Abstract
Background:
In addition to the great challenge of early diagnosis and prognosis in breast cancer (BC), the role of gene promoters in BC remains largely unexplored. This study aimed to evaluate aldo-keto reductase family 1 member B1 (AKR1B1) methylation as noninvasive biomarker for early BC diagnosis.
Methods:
A total of 200 (120 with BC, 40 with benign breast diseases, 40 healthy) Egyptian women were enrolled. AKR1B1 methylation level was determined using EpiTect Methyl II QPCR assay quantitative polymerase chain reaction.
Results:
Findings revealed that hypermethylation AKR1B1 was reported to be associated (P < .0001) with BC cases (93.2 [75.4-98.6]) compared with benign (23.9 [22.6-48.3]) or healthy (15.5 [10.6-16]) controls. It had a great diagnostic power (area under the curve [AUC] = 0.909) that was superior to cancer antigen (CA) 15-3 (AUC = 0.681) and carcinoembryonic antigen (CEA) (AUC = 0.539). Interestingly, AKR1B1 hypermethylation was reported to be significant in identifying BC early stages (AUC = 0.899) and grades (AUC = 0.903). Independent to hormonal status and HER2neu expression, AKR1B1 hypermethylation was related to some tumor severity features, including advanced stages, high histological grades, and lymph node invasion. Also, AKR1B1 high degrees of methylation were significantly correlated with the increase in CEA (r = .195; P = .027), CA-15.3 (r = .351; P = .0001) and tumor stages (r = .274; P = .014), grades (r = .253; P = .024), and lymph node invasion (r = .275; P = .014).
Conclusions:
This study revealed that aberrant AKR1B1 methylation could facilitate early BC detection from benign br0east disorders. Hypermethylated AKR1B1 was related to BC aggressiveness suggesting its potential role as diagnostic and prognostic BC biomarker.
Keywords: Breast cancer, AKR1B1 methylation, diagnosis, epigenetics, biomarker
Introduction
Worldwide, breast cancer (BC) stills a prevalent and complex health concern affecting millions of patients and one of the commonly malignant cancers affecting women.1,2 It is developed and obtained as a result of many external and internal factors. 1 It has been suggested that 20% to 30% of BCs can be associated with modifiable factors and about 5% to 10% of BCs can be associated with family history and genetic mutations. 3 Declined and unsatisfactory BC survival rates, particularly in developing countries, are mainly due to the insufficiency of early detection programs, the lack of adequate treatment and diagnosis facilities, delays related to treatment, and the consequence high percentage of women presenting with late-stage BC. 4 Regarding BC early detection, there is a great challenge due to some noted limitations of the varied available approaches, especially mammography, including low sensitivity and high false-positive rates. 5 Also, carcinoembryonic antigen (CEA) and cancer antigen (CA) 15-3 are the most relevant tumor markers in BC and both are correlated with tumor nodal involvement and tumor size.6,7 Despite that, they cannot be recommended for BC screening due to their low sensitivity at primary diagnosis and they are useful tools only in therapy monitoring and follow-up.6,7
So, there is a significant requirement for evaluating novel and reliable biomarkers to aid in BC early detection, facilitate targeted therapeutic approaches development, enable precise disease behavior prediction and enhance prognostic accuracy. 8 Accumulating evidence demonstrated that DNA methylation may play important role for BC progression and development. 9 Within a cytosine guanine (CpG) dinucleotide, DNA methylation involves the methyl group addition to carbon 5-position of cytosine. 10 This molecular event is definitive for many critical cellular mechanisms, including genomic stability and imprinting, X-chromosome inactivation, gene expression regulation, and embryonic development. 11 Aberrant DNA hypermethylation and hypomethylation patterns have been reported as critical players in tumorigenesis, promoting tumor suppressor genes silencing and oncogenes expression. 12 Thus, as a cancer-related biomarker, abnormal DNA methylation could be aid in tumor, including BC, prognosis, and early detection. 11
Aldo-keto reductase family 1 (AKR1) is one of AKR 16 family families which are divided into members and subfamilies according to their amino acid sequence identity. 13 Subfamily B of AKR1 is comprises 3 members: AKR1B1, AKR1B10, and AKR1B15. 14 AKR1B1 gene (18 kb long) is located on chromosome 7q33 and its coding transcript involves 10 exons. 15 Biologically, AKR1B1 catalyzes many aldehydes to alcohols consuming NADPH. However, its ability to reduce different substrates renders difficult to completely ensure its biological role. 16 In several tumors such as BC, rectal, cervical, and ovarian cancers, AKR1B1 overexpression was indicated using immunoblotting. 17 AKR1B1 upregulation was reported in BC cell lines basal subtype and in triple-negative BC (TNBC). 18 Although reports assessing AKR1B1 expression could not clearly highlight its effect on BC, great evidence demonstrated that AKR1B1 could play an important role in BC tumorigenesis. 19
Till now, most studies have evaluated small number of genes in BC. 20 Furthermore, few studies have evaluated regarding AKR1B1 promoter gene methylation among other genes in BC,21,22 regardless BC clinicopathological data, including tumor subtypes, stage, grade, lymph node invasion, estrogen receptor, progesterone receptor, and HER2 protein status. Thus, this research aimed to evaluate the aberrant AKR1B1 methylation patterns among BC Egyptian cases in comparison with other noncancer participants (with benign breast diseases and healthy controls). Also, we aimed to evaluate its diagnostic ability compared with established tumor markers (CEA and CA15.3) and to evaluate the association of AKR1B1 methylation patterns and the disease clinicopathological features.
