Abstract
Objectives
We report prolonged mpox (>14 weeks) in a patient with HIV complicated by deep tissue MPXV infection despite two courses of tecovirimat treatment.
Methods
MPXV-DNA levels in lesional swabs, blood and tissue were quantified by qPCR. Anti-MPXV antibodies were analyzed by IF and VNT. Infectivity was assessed by virus isolation. Sequencing was performed to assess for tecovirimat resistance mutations and quantitative results were obtained by digital SNP PCR (A288P).
Results
The patient's clinical condition improved significantly during both tecovirimat treatment courses (each 14 days), yet we observed persistent MPXV-DNA in lesions accompanied by viremia (mean 1.4 × 104 copies/ml) for >14 weeks. A deep tissue infection driven by MPXV complicated the clinical course (week 9). Presence of infectious virus within the tissue and high infectious titers (>106 PFU/ml) were observed. The VP37 protein sequence revealed A288P substitutions. Digital PCR showed 1 % and less abundance (A288P) during first treatment course (blood and swabs), with increasing proportion during second course (week 8–9; 28 % in blood and swabs), however the mutation was absent in samples from deep tissue infection and MPXV isolates (week 9) indicating compartimentalization. Morphological fully enveloped MPXV partices visualized by TEM in necrotic areas suggesting tecovirimat treatment failure in the deep tissue compartment.
Conclusion
Our data provide evidence that Tecovirimat treatment selects for compartimentalized viral mutations (A288P). While the patient clinically benefited from repeated tecovirimat course, emergence of viral muations and deep tissue infection emphasizes the challenge and importance of infectious disease monitoring in mpox patient management.
Keywords: Mpox, MPXV, HIV, Tecovirimat
1. Introduction
The 2022/23 mpox (formely known as monkeypox) outbreak was unprecedented in its scale and spread, and although case numbers have fallen sharply, increased incidence continues to be reported [[1], [2], [3]]. In addition, an ongoing clade I monkeypoxvirus (MPXV) related outbreak in africa (Democratic Republic of the Congo) is a cause for concern [4].
Immunocompromised patients are at risk of severe and prolonged mpox with increased mortality [[5], [6], [7]]. It is therefore being discussed whether mpox should be considered an opportunistic, AIDS-defining infection [8,9]. Emergency treatment with tecovirimat is approved for these patients (under exceptional circumstances) and was widely used during the outbreak [7,10,11]. Tecovirimat treatment failure has been described with the emergence of both known and new VP37 resistance mutations [12,13]. We report here severe and persistent mpox in an HIV-positive patient with low CD4 count, who failed MPXV clearance despite two tecovirimat treatment courses. The aim of this study was to decipher factors/mechanisms that contributed to the overall treatment failure through a virological, histopathological and genetic analysis. We conclude that beside sporadical abundance of VP37 resistance mutations, additional factors such as a weak immune response and/or insufficient tecovirimat concentration at the site of deep tissue infection may have contributed to MPXV therapy escape.
2. Methods
2.1. Patient and ethics
A 31-year-old male patient living with HIV-1 (first diagnosed 3 months before initial presentation, CDC C3) was hospitalized for mpox with ubiquitous lesions. At initial consultation, the patient presented with predominantly facial lesions, fever, malaise and lymphadenopathy that reportedly developed over the last 20 days. Brief details of its clinical presentation and disease course were published previously [10,14,15]. The patient gave written informed consent to the use of the samples and to the publication of the analysis results.
2.2. Samples and molecular diagnostic
Lesional swabs and EDTA blood samples were obtained during the clinical routine. MPXV DNA was detected and quantified as described previously [16,17]. Native tissue samples were processed as described [18] and viral DNA was extracted using an automated system (MagNA Pure, Roche, USA) prior to qPCR (Light Cycler 480 II, Roche, USA).
2.3. MPXV culture, virus titration and molecular detection
MPXV isolation was performed on Vero 76 cells (ATCC CRL1587) [16]. For virus titration from homogenized original samples, adsorption of serial virus dilutions was performed on Vero 76 cells seeded to confluency. After incubation for 1h at 37 °C the supernatant was replaced by methylcellulose-overlay medium (methylcellulose mixed 1:3 with DMEM). Cells were incubated for 4 days at 37 °C prior to fixation in 4 % Formaldehyde and plaques were vizualized by staining with crystal violet solution.
