Abstract
Hepatitis B viruses replicate through reverse transcription of an RNA intermediate, the pregenomic RNA (pgRNA). Replication is initiated de novo and requires formation of a ribonucleoprotein complex comprising the viral reverse transcriptase (P protein), an RNA stem-loop structure (ɛ) on the pgRNA, and cellular proteins, including the heat shock protein Hsp90, the cochaperone p23, and additional, as yet unknown, factors. Functional complexes catalyze the synthesis of a short DNA primer that is templated by ɛ and covalently linked to the terminal protein (TP) domain of P protein. Currently, the only system for generating such complexes in the test tube is in vitro translation of duck hepatitis B virus (DHBV) P protein in rabbit reticulocyte lysate (RRL), which also provides the necessary factors. However, its limited translation capacity precludes a closer analysis of the complex. To overcome this restriction we sought to produce larger amounts of DHBV P protein by expression in Escherichia coli, followed by complex reconstitution in RRL. Because previous attempts to generate full-length P protein in bacteria have failed we investigated whether separate expression of the TP and reverse transcriptase-RNase H (RT-RH) domains would allow higher yields and whether these domains could trans complement each other. Indeed, TP and, after minor C-terminal modifications, also RT-RH could be expressed in substantial amounts, and when added to RRL, they were capable of ɛ-dependent DNA primer synthesis, demonstrating posttranslational activation. This reconstitution system should pave the way for a detailed understanding of the unique hepadnaviral replication initiation mechanism.
Hepatitis B viruses, or hepadnaviruses, are small, hepatotropic DNA-containing viruses that replicate through an RNA intermediate. Their type member, hepatitis B virus (HBV), is an important human pathogen that causes acute and chronic hepatitis B (6, 33). Because only human and chimpanzee hepatocytes are susceptible to efficient HBV infection, the related animal viruses of woodchucks (woodchuck hepatitis B virus) and of ducks (duck hepatitis B virus [DHBV]) have become important model systems. In all hepadnaviruses, a terminally redundant transcript, the pregenomic RNA (pgRNA), acts as mRNA for core protein and P protein, the reverse transcriptase (RT). Furthermore, this RNA is selectively packaged into viral capsids and reverse transcribed into partially double-stranded circular DNA (22, 23). Both nucleocapsid assembly and replication initiation are crucially dependent on the binding of P protein to a stem-loop structure, ɛ, close to the 5′ end of the pgRNA (5, 16, 25); a second copy of ɛ, in the 3′-terminal redundancy, is functionally silent in vivo (27). In contrast to that of retroviruses, hepadnavirus replication is initiated de novo, i.e., without a nucleic acid primer, within a bulged region of ɛ (Fig. 1B), resulting in the synthesis of a 3- or 4-nucleotide (nt) DNA primer (24, 37, 38) whose 5′ end becomes covalently linked to P protein (20, 41, 44). The primer-P protein complex is subsequently translocated to a 3′-proximal RNA element, DR1*, for minus-strand DNA elongation.
FIG. 1.
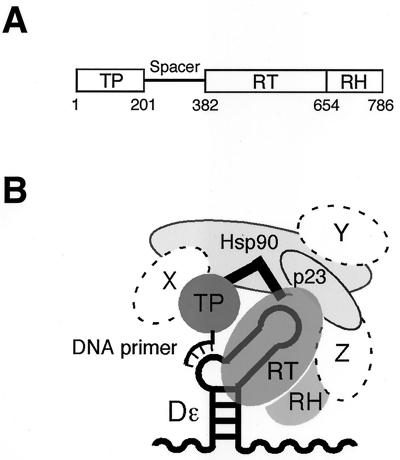
(A) Domain organization of hepadnaviral P proteins. Numbers indicate amino acid positions of DHBV P. (B) Model of the DHBV replication initiation complex. P protein with its TP and RT-RH domains connected via the spacer (black angled bar) is bound to the RNA stem-loop Dɛ. A bulged region in Dɛ serves as a template for the synthesis of a short DNA primer that is covalently linked to a Tyr residue in the TP domain. Binding to Dɛ requires P protein to be present in a multicomponent complex composed of Hsp90, p23 (light gray objects), and, most likely, additional, as yet unknown, factors (designated X, Y, and Z).
All hepadnaviral P proteins are about 90 kDa in size and composed of four domains (Fig. 1A): the terminal protein (TP), the spacer region, the reverse transcriptase (RT), and the RNase H (RH). TP is unique to hepadnaviruses and contains a Tyr residue to which the DNA primer is linked (20, 41, 44). The spacer region is not essential for P protein function (26) and is rather divergent in sequence between different hepadnaviruses. The RT and RH domains, by contrast, contain highly conserved sequence motifs that occur in all RTs (42).
A general outline of hepadnaviral replication has been worked out genetically, but mechanistic questions have become tractable only after the establishment of a system allowing reconstitution of replication initiation in a cell-free system (39). The P protein of DHBV, but for unknown reasons not that of HBV, when generated by in vitro translation in rabbit reticulocyte lysate (RRL) is able to perform the authentic DHBV ɛ (Dɛ)-dependent (−)-DNA priming reaction. Dɛ may be present at either end of the P mRNA used to program the lysate (cis priming) or be added as a separate, short RNA molecule (trans priming) (40). This relaxed position specificity appears to be the only obvious difference to genuine replication initiation. The system has therefore extensively been used to define critical determinants of the P-ɛ interaction using mutant P (32, 36) and Dɛ (2–5, 29, 36) molecules. An additional unique aspect of hepadnaviral replication is the dependence of Dɛ-P ribonucleoprotein (RNP) complex formation on cellular chaperones which are provided by RRL. Hsp90 and the small cochaperone p23 appear to be essential components (13, 14); by analogy to steroid hormone receptor complexes (34) and based on ultracentrifugation and gel filtration data (12, 29), it is highly likely that additional, as yet unknown, cellular proteins are present in the complex. The major obstacle for their identification is the limited amount of P protein, and consequently P protein RNPs, obtainable by in vitro translation (in the range of 1 ng/μl). The problem is potentiated by the very high abundance in RRL of several of the known and suspected chaperones (31) involved in RNP formation (in the range of 100 ng/μl), making it very difficult to completely separate these from their P protein-associated counterparts.