Materials and Methods
Patients
A retrospective study was performed in a total of 120 newly diagnosed Egyptian patients with primary BC. Moreover, as controls, 40 age-matched female cases with breast benign disorders and 40 age-matched healthy females were included. Patients were radiologically, clinically, and if available, pathologically diagnosed for BC at Mansoura University Oncology Center, Egypt from January 2022 to January 2023. Patients with any other tumors and/or received any type of therapy were excluded. Medical reports of cases were reviewed and clinicopathological data, including age, TNM stage, 23 her2neu expression, progesterone and estrogen receptors status, and histological grade, were obtained. The study protocol was ethically approved by the Institutional Research Board of Mansoura University.
Extraction of DNA
Before any interventions, venous blood (5 mL) was collected from all participants, into sterile plain tubes. Serum was separated (by centrifugation [4000 rpm, 10 minutes]) and stored at −20°C until its use for quantifying AKR1B1 promoter gene methylation and detecting tumor markers. Commercial DNA Min kit (Qiagen, Germany, Cat 51104) was used for DNA extraction based on spin column and according to manufacturer’s recommendations. The concentration and purity of the produced DNA were detected using Nano-drop spectrophotometer (Quawell, Scribner, USA) then stored (−20°C) till further analysis.
Methylation pattern
AKR1B1 methylation profile was assessed using EpiTect Methyl II quantitative polymerase chain reaction (qPCR) System (Qiagen, Germany). In step 1, 4 equal genomic DNA aliquots were subjected to 4 different tubes, mock (M0), methylation-sensitive dependent (Msd), methylation-dependent (Md), and methylation-sensitive (Ms) enzymes. Using thermal cycler (SureCycler 8800, Santa Clara, California), all tubes were incubated for 6 hours at 37°C and for 20 minutes at 65°C. In step 2, system of Max3005P QPCR (Stratagene, Agilent Technologies, Santa Clara, California) was used and directly the enzyme reactions were mixed with qPCR master mix (RT2 qPCR SYBR Green/ROX Master Mix) and were dispensed into a plate containing pre-aliquoted primer mixes (EpiTect Methyl II qPCR Primer Assay). Cycle conditions were 95°C for 10 minutes (1 cycle), 99°C for 30 seconds and 72°C for 1 minute (3 cycles), and finally 97°C for 15 seconds and 72°C for 1 minute (40 cycles). Relative quantities of unmethylated and methylated DNA were automatically calculated by pasting raw ΔCT values into pre-performed data analysis spreadsheet (EpiTect Methyl II PCR Array Microsoft Excel based data analysis template).
BC markers
Based on manufacturer’s instructions, all cases and controls, after diagnosis and sample collection, were tested for CA 15.3 and CEA using commercial enzyme-linked immunosorbent assay (ELISA) kit (MyBioSource, San Diego, California).
