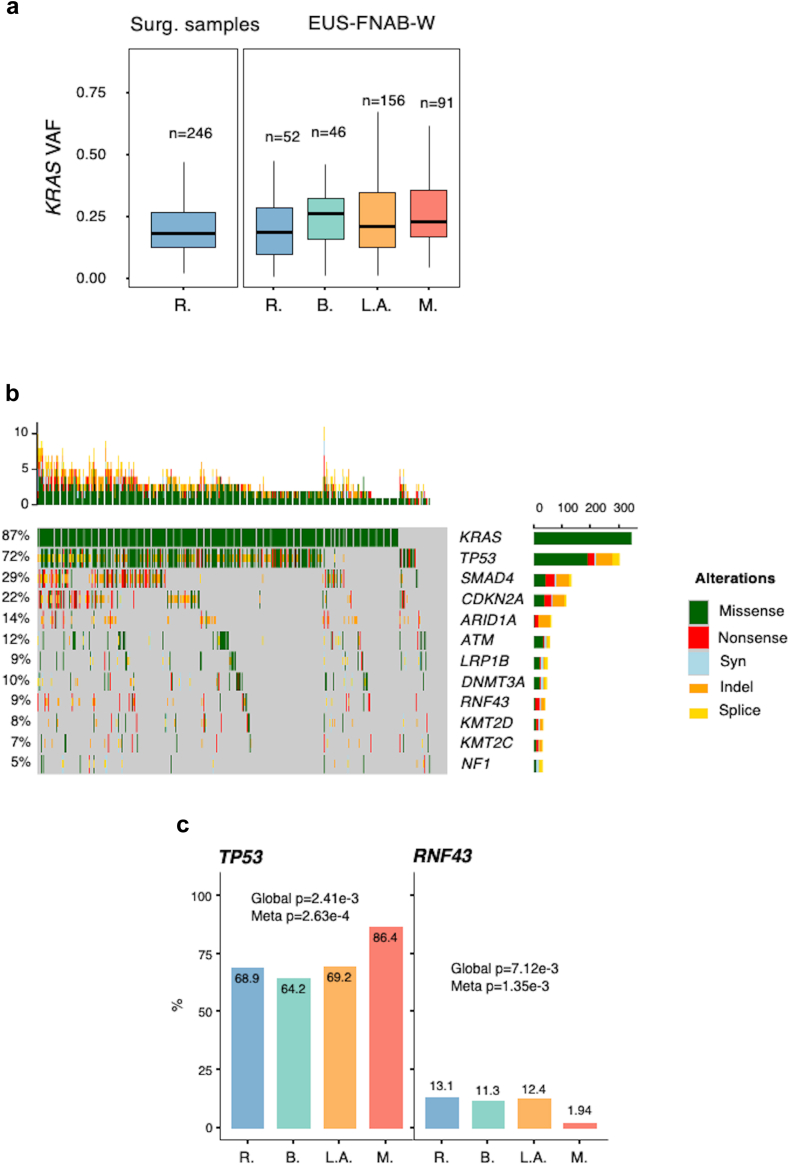
One crucial aspect in assessing the clinical relevance of EUS-FNAB-W samples is estimating tumour cellularity from the complex mixture composed of adenocarcinoma cells, stroma cells, and blood cells, among others. As a surrogate marker of PDAC cellularity, we chose the variant allele frequency (VAF) of the KRAS oncogene hotspots mutation. EUS-FNAB-W-derived KRAS VAF was compared to a reference VAF obtained from resected tumours for which the DNA was extracted using a punch biopsy in an area of high cellularity, localised by anatomopathological analysis. Fig. 2a shows the distribution of KRAS VAF in the punched resected tumours (n = 246) versus those obtained from EUS-FNAB-W from various disease stages (total n = 345). Mean VAF in the reference surgical group was 0.18 ± 0.27 (left panel), and ranged from 0.18 to 0.25 in EUS-FNAB-W material (right panel). These results indicate that tumour cells cellularity measured using KRAS VAF analysis in EUS-FNAB-W is representative of surgical specimens of PDAC tumour cells content, thus validating the clinical relevance of our approach.
Fig. 2.
Landscape of DNA mutations from EUS-FNAB-W. a: Distributions of the Variant Allele Frequency (VAF) of KRAS mutations in a retrospective series of punched surgical samples (left panel) and in each disease stage of EUS-FNAB-W from the BACAP protocol (right panel). VAF analysis was performed in a total of 345 patients (86.9% of cases). b: Oncoplot of the main mutations found by EUS-FNAB-W of the BACAP protocol. DNA sequencing was performed in a total of 390 patients (98.2% of cases). c: Proportion of patients presenting mutations in TP53 and RNF43 genes in each disease stage (R: resected and/or resectable; B: borderline; L.A.: locally advanced; M: metastatic). (Fisher’s exact test).
DNA mutational profile
We next performed targeted DNA sequencing from EUS-FNAB-W material. Results shown in Fig. 2b indicate the prevalence and the nature of the mutation events within the 12 most altered genes. These mutations correspond to the widely acknowledged mutational profile of PDAC tumours, with frequent alterations in the top 4 main driver genes for PDAC (i.e., KRAS, TP53, SMAD4 and CDKN2A). These findings validate at a large scale the use of EUS-FNAB-W material for targeted DNA sequencing.
We further explored the translational value of this strategy. Thus, the association between the mutation profile and disease stage was tested for every gene that was found to be altered in at least 5% of the samples. Using this approach, only two genes showed a significant association (False Discovery Rate, FDR, <5%) with disease stage, especially when comparing metastatic tumours to other tumour stages. Indeed, we found that TP53 mutations are significantly more frequent in metastatic tumours (86.4% versus 68.9%, 64.2% and 69.2% in resected, borderline and locally advanced tumours, respectively, p = 2.63e-4), whereas RNF43 is significantly less frequently mutated in the same metastatic tumours (1.94% versus 13.1%, 11.3%, 12.4% in resected, borderline and locally advanced tumours respectively p = 1.35e-3) (Fisher’s exact test) (Fig. 2c). This result suggests that primary tumours that have metastasised are genetically distinct from other PDAC tumour stages. We have also checked for the age of the patients depending on the mutational status of KRAS gene: there is no statistical difference between subgroup of patients with KRAS mutation versus those with wild-type KRAS (p = 0.854, Mann–Whitney test).