A possible solution would be to exogenously add to RRL, in larger amounts, P protein from a heterologous source which then may interact with RRL-contained factors to produce functional RNPs. However, heterologous expression of full-length P protein polypeptides capable of authentic ɛ-dependent priming has not yet been achieved. HBV P protein produced from recombinant baculoviruses in insect cells exerts some DNA synthesis activity that is, however, not ɛ dependent (19, 20). DHBV P protein, in that system, is subject to heavy proteolytic degradation (J. Römmelt and M. Nassal, unpublished data). An enzymatically active DHBV P fusion protein with the Gag protein of the Ty1 retrotransposon has been described, but it appears to be stable and active only inside the generated virus-like particles. Escherichia coli would offer many advantages as an expression system. However, virtually all previous attempts to generate full-length priming-competent P protein in bacteria have failed. Very recently the synthesis of DHBV P protein fusions with priming activity has been reported (12). However, the P protein parts were substantially truncated, and the yields were apparently low; hence, it remains to be determined whether this system is suitable for generating authentic replication complexes at a larger scale.
Here we took advantage of the multidomain nature of the hepadnaviral P protein and explored whether individual DHBV P domains could be expressed in E. coli more efficiently than the full-length protein and whether these separate domains are able to trans complement each other to form functional initiation complexes. The principal possibility for such a complementation has been demonstrated with insect cell-derived HBV P protein TP and RT-RH domains. However, activity was restricted to lysates from coexpressing cells and was not found after mixing either lysates from separately infected cells or the isolated proteins (20). Because DHBV P protein translated in RRL exerts a clearly Dɛ-dependent priming activity, we first used in vitro cotranslation of DHBV TP and RT-RH to establish that trans complementation is possible. We then investigated expression of the two domains in E. coli and found that TP and, after relatively minor deletions in the C-terminal part, also RT-RH polypeptides could be obtained in substantial amounts. Importantly, the purified domains produced in E. coli could be reconstituted with RRL into functional, Dɛ-dependent priming complexes.
MATERIALS AND METHODS
Plasmid constructs.
The parental plasmid for in vitro translations was pT7AMVpol16 (41), which contains the T7 promoter followed by nt 170 to 2843 of DHBV type 16, i.e., the complete P protein open reading frame (ORF) plus the 3′ copy of Dɛ. The TP and RT constructs were obtained by appropriate restriction digestions and/or PCR-mediated mutagenesis (details are available from the authors upon request). N-terminal His tags were introduced between amino acids (aa) 2 and 3 of the TP constructs and at the C terminus of the RT constructs between aa 785 and 786. The plasmid names indicate the P protein amino acid sequence present in the translation product. pT7-TP1-220 and pT7-RT209-786 contain aa 1 to 220 and aa 209 to 786, respectively. pT7-TP1-220ɛ+ and pT7-RT209-786ɛ+ contain in addition a copy of Dɛ downstream of the TP or RT-RH ORF, respectively. For expression in E. coli the corresponding DHBV sequences were inserted into the vector p30a(+) (Novagen). pET-TP1-220 for expression of TP1-220 was generated by inserting a restriction fragment from pT7-TP1-220 comprising the complete ORF immediately downstream of the Shine-Dalgarno sequence of pET-30a(+). The encoded polypeptide is identical to that of the in vitro translation construct. The pET-RT series of constructs for expression of RT-RH domains were constructed by inserting the corresponding DHBV sequence using PCR-mediated mutagenesis into the polylinker of pET-30a(+). The encoded proteins start with a vector-derived sequence of 50 aa, including a His tag for purification followed by the DHBV P protein sequence specified by the numbers. pET-RT349-729, pET-RT349-753, pET-RT349-761, and pET-RT349-775 additionally contain the vector-derived sequence Val Asp Lys Leu Ala Ala Ala Leu Glu His His His His His His at the C terminus. The plasmid pET-RT349-786co differs from pET-RT349-786 by several silent mutations in the DHBV sequence from nt 2426 to 2527 (codons 753 to 786) which do not affect the amino acid sequence. pET-RT349-786Pol− carries a single nucleotide substitution at nt 1706 that changes Asp 513 in the catalytic core of the RT domain to His. This mutation completely abolishes polymerase activity (41). All of the pET constructs lack the Dɛ sequence. Dɛ-RNA was obtained by in vitro transcription from plasmid pDɛ1 (2). The transcript of 76 nt contains DHBV sequence from nt 2557 to 2624.