Statistical analyses
Based on normality distribution, variables were descriped as absolute numbers, mean ± SD or median (interquartile range, IQR), appropriately. Difference between different groups was assessed using analysis of variance (ANOVA) and Kruskal-Wallis for normally and non-normally distributed values, respectively, and Fisher least significant difference as post hoc test. P value < .05 was significant. The degree of AKR1B1 methylation and BC biomarkers were subjected to receiver operating characteristic (ROC) curve 24 to identify their diagnostic power. For determining the correlation of AKR1B1 methylation degree with different variables, Pearson and Spearman correlations were assessed, appropriately. All results were analyzed using GraphPad prism and SPSS (Chicago, Illinois) programs.
Results
Characteristics of participants
Cases and controls data are summarized in Table 1. At time of diagnosis, BC cases, patients with benign breast diseases, and healthy controls were age-matched (P = .902). Pathologically, breast benign diseases in this study included fibrocystic changes, intraductal papillomatosis, and follicular hyperplasia. Most of participants were premenopausal women. Also, data about tumor invasion, stage, grade, lymph node invasion, hormones (estrogen and progesterone) receptors status, and HER-2 protein expression are shown in Table 1.
Table 1.
Characteristics of participants.
| Parameter | Breast cancer | Benign | Healthy | P value |
|---|---|---|---|---|
| Number | 120 | 40 | 40 | — |
| Age, y | 52.3 ± 9.1 | 51.9 ± 9.2 | 51.45 ± 9.0 | .902 |
| Menopause (pre-/post-menopausal) | 75/45 | 27/13 | 26/14 | .923 |
| CEA (ng/mL) | 12 (8.1-16.8) | 14.3 (8.9-15.2) | 7.9 (5.5-10.7) | .016 |
| CA-15.3 (U/mL) | 22.1 (14.1-24.1) | 16.9 (12.8-21.9) | 11.9 (10.9-14) | .0020 |
| Methylated AKR1B1 | 93.2 (75.4-98.6) | 23.9 (22.6-48.3) | 15.5 (10.6-16) | <.0001 |
| Invasion (Insitu/Invasive) | 48/72 | — | — | — |
| Tumor depth (T ⩽ 2/T > 2) | 51/69 | — | — | — |
| Tumor grade (G1/G2-3) | 45/75 | — | — | — |
| Lymph node invasion (negative/positive) | 59/61 | — | — | — |
| Estrogen receptor (negative/positive) | 60/60 | — | — | — |
| Progesterone receptor (negative/positive) | 42/78 | — | — | — |
| HER-2/neu (negative/positive) | 42/78 | — | — | — |
| Luminal (ER/PR+) A (HER2−)/B(HER2+) | 12/78 | — | — | — |
| Non-luminal HER2+ (ER-/PR-/HER2+) | 0 | — | — | — |
| Triple negative (ER-/PR-/HER2-) | 30 | — | — | — |
Differences between groups were established by ANOVA test or χ2 test appropriately. P < .05 is significant.
AKR1B1 methylation was associated with BC development
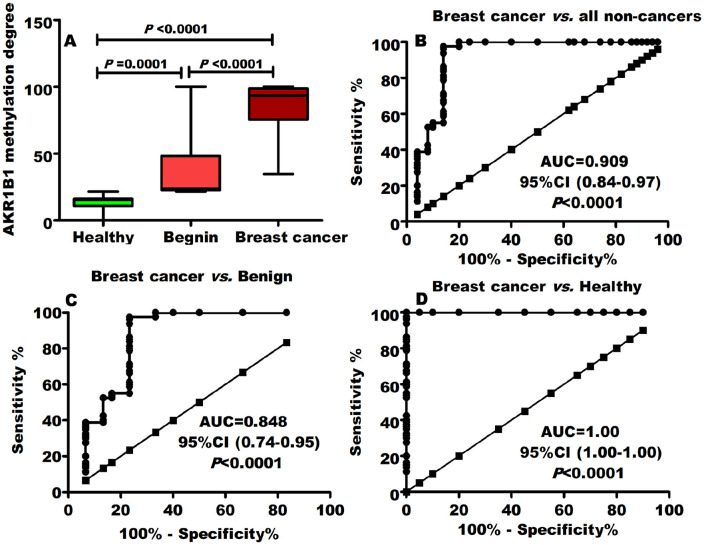
Despite BC tumor markers CEA and CA15.3 (Table 1), AKR1B1 methylation pattern was distinctly (P < .0001) associated with patients with BC. As expressed by median (IQR), BC (93.2 [75.4-98.6]) cases displayed significantly (P < .0001) greater degree of AKR1B1 methylation in comparison with benign (23.9 [22.6-48.3]) breast disorders and healthy (15.5 [10.6-16]) females (Figure 1A).