RNA-seq marker-based deconvolution for quantitative cell composition estimation
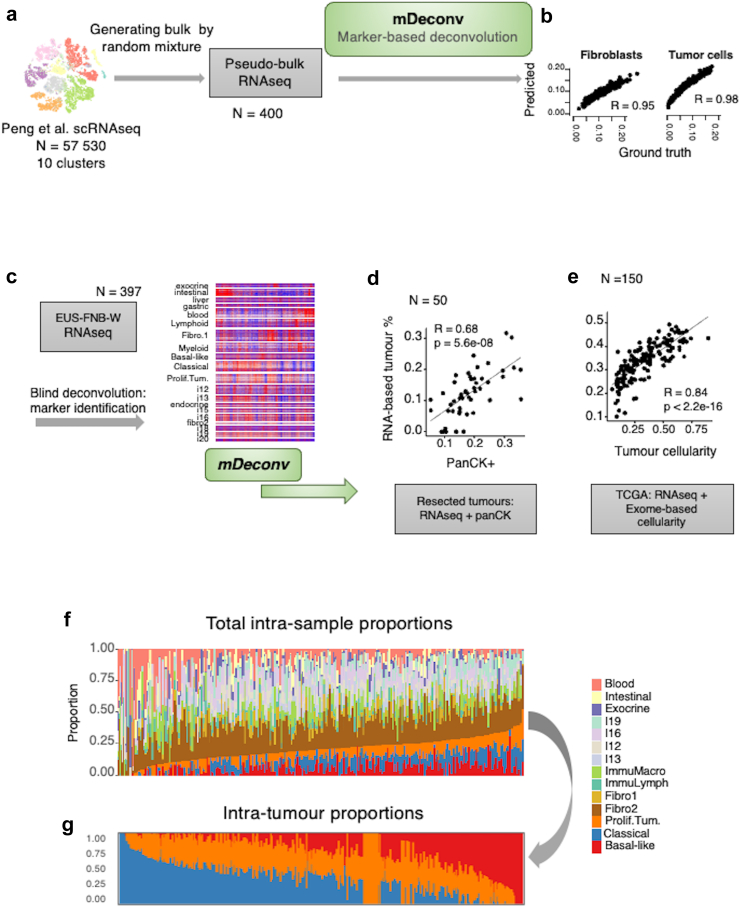
Given the high heterogeneity of PDAC samples and the diversity of potential contaminants in EUS-FNAB-W, we developed a robust deconvolution approach that can be applied to any type of tissue and sample. We propose a semi-supervised approach of marker-based deconvolution, termed mDeconv, which aims to quantify cellular components based on a set of marker genes defined to be specifically over-expressed in each tumour component that is to be quantified. mDeconv internally uses a non-negative matrix factorisation approach which constrains each factor to be specific to their designated marker set. mDeconv was first applied to a pseudo-bulk dataset built from 400 mixtures of 10 clusters of PDAC single-cell RNA-seq profiles27 (Fig. 3a). Using 10 independent components to blindly extract 10 sets of marker genes, mDeconv quantified each cellular component with an accuracy (Spearman’s correlation) ranging from 0.95 to 0.99 (Fig. 3b). Following this, 20 marker sets were recovered from an independent component analysis of the EUS-FNAB-W transcriptome profiles (n = 397, Fig. 3c), encompassing normal tissues (exocrine parenchyma, intestinal, or gastric tissues, whole blood), general immune populations (lymphoid or myeloid), two fibroblastic components, three tumour components, as well as undetermined components. mDeconv was then applied to two different PDAC transcriptome datasets using these marker sets. First, on a series of 50 resected tumours, tumour cellularity was estimated using the area of positive pan-cytokeratin immunohistochemistry staining (panCK).29 The sum of the three tumour components quantified by mDeconv correlated with the panCK-estimated cellularity (Spearman’s R = 0.68, Fig. 3d). Second, on the 150 resected tumours of the TCGA,28 RNAseq-based tumour cellularity quantification recovered by mDeconv correlated with the exome-based cellularity estimate (Spearman’s R = 0.84, Fig. 3e). The mDeconv-based cellular component quantification on the present EUS-FNAB-W cohort is shown in Fig. 3f, and the intra-tumour quantification of the three tumour components in Fig. 3g: basal-like profile (in red), classical (in blue) and a more generic tumour component associated to proliferating transformed epithelial cells referred to as “proliferating tumour” (in orange). The “proliferating tumour” components described in our study may be related and considered as tumour components without typical differentiation markers, and in that aspect may relate to the intermediary phenotype recently described33,34 in studies made on tissue specimens. In this case, 12 tumours had no quantified tumour components, suggesting either that there were too few tumour cells to be detected or that an undetermined tumour phenotype was present. These tumours also tended to be more frequently KRAS wild-type (odds ratio = 7.68, Fisher’s exact test p = 0.00152). Collectively, we successfully assessed tumour heterogeneity at the transcriptional level using EUS-FNAB-W material analysis and a bioinformatic pipeline. We identified 3 main tumour components (basal-like, classical, and proliferating) in most tumours, with the exception of 12 samples (3%), in which KRAS wild-type status tended to be overrepresented/dominant.
Fig. 3.
RNAseq deconvolution. a: Schematic diagram of the deconvolution method mDeconv on a synthetic dataset. Ten cell types obtained from the clustering of 57,530 single cell RNA-seq were randomly sampled and summed to generate 400 synthetic pseudo-bulk RNA-seq. mDeconv was applied blindly to the synthetic dataset and proportions were estimated from the pseudo-bulk RNA-seq. b: Spearman’s correlations are shown, comparing estimated and true mixture proportions for each of the cell types (fibroblasts and tumour cells). c: Schematic diagram of the application of mDeconv on the BACAP EUS-FNAB-W RNA-seq, identifying 20 independent component-based marker sets. d: Application of mDeconv on a dataset of 50 resected tumours with RNA-seq and corresponding pan-cytokeratin-positive area (panCK+). Scatter plot shows the correlation between the panCK+ and the sum of the estimated proportions of the three RNA-seq-based tumour components. e: Application of mDeconv on the TCGA dataset (150 resected primary tumours). Scatter plot shows the correlation between the exome-based and the RNA-seq estimations of tumour proportions. f: Intra-sample proportions of all the relevant RNA components and g: of tumour components only. The results are from the samples from all the 397 patients.