E. coli protein expression and purification.
Expression constructs were transformed into E. coli strain BL21-CodonPlus-RIL (Stratagene). Bacterial cultures, grown at 37°C to an optical density at 600 nm of 0.5 to 1.0, were induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) and harvested after 2 to 16 h at room temperature. For purification of TP, the cells were lysed in buffer N1 (50 mM sodium phosphate, pH 7.5; 300 mM NaCl; 20 mM imidazole; 0.1% NP-40) containing lysozyme (1 mg/ml) followed by sonication. Cleared lysates were subjected to immobilized metal affinity chromatography (IMAC) using a batch procedure. For native purification the supernatant was mixed with Ni-nitrilotriaacetic acid (NTA)-agarose (Qiagen) and incubated for 1 h at 4°C. The beads were washed twice with buffer N1, once with buffer N1 containing 1 M NaCl, and once again with buffer N1. Elution was performed with buffer N1 supplemented with 200 mM imidazole. Peak fractions were dialyzed against dialysis buffer 1 (50 mM Tris HCl, pH 7.5; 100 mM KCl; 1 mM dithiothreitol [DTT]) at 4°C, adjusted to 10% glycerol (vol/vol) and stored at −80°C. The insoluble fraction of the lysate was dissolved in buffer D1 (8 M urea, 100 mM sodium phosphate [pH 8.0]), and the TP protein was purified using Ni-NTA-agarose according to the manufacturer's instructions. For renaturation, TP in the eluate was dialyzed against dialysis buffer 2 (20 mM HEPES, 100 mM potassium acetate, 1 mM DTT, 0.1% NP-40 [pH 7.4]), containing 3 M urea in the first step and 1 M urea in the second step. After centrifugation the clear supernatant was adjusted to 10% glycerol and stored at −80°C. RT349-761 was purified similarly but with the some modifications. Buffer N1 was replaced with buffer N2 (100 mM potassium phosphate [pH 7.5], 500 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.5 mM DTT, EDTA-free protease inhibitor cocktail [“complete”; one tablet per 100 ml of buffer; Roche]), and poly-His protein purification resin (Roche) was used instead of Ni-NTA agarose. The beads were washed twice with buffer N2 containing 0.1% Triton X-100, and the RT protein was eluted from the beads in multiple fractions with the same buffer containing increasing concentrations of imidazole from 10 to 200 mM. Pooled peak fractions were dialyzed against dialysis buffer 3 (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, 1 mM DTT) in the presence of protease inhibitor cocktail. The cleared dialysate was adjusted to 10% glycerol and stored at −80°C. Protein concentrations were estimated by comparison of Coomassie blue-stained band intensities with a bovine serum albumin standard.
In vitro translation and trans-complementation priming assay.
In vitro translations were performed in an RRL-coupled in vitro transcription and translation system (TNT T7 Quick Coupled Transcription/Translation System; Promega, Madison, Wis.) according to the manufacturer's recommendations. Usually translation reactions were set up in a total volume of 20 μl programmed with 0.4 μg of template DNA and incubated at 30° C for 2 h. When Dɛ was provided in trans, the in vitro-transcribed 76-nt Dɛ-RNA was added at the beginning of the translation reaction. Priming was performed by adding 5 μCi of [α-32P]dATP (3,000 Ci/mmol) in 20 μl of 2× priming buffer (41) followed by incubation for 1 h at 37°C. Reactions were stopped by addition of sodium dodecyl sulfate (SDS) sample buffer, and aliquots were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Labeled P protein was visualized with a phosphoimaging system (Fuji). For calculation of relative priming efficiencies the intensities of the 32P-labeled bands were measured with the MacBas software (Fuji) and normalized against the amount of translated protein which was quantified from [35S]Met-labeled in vitro translation reactions carried out in parallel.
Priming complex reconstitution from E. coli-expressed P protein domains.
In the priming complex reconstitution assay 0.5 μl of purified TP1-220 (0.5 mg/ml) was incubated with 1 μl of purified RT349-761 (0.1 mg/ml), 0.2 μl of in vitro-transcribed Dɛ-RNA (50 μM), and 5 μl of RRL (Promega) and adjusted to a final volume of 10 μl with H2O. In control reactions lacking TP or RT, a corresponding volume of 100 mM NaCl was added to keep the concentration of monovalent cations constant. In the RRL-minus control RRL was replaced with 5 μl of buffer (50 mM Tris HCl [pH 7.5], 100 mM KCl, 5 mM MgCl2). The translation-minus control contained 0.5 μl of cycloheximide (400 μg/ml). All samples were incubated for 1 h at 30 C to allow for priming complex formation. Subsequent priming assays were performed as described above for in vitro-translated P protein.
RESULTS
Separate DHBV TP and RT-RH domains in vitro translated in RRL can trans complement each other to form a functional priming complex.
An essential prerequisite for our approach was to first prove, in an established system, that the separate TP and RT-RH domains can indeed assemble into RNPs with authentic Dɛ-dependent priming activity. We therefore used, initially, in vitro translation in RRL to test for trans complementation. We started out with two T7 promoter-controlled constructs comprising aa 1 to 220 of P protein, i.e., the entire TP domain, and aa 209 to 786, i.e., the entire RT-RH domain preceded by the spacer. To address Dɛ dependence, each construct was generated in two versions, one with and one without a Dɛ element in the 3′ part of the RNA (TP1-220 and TP1-220ɛ+ and RT209-786 and RT209-786ɛ+, respectively [Fig. 2A]). Covalent labeling by [32P]deoxynucleoside monophospate of the TP protein during priming reactions with the corresponding [α-32P]deoxynucleoside triphosphate would indicate productive trans complementation, and specificity would be proven by its dependence on the presence of RT-RH and Dɛ.
FIG. 2.
