Abstract
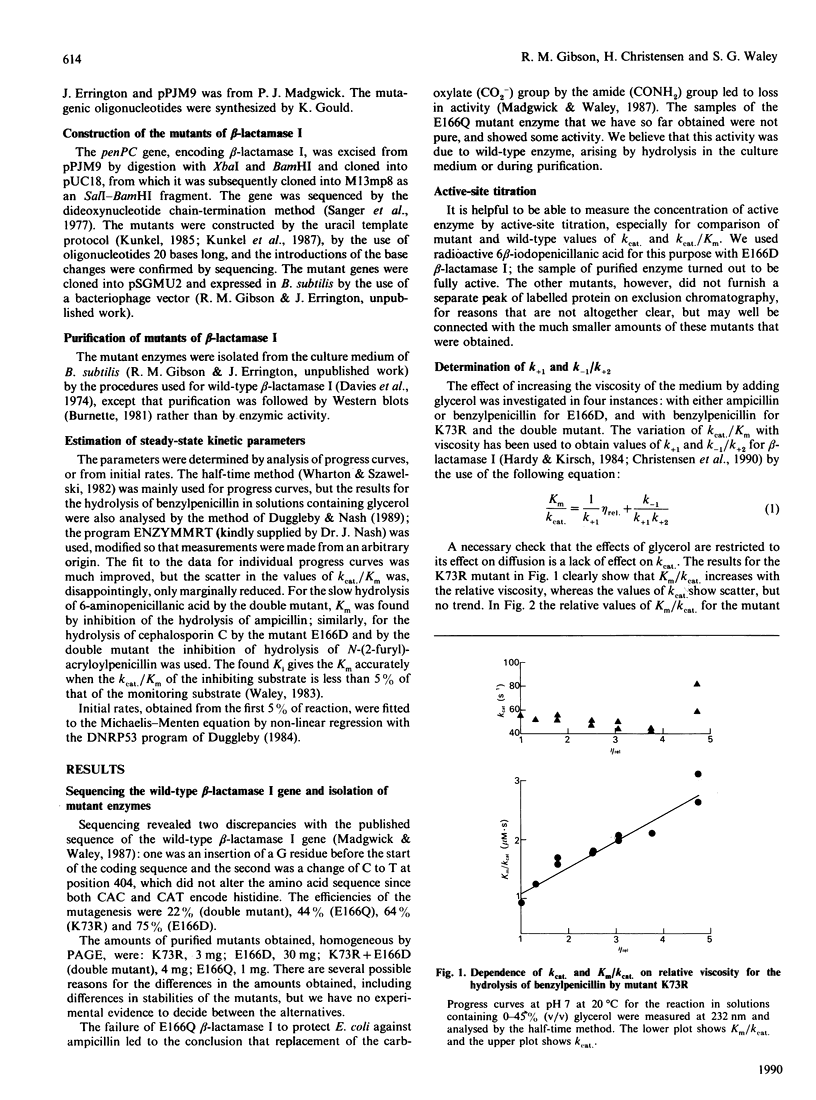
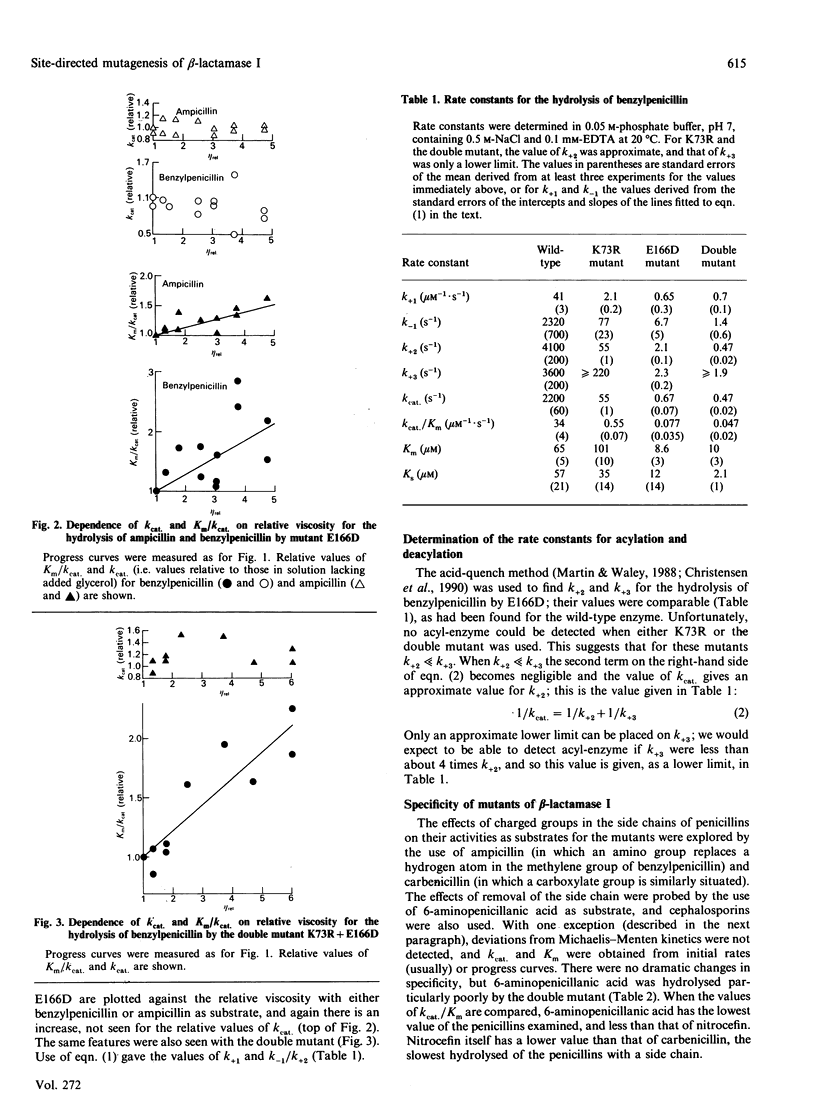
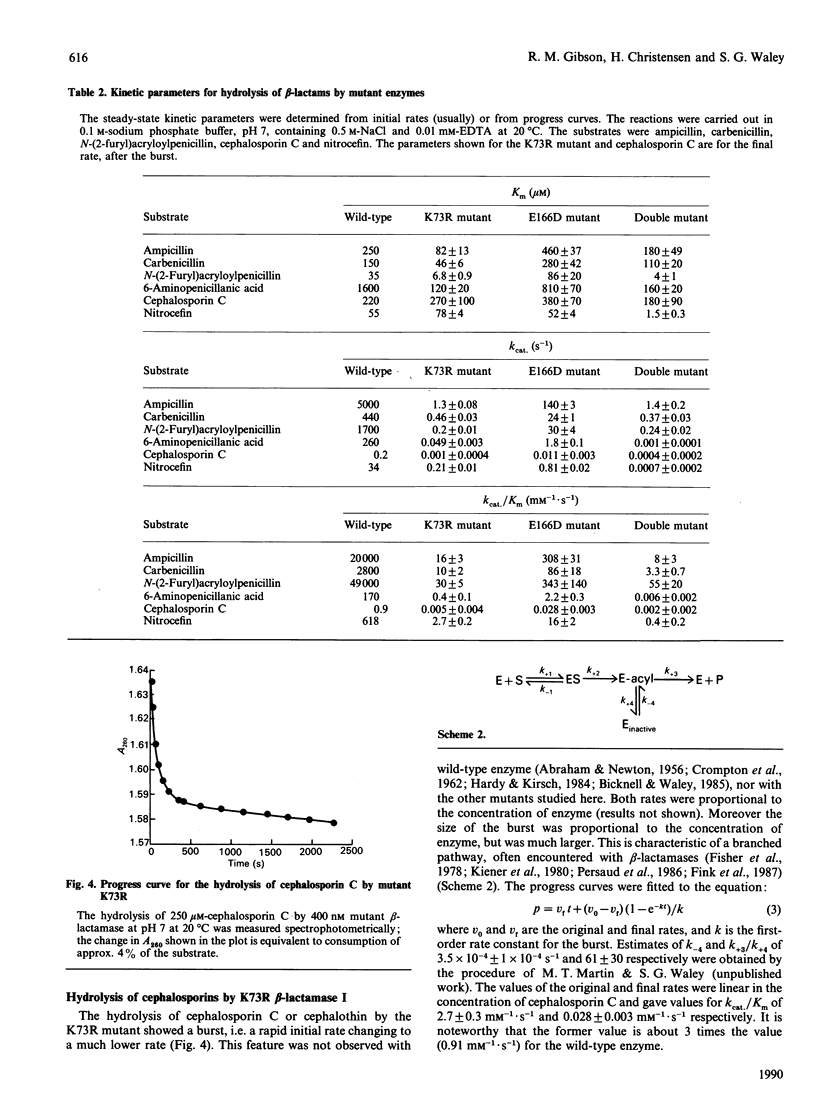
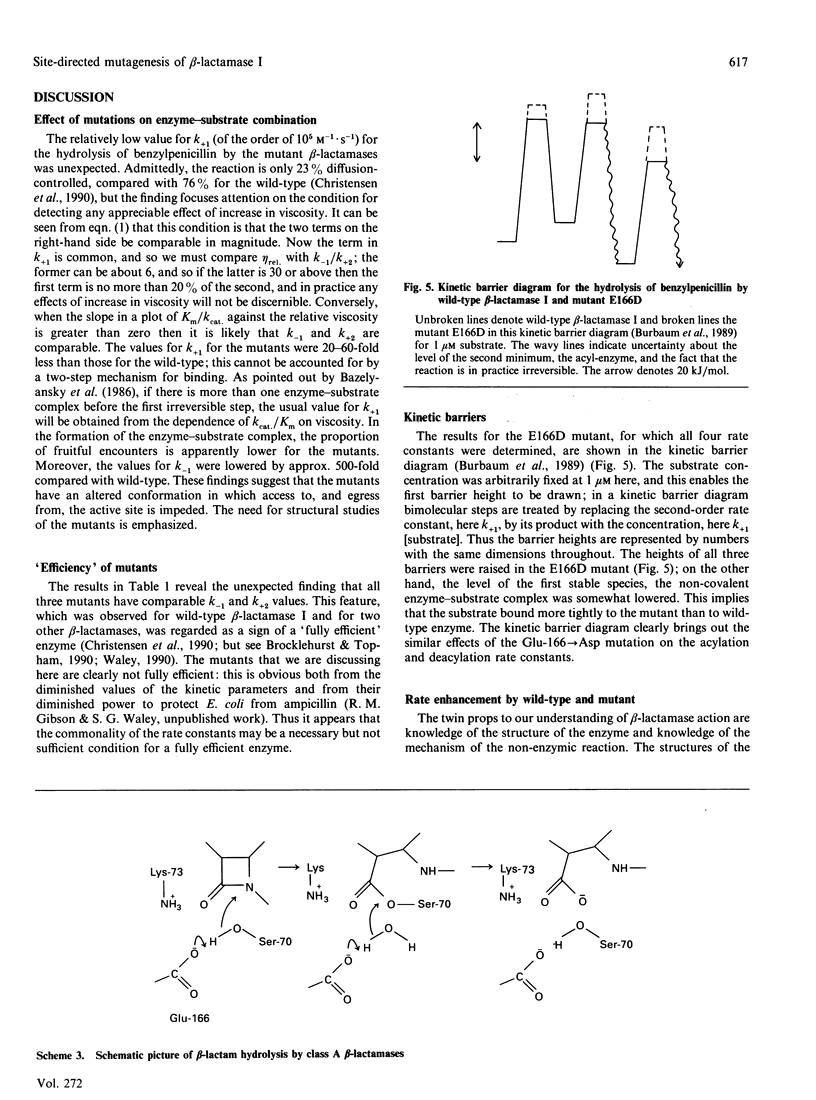
Two single mutants and the corresponding double mutant of beta-lactamase I from Bacillus cereus 569/H were constructed and their kinetics investigated. The mutants have Lys-73 replaced by arginine (K73R), or Glu-166 replaced by aspartic acid (E166D), or both (K73R + E166D). All four rate constants in the acyl-enzyme mechanism were determined for the E166D mutant by the methods described by Christensen, Martin & Waley [(1990) Biochem. J. 266, 853-861]. Both the rate constants for acylation and deacylation for the hydrolysis of benzylpenicillin were decreased about 2000-fold in this mutant. In the K73R mutant, and in the double mutant, the rate constants for acylation were decreased about 100-fold and 10,000-fold respectively. All three mutants also had lowered values for the rate constants for the formation and dissociation of the non-covalent enzyme-substrate complex. The specificities of the mutants did not differ greatly from those of wild-type beta-lactamase, but the hydrolysis of cephalosporin C by the K73R mutant gave 'burst' kinetics.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ABRAHAM E. P., NEWTON G. G. A comparison of the action of penicillinase on benzylpenicillin and cephalosporin N and the competitive inhibition of penicillinase by cephalosporin C. Biochem J. 1956 Aug;63(4):628–634. doi: 10.1042/bj0630628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber T. C., Davenport R. C., Jr, Giammona D. A., Lolis E., Petsko G. A., Ringe D. Crystallography and site-directed mutagenesis of yeast triosephosphate isomerase: what can we learn about catalysis from a "simple" enzyme? Cold Spring Harb Symp Quant Biol. 1987;52:603–613. doi: 10.1101/sqb.1987.052.01.069. [DOI] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Bazelyansky M., Robey E., Kirsch J. F. Fractional diffusion-limited component of reactions catalyzed by acetylcholinesterase. Biochemistry. 1986 Jan 14;25(1):125–130. doi: 10.1021/bi00349a019. [DOI] [PubMed] [Google Scholar]