RNA expression profile of PDAC tumours samples using EUS-FNAB-W
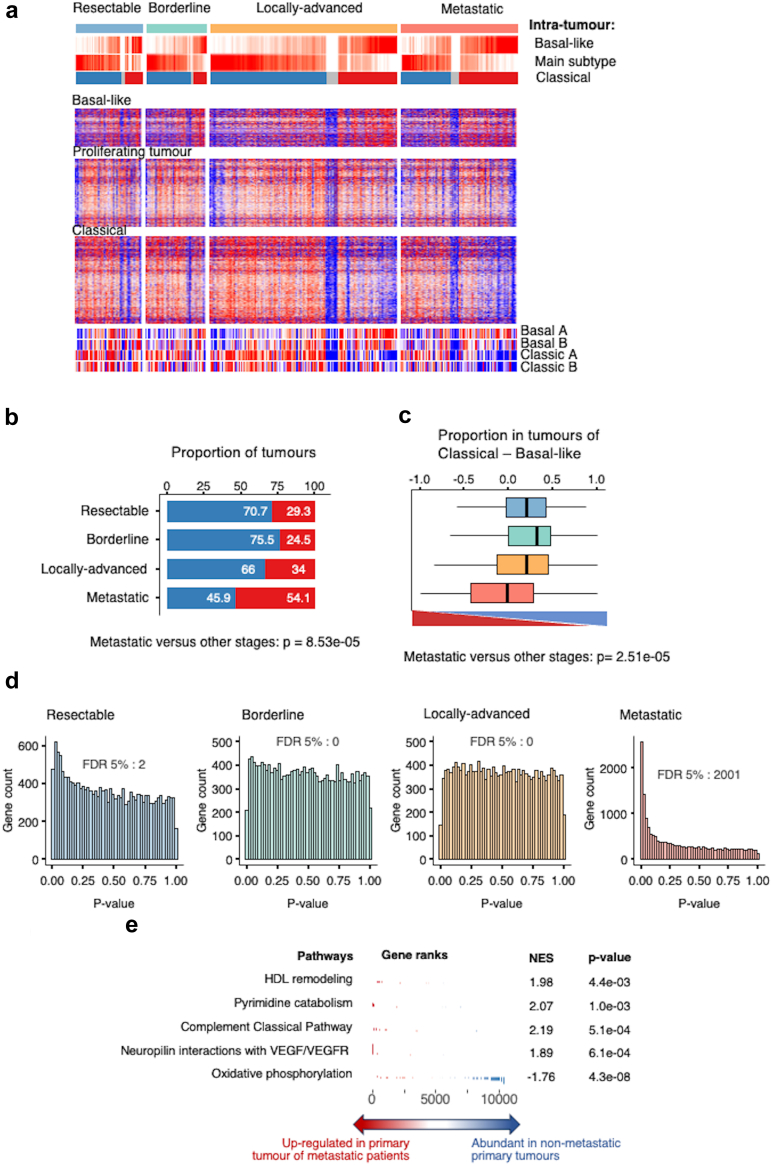
While anatomical stages of disease have major implications in PDAC management, in particular regarding surgery, it is still unclear to what extent these differ biologically. Focusing on the three tumour components (i.e., classical, basal-like, and proliferating tumour), the expression patterns were globally similar across the four stages (Fig. 4a). However, the basal-like tumour component (in red) was more prevalent in tumours at the metastatic stage (54.1%, Fisher’s exact test versus other stages: p = 8.53e-5) (Fig. 4b). This finding was confirmed when considering the intra-tumour composition by calculating the difference between the basal-like and classical component quantities. Here again, primary tumours at the metastatic stage display significantly less classical (blue) and more basal-like (red) component, when compared to other tumour stages (Fig. 4c) (p = 0.00019). Finally, the transcriptomes of each stage were compared with one another. Fig. 4d shows the distribution of Mann–Whitney’s test p-values, demonstrating little to no differentially expressed genes between resectable, borderline, and locally advanced primary tumours, while metastatic PDAC is highly distinct in its transcriptomic profile with 2001 differentially expressed genes (FDR <5%). During this analysis, normality was checked for sequenced normalised RNA counts, and similarly to previous observations, genes in general were not found to follow a gaussian distribution.
Fig. 4.
RNAseq landscape by disease stage. a: Heatmap of the 392 patients with known disease stage. Patients are arranged according to the difference between the classical and basal-like intra-tumour proportions. Individual gene expression levels from each of the three tumour components are shown. b: Proportion of tumours of each subtype, defined by the main one between basal-like (red) and classical (blue), at each disease stage. Fisher’s exact test indicated that basal-like component is significantly expressed within metastatic primary tumours versus other stages. c: Distribution of the difference between the classical and basal-like intra-tumour proportions in each disease stage (Blue: resectable; green: borderline; orange: locally advanced; light red: metastatic) (bottom of the figure: triangle red “basal-Like”; triangle blue “classical”). The Kruskal–Wallis test was significant with p = 0.00019 and a significant difference between metastatic subgroup versus other stages regarding the presence of “basal-like” component (Mann–Whitney test). d: Mann–Whitney test p-value histograms comparing the expression of 18,220 genes between each disease stage against the rest. The number of genes with an FDR <5% is shown at the bottom. e: Gene set enrichment analysis of the differential gene expression between primary tumour samples from metastatic versus non-metastatic patients.
Gene set enrichment analysis (GSEA) indicates that primary tumours at the metastatic stage show enrichment in HDL remodelling, pyrimidine catabolism, complement classical pathway, and neuropilin interactions with VEGF/VEGFR signatures, while the signature for oxidative phosphorylation is repressed, as compared to other tumour stages (Fig. 4e). Collectively, transcriptomic analysis confirmed that primary tumours at a metastatic stage molecularly differ from PDAC tumours at different stages.