Figure 1.
AKR1B1 methylation and breast cancer development. (A) Patients with breast cancer were significantly related to hypermethylated AKR1B1. The ROC curve revealed that AKR1B1 methylation had a great diagnostic power for separating breast cancer from (B) all noncancer individuals (benign, healthy combined), (C) benign disorders, and (D) healthy controls.
Accuracy of AKR1B1 methylation in BC diagnosis
To determine its diagnostic ability, ROC curve analysis was performed for AKR1B1 DNA methylation and the optimal cutoff value was assessed. Despite CA 15-3 (AUC = 0.681; P = .010) and CEA (AUC = 0.539; P = .465) (Table 2), AKR1B1 DNA methylation had superior (AUC = 0.909; P < .0001; Figure 1B) diagnostic ability for diagnosing BC from all noncancers (benign and healthy combined). When comparing BC with only benign breast disorders, AKR1B1 DNA methylation ability to detect BC did not markedly alter (Figure 1C) indicating its cancer specification. Furthermore, this power rises to absolute value AUC = 1.00 (P < .0001) when comparing BC with only healthy controls (Figure 1D). Interestingly, AKR1B1 methylation reported to be significant in identifying BC early stages and grades (Table 2).
Table 2.
Diagnostic power of AKR1B1 methylation against CEA and CA-15.3.
| Marker | AUC (95% CI) | P value | Cutoff | Sen. (%) | Sp. (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|---|
| Breast cancer vs all noncancers | ||||||||
| CEA | 0.539 0.44-0.64 |
.465 | >9 | 70 | 40 | 65.1 | 45.5 | 58.5 |
| CA-15.3 | 0.681 0.60-0.78 |
.010 | >13 | 75 | 48 | 69.8 | 54.5 | 64.6 |
| AKR1B1 methylation | 0.909 0.84-0.97 |
<.0001 | >45 | 98 | 86 | 91 | 96 | 93 |
| Breast cancer vs. healthy females | ||||||||
| CEA | 0.669 0.56-0.78 |
.020 | >9 | 70 | 65 | 88.9 | 35.1 | 69 |
| CA-15.3 | 0.686 0.60-0.78 |
<.001 | >13 | 75 | 65 | 89.6 | 39.4 | 73 |
| AKR1B1 methylation | 1.00 1.0-1.0 |
<.0001 | >45 | 98 | 100 | 100 | 96 | 99 |
| Early stages from all noncancers | ||||||||
| CEA | 0.469 0.33-0.60 |
.632 | >9 | 60 | 40 | 60 | 40 | 52 |
| CA-15.3 | 0.671 0.58-0.76 |
.003 | >13 | 70 | 48 | 67 | 51 | 61 |
| AKR1B1 methylation | 0.899 0.83-0.97 |
<.0001 | >45 | 100 | 86 | 92 | 100 | 95 |
| Low grades from all noncancers | ||||||||
| CEA | 0.418 0.28-0.56 |
.222 | >9 | 60 | 40 | 60 | 40 | 52 |
| CA-15.3 | 0.675 0.58-0.77 |
.003 | >13 | 70 | 48 | 67 | 51 | 61 |
| AKR1B1 methylation | 0.903 0.83-0.97 |
<.0001 | >45 | 100 | 86 | 92 | 100 | 95 |
Abbreviations: AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value; Sen, sensitivity; Sp, specificity.