- Bicknell R., Waley S. G. Single-turnover and steady-state kinetics of hydrolysis of cephalosporins by beta-lactamase I from Bacillus cereus. Biochem J. 1985 Oct 1;231(1):83–88. doi: 10.1042/bj2310083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklow S. C., Raines R. T., Lim W. A., Zamore P. D., Knowles J. R. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: Analysis of triose phosphate equilibria in aqueous solution by 31P NMR. Biochemistry. 1988 Feb 23;27(4):1158–1167. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- Brocklehurst K., Topham C. M. Kinetic parameters of the acyl-enzyme mechanism and conditions for quasi-equilibrium and for optimal catalytic characteristics. Biochem J. 1990 Sep 1;270(2):561–563. doi: 10.1042/bj2700561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbaum J. J., Raines R. T., Albery W. J., Knowles J. R. Evolutionary optimization of the catalytic effectiveness of an enzyme. Biochemistry. 1989 Nov 28;28(24):9293–9305. doi: 10.1021/bi00450a009. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- CROMPTON B., JAGO M., CRAWFORD K., NEWTON G. G., ABRAHAM E. P. Behaviour of some derivatives of 7-aminocephalosporanic acid and 6-aminopenicillanic acidas substrates, inhibitors and inducers of penicillinases. Biochem J. 1962 Apr;83:52–63. doi: 10.1042/bj0830052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen H., Martin M. T., Waley S. G. Beta-lactamases as fully efficient enzymes. Determination of all the rate constants in the acyl-enzyme mechanism. Biochem J. 1990 Mar 15;266(3):853–861. [PMC free article] [PubMed] [Google Scholar]
- Citri N., Samuni A., Zyk N. Acquisition of substrate-specific parameters during the catalytic reaction of penicillinase. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1048–1052. doi: 10.1073/pnas.73.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corran P. H., Waley S. G. The amino acid sequence of rabbit muscle triose phosphate isomerase. FEBS Lett. 1973 Feb 15;30(1):97–99. doi: 10.1016/0014-5793(73)80627-1. [DOI] [PubMed] [Google Scholar]