Role of DNA mutational profile in the prediction of the response to treatment
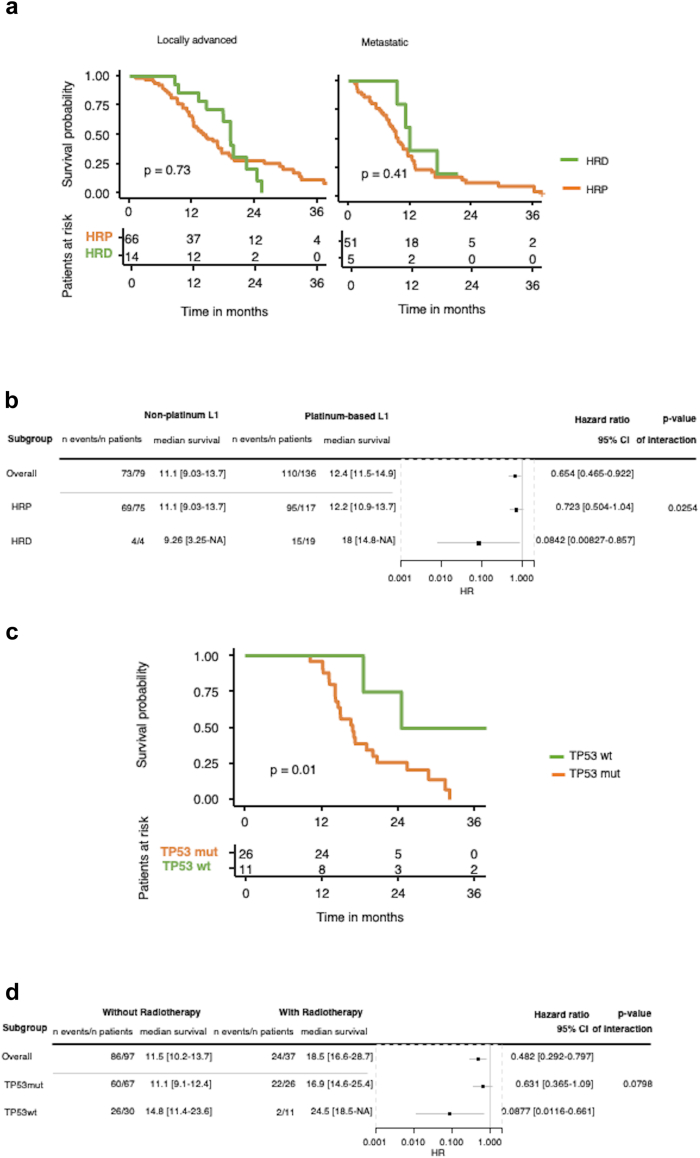
To gain further insights into the translational potential of the genomic analysis of EUS-FNAB-W material, we first investigated the influence of mutation in homologous recombination deficiency genes on the survival of patients receiving platinum-derived chemotherapy as a first-line treatment. Thus, we analysed the influence of the presence of mutation(s) of at least one HRD gene on the overall survival (OS) in the sub-group of patients with unresectable tumours (i.e., metastatic and/or locally advanced with no subsequent resection) who had received at least one cycle of chemotherapy containing platinum-derived chemotherapy (mainly oxaliplatin in FOLFIRINOX or FOLFOX protocols) as first-line regimen. These patients were compared to sub-groups of patients without mutations (HRP group for Homologous Recombination Proficient genes, i.e., wild-type). Fig. 5a shows the survival curves of patients with locally advanced (left panel) and metastatic (right panel) PDAC. Detection of HRD gene mutation(s) in the EUS-FNAB-W material of primary tumours was associated with a better OS but was not statistically significant (locally advanced PDAC p = 0.73; metastatic PDAC p = 0.41) (Log rank test).
Fig. 5.
DNA mutational profile and response to treatment. a: Kaplan–Meier curve for patients with locally advanced (left) or metastatic (right) pancreatic cancer who received a platinum-based regimen as first-line treatment (L1), stratified by the mutation status of genes involved in homologous repair (HRD: mutated; HRP: wild type). b: Forest plot of the interaction between platinum-based L1 and HR status among patients with advanced diseases (HRD: mutated; HRP: wild-type) (cox proportional hazards regression model). c: Kaplan–Meier curve for patients with locally advanced pancreatic adenocarcinoma who received a radio-chemotherapy stratified by TP53 mutation status. d: Forest plot of the interaction between radio-chemotherapy and TP53 mutations. All Forest plots and Cox proportional hazards regressions are stratified by disease stage and performance status. (HRD: homologous repair deficient genes; HRP: homologous recombination proficient genes). List of the HR genes is as follow: BRCA2, BRCA1, PALB2, BARD1, BLM, BRIP1, CDK12, CHEK1, CHEK2, FANCC, FANCD2, FANCE, FANCF, FANCI, FANCL, FANCM, MRE11, NBN, RAD51, RAD51B, RAD51C, RAD51D, RAD52, RAD54L, RPA1.
However, we found that HRD gene mutation(s) in patients with both locally advanced and metastatic PDAC associated with a significant benefit in OS following treatment (Hazard Ratio: 0.084; p = 0.025) (Fig. 5b) (Cox proportional hazard regression model).
We then assessed the influence of TP53 mutations on the survival of patients with locally advanced PDAC who received radio-chemotherapy as a first-line treatment. As shown in Fig. 5c and d, the presence of wild-type TP53 within the EUS-FNAB-W material both positively and significantly influenced (i.e., improved) the patients’ OS (p = 0.01), with a Hazard Ratio of 0.087 (p = 0.079) (Cox proportional hazard regression model).
Collectively, these results show that genetic interrogation of EUS-FNAB-W material shows promise to predict response to treatment in patients with PDAC.