Methylation degree was positively correlated with disease severity
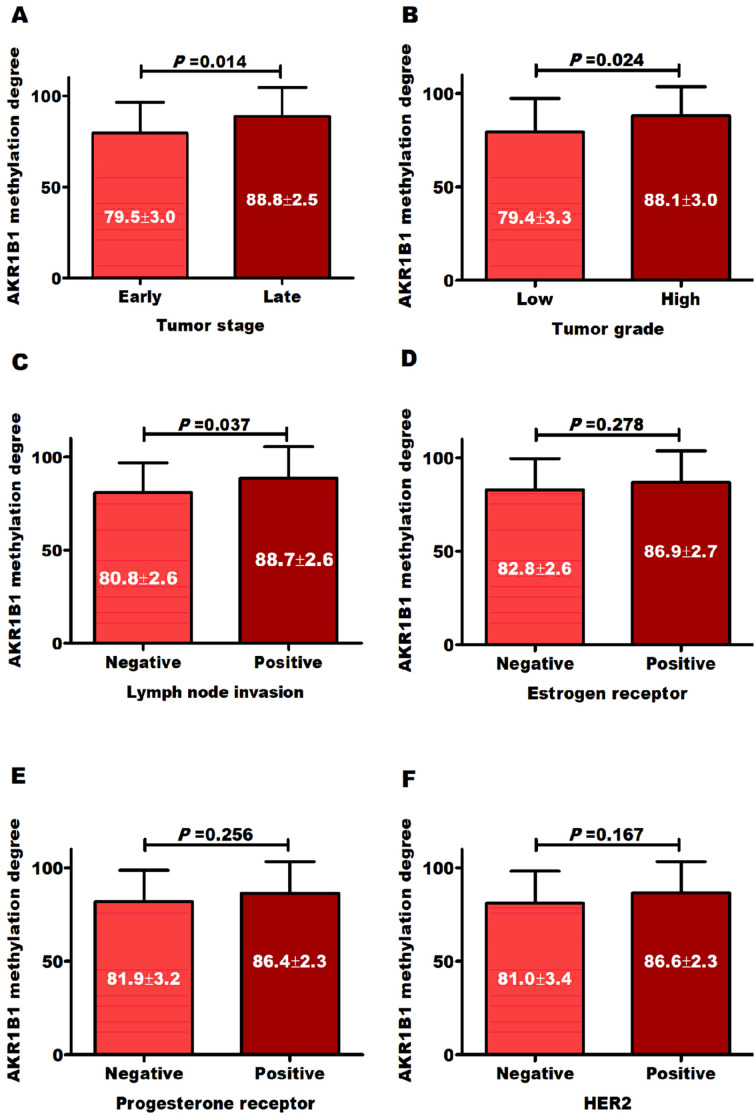
AKR1B1 hypermethylation was related to BC advanced stages (Figure 2A), high histological grades (Figure 2B), and lymph node invasion (Figure 2C), and this is independent to hormonal status and HER2neu expression (Figure 2D to F). Also, AKR1B1 high degrees of methylation were significantly correlated with the increase in CEA, CA-15.3 and tumor stages, grades, and lymph node invasion (Table 3). Interestingly, regarding BC molecular subtypes, TNBC (which is particularly difficult to treat) was significantly (P = .016) associated with AKR1B1 high degrees of methylation (Figure 3) compared with luminal subtypes.
Figure 2.
Distribution of AKR1B1 methylation degree according to tumor (A) stages, (B) grades, (C) lymph node invasion, (D) estrogen and (E) progesterone receptors, and (F) HER2 expression.
Table 3.
Correlation between methylated AKR1B1 and other parameters.
| Factor correlated with AKR1B1 | Correlation coefficient (r) | P value |
|---|---|---|
| Age | −.089 | .315 |
| Menopause | −.114 | .198 |
| CEA | .195 | .027 |
| CA-15.3 | .351 | .0001 |
| Tumor stage | .274 | .014 |
| Tumor grade | .253 | .024 |
| Lymph node | .275 | .014 |
| Estrogen receptor | .123 | .278 |
| Progesterone receptor | .128 | .265 |
| HER2 | .144 | .202 |
Pearson correlation was used for variables with interval scale, whereas Spearman correlation was used for variables with ordinal scales.
Figure 3.