- Coulson A. Beta-lactamases: molecular studies. Biotechnol Genet Eng Rev. 1985;3:219–253. doi: 10.1080/02648725.1985.10647814. [DOI] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Cohen L. W., Riggs A. D., Morin C., Itakura K., Richards J. H. Oligonucleotide-directed mutagenesis as a general and powerful method for studies of protein function. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6409–6413. doi: 10.1073/pnas.79.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. B., Abraham E. P. Separation, purification and properties of beta-lactamase I and beta-lactamase II from Bacillus cereus 569/H/9. Biochem J. 1974 Oct;143(1):115–127. doi: 10.1042/bj1430115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Mare S., Coulson A. F., Knowles J. R., Priddle J. D., Offord R. E. Active-site labelling of triose phosphate isomerase. The reaction of bromohydroxyacetone phosphate with a unique glutamic acid residue and the migration of the label to tyrosine. Biochem J. 1972 Sep;129(2):321–331. doi: 10.1042/bj1290321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Wéry J. P., Dehottay P., Dusart J., Erpicum T., Frère J. M., Ghuysen J. M. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem J. 1987 Aug 1;245(3):911–913. doi: 10.1042/bj2450911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggleby R. G., Nash J. C. A single-parameter family of adjustments for fitting enzyme kinetic models to progress-curve data. Biochem J. 1989 Jan 1;257(1):57–64. doi: 10.1042/bj2570057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggleby R. G. Regression analysis of nonlinear Arrhenius plots: an empirical model and a computer program. Comput Biol Med. 1984;14(4):447–455. doi: 10.1016/0010-4825(84)90045-3. [DOI] [PubMed] [Google Scholar]
- Fink A. L., Behner K. M., Tan A. K. Kinetic and structural characterization of reversibly inactivated beta-lactamase. Biochemistry. 1987 Jul 14;26(14):4248–4258. doi: 10.1021/bi00388a011. [DOI] [PubMed] [Google Scholar]
- Fisher J., Belasco J. G., Khosla S., Knowles J. R. beta-Lactamase proceeds via an acyl-enzyme intermediate. Interaction of the Escherichia coli RTEM enzyme with cefoxitin. Biochemistry. 1980 Jun 24;19(13):2895–2901. doi: 10.1021/bi00554a012. [DOI] [PubMed] [Google Scholar]
- Fisher J., Charnas R. L., Knowles J. R. Kinetic studies on the inactivation of Escherichia coli RTEM beta-lactamase by clavulanic acid. Biochemistry. 1978 May 30;17(11):2180–2184. doi: 10.1021/bi00604a024. [DOI] [PubMed] [Google Scholar]