MPXV DNA loads in supernatants were quantified by qPCR as described above [17]. VP37 was amplified as described (primers 5′-CCTGTTTAACATGATGGCGTT-3′ and 5′-AGGTTGTGATGTCGACTTTGA-3′ [19]) and sequenced.
2.4. SNP analysis and digital PCR
The presence of the A288P mutation in the patient samples or a possible selection of this mutation was quantified by digital PCR using a newly designed SNP specific LNA probes as described [20]. Primers mpxv-op57A-F 5′-GGAGTTAAGATCAGACTTCTAGTT-3′and mpxv-op57A-R 5′-ACACACAACGCATCTAGACTT-3′ were used in the PCR reaction and combined with a probes mpxv-op57A-WT (FAM- ATT CT(+A) T(+G)(+G) (+C)A(+A) CCG -BHQ1) and mpxv-op57A-A288P (HEX- ATT CT(+A) T(+G)(+C) (+C)A(+A) CCG -BHQ1), the latter being specific for the A288P SNP (G→C). Digital PCR was performed and analyzed using the performed QIAcuity (Qiagen) platform.
2.5. Immunohistochemistry
Paraffin embedded patients tissue from surgical procedure was cut at 2 μm and used for eosin hematoxylin (H&E) staining according to standard procedures. For the detection of MPXV protein in the tissue, immunohistochemical staining was performed. For this, the Ventana Benchmark XT machine (Ventana, Tuscon, Arizona, USA) was used including automated inactivation of endogenous peroxidases (PBS/3 % hydrogen peroxide) and antibody-specific antigen retrieval. After blocking, sections were incubated with the primary rabbit polyclonal antibody against B5R VACV (homolog of MPXV B6R) #NR-629 (provided by BEI Resources). For detection of specific primary antibody binding, the Ultra View Universal 3,3′-Diaminobenzidine (DAB) Detection Kit (#760-500, Ventana, Roche) was used, which contains secondary antibodies, DAB stain, and counterstaining reagent for detection of nuclei. Stained sections were evaluated by experienced pathologists and representative images were taken with a Leica DMD108 digital microscope.
2.6. Indirect immunofluorescence
For indirect immunofluorescence test (IIFT), MPXV-infected Vero cells were spotted on glass slides as described [21]. Briefly, serial dilutions of patient sera were incubated on fixed and dried cells for 1h, and after washing FITC-labeled secondary antibody (anti-human IgG) was added. IIF was performed under BSL-3 conditions.
2.7. Transmission electron microscopy
Patients tissue was fixed in a mixture of 4 % paraformaldehyde and 1 % glutaraldehyde in 0.1 M PBS, pH 7.2 overnight. After washing in PBS for three times, the tissue was rinsed three times in 0.1 M sodium cacodylate buffer (pH 7.2–7.4), and osmicated using 1 % osmium tetroxide in cacodylate buffer. Then, the tissue was dehydrated in ascending ethyl alcohol concentration steps. After two rinses in propylene oxide, infiltration of the embedding medium was performed by immersing the samples in a 1:1 mixture of propylene oxide and Epon and finally in neat Epon. Polymerization was carried out at 60 °C. For light microscopy, semithin sections (0.5 μm) were prepared by staining for 1 minute with 1 % Toluidine blue and mounting on glass slides. Ultrathin sections (60 nm) were examined in a JEM2100Plus Transmission Electron Microscope (Jeol). Pictures were acquired with the Xarosa CMOS camera (EMSIS, Germany).
3. Results
3.1. Clinical course and MPXV kinetics
At initial clinical presentation, low CD4 count (30/μl) and high HIV-1 RNA loads (1.29 × 106 copies/ml) were observed despite 3 months cART (combination of bictegravir/entricitabine/tenofovir (Bictavy)), suggesting non-adherence. No details were available on the diagnosis and course of the HIV infection.
High MPXV DNA loads were detected in facial, oral, genital, and plantar lesions (≤9.25 × 107 copies/ml) and the patient proved viremic (≤1.03 × 105 copies/ml) (Fig. 1A). Given the patient's poor condition with >30 severely painful, itchy and necrotizing lesions, a 14day treatment course with tecovirimat was started (2 × 600 mg daily), with resumption of cART and initiation of trimethoprim/sulfamethoxazole prophylaxis. Tecovirimat was administered with a fatty meal to ensure optimal absorption and intake was supervised to ensure compliance.