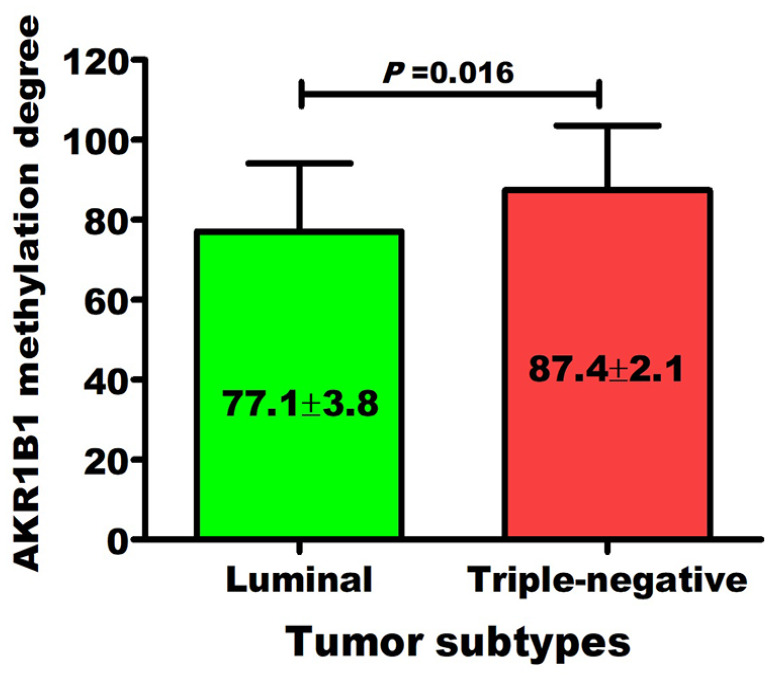
Distribution of AKR1B1 methylation degree among breast cancer subtypes.
Discussion
Screening methods, particularly mammography, can help reduce BC-related mortality rate by about 28% to 45%. However, in young cases and patients with small breasts, its use is limited by false-negative diagnosis possibility, dense breast lesions poor discrimination, and high radiation.25,26 Regarding early detection, there is a great challenge in the light of the limitations of available prognostic and diagnostic techniques, including false-positive results and low sensitivity. 5 In this study, we analyzed and evaluated AKR1B1 methylation profile in BC Egyptian patients as an efficient genetic marker for BC. AKR1B1 methylation showed high specificity and sensitivity for BC early diagnosis.
There were some studies that demonstrated a potential association of AKR1B1 DNA methylation and BC development.21,22,27 However, these studies did not include sufficient number of samples, almost did not include benign breast diseases, there is almost no study focusing on the association of AKR1B1 hypermethylation or hypomethylation and the disease severity. Here, AKR1B1 methylation was distinctly (P < .0001) related to BC (93.2 [75.4-98.6]) development as it displayed greater degree of hypermethylation compared with benign disorders (23.9 [22.6-48.3]) and healthy (15.5 [10.6-16]) females. Receiver operating characteristic curve revealed a great diagnostic power for AKR1B1 methylation (AUC = 0.909) that was superior to CA 15-3 (AUC = 0.681) and CEA (AUC = 0.539). When comparing BC with only benign breast disorders, AKR1B1 methylation ability to detect BC did not markedly change and raised to absolute value AUC = 1.00 comparing BC with only healthy controls. Interestingly, AKR1B1 hypermethylation was reported to be significant in identifying BC early stages (AUC = 0.899) and grades (AUC = 0.903).
In a trial to obtain a comprehensive gene methylation signature of HER2-positive BC, Lindqvist et al 21 reported AKR1B1-specific gene methylation. Among a panel of 19 candidate genes, de Groot et al found that AKR1B1 promoters were significantly differentially methylated in BC versus normal tissues. They found that AKR1B1 and TM6SF1 could diagnosed BC (AUC = 0.986) efficiently. 22 In a range of BC cell lines, Le et al 27 using methylation-sensitive high-resolution melting found that AKR1B1 methylation was specific for epithelial BC cell lines.
In this study, independent to hormonal status and HER2neu expression, AKR1B1 hypermethylation was related to BC advanced stages, high histological grades, and lymph node invasion. Moreover, high methylation degrees were significantly correlated with the increase in CEA (r = .195; P = .027), CA-15.3 (r = .351; P = .0001) and tumor stages (r = .274; P = .014), grades (r = .253; P = .024), and lymph node invasion (r = .275; P = .014).
This result may align with findings of previous studies. Among a total of 21 491 identified differentially methylated regions, Luo et al 28 found that the promoter methylation levels of AKR1B1 was increased in positive lymph node compared with the LN-negative BC. Also, highly methylated AKR1B1 gene promoters were also found in ER-positive and HER2-negative BC with axillary lymph node metastasis. 28 Benezeder et al 29 assessed multigene methylation analysis of enriched circulating tumor cells (CTCs) and they found that there was an association of these genes, including AKR1B1, methylation, and poor progression-free survival in patients with metastatic BC. Patients with CTCs unmethylated genes exhibited significantly longer progression-free survival compared with cases with methylated CTCs. 29 Recently, an increase in AKR1B1 methylation during treatment was reported to be correlated with a higher residual cancer burden and a decrease in this marker was found in cases who responded to treatment, but not in cases who did not respond. 30 All of these indicating that AKR1B1 DNA methylation may have a potential role in BC aggressiveness and its prognostic use in BC may need further investigations.