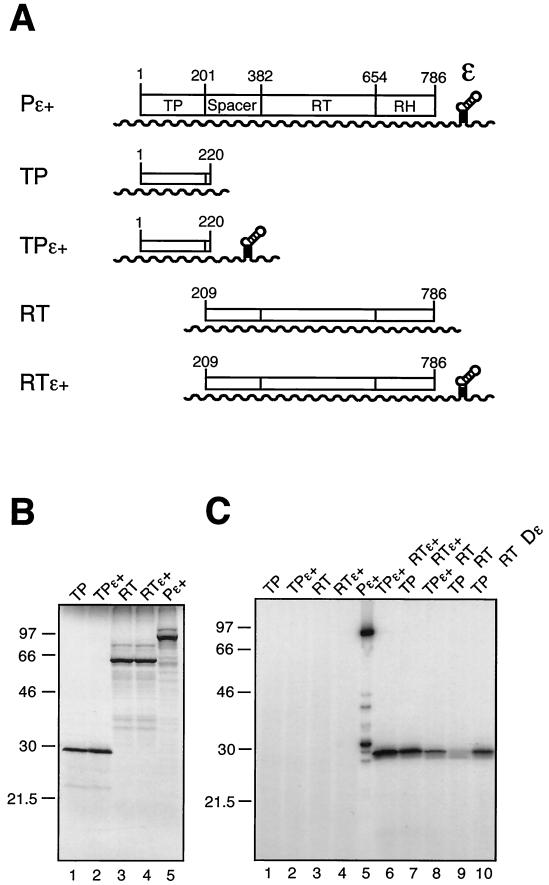
Functional trans-complementation of DHBV P protein domains coexpressed in RRL. (A) Schematic drawing of plasmid constructs and RNA transcripts used for in vitro translation. ORFs are given as boxes, and T7 promoter-driven transcripts are shown as wavy lines. Plasmid pT7AMVpol (Pɛ+) contains the complete 786-aa P protein ORF; the approximate borders of the four P protein domains are indicated by amino acid positions. Constructs Pɛ+, pT7-TP1-220ɛ+ (TPɛ+), and pT7-RT209-786ɛ+ (RTɛ+) carry a cis Dɛ element downstream of the coding region, whereas pT7-TP1-220 (TP) and pT7-RT209-786 (RT) are Dɛ deficient. (B) [35S]Met-labeled in vitro translation products of the constructs shown in panel A. Proteins were analyzed by SDS-PAGE and autoradiography. Molecular size markers are given in kilodaltons. (C) In vitro cotranslation and protein priming assay in the presence of [α-32P]dATP. Priming assays were performed as described in Materials and Methods. In the sample shown in lane 10 a 76-nt Dɛ RNA in vitro transcript was supplied in trans at a 1 μM concentration. An equal volume of each sample (except for Pɛ+ [20%]) was analyzed by SDS-PAGE and autoradiography. The band of about 32 kDa in the Pɛ+ sample corresponds most likely to a degradation product of the 32P-primed full-length P protein and did not occur as prominently in similar experiments.
For a semiquantitative estimate of the protein concentrations, the individual polypeptides were first separately translated in the presence of [35S]Met (Fig. 2B). Each of the TP and RT-RH constructs yielded in similar amounts a labeled protein of about 30 and 60 kDa, respectively. Next the two domains were cotranslated and subjected to priming assays with [α-32P]dATP. When no Dɛ was present in cis, the priming reactions were supplemented with in vitro-transcribed Dɛ RNA at a 1 μM concentration in trans (2). No signal was seen in the absence of RT-RH or TP; when Dɛ was omitted, a weak background signal was observed at the TP position (Fig. 2C). This may reflect a low level of Dɛ-independent priming activity as previously reported for the full-length protein (5). Importantly, however, much stronger signals were obtained when all three components were present. The most intense TP labeling, compared to that observed in a standard trans-priming assay with full-length P protein, occurred with Dɛ being present in cis on both RNAs or on the RT-RH RNA. However, specific labeling was also easily detected with Dɛ only on the TP mRNA or entirely provided as separate RNA in trans. These data suggested rather efficient RNP formation.
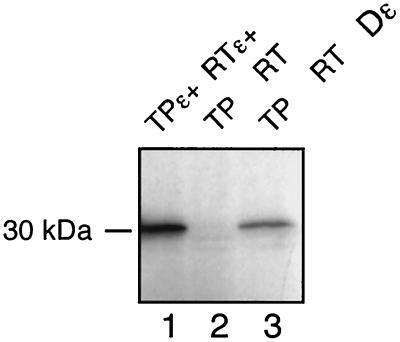
In a second set of experiments, TP and RT-RH were expressed in separate translation reactions, translation was stopped with cycloheximide, and then the lysates were mixed. To allow for complex formation, the mixtures were incubated for 1 h at 30°C and subsequently adjusted to priming conditions by the addition of priming buffer and [α-32P]dATP. The products were analyzed on the same gel as those presented in Fig. 2C. Because the signals were generally weaker, a fourfold longer exposure is shown (Fig. 3). Virtually no signals were observed in the absence of Dɛ, while 32P labeling was easily detected when Dɛ was present in either cis or trans. A semiquantitative evaluation by phosphorimager analysis indicated that posttranslational priming in cis and priming in trans were about 20 to 30% and about 10% as efficient as in the cotranslational setting, respectively. Hence, cotranslational events involving the P protein sequence as a whole are not essential for TP–RT-RH complex formation, although they may contribute to its efficiency. Also, it was still possible that some essential factors from the RRL were cotranslationally loaded onto the TP or the RT-RH domains or both (see below).
FIG. 3.