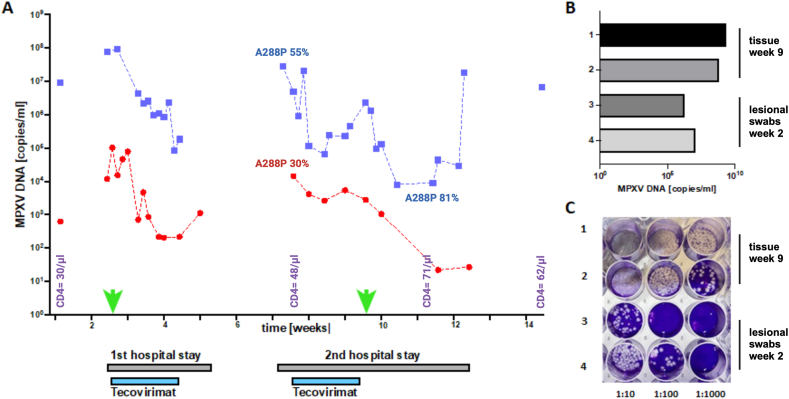
Fig. 1.
MPXV kinetics and detection of infectious virus in the patient during the clinical course indicate persistence of MPXV. (A) The kinetics of MPXV DNA levels in lesional swabs (blue squares) and blood (red circles) are shown. In week 1, one swab and one blood sample were obtained on an outpatient ward, values obtained during inpatient stays (weeks 2–4, weeks 7–9) are connected by dashed lines. The time period of tecovirimat therapy is highlighted in grey. For virus isolation experiments, 2 swab samples (week 2) and 2 of the tissue samples (week 9) were selected (green arrows). The CD4 count over time is shown in purple, A288P prevalence as determinded by digital PCR is given for blood and swab samples (B) MPXV DNA levels as quantified by qPCR in the 4 samples (1, 2 = tissue samples, 3,4 = swabs). To assess for presence of infectious virus in the samples, plaque assay using serial sample dilutions of the original tissue samples for infection was performed (appendix). Clearly visible plaques prove the infectivity of the samples (C).
The lesions were treated topically (zinc-shake mixture), and analgesics (opiates and/or metamizol) and antihistamines (for pruritus, clemastine) were administered as required. The treatment resulted in significant clinical improvement, with rapid decrease of HIV RNA load to <300 copies/ml within 2 weeks. The patient could be discharged at week 4. During week 7 he was readmitted due to clinical deterioration with newly emerged painful lesions. High MPXV DNA levels rebounded and were again detected in lesions (≤2.85 × 107 copies/ml) and the patient was viremic (≤1.44 × 104 copies/ml) (week 7, Fig. 1A). A second 14d treatment course with Tecovirimat was carried out resulting in clinical improvement and decrease of lesional swab and blood MPXV DNA levels. Yet, shortly after completion of this treatment (week 9, Fig. 1A), the patient complained of a very painful swelling in the area of the lower leg, which appeared abscess-like in magnetic resonance imaging (MRI) [14] and required surgical intervention. After debridement and drainage, the patient improved rapidly and could be discharged at week 12. At re-presentation in our outpatient clinic (week 14), the patient still had lesions with MPXV DNA detectable (≤6.75 × 106 copies/ml), yet appeared clinically stable with sustained low HIV RNA load (5.9 × 101 copies/ml) and low CD4 counts (62/μl). After week 14, the patient was lost to follow-up.
3.2. Humoral immune response
While we observed a clear IgG seroconversion with anti-MPXV IgG titers of 1:320 (week 2) raised to 1: 5120 in week 14, (IIFT), no neutralizing activity was detected in virus neutralization assay throughout the entire observation period (appendix figure S1) in line with failure of virus clearance.