AKR1B1 role in cancer is not totally clear but increasing evidence is demonstrating to have a great impact on tumor progression. It could involve in a complicated network of miRNAs, proteins, and signaling pathways mediating mechanisms such as epithelial to mesenchymal transition, cell cycle, inflammatory responses, cell apoptosis, and survival. 19 In cancer, AKR1B1 overexpression has been related to inflammatory mediators, cell cycle mediators, survival proteins and pathways such as protein kinase B or Akt and mammalian target of rapamycin (mTOR), and other regulatory factors in response to prostaglandin and reactive oxygen species synthesis. 19 Despite that the exact mechanism linked hypermethylated AKR1B1 with BC progression remains elusive and there is a need to conduct more clinical studies to reveal this association.
Conclusions
Our study results revealed that AKR1B1 DNA hypermethylation was related to early BC development. From benign pre-malignant breast diseases, high methylation degrees could accurately predict BC. So, AKR1B1 methylation could specifically facilitate early BC screening so as to give accurate and timely decisions. This study is limited because of it include a single-center cohort and is retrospective in nature. Thus, there is an urgent need for future more multicentric comprehensive researches to determine AKR1B1 methylation-specific role in BC pathogenesis and aggressiveness.
Acknowledgments
The authors are indebted to the stuff of Mansoura Oncology Centre, Mansoura University, Egypt for their help in pathological reports for the enrolled individuals. Also, authors are grateful to the Science Technology & Innovation Funding Authority (STDF), Egypt as the instruments used in this work were purchased through their Capacity Building Grant.
Footnotes
Author Contributions: ME, MS, and MAA designed the study. MAA and BMF contributed to the experimental work. AA provide samples and diagnosis. MAA drafted and conceived the manuscript. All authors approved and revised the final manuscript.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Consent for Publication: Not applicable
Data Availability: Data related to the study are available under request.
Ethical Approval: The study protocol was ethically approved by the Institutional Research Board of Mansoura University and National Research Centre Medical Ethical Committee (ID: 15170) and was established based on the Helsinki Declaration ethical guidelines. Informed consent was obtained from each participant to be involved in this work.
ORCID iD: Mohamed A Abdelrazek  https://orcid.org/0000-0002-4111-0682
https://orcid.org/0000-0002-4111-0682
References
- 1. Obeagu EI, Obeagu GU. Breast cancer: a review of risk factors and diagnosis. Medicine (Baltimore). 2024;103:e36905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12-49. [DOI] [PubMed] [Google Scholar]
- 3. Sun YS, Zhao Z, Yang ZN, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13:1387-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rivera-Franco MM, Leon-Rodriguez E. Delays in breast cancer detection and treatment in developing countries. Breast Cancer (Auckl). 2018;12:1178223417752677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdelrazek MA, Nageb A, Barakat LA, Abouzid A, Elbaz R. BC-DETECT: combined detection of serum HE4 and TFF3 improves breast cancer diagnostic efficacy. Breast Cancer. 2022;29:507-515. [DOI] [PubMed] [Google Scholar]
- 6. Laessig D, Nagel D, Heinemann V, Untch M, et al. Importance of CEA and CA 15-3 during disease progression in metastatic breast cancer patients. Anticancer Res. 2007;27:1963-1968. [PubMed] [Google Scholar]
- 7. Ryu JM, Kang D, Cho J, et al. Prognostic impact of elevation of cancer antigen 15-3 (CA15-3) in patients with early breast cancer with normal serum CA15-3 level. J Breast Cancer. 2023;26:126-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Golestan A, Tahmasebi A, Maghsoodi N, Faraji SN, Irajie C, Ramezani A. Unveiling promising breast cancer biomarkers: an integrative approach combining bioinformatics analysis and experimental verification. BMC Cancer. 2024;24:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Downs BM, Mercado-Rodriguez C, Cimino-Mathews A, et al. DNA methylation markers for breast cancer detection in the developing world. Clin Cancer Res. 2019;25:6357-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15:459-466. [DOI] [PubMed] [Google Scholar]