Role of transcriptomic analysis in the prediction of response to gemcitabine treatment
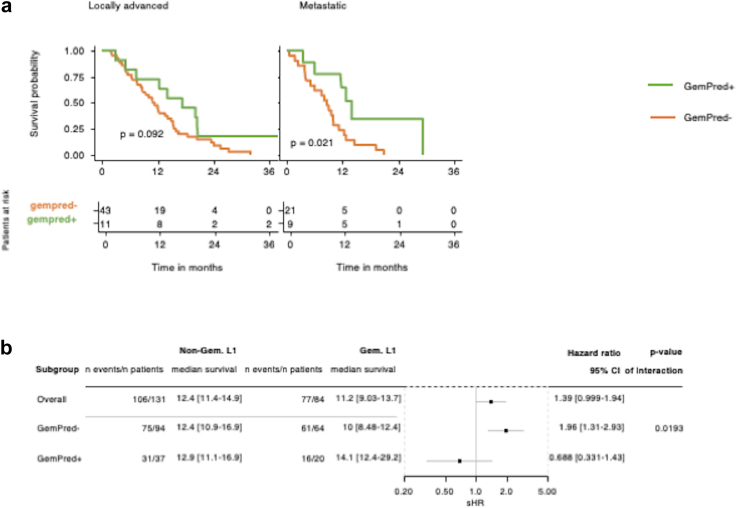
On the same line, we explored whether the transcriptomic profile of PDAC tumours obtained following EUS-FNAB-W may help guide therapeutic decisions. Thus, we explored the correlation between the presence of the GemPred signature (that was previously found to be associated with gemcitabine response),30,31 and OS in patients with locally advanced or metastatic PDAC treated with gemcitabine-based therapy as a first-line option. As shown in Fig. 6a, the presence of the Gem-predictive mRNA profile in the EUS-FNAB-W material of primary PDAC is significantly associated with a better survival in patients with metastatic PDAC (p = 0.021) (Log rank test) and tends to improve OS in patients with locally advanced PDAC (p = 0.092) treated by gemcitabine (Log rank test). When pooling both tumour types, we found that enrichment of the GemPred signature is associated with a significantly higher survival rate (Hazard Ratio: 0.68; p = 0.019) (Fig. 6b) (Cox proportional hazard regression model). This result argues for transcriptomic exploration of EUS-FNAB-W material for treatment response prediction.
Fig. 6.
Transcriptomic profile and response to treatment. a: Kaplan–Meier curve for patients with locally advanced (left) or metastatic (right) pancreatic cancer who received a gemcitabine-based regimen as first-line treatment (L1), stratified by the transcriptome-based GemPred signature. b: Forest plot of the interaction between gemcitabine-based L1 and GemPred among all patients with advanced diseases (i.e., locally advanced plus metastatic). The Forest plot and Cox proportional hazards regression is stratified by disease stage and performance status.
Influence of sex and age on prognosis and response to treatment
In the present cohort we also included the sex and age of patients in our analysis (Cox proportional hazard regression model). Sex was not associated with survival whatever the subgroup of patients. Age was not associated with survival in metastatic patients. However, among patients with locally advanced disease, older patients tended to have a shorter overall survival (statistically not significant). However, age had no significant association with survival in a multivariate model in locally advanced diseases which also included treatment modalities (radiotherapy, surgery and FOLFIRINOX), suggesting that the detrimental effect of older age was caused by a less frequent use of aggressive therapeutic options. We evaluated several models with Gempred stratification, stratified by sex, age and/or radiotherapy and found no statistically significant difference.
Discussion
The present study is a real-life, prospective and multicentric investigation of the feasibility and of the translational potential of DNA and RNA analysis of primary pancreatic tumours of all stages naïve of treatment. We found that genomic and transcriptomic exploration of EUS-FNAB-W material segregates primary tumours at the metastatic stage from other tumour stages. In addition, we validated gene mutations and RNA expression profiles as biomarkers for prediction of patients’ responses to chemotherapy or radiotherapy, demonstrating the translational value of the approach.
The multicentric and prospective BACAP cohort is so far the largest clinical and biological repository dedicated to patients with PDAC from all stages worldwide.24,25 In addition, this cohort can be considered representative of the current practice as median survival of patients are similar to those observed in “real-life studies”.35, 36, 37 Moreover, the percentage of KRAS mutations and that of the three other main driver genes are similar to the percentages currently observed in PDAC.18,33,34
However, our study highlights some limitations. Indeed, 57 samples (6.4%) were negative for nucleic acids in the EUS-FNAB-W, while the core biopsy was of acceptable quality. In addition, 45% of the samples did not reach the threshold in RNA or DNA material recommended by subcontractors (i.e., >150 ng per sample).
Besides the intratumoural environment (fibrosis or not, stroma reaction or not, apoptosis, etc.) there are many reasons for explaining the difference in quantities of DNA and/or RNA within the EUS-FNAB wash materials, such as various needle types used for sampling, multiple operators in some centres, size and localisation of the tumour, etc. Thus, it is difficult to assess some of these parameters as well as histology, taking into account the small size of micro biopsies and sometimes the presence of only cytological materials. However, we have checked for several parameters that might account for these two subgroups. First, the survival rates, stage to stage, of patients with insufficient biopsy quality were not significantly different from those of patients with sufficient biopsy quality (data not shown), reflecting that the biological behaviour of the tumour seems to be equivalent among these two subgroups. Following this, we checked for the influence of the multicentre design of our study. There is no significant difference between any two pairs of centres in terms of DNA quantities extracted, KRAS mutation rate as well as SMAD4, CDKN2A, and TP53 mutation rate (data not shown).
Recent advancements in sample preparation and sequencing efficiency have, however, lowered the amount of biological material required to around 10–30 ng, indicating that more than 90% of samples from this study would become eligible for analysis of DNA and/or RNA.38
By performing different sets of molecular analysis (tumour cellularity, mutation and transcriptional profiles), we established here that the molecular analysis of tumour material recovered from EUS-FNAB-W is not of secondary importance to data obtained from PDAC surgical specimen tissues, demonstrating that EUS-FNAB-W material is relevant to molecular analysis in clinical practice.
We highlight here that specific mutation profiles may be of interest for tumour classification and prognosis.39 Indeed, primary tumours at the metastatic stage differs significantly from other tumours stages due to enrichment in mutated TP53 and low proportion of RNF43 gene mutations. Just like KRAS mutations,18,22 the presence of mutated TP53 in tumours is associated with poorer prognosis,40 therefore, it is no surprise that the primary metastatic tumours from this study show a high prevalence of TP53 mutations.39 On the other hand, RNF43 (ring finger protein 43) is an E3 ubiquitin ligase that is frequently mutated in PDAC and its precursors.41 However, the impact of RNF43 mutations on PDAC biology was poorly understood until recently, when Hossein et al. described RNF43 as a novel tumour suppressor gene of PDAC.42 A recent publication from Dreyer et al.43 indicated that RNF43 mutations are frequently found in primary resected tumours of patients with low recurrence. They also found a low mutational burden of RNF43 in hepatic metastasis.