3.3. MPXV tissue infection
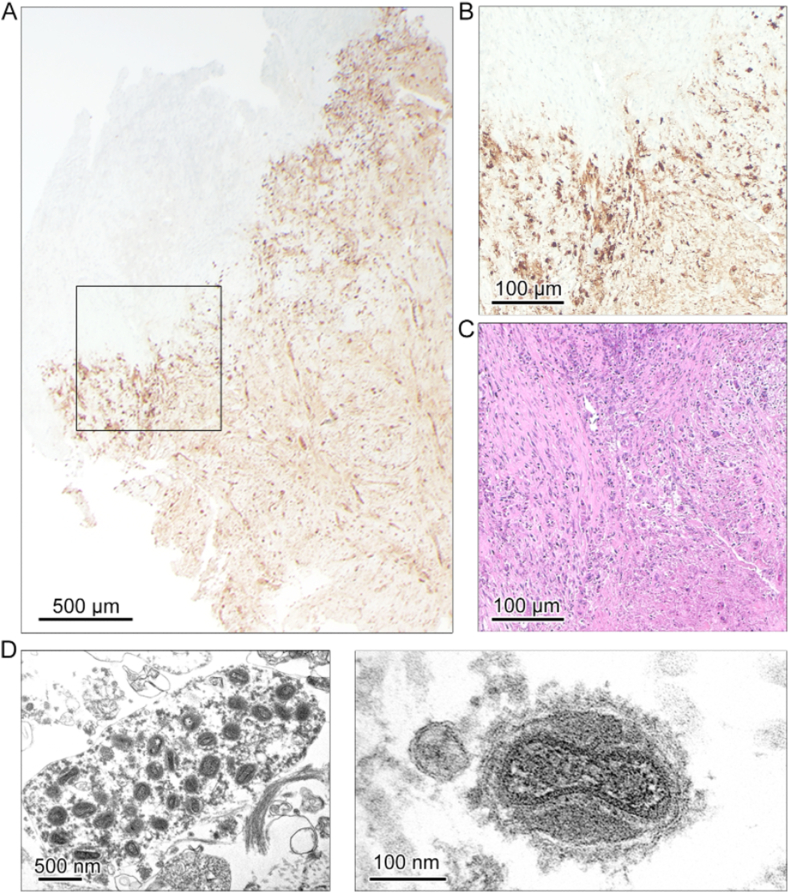
In the surgically removed tissue samples (n = 2) 1.2 × 109 and 1.8 × 109 copies/ml of MPXV DNA were detected (Fig. 1B). Virus isolation on Vero cells (ATCC CRL1587) [16] and MPXV titration from the original samples proved presence of infectious virus with titers of >1 × 106 PFU/ml (Fig. 1C). Routine microbiological screening comprising aerobic and anaerobic bacterial cultures, pan-bacterial (16-S-rRNA) and pan-fungal (18-S-rRNA) PCR were sterile and detected no bacterial or fungal pathogen, thus the deep tissue infection represented most likely a purely MPXV-driven process. Histopathological analysis revealed central necrosis with a high rate of tissue destruction and presence of MPXV-proteins in the majority of cells (Fig. 2A–C). The abundance of MPXV particles in the necrotic tissue lesion core was confirmed by TEM including the detection of enveloped particles suggesting tecovirimat treatment failure (Fig. 2D).
Fig. 2.
Widespread distribution of virus-positive cells in the tissue lesion of the patient. (A) Representative overview image of a tissue section from the abscess region stained for MPXV B6R showing widespread distribution of the MPXV B6R protein in 25× magnification. (B) Close-up of boxed region in A at 100× magnification (C) a consecutive section stained for H&E displaying the same region as in B showing severe necrosis and pronounced tissue destruction in virus-protein positive areas. (D) Transmission electron microscopy of the necrotic lesion reveils intracellular mature virus particles and single fully enveloped virus particles.
3.4. Tecovirimat resistance mutations
To screen for potential Tecovirimat resistance mutations, Sanger sequencing of VP37 encoding gene [19] was performed. The only SNP besides the E353K substitution (indicative for the outbreak) was a compartmentalized A288P substitution (GCA- > CCA) [22], (Table 1). Digital PCR with SNP and reference sequence specific probes confirmed A288P compartmentalisation and heterogeneity over time (Table 1).
Table 1.