In vitro-translated DHBV TP and RT-RH domains interact posttranslationally in a Dɛ-dependent manner. The indicated constructs were separately expressed in RRL, and translation was stopped by adding cycloheximide to a final concentration of 20 μg/ml. The TP- and RT-containing lysates were mixed and incubated together with Dɛ RNA (only lane 3; final concentration, 1 μM) for 1 h at 30° C. The samples were subjected to priming assays in the presence of [α-32P]dATP and analyzed by SDS-PAGE on the same gel as those in Fig. 2C; the exposure shown here is four times longer. See the legend to Fig. 2A for details about constructs.
E. coli-derived TP can functionally interact with RT-RH expressed in RRL.
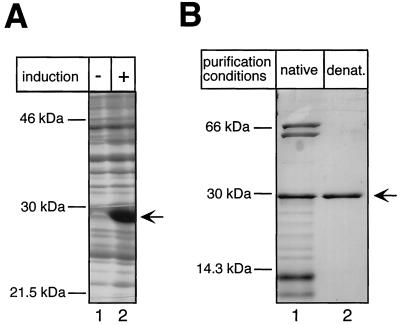
Next we tested whether the separate TP and RT-RH domains, as opposed to the full-length protein, could efficiently be expressed in E. coli. First we focused on TP. As shown in Fig. 4A, E. coli BL21-CodonPlus cells transformed with pET-TP1-220, upon induction, produced large amounts of the TP protein with an apparent molecular mass of 29 kDa. While a larger proportion of the protein was present as insoluble inclusion bodies, a substantial fraction remained soluble. Using the terminal His tag, the protein was purified by IMAC on Ni2+-agarose. This native preparation yielded about 100 μg of TP per 100 ml of bacterial culture. SDS-PAGE revealed, in addition to TP, three distinct bands of copurifying E. coli proteins with apparent molecular masses of about 70, 60, and 14 kDa (Fig. 4B). According to Western blots with specific antibodies (kindly provided by B. Bukau) the two larger proteins corresponded to the E. coli chaperones DnaK and GroEL (data not shown). Because these prokaryotic chaperones might have influenced the subsequent reconstitution experiments, we also purified the insoluble TP fraction by solubilization and purification in the presence of 8 M urea (Fig. 4B). As expected, no major contamination with E. coli proteins was detectable after this treatment. For renaturation, the purified protein was dialyzed against buffers containing 3 and 1 M urea, yielding about 2 mg of TP per 100 ml of bacterial culture. Further decreasing the urea concentration led to increased protein precipitation, but the 1 M urea solution turned out to be fully compatible with the subsequent functional assays unless it exceeded 10% of the total reaction volume.
FIG. 4.
Bacterial expression and purification of the DHBV TP domain. (A) Expression of TP1-220 in E. coli BL21-CodonPlus-RIL cells. E. coli cultures transformed with plasmid pET-TP1-220 were grown in the presence (+) or absence (−) of IPTG, and the pelleted cells were lysed by boiling in SDS sample buffer. Total lysates were analyzed by SDS-PAGE and Coomassie blue staining. (B) Purification of TP1-220. The soluble fraction of TP1-220 was subjected to IMAC and subsequent dialysis against a physiological buffer. The insoluble faction was solubilized in 8 M urea and purified by IMAC under denaturing (denat.) conditions. The protein was refolded by two-step dialysis against buffers containing 3 and 1 M urea, respectively. Aliquots of both purified fractions were resolved by SDS-PAGE and stained with Coomassie blue. Note the presence in the natively purified fraction (lane 1) of additional proteins of about 70 , 60, and <14 kDa. The former two correspond to E. coli DnaK and GroEL, as indicated by Western blotting with specific antibodies (not shown).
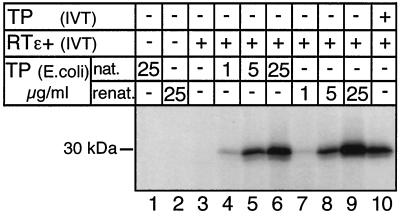
Various amounts of the native and of the denatured and renatured TP preparation, ranging from 10 to 250 ng per 10 μl of reaction mixture, were then added to RRL which had been programmed with pT7-RT209-786ɛ+ and subjected to priming assays. For a relative estimate of the fraction of active heterologously expressed TP molecules, an RRL cotranslation experiment with TP1-220 and RT209-786ɛ+ was run in parallel. As shown in Fig. 5, both TP preparations yielded specific signals in the presence of RT-RH and Dɛ; signal intensities increased proportionally to the dose of exogenously added TP. At the highest concentration TP labeling was about two- to threefold stronger than in the cotranslation experiment which, according to [35S]Met labeling, contained approximately 10 ng of TP. No significant difference was observed between TP prepared under native conditions and TP prepared under denaturing conditions. Hence, both preparations had at least 1/10 the specific activity of that of the in vitro-translated protein. The result also suggested that the copurified E. coli proteins in the native preparation had no influence on complex formation and/or priming activity. Hence, pure TP1-220 produced in E. coli is able to reconstitute the hepadnaviral reverse transcription initiation complex and can act as a protein primer. This activity does not require cotranslational events.
FIG. 5.
In vitro reconstitution of functional priming complexes from in vitro-translated RT-RH and E. coli-expressed TP. RT209-786 (RTɛ+) was expressed in RRL from construct pT7-RT209-786ɛ+ in the presence of the indicated amounts of E. coli-derived TP1-220 (TP) purified under native conditions (nat.) (lanes 4 to 6), or purified under denaturing conditions followed by renaturation (renat.) (lanes 7 to 9), or without TP1-220 (lane 3). As negative controls, the TP fractions were incubated in RRL in the absence of RT209-786 (lanes 1 and 2). For comparison an in vitro cotranslation of TP1-220 and RT209-786 was performed (lane 10). After in vitro translation the samples were assayed for priming with [α-32P]dATP. 32P-labeled TP was detected by SDS-PAGE and autoradiography.