- Frère J. M., Joris B. Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit Rev Microbiol. 1985;11(4):299–396. doi: 10.3109/10408418409105906. [DOI] [PubMed] [Google Scholar]
- Hall A., Knowles J. R. Directed selective pressure on a beta-lactamase to analyse molecular changes involved in development of enzyme function. Nature. 1976 Dec 23;264(5588):803–804. doi: 10.1038/264803a0. [DOI] [PubMed] [Google Scholar]
- Hardy L. W., Kirsch J. F. Diffusion-limited component of reactions catalyzed by Bacillus cereus beta-lactamase I. Biochemistry. 1984 Mar;23(6):1275–1282. doi: 10.1021/bi00301a040. [DOI] [PubMed] [Google Scholar]
- Healey W. J., Labgold M. R., Richards J. H. Substrate specificities in class A beta-lactamases: preference for penams vs. cephems. The role of residue 237. Proteins. 1989;6(3):275–283. doi: 10.1002/prot.340060310. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science. 1987 May 8;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- Hibler D. W., Stolowich N. J., Reynolds M. A., Gerlt J. A., Wilde J. A., Bolton P. H. Site-directed mutants of staphylococcal nuclease. Detection and localization by 1H NMR spectroscopy of conformational changes accompanying substitutions for glutamic acid-43. Biochemistry. 1987 Sep 22;26(19):6278–6286. doi: 10.1021/bi00393a048. [DOI] [PubMed] [Google Scholar]
- Jacob F., Joris B., Dideberg O., Dusart J., Ghuysen J. M., Frère J. M. Engineering a novel beta-lactamase by a single point mutation. Protein Eng. 1990 Oct;4(1):79–86. doi: 10.1093/protein/4.1.79. [DOI] [PubMed] [Google Scholar]
- Jaurin B., Grundström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. A., Dideberg O., Charlier P., Wery J. P., Libert M., Moews P. C., Knox J. R., Duez C., Fraipont C., Joris B. On the origin of bacterial resistance to penicillin: comparison of a beta-lactamase and a penicillin target. Science. 1986 Mar 21;231(4744):1429–1431. doi: 10.1126/science.3082007. [DOI] [PubMed] [Google Scholar]
- Kelly J. A., Knox J. R., Zhao H., Frère J. M., Ghaysen J. M. Crystallographic mapping of beta-lactams bound to a D-alanyl-D-alanine peptidase target enzyme. J Mol Biol. 1989 Sep 20;209(2):281–295. doi: 10.1016/0022-2836(89)90277-5. [DOI] [PubMed] [Google Scholar]