Abundance of SNP A288P quantified by digital PCR (dPCR) and detection by SANGER-Sequencing.
| material | week | Target | A288P fraction | Partitions (valid) | Partitions positive | Partitions negative | A288P (SANGER) |
|---|---|---|---|---|---|---|---|
| Lesional swab | 2 | A288P | 0.00 % | 25405 | 0 | 25405 | Not detected |
| WT | – | 25405 | 25405 | 0 | |||
| Lesional swab | 4 | A288P | 0.92 % | 25498 | 10 | 25488 | Not detected |
| WT | – | 25494 | 1049 | 24445 | |||
| Lesional swab | 7 | A288P | 0.12 % | 25496 | 212 | 25284 | Not detected |
| WT | – | 25496 | 25474 | 22 | |||
| blood | 8 | A288P | 28.78 % | 25391 | 19 | 25372 | Detected |
| WT | – | 25391 | 47 | 25344 | |||
| Lesional swab | 8 | A288P | 54.60 % | 25470 | 490 | 24980 | Detected |
| WT | – | 25468 | 408 | 25060 | |||
| Lesional swab | 9 | A288P | 0.00 % | 25349 | 3 | 25346 | Not detected |
| WT | – | 25349 | 24482 | 867 | |||
| Tissue biopsie, sample 1 | 9 | A288P | 0.00%o | 25481 | 0 | 25481 | Not detected |
| WT | – | 25481 | 25481 | 0 | |||
| Tissue biopsie, sample 2 | 9 | A288P | 0.00 % | 25466 | 0 | 25466 | Not detected |
| WT | – | 25466 | 25466 | 0 | |||
| Lesional swab | 11 | A288P | 80.90 % | 25479 | 249 | 25230 | Sequencing failed |
| WT | 25479 | 59 | 25420 | ||||
| MPXV tissue derived isolate 1 | 9 | A288P | 0.00 % | 25418 | 0 | 25418 | Not detected |
| WT | – | 21637 | 18731 | 2906 | |||
| MPXV tissue derived isolate 2 | 9 | A288P | 0.02 % | 25497 | 3 | 25494 | Not detected |
| WT | – | 25497 | 13339 | 12158 | |||
| MPXV tissue derived isolate 3 | 9 | A288P | 0.00 % | 25380 | 1 | 25379 | Not detected |
| WT | – | 25380 | 14803 | 10577 | |||
| MPXV tissue derived isolate 4 | 9 | A288P | 0.01 % | 25466 | 2 | 25464 | Not detected |
| WT | – | 25309 | 18910 | 6399 | |||
| MPXV tissue derived isolate 5 | 9 | A288P | 0.01 % | 25340 | 2 | 25338 | nd |
| WT | – | 25340 | 19580 | 5760 | |||
| MPXV tissue derived isolate 6 | 9 | A288P | 0.01 % | 25340 | 2 | 25338 | nd |
| WT | – | 25340 | 19580 | 5760 | |||
| MPXV tissue derived isolate 7 | 9 | A288P | 0.01 % | 25488 | 1 | 25487 | nd |
| WT | – | 25488 | 11192 | 14296 | |||
| MPXV tissue derived isolate 8 | 9 | A288P | 0.00 % | 25471 | 0 | 25471 | nd |
| WT | – | 25471 | 22452 | 3019 | |||
| MPXV tissue derived isolate 9 | 9 | A288P | 0.00 % | 25472 | 0 | 25472 | nd |
| WT | – | 25472 | 18573 | 6899 | |||
| MPXV tissue derived isolate 10 | 9 | A288P | 0.00 % | 25475 | 0 | 25475 | nd |
| WT | – | 25475 | 18383 | 7092 | |||
| Lesional swab (patient B) | na | A288P | 0.00 % | 25459 | 0 | 25459 | Not detected |
| WT | – | 25082 | 8293 | 16789 | |||
| Negative control | na | A288P | 0.00 % | 25451 | 0 | 0 | nd |
| WT | – | 25451 | 0 | 0 |
Table 1: By applying dPCR, the relative proportion of the VP37 A288P SNP compared to ancestral epidemic MPXV sequence was assessed in clinical samples and 10 of the MPXV isolates (MPXV tissue derived isolates 1–10, all rescued from tissue obtained during chirurgical intervention in week 9), therein those used for the in vitro tests. A newly designed SNP-specific and a reference sequence-specific probe were used and dPCR was performed on the QIAcuity platform. As controls, a different patient (patient B) and a negative sample are included.
The table lists the type of material, in the case of patient samples the week of the observation period, gives the share of the A288P SNP compared to the reference sequence (percentage), indicates the overall number of positive partitions per sample (e.g. partitions with MPXV DNA detected by either probe) and gives the share of target specific partitions. Additionally, A288P detection in SANGER-Sequencing is indicated (A288P SANGER).
Nd = not done.