A short C-terminal sequence in the RH domain interferes with expression of RT-RH constructs in E. coli.
Next we focused on expressing the RT-RH domain in E. coli. The initially used construct pET-RT209-786 did not yield detectable amounts of the 71-kDa full-length protein (data not shown). We therefore constructed a series of terminal deletion variants to define N- and C-terminal borders which would allow a higher level of production in E. coli while preserving P protein enzymatic activity.
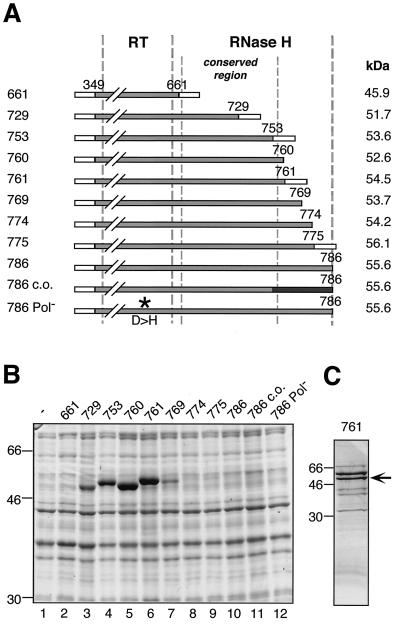
First, the N-terminal spacer region was deleted (construct RT349-786). When cotranslated with TP1-220 in RRL this protein produced a priming-competent complex, demonstrating that the spacer region is required neither for complex formation nor for priming (data not shown). However, this C-terminally full-length protein of nominally 56 kDa was poorly expressed in E. coli (Fig. 6B). Therefore, a series of C-terminal deletion variants was constructed (Fig. 6A), most of them still containing the conserved part of the RH domain. SDS-PAGE of the corresponding bacterial lysates followed by Coomassie blue staining revealed massive variations in the expression levels (Fig. 6B). Whereas deleting up to 12 aa (RT349-774, RT349-775) had no significant effect, protein RT349-769 with a 17-aa deletion gave a clearly visible band. Deleting 25 to 33 aa (RT349-761, RT349-760, RT349-753) strongly enhanced expression. Even larger deletions (RT349-729, RT349-661), by contrast, resulted again in reduced expression levels. The presence in some of the constructs of short vector-derived C-terminal sequences had no significant influence. These data indicated that aa 762 to 786 strongly contributed to the low yield of RT349-786.
FIG. 6.
Bacterial expression and purification of the RT-RH domain. (A) Schematic drawing of C-terminally truncated RT-RH constructs analyzed for protein expression in E. coli. Each construct starts with 50 vector-derived aa (open boxes at left) followed by DHBV P sequence from aa 349 to the indicated C-terminal position. Some constructs contain short vector-encoded C-terminal tags (open boxes at right). Numbers on the left represent abbreviations of the construct names; e.g., “661” refers to plasmid pET-RT349-661. Construct pET-RT349-786 c.o. is codon optimized for E. coli in the C-terminal region (black box). Construct pET-RT349-786Pol− contains a D-to-H amino acid substitution in the YMDD motif of the polymerase active site (asterisk). Borders of the RT and RH domains are indicated by thick vertical broken lines. The conserved region of the RH domain from aa 660 to 759 (9) is labeled by thin broken lines. (B) Expression of RT-RH constructs in E. coli strain BL21-CodonPlus-RIL. Total cell lysates of induced bacterial cultures were analyzed by SDS-PAGE and Coomassie blue staining; molecular mass markers are given in kilodaltons. (C) Purification of bacterially expressed RT349-761. The soluble fraction of RT349-761 was purified by IMAC and subsequent dialysis against a physiological buffer. The purified protein fraction was analyzed by SDS-PAGE and Coomassie blue staining. The nominally 54-kDa RT349-761 protein is marked by an arrow. Additional bands of 60 kDa (strong) and 70 kDa (weak) were identified by immunoblotting as E. coli GroEL and DnaK.
Two possible explanations were explicitly tested. First, we introduced silent mutations to optimize codons with respect to codon usage in E. coli (RT349-786co) because this region contains several codons that are very infrequently used in E. coli (15). Second, the polymerase active site was mutated to abolish potential RT activity (RT349-786Pol-). However, neither mutation significantly improved the protein yields (Fig. 6B), suggesting that the poor expression of RT-RH proteins extending beyond amino acid 762 was caused neither by scarcity of certain tRNAs nor by polymerase activity.
Because RT349-761 was the least truncated of the highly expressed constructs it was chosen for purification and subsequent priming complex reconstitution experiments. After lysis of the bacteria, a larger fraction of the nominally 55-kDa RT349-761 protein was present in inclusion bodies. However, significant amounts, i.e., approximately 100 μg per 100 ml of culture, remained soluble and were purified via the terminal His tags, by IMAC under native conditions (Fig. 6C). As with TP1-220, several distinct copurifying E. coli proteins were present, two of which, according to Western blotting, corresponded again to DnaK and GroEL (data not shown).