Regarding KRAS mutation status, there is no significant difference between the age of patients bearing wild-type KRAS and that of patients with mutated KRAS, which is in line with recent studies.44 In addition, the analysis of point mutations showed no statistical enrichment of any other mutation in KRAS wild-type tumours. However, the only BRAF V600E mutation was found in a KRAS wild-type tumour.
The differences between primary tumours at a metastatic stage and localised tumours are also highlighted by the transcriptomic analysis that we performed on the EUS-FNAB-W materials. This analysis was only made possible by accessing these clinically highly different tumours using a homogeneous sampling and profiling process. The deconvolution of transcriptome profiles presents a challenge that has been successfully tackled, as previously demonstrated in a supervised way, to quantify intra-sample immune populations.45 This deconvolution allowed us to map the many tumour components, to compare the intra-tumour diversity of tumour at different stages46 while minimising the impact of EUS-FNAB-W-related contaminants.
We found that primary tumours with metastases, which do not differ significantly in size from locally advanced tumours (see Table 1), are enriched in a basal-like profile that is significantly more pronounced than the classical profile compared to other primary tumours from different stages with the same profile. These results are consistent with the poor prognosis of these tumours.3,10,11,29,37 In addition, these findings might be helpful to make predictions to help anticipate early tumour dissemination, regardless of tumour size. For this, it would be interesting to derive a signature from the 2001 genes that are significantly and differentially expressed in these tumours to detect a possible “metastatic” profile early in the tumoural progression. This may have a significant impact on the management of patients with resectable tumours, as it may help avoiding unnecessary resections, or advocate for a more aggressive neoadjuvant approach if the decision to proceed with surgery is still made. Noteworthy is that the so-called “proliferating tumour components” that we described in our study may be related and considered as the tumour component without typical differentiation markers, and in that aspect may relate to the intermediary phenotype recently described33,34 in studies that were performed on tissue specimens.
Our study also highlights the potential therapeutic value of the molecular analysis of the primary tumour, regardless of the disease stage alone. The DNA mutation status of the HRD genes is positively associated with the overall survival of patients who have received first-line platinum-based chemotherapy (mainly FOLFIRINOX and FOLFOX protocols),2,3 the more representative being BRCA1, BRCA2 and PALB2. It is widely accepted that patients with a germline mutation of the BRAC1 and BRCA2 genes within tumours will be more sensitive to platinum and even to PARP inhibitors in second-line treatment.2, 3, 4, 5,12 Our study contributes the growing body of molecular evidence showing that somatic mutations in such genes, but also within in other genes contributing to the HRD status that we describe in this work, also have prognostic value.
We also report here that the wild type status of the TP53 gene significantly correlates with better overall survival of patients who have received radio-chemotherapy, even if they may have had other second or even third-line treatments. Even if the place of radio-chemotherapy is still debated in the therapeutic arsenal used in treating PDAC, such molecular approaches based on P53 mutation status analysis may help clinicians select patients who are likely to have a better response.47 Hence, in the current era of theragnostic approaches, our strategy allows to conduct a pre-therapeutic search within the primary tumour for gene mutations that can be actionable, such as involving KRAS G12C, KRAS G12D, pan RAS, SOS or NTRK inhibitors.2,7, 8, 9,15 Beside DNA analysis, we found that transcriptomic characterisation of the GemPred profile in biopsy material is feasible and may have clinical applications, as we confirmed the prognostic value of response to gemcitabine not only in the material of resected tumours, but also within locally advanced and metastatic primary tumours. Taking the best of both worlds, RNA analysis coupled with DNA analysis could therefore be combined to drive therapeutic decision not only on the choice of first-line treatment (chemotherapy or radio-chemotherapy) but also depending on the enrichment of the GemPred signature and/or on the mutational profile of the tumour DNA.30,31,48
In summary, in this largest prospective, multicentre and clinically compliant study conducted to date, we confirm the feasibility of combined DNA and RNA molecular analyses using samples obtained through endoscopic ultrasound. We were able to comparatively characterise the primary tumours of metastatic, locally advanced, and resectable pancreatic cancers. We found that primary tumours in metastatic forms exhibit a transcriptomic profile associated with a poorer prognosis compared to other primary tumours, indicating that their molecular portrait could be crucial in the spread of cancer cells beyond the pancreas. Analysing both DNA and RNA levels helps to predict therapeutic response, impacting the patient survival outcomes for platinum-based or gemcitabine-based chemotherapy, as well as for radio-chemotherapy. This molecular approach would also likely benefit the field of targeted treatments, paving the way for predictive medicine for patients with pancreatic cancer.
Contributors
BL, NR, BB designed the study, conceived the study, conducted experimental work, acquired and analysed data, wrote the first manuscript draft and the revised manuscript; TJ, AM, CP, GS designed the study, analysed data, wrote the first manuscript draft and revised manuscript; PC, CJ, SJ, DN analysed data and revised manuscript; PL, NB, BL, BB, CC contributed to patient recruitment, the collection of biomaterials and clinical data, and revised the manuscript. All authors reviewed, edited and approved the final version of the manuscript. The members of the BACAP consortium have included and followed the included patients in their respective centres and some of them have performed EUS-FNAB. Remy Nicolle, Cindy Canivet, Camille Pignolet, Sophie Gourgou, Louis Buscail, and Barbara Bournet have accessed and verified the underlying data.
Data sharing statement
Both clinical and molecular data are available on Gene Expression Omnibus: GSE253260.
Declaration of interests
Nelson Dusetti is co-owner and inventor for the patent (PCT/EP2022/065222.462) dedicated to predictive medicine for pancreatic cancer. None of the other authors has competing interest in relation to the present work.
Investigators implicated in the BACAP consortia received a financial compensation for patient inclusion and data collection from Toulouse Hospital as the sponsor of BACAP cohort funded by a grant from INCa.
Acknowledgements
Writing assistance was provided by “The Language Room” company, Lyon, France. Funding was provided by Institut National Du Cancer (BCB INCa_7294) France, CHU of Toulouse France, Inserm and Ligue Nationale Contre le Cancer (CIT program), France.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105373.