We firstly detected A288P in a lesional swab as a minority variant (0.92 %) at the beginning of the first tecovirimat treatment course, but its relative proportion remained low through week 7 (0.12 %). Proportion of the SNP increased during the course (28.78 % in the blood and 54.6 % in a swab at week 8), but disappeared after completion of the 2nd tecovirimat treatment course (A288P undetectable in swabs and tissue samples obtained in week 9). Although in a later sample (week 11) A288P abundance appeared to be high (81 %), the significance of this result is unclear because overall MPXV DNA levels in that sample were low (Table 1). Notably, none of the MPXV isolates (neither from the deep tissue infection nor from lesional swabs) harbored the A288P SNP (abundance ≤0.02 % at first cell culture passage) (Table 1).
4. Discussion
We describe a severe and unusual mpox course in an HIV-1-positive patient. While our patient mirrors the mpox course in HIV patients in many aspects [5,6,11,12], the salient aspect of this case is the development of a MPXV deep tissue infection despite two 14d Tecovirimat treatment courses. The disease course was suggestive for VP37 resistance mutation selection, as described in preovious reports [5,9,10] [13,22], yet by combination of SANGER-Sequencing and digital PCR we only inconstantly detected a single SNP (A288P) in different sample types over the course of the disease indicating compartimentalization of the mutation.
However, A288P was present in blood and superficial lesion samples at an early stage. It is conceivable that although that SNP was not dominant in most of the samples, the resistant MPXV subpopulation led to resurgence of infection after completion of tecovirimat treatment. Under this assumption, it is noteworthy that the patient benefited and improved significantly during both treatment courses. This might indicate that abundance of a resistant MPXV subpopulation does not necessarily preclude tecovirimat treatment.
Notably, A288P was not detected in tissue samples, yet fully enveloped virus particles were vizualized in the necrotic lesion core which might point to ineffective drug concentrations and together with the patient's poor immune status may have been decisive for failure of virus clearance in this compartment.
The case presented here emphasizes the challenge and importance of infectious disease monitoring in mpox patient management and also of immune reconstitution in combination with tecovirimat treatment. Although we investigated only one patient, the in-dephts virological and immunohistochemical evaluation sheds new light in the understanding of treatment failure particularly in cases of severe or fulminant mpox.
Data and code availability statement
Data will be made available on request.
CRediT authorship contribution statement
Susanne Pfefferle: Writing – original draft, Visualization, Validation, Investigation, Formal analysis, Data curation, Conceptualization. Michaela Schweizer: Visualization, Methodology. Kristin Hartmann: Methodology, Investigation. Julia Berger: Methodology, Investigation. Dominik Nörz: Methodology, Investigation. Petra Emmerich: Validation, Methodology, Investigation. Ronald von Possel: Methodology, Investigation. Katja Giersch: Methodology, Investigation. Lisa Sophie Pflüger: Methodology, Investigation. Christian Bernreuther: Methodology, Investigation. Markus Glatzel: Supervision, Resources. Susanne Krasemann: Writing – review & editing, Methodology, Investigation, Data curation. Thomas Theo Brehm: Investigation. Julian Schulze zur Wisch: Investigation. Nicole Fischer: Writing – review & editing, Validation, Methodology, Investigation. Stefan Schmiedel: Writing – review & editing, Investigation. Martin Aepfelbacher: Writing – review & editing, Supervision, Resources. Marc Lütgehetmann: Writing – review & editing, Validation, Supervision, Resources, Investigation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Chudamani Raithore and Emanuela Szpotowicz for technical assistance. The following reagents were obtained through BEI Resources, NIAID, NIH: Polyclonal Anti-Vaccinia Virus (WR) B5R Protein, (antiserum, Rabbit), NR-629.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e38873.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Endo A., Jung S.M., Miura F. Mpox emergence in Japan: ongoing risk of establishment in Asia. Lancet. 2023;401(10392):1923–1924. doi: 10.1016/S0140-6736(23)00766-3. [DOI] [PubMed] [Google Scholar]