E. coli-derived TP and RT-RH can assemble into a functional initiation complex in RRL.
To test for functional RNP formation from both TP and RT-RH produced in E. coli, a four-component reconstitution system was set up. Purified TP1-220 and RT349-761 were incubated with in vitro-transcribed Dɛ-RNA and RRL. In this setting, RRL served only as a source for the host factors required for RNP formation. Initiation complex assembly was monitored by [α-32P]dATP priming assays (Fig. 7). Indeed, a strong labeling of the 30-kDa TP1-220 was observed (Fig. 7, lane 1) when all three other components were also present. Leaving out either TP, RT, Dɛ, or RRL abolished the signal completely (Fig. 7, lanes 2 to 5), strongly suggesting that the reconstituted complex is capable of authentic priming. Likewise, no priming activity was observed when the reaction components were immediately mixed in priming buffer without prior incubation (Fig. 7, lane 7). This resembles the previous observation that RNP complex formation of in vitro-translated full-length P protein with Dɛ-RNA requires the buffer conditions of RRL and does not occur in priming buffer (J. Beck and M. Nassal, unpublished data.). Though unlikely, it was formally possible that some TP- and/or RT-RH-encoding RNA was copurified with the bacterial protein preparations and then was in vitro translated in the RRL, generating the actual protein substrates for trans complementation. This possibility was explicitly excluded by blocking translation in RRL by cycloheximide, with no negative effect on priming (Fig. 7, lane 6). Hence, bacterially expressed TP and RT-RH can be reconstituted in RRL into a complex with genuine Dɛ-dependent priming activity.
FIG. 7.
In vitro reconstitution of functional priming complexes from E. coli-derived TP and RT-RH domains in RRL. TP, RT, RRL, and Dɛ RNA were mixed, incubated for 1 h at 30°C to allow RNP formation, and subsequently assayed for protein priming activity in the presence of [α-32P]dATP (for details, see Materials and Methods). The reaction products were resolved by SDS-PAGE, and 32P-labeled TP protein was detected by autoradiography. Abbreviations: TP, bacterially expressed TP1-220 (25 ng/μl); RT, bacterially expressed RT349-761 (10 ng/μl); Dɛ, in vitro-transcribed Dɛ RNA (1 μM); RRL, rabbit reticulocyte lysate; +CHX, sample supplemented with cycloheximide (20 μg/ml) at the beginning of the incubation period (lane 6); -binding, the indicated components were mixed and subjected to priming conditions without prior incubation at 30°C (lane 7). For comparison, a priming assay with in vitro-cotranslated TP1-220 and RT349-761 proteins from Dɛ-deficient constructs, supplemented with 1 μM Dɛ RNA in trans, was performed in parallel (lane 8).
An estimate for the relative priming efficiencies of the bacterially produced proteins was obtained by comparison with the TP signal generated in a cotranslational trans-priming assay with in vitro-translated TP and RT-RH (Fig. 7, lane 8). According to results of [35S]Met labeling, this reaction contained about 10 and 15 ng of TP and RT, respectively, and it produced a signal about 30% weaker than that with 250 and 100 ng of E. coli-derived TP and RT-RH, respectively (Fig. 7, lane 1); hence, the specific priming activity was roughly sevenfold higher. This ratio is, however, very similar to that previously seen for cotranslational versus posttranslational trans priming with the separately in vitro-translated domains (see above), indicating a similar degree of priming competence for the E. coli-expressed proteins.
DISCUSSION
Reverse transcription of the hepadnaviral pgRNA follows an intricate mechanism that, in several aspects, differs fundamentally from retroviral replication (22, 23). Two hallmarks are site-specific initiation without a nucleic acid primer and the dependence of RT activity on cellular factors. Proper synthesis of the first few nucleotides of minus-strand DNA requires that the Tyr residue in TP and the active site of RT be exactly positioned over the ɛ RNA template. Preparing ɛ for copying involves major structural rearrangements in the upper part of the stem-loop (3), and interactions of the protein with the stem underlying the initiation site might be involved in arresting synthesis after 3 or 4 nt (30). Also, P protein displays distinct differences in protease sensitivity during different stages of initiation (35, 36). These data imply dynamic and ordered changes in the RNA and the protein which, conceivably, are mediated by cellular factors such as the chaperones (12). However, a true mechanistic understanding will depend on identifying all elements comprising a functional complex and, eventually, on reconstituting this complex from purified components. All necessary factors for DHBV, but not HBV, P protein activation are obviously contained in RRL. Our data demonstrate that the limited translational capacity of RRL can be overcome by provision of separate TP and RT-RH domains from an exogenous source. The cell-free reconstitution system reported here should hence be suitable for further elucidating the hepadnaviral replication mechanism.
E. coli expression of DHBV P protein domains.
Whereas various RTs have successfully been expressed in E. coli, most such experiments with hepadnaviral P proteins have failed. Possible explanations include instability of the mRNA or the protein, inefficient translation due to suboptimal codon usage, or toxicity caused by enzymatic activities. The likelihood for these potentially deleterious factors generally increases with protein size. We therefore reasoned that breaking up the 90-kDa P protein into separate domains might allow for more efficient expression. Indeed, the 220-aa TP was highly expressed in BL21-CodonPlus cells, which carry an extra plasmid encoding rare E. coli tRNAs. Since expression was very poor in the same strain lacking this plasmid, codon usage was indeed a crucial factor even with the relatively small TP domain. As with many overexpressed proteins, a major proportion of TP was present in inclusion bodies. However, substantial amounts remained soluble. The corresponding IMAC fractions contained, in addition, a few distinct E. coli proteins, most prominently the E. coli chaperones DnaK and GroEL, homologues of the eukaryotic Hsp70 and Hsp60, respectively. Whether this reflects an adventitious or a physiological role is not clear at present (see below). To exclude any influence of the bacterial chaperones on the subsequent reconstitution experiments we also purified the insoluble fraction of TP under denaturing conditions. After renaturation we obtained milligram amounts of essentially pure TP which, in the subsequent priming assays, was as active as the native preparation. Hence, the E. coli chaperones were not important for activity. With further optimization, this procedure may allow the preparation of TP in amounts sufficient for structural studies.