- Kiener P. A., Knott-Hunziker V., Petursson S., Waley S. G. Mechanism of substrate-induced inactivation of beta-lactamase I. Eur J Biochem. 1980 Aug;109(2):575–580. doi: 10.1111/j.1432-1033.1980.tb04830.x. [DOI] [PubMed] [Google Scholar]
- Klemes Y., Citri N. Catalytic and conformational properties of cross-linked derivatives of penicillinase. Biochim Biophys Acta. 1979 Apr 12;567(2):401–409. doi: 10.1016/0005-2744(79)90126-8. [DOI] [PubMed] [Google Scholar]
- Knott-Hunziker V., Petursson S., Waley S. G., Jaurin B., Grundström T. The acyl-enzyme mechanism of beta-lactamase action. The evidence for class C Beta-lactamases. Biochem J. 1982 Nov 1;207(2):315–322. doi: 10.1042/bj2070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Madgwick P. J., Waley S. G. beta-lactamase I from Bacillus cereus. Structure and site-directed mutagenesis. Biochem J. 1987 Dec 15;248(3):657–662. doi: 10.1042/bj2480657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. T., Waley S. G. Kinetic characterization of the acyl-enzyme mechanism for beta-lactamase I. Biochem J. 1988 Sep 15;254(3):923–925. doi: 10.1042/bj2540923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moews P. C., Knox J. R., Dideberg O., Charlier P., Frère J. M. Beta-lactamase of Bacillus licheniformis 749/C at 2 A resolution. Proteins. 1990;7(2):156–171. doi: 10.1002/prot.340070205. [DOI] [PubMed] [Google Scholar]
- Mézes P. S., Blacher R. W., Lampen J. O. Processing of Bacillus cereus 569/H beta-lactamase I in Escherichia coli and Bacillus subtilis. J Biol Chem. 1985 Jan 25;260(2):1218–1223. [PubMed] [Google Scholar]
- Oefner C., D'Arcy A., Daly J. J., Gubernator K., Charnas R. L., Heinze I., Hubschwerlen C., Winkler F. K. Refined crystal structure of beta-lactamase from Citrobacter freundii indicates a mechanism for beta-lactam hydrolysis. Nature. 1990 Jan 18;343(6255):284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- Persaud K. C., Pain R. H., Virden R. Reversible deactivation of beta-lactamase by quinacillin. Extent of the conformational change in the isolated transitory complex. Biochem J. 1986 Aug 1;237(3):723–730. doi: 10.1042/bj2370723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock M. R. The function and evolution of penicillinase. Proc R Soc Lond B Biol Sci. 1971 Dec 31;179(1057):385–401. doi: 10.1098/rspb.1971.0104. [DOI] [PubMed] [Google Scholar]