- 2.Zhao T., Wu Z. Prevention of a potential mpox outbreak in China. Lancet. 2023;402(10407):1038–1039. doi: 10.1016/S0140-6736(23)01793-2. [DOI] [PubMed] [Google Scholar]
- 3.Moschese D., et al., Surge of mpox cases in lombardy region, Italy, october 2023 - january 2024, Clin. Infect. Dis. (2024), online ahead of print. [DOI] [PubMed]
- 4.Kibungu E.M., et al. Clade I-associated mpox cases associated with sexual contact, the democratic republic of the Congo. Emerg. Infect. Dis. 2024;30(1):172–176. doi: 10.3201/eid3001.231164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitja O., et al. Mpox in people with advanced HIV infection: a global case series. Lancet. 2023;401(10380):939–949. doi: 10.1016/S0140-6736(23)00273-8. [DOI] [PubMed] [Google Scholar]
- 6.Riser A.P., et al. Epidemiologic and clinical features of mpox-associated deaths - United States, may 10, 2022-March 7, 2023. MMWR Morb. Mortal. Wkly. Rep. 2023;72(15):404–410. doi: 10.15585/mmwr.mm7215a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippov E., et al. Treatment failure in patient with severe mpox and Untreated HIV, Maryland, USA. Emerg. Infect. Dis. 2023;29(6) doi: 10.3201/eid2906.230059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitja O., et al. Classifying necrotising mpox as an AIDS-defining condition. Lancet. 2023;402(10414):1751–1752. doi: 10.1016/S0140-6736(23)01896-2. [DOI] [PubMed] [Google Scholar]
- 9.Pinnetti C., et al. Mpox as AIDS-defining event with a severe and protracted course: clinical, immunological, and virological implications. Lancet Infect Dis. 2024;24(2):e127–e135. doi: 10.1016/S1473-3099(23)00482-6. [DOI] [PubMed] [Google Scholar]
- 10.Hermanussen L., et al. Tecovirimat therapy for severe monkeypox infection: longitudinal assessment of viral titers and clinical response pattern-A first case-series experience. J. Med. Virol. 2023;95(1) doi: 10.1002/jmv.28181. [DOI] [PubMed] [Google Scholar]
- 11.McLean J., et al. Tecovirimat treatment of people with HIV during the 2022 mpox outbreak : a retrospective cohort study. Ann. Intern. Med. 2023;176(5):642–648. doi: 10.7326/M22-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contag C.A., et al. Treatment of mpox with suspected tecovirimat resistance in immunocompromised patient, United States, 2022. Emerg. Infect. Dis. 2023;29(12):2520–2523. doi: 10.3201/eid2912.230849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith T.G., et al. Tecovirimat resistance in mpox patients, United States, 2022-2023. Emerg. Infect. Dis. 2023;29(12):2426–2432. doi: 10.3201/eid2912.231146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brehm T.T., Hermanussen L., Schmiedel S. Orthopox simiae muscle abscess. Infection. 2023;51(3):799–800.. doi: 10.1007/s15010-022-01961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermanussen L., et al. Tecovirimat for the treatment of severe Mpox in Germany. Infection. 2023:1–6. doi: 10.1007/s15010-023-02049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norz D., et al. Evidence of surface contamination in hospital rooms occupied by patients infected with monkeypox, Germany, June 2022. Euro Surveill. 2022;27(26) doi: 10.2807/1560-7917.ES.2022.27.26.2200477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norz D., et al. Rapid adaptation of established high-throughput molecular testing infrastructure for monkeypox virus detection. Emerg. Infect. Dis. 2022;28(9):1765–1769. doi: 10.3201/eid2809.220917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wichmann D., et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann. Intern. Med. 2020;173(4):268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pires M.A., et al. In vitro susceptibility to ST-246 and Cidofovir corroborates the phylogenetic separation of Brazilian Vaccinia virus into two clades. Antivir. Res. 2018;152:36–44. doi: 10.1016/j.antiviral.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Norz D., et al. Rapid automated screening for SARS-CoV-2 B.1.617 lineage variants (Delta/Kappa) through a versatile toolset of qPCR-based SNP detection. Diagnostics. 2021;11(10) doi: 10.3390/diagnostics11101818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emmerich P., et al. Longitudinal detection of SARS-CoV-2-specific antibody responses with different serological methods. J. Med. Virol. 2021;93(10):5816–5824. doi: 10.1002/jmv.27113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrigues J.M., et al. Identification of tecovirimat resistance-associated mutations in human monkeypox virus - Los Angeles county. Antimicrob. Agents Chemother. 2023;67(7) doi: 10.1128/aac.00568-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.