- 11. Mao XH, Ye Q, Zhang GB, et al. Identification of differentially methylated genes as diagnostic and prognostic biomarkers of breast cancer. World J Surg Oncol. 2021;19:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong S, Li W, Wang L, et al. Histone-related genes are hypermethylated in lung cancer and hypermethylated HIST1H4F could serve as a pan-cancer biomarker. Cancer Res. 2019;79:6101-6112. [DOI] [PubMed] [Google Scholar]
- 13. Satoshi E, Matsunaga T, Hara A. Characterization of aldo-keto reductase 1C subfamily members encoded in two rat genes (akr1c19 and RGD1564865). Relationship to 9-hydroxyprostaglandin dehydrogenase. Arch Biochem Biophys. 2021;700:108755. [DOI] [PubMed] [Google Scholar]
- 14. Hyndman D, Bauman DR, Heredia VV, Penning TM. The aldo-keto reductase superfamily homepage. Chem Biol Interact. 2003;143-144:621-631. [DOI] [PubMed] [Google Scholar]
- 15. Graham A, Heath P, Morten JE, Markham AF. The human aldose reductase gene maps to chromosome region 7q35. Hum Genet. 1991;86:509-514. [DOI] [PubMed] [Google Scholar]
- 16. Di Benedetto C, Borini Etichetti C, Cocordano N, et al. The p53 tumor suppressor regulates AKR1B1 expression, a metastasis-promoting gene in breast cancer. Front Mol Biosci. 2023;10:1145279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saraswat M, Mrudula T, Kumar PU, et al. Overexpression of aldose reductase in human cancer tissues. Med Sci Monit. 2006;12:CR525-CR529. [PubMed] [Google Scholar]
- 18. Wu X, Li X, Fu Q, et al. AKR1B1 promotes basal-like breast cancer progression by a positive feedback loop that activates the EMT program. J Exp Med. 2017;214:1065-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khayami R, Hashemi SR, Kerachian MA. Role of aldo-keto reductase family 1 member B1 (AKR1B1) in the cancer process and its therapeutic potential. J Cell Mol Med. 2020;24:8890-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Almeida BP, Apolónio JD, Binnie A, Castelo-Branco P. Roadmap of DNA methylation in breast cancer identifies novel prognostic biomarkers. BMC Cancer. 2019;19:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindqvist BM, Wingren S, Motlagh PB, Nilsson TK. Whole genome DNA methylation signature of HER2-positive breast cancer. Epigenetics. 2014;9:1149-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Groot JS, Pan X, Meeldijk J, van der Wall E, van Diest PJ, Moelans CB. Validation of DNA promoter hypermethylation biomarkers in breast cancer—a short report. Cell Oncol (Dordr). 2014;37:297-303. [DOI] [PubMed] [Google Scholar]
- 23. O’Sullivan B, Brierley J, Byrd D, et al. The TNM classification of malignant tumours-towards common understanding and reasonable expectations. Lancet Oncol. 2017;18:849-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5:1315-1316. [DOI] [PubMed] [Google Scholar]
- 25. Warner E. Clinical practice. Breast-cancer screening. N Engl J Med. 2011;365:1025-1032. [DOI] [PubMed] [Google Scholar]
- 26. Wang T, Li P, Qi Q, et al. A multiplex blood-based assay targeting DNA methylation in PBMCs enables early detection of breast cancer. Nat Commun. 2023;14:4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Le AV, Szaumkessel M, Tan TZ, et al. DNA methylation profiling of breast cancer cell lines along the epithelial mesenchymal spectrum-implications for the choice of circulating tumour DNA methylation markers. Int J Mol Sci. 2018;19:2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo J, Chen S, Chen J, Zhou Y, He F, Wang E. Identification and validation of DNA methylation markers to predict axillary lymph node metastasis of breast cancer. PLoS ONE. 2022;17:e0278270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Benezeder T, Tiran V, Treitler AAN, et al. Multigene methylation analysis of enriched circulating tumor cells associates with poor progression-free survival in metastatic breast cancer patients. Oncotarget. 2017;8:92483-92496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janssen LM, Janse MHA, Penning de, Vries BBL, et al. Predicting response to neoadjuvant chemotherapy with liquid biopsies and multiparametric MRI in patients with breast cancer. NPJ Breast Cancer. 2024;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]