Contributor Information
Louis Buscail, Email: buscail.l@chu-toulouse.fr.
the BACAP Consortium:
Barbara Bournet, Cindy Canivet, Louis Buscail, Nicolas Carrère, Fabrice Muscari, Bertrand Suc, Rosine Guimbaud, Corinne Couteau, Marion Deslandres, Pascale Rivera, Emily Alouany, Nadim Fares, Karl Barange, Janick Selves, Anne Gomez-Brouchet, Adrian Culetto, Guillaume Le Cosquer, Marion Jaffrelot, Bertrand Napoléon, Bertrand Pujol, Fabien Fumex, Jérôme Desrame, Christine Lefort, Vincent Lepilliez, Rodica Gincul, Pascal Artru, Léa Clavel, Anne-Isabelle Lemaistre, Laurent Palazzo, Jérôme Cros, Sarah Tubiana, Nicolas Flori, Pierre Senesse, Pierre-Emmanuel Colombo, Emmanuelle Samalin-Scalzi, Fabienne Portales, Sophie Gourgou, Lise Roca, Claire Honfo Ga, Carinne Plassot, Marc Ychou, Pierre Guibert, Christelle De La Fouchardière, Mathieu Sarabi, Patrice Peyrat, Séverine Tabone-Eglinger, Caroline Renard, Guillaume Piessen, Stéphanie Truant, Alain Saudemont, Guillaume Millet, Florence Renaud, Emmanuelle Leteurtre, Patrick Gelé, Eric Assenat, Jean-Michel Fabre, François-Régis Souche, Marie Dupuy, Anne-Marie Gorce-Dupuy, Jeanne Ramos, Jean-François Seitz, Jean Hardwigsen, Emmanuelle Norguet-Monnereau, Philippe Grandval, Muriel Duluc, Dominique Figarella-Branger, Véronique Vendrely, Clément Subtil, Eric Terrebonne, Jean-Frédéric Blanc, Jean-Philippe Merlio, Dominique Farges-Bancel, Jean-Marc Gornet, Daniela Geromin, Geoffroy Vanbiervliet, Anne-Claire Frin, Delphine Ouvrier, Marie-Christine Saint-Paul, Philippe Berthélémy, Chelbabi Fouad, Stéphane Garcia, Nathalie Lesavre, Mohamed Gasmi, Marc Barthet, Vanessa Cottet, and Cyrille Delpierre
Appendix ASupplementary data
Supplementary Fig. S1.
Supplementary Figure 1: Survival curves of the 397 patients with pancreatic cancer included in the sequenced series of DNA and RNA on endoscopic ultrasound-guided fine needle aspiration biopsy wash of material from primary tumours. a: Progression-free survival. b: Overall survival.
References
- 1.Ferlay J., Partensky C., Bray F. More deaths from pancreatic cancer than breast cancer in the EU by 2017. Acta Oncol. 2016;55:1158–1160. doi: 10.1080/0284186X.2016.1197419. [DOI] [PubMed] [Google Scholar]
- 2.Hu Z.I., O’Reilly E.M. Therapeutic developments in pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2024;21:7–24. doi: 10.1038/s41575-023-00840-w. [DOI] [PubMed] [Google Scholar]
- 3.Neoptolemos J.P., Kleeff J., Michl P., Costello E., Greenhalf W., Palmer D.H. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15:333–348. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
- 4.Kindler H.L., Hammel P., Reni M., et al. Overall survival results from the polo trial: a phase III study of active maintenance olaparib versus placebo for germline BRCA-mutated metastatic pancreatic cancer. J Clin Oncol. 2022;40:3929–3939. doi: 10.1200/JCO.21.01604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Reilly E.M., Lee J.W., Zalupski M., et al. Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J Clin Oncol. 2020;38:1378–1388. doi: 10.1200/JCO.19.02931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philip P.A., Azar I., Xiu J., et al. Molecular characterization of KRAS wild-type tumours in patients with pancreatic adenocarcinoma. Clin Cancer Res. 2022;28:2704–2714. doi: 10.1158/1078-0432.CCR-21-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strickler J.H., Satake H., George T.J., et al. Sotorasib in KRAS p.G12C-mutated advanced pancreatic cancer. N Engl J Med. 2023;388:33–43. doi: 10.1056/NEJMoa2208470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D., Herdeis L., Rudolph D., et al. Pan-KRAS inhibitor disables oncogenic signalling and tumour growth. Nature. 2023;619:160–166. doi: 10.1038/s41586-023-06123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drilon A., Laetsch T.W., Kummar S., et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collisson E.A., Sadanandam A., Olson P., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moffitt R.A., Marayati R., Flate E.L., et al. Virtual microdissection identifies distinct tumour- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waddell N., Pajic M., Patch A.M., et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey P., Chang D.K., Nones K., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 14.Pishvaian M.J., Blais E.M., Brody J.R., et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the Know Your Tumour registry trial. Lancet Oncol. 2020;21:508–518. doi: 10.1016/S1470-2045(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S., Zheng Y., Yang F., et al. The molecular biology of pancreatic adenocarcinoma: translational challenges and clinical perspectives. Signal Transduct Target Ther. 2021;6:249. doi: 10.1038/s41392-021-00659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalimuthu S N., Wilson G.W., Grant R.C., et al. Morphological classification of pancreatic ductal adenocarcinoma that predicts molecular subtypes and correlates with clinical outcome. Gut. 2020;69:317–328. doi: 10.1136/gutjnl-2019-318217. [DOI] [PubMed] [Google Scholar]
- 17.Nicolle R., Blum Y., Duconseil P., et al. Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EBioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buscail L., Bournet B., Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17:153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 19.Fuccio L., Hassan C., Laterza L., et al. The role of K-ras gene mutation analysis in EUS-guided FNA cytology specimens for the differential diagnosis of pancreatic solid masses: a meta-analysis of prospective studies. Gastrointest Endosc. 2013;78:596–608. doi: 10.1016/j.gie.2013.04.162. [DOI] [PubMed] [Google Scholar]
- 20.Bournet B., Souque A., Senesse P., et al. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with KRAS mutation assay to distinguish pancreatic cancer from pseudotumoural chronic pancreatitis. Endoscopy. 2009;41:552–557. doi: 10.1055/s-0029-1214717. [DOI] [PubMed] [Google Scholar]
- 21.Bournet B., Selves J., Grand D., et al. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with a KRAS mutation assay using allelic discrimination improves the diagnosis of pancreatic cancer. J Clin Gastroenterol. 2015;49:50–56. doi: 10.1097/MCG.0000000000000053. [DOI] [PubMed] [Google Scholar]