Efficient expression of RT-RH required sequence modifications, in particular in the C-terminal region of the RH domain. Deleting the N-proximal spacer region did not affect activity, in accord with previous studies (12, 19, 20, 26), but also did not increase expression levels. By contrast, short C-terminal deletions led to a drastic increase in protein yield. Codon usage in general was again important because most RT-RH variants remained nearly undetectable when expressed in cells lacking the tRNA plasmid. However, optimizing codon usage for E. coli in the very C-terminal RH part did not improve expression of the full-length protein, and neither did knocking out the RT active site. Also, further truncations upstream of aa 729 led to decreased protein levels. This argues against the presence, in the wild-type mRNA, of a specific instability determinant that leads to rapid RNA degradation. Possibly, therefore, the relatively large amounts of RT349-761 and similarly truncated variants are related to protein folding.
The amounts of protein detectable after induction reflect the rates of synthesis versus degradation. Two opposite views, which may only be distinguished by experimentally determining these rates for the different variants, are that the very C-terminal region of RH is required for proper folding; its deletion may then lead to rapid aggregation and accumulation of the truncated, nonnatively folded proteins in protease-resistant inclusion bodies. Alternatively, the C terminus might interfere with folding in E. coli, e.g., because its incorporation into the entire structure requires interacting factors present only in eukaryotic cells; this would result in more rapid degradation of P proteins with a complete C terminus. More practically important, however, is that the well-expressed RT349-761 appears to be a suitable surrogate for the full-length protein. First and foremost, the soluble fraction of the protein could be assembled into complexes competent for Dɛ-dependent priming (see below). Second, the primary sequence of RT–RH349-761 contains all RH residues that are highly conserved among hepadnaviral P proteins from different species (Fig. 8), including those important for RH activity in DHBV P protein (8–10) as well as in the RH proteins from human immunodeficiency virus type 1 (HIV-1) and E. coli (11, 43), arguing again that RT349-761 comprises all the sequence required for folding and nucleolytic activity of RH enzymes (Fig. 8). Finally, a glutathione S-transferase fusion of an even further truncated DHBV P protein variant ending at aa 734 exhibited priming activity (12).
FIG. 8.
Alignment of RNase H domains of DHBV, HBV, and HIV-1 RTs. The alignment is based on a previously published version (9). Amino acid residues conserved between DHBV and HBV are boxed. Residues conserved between hepadnaviral and retroviral RH domains are shaded. Invariant Asp and Glu residues implicated in catalysis are marked with asterisks. Numbers above and below the sequence correspond to amino acid positions of DHBV P and HIV-1 RT, respectively. The horizontal bars and boxes below the sequence indicate β-sheets and α-helices of the RH domain in HIV-1 RT as determined by X-ray crystallography (11). Helix αE is unlikely to exist in hepadnaviral RHs due to multiple Pro residues in this region. Note that DHBV residue 759 represents the C-terminal border of the homologous region.
As for TP, the E. coli chaperones DnaK and GroEL copurified with RT349-761 during IMAC under native conditions. This may suggest a specific (12) but not necessarily physiological association. Chaperones interact with a whole variety of hydrophobic protein regions if they are exposed to the solvent, for instance upon heat shock or some other stress, including overexpression (7, 21); notably, potential DnaK binding motifs occur statistically every 36 residues (28). Hence, it is difficult to distinguish between an association caused by nonnative folding and one that reflects the authentic exposure of such regions on a natively folded chaperone target protein. We also noted that, even in the absence of any P protein peptide, a well-detectable fraction of DnaK and GroEL eluted at higher imidazole concentrations from the Ni2+ agarose beads than the bulk of E. coli proteins. Because chaperone association is per se affected by salt concentrations (12, 17), thoroughly addressing this question will require the use of strictly controlled variations in buffer conditions. At any rate, the E. coli chaperones appear to enhance the solubility of RT-RH, and probably TP; their coexpression may hence be a means to further increase the fraction of soluble TP and RT-RH. Even now, however, up to 100 μg per 100 ml of E. coli culture of TP and RT-RH was obtained under native conditions, exceeding by several logs the standard yields of in vitro translation.
An attempt to refold the insoluble fraction of RT-RH under the same conditions as used for TP did not yield priming active protein. In view of its substantially larger size and two-domain structure, this may not be too surprising. Formally, therefore, some effect on the priming reaction of the E. coli chaperones present in the native RT-RH preparation cannot be ruled out. However, we have very recently generated an RT-RH fusion protein containing a heterologous domain that is more soluble and can be prepared with only little contamination by E. coli proteins. Preliminary experiments show that this chimeric protein is priming competent, arguing against such a role for the bacterial chaperones.
Efficient trans complementation between separate DHBV P protein TP and RT-RH domains.
The principal possibility of trans complementation between hepadnaviral TP and RT-RH domains has been demonstrated using insect cell-derived HBV P protein domains (18, 20). Even though, in this setting, initiation of DNA synthesis was ɛ dependent, activity was restricted to lysates from coexpressing cells but was undetectable upon mixing separate lysates or isolated TP and RT-RH. This suggested an important role for cotranslational events in initiation complex formation.
Our initial experiments with both TP and RT-RH expressed in RRL showed that the separate DHBV P protein domains can likewise assemble into complexes with Dɛ-dependent priming activity. With the cis constructs, TP labeling was stronger with Dɛ on the RT-RH RNA than on the TP RNA, suggesting that RT-RH maintains the cis preference