- Raines R. T., Sutton E. L., Straus D. R., Gilbert W., Knowles J. R. Reaction energetics of a mutant triosephosphate isomerase in which the active-site glutamate has been changed to aspartate. Biochemistry. 1986 Nov 4;25(22):7142–7154. doi: 10.1021/bi00370a057. [DOI] [PubMed] [Google Scholar]
- Samraoui B., Sutton B. J., Todd R. J., Artymiuk P. J., Waley S. G., Phillips D. C. Tertiary structural similarity between a class A beta-lactamase and a penicillin-sensitive D-alanyl carboxypeptidase-transpeptidase. 1986 Mar 27-Apr 2Nature. 320(6060):378–380. doi: 10.1038/320378a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., DeGrado W. F., Thomas B. J., Petteway S. R., Jr Purification and properties of thiol beta-lactamase. A mutant of pBR322 beta-lactamase in which the active site serine has been replaced with cysteine. J Biol Chem. 1984 Apr 25;259(8):5327–5332. [PubMed] [Google Scholar]
- Tsukamoto K., Tachibana K., Yamazaki N., Ishii Y., Ujiie K., Nishida N., Sawai T. Role of lysine-67 in the active site of class C beta-lactamase from Citrobacter freundii GN346. Eur J Biochem. 1990 Feb 22;188(1):15–22. doi: 10.1111/j.1432-1033.1990.tb15365.x. [DOI] [PubMed] [Google Scholar]
- Waley S. G. Beta-lactamases: a major cause of antibiotic resistance. Sci Prog. 1988;72(288 Pt 4):579–597. [PubMed] [Google Scholar]
- Waley S. G. Kinetic parameters from progress curves of competing substrates. Application to beta-lactamases. Biochem J. 1983 May 1;211(2):511–513. doi: 10.1042/bj2110511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waley S. G., Miller J. C., Rose I. A., O'Connell E. L. Identification of site in triose phosphate isomerase labelled by glycidol phosphate. Nature. 1970 Jul 11;227(5254):181–181. doi: 10.1038/227181a0. [DOI] [PubMed] [Google Scholar]
- Wharton C. W., Szawelski R. J. Half-time analysis of the integrated Michaelis equation. Simulation and use of the half-time plot and its direct linear variant in the analysis of some alpha-chymotrypsin, papain- and fumarase-catalysed reactions. Biochem J. 1982 May 1;203(2):351–360. doi: 10.1042/bj2030351. [DOI] [PMC free article] [PubMed] [Google Scholar]