- 22.Bournet B., Muscari F., Buscail C., et al. KRAS G12D mutation subtype is A prognostic factor for advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol. 2016;7 doi: 10.1038/ctg.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bournet B., Pointreau A., Souque A., et al. Gene expression signature of advanced pancreatic ductal adenocarcinoma using low density array on endoscopic ultrasound-guided fine needle aspiration samples. Pancreatology. 2012;12:27–34. doi: 10.1016/j.pan.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Canivet C., Gourgou-Bourgade S., Napoléon B., et al. A prospective clinical and biological database for pancreatic adenocarcinoma: the BACAP cohort. BMC Cancer. 2018;18:986. doi: 10.1186/s12885-018-4906-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frere C., Bournet B., Gourgou S., et al. Incidence of venous thromboembolism in patients with newly diagnosed pancreatic cancer and factors associated with outcomes. Gastroenterology. 2020;158:1346–1358.e4. doi: 10.1053/j.gastro.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Neuzillet C., Nicolle R., Raffenne J., et al. Periostin- and podoplanin-positive cancer-associated fibroblast subtypes cooperate to shape the inflamed tumour microenvironment in aggressive pancreatic adenocarcinoma. J Pathol. 2022;258:408–425. doi: 10.1002/path.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng J., Sun B.F., Chen C.Y., et al. Single-cell RNA-seq highlights intra-tumoural heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Cancer Genome Atlas Research Network Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puleo F., Nicolle R., Blum Y., et al. Stratification of pancreatic ductal adenocarcinomas based on tumour and microenvironment features. Gastroenterology. 2018;155:1999–2013.e3. doi: 10.1053/j.gastro.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 30.Nicolle R., Gayet O., Duconseil P., et al. A transcriptomic signature to predict adjuvant gemcitabine sensitivity in pancreatic adenocarcinoma. Ann Oncol. 2021;32:250–260. doi: 10.1016/j.annonc.2020.10.601. [DOI] [PubMed] [Google Scholar]
- 31.Nicolle R., Gayet O., Bigonnet M., et al. Relevance of biopsy-derived pancreatic organoids in the development of efficient transcriptomic signatures to predict adjuvant chemosensitivity in pancreatic cancer. Transl Oncol. 2022;16 doi: 10.1016/j.tranon.2021.101315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moncada R., Barkley D., Wagner F., et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 33.Williams H.L., Dias Costa A., Zhang J., et al. Spatially resolved single-cell assessment of pancreatic cancer expression subtypes reveals Co-expressor phenotypes and extensive intratumoral heterogeneity. Cancer Res. 2023;83:441–455. doi: 10.1158/0008-5472.CAN-22-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saillard C., Delecourt F., Schmauch B., et al. Pacpaint: a histology-based deep learning model uncovers the extensive intratumor molecular heterogeneity of pancreatic adenocarcinoma. Nat Commun. 2023;14:3459. doi: 10.1038/s41467-023-39026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chikhladze S., Lederer A.K., Kousoulas L., et al. Adjuvant chemotherapy after surgery for pancreatic ductal adenocarcinoma: retrospective real-life data. World J Surg Oncol. 2019;17:185. doi: 10.1186/s12957-019-1732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auclin E., Marthey L., Abdallah R., et al. Role of FOLFIRINOX and chemoradiotherapy in locally advanced and borderline resectable pancreatic adenocarcinoma: update of the AGEO cohort. Br J Cancer. 2021;124:1941–1948. doi: 10.1038/s41416-021-01341-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marschner N., Hegewisch-Becker S., Reiser M., et al. FOLFIRINOX or gemcitabine/nab-paclitaxel in advanced pancreatic adenocarcinoma: a novel validated prognostic score to facilitate treatment decision-making in real- world. Int J Cancer. 2023;152:458–469. doi: 10.1002/ijc.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hilmi M., Armenoult L., Ayadi M., Nicolle R. Whole-transcriptome profiling on small FFPE samples: which sequencing kit should Be used? Curr Issues Mol Biol. 2022;44:2186–2193. doi: 10.3390/cimb44050148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du J., Qiu X., Lu C., et al. Molecular landscape and prognostic biomarker analysis of advanced pancreatic cancer and predictors of treatment efficacy of AG chemotherapy. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.844527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., Chen B., Chen J., Sun S. A novel tp53-associated nomogram to predict the overall survival in patients with pancreatic cancer. BMC Cancer. 2021;21:335. doi: 10.1186/s12885-021-08066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinhart Z., Pavlovic Z., Chandrashekhar M., et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumours. Nat Med. 2017;23:60–68. doi: 10.1038/nm.4219. [DOI] [PubMed] [Google Scholar]
- 42.Hosein A.N., Dangol G., Okumura T., et al. Loss of Rnf43 accelerates kras-mediated neoplasia and remodels the tumour immune microenvironment in pancreatic adenocarcinoma. Gastroenterology. 2022;162:1303–1318.e18. doi: 10.1053/j.gastro.2021.12.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dreyer S.B., Upstill-Goddard R., Legrini A., et al. Genomic and molecular analyses identify molecular subtypes of pancreatic cancer recurrence. Gastroenterology. 2022;162:320–324.e4. doi: 10.1053/j.gastro.2021.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Ammar I., Rousseau A., Nicolle R., et al. Precision medicine for KRAS wild-type pancreatic adenocarcinomas. Eur J Cancer. 2024;197 doi: 10.1016/j.ejca.2023.113497. [DOI] [PubMed] [Google Scholar]
- 45.Sturm G., Finotello F., Petitprez F., et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–i445. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayashi A., Fan J., Chen R., et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat Cancer. 2020;1:59–74. doi: 10.1038/s43018-019-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong X., Yu D., Wang Z., Li S. Relationship between p53 status and the bioeffect of ionizing radiation. Oncol Lett. 2021;22:661. doi: 10.3892/ol.2021.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraunhoffer N., Chanez B., Teyssedou C., et al. A transcriptomic-based tool to predict gemcitabine sensitivity in advanced pancreatic adenocarcinoma. Gastroenterology. 2023;164:476–480.e4. doi: 10.1053/j.gastro.2022.11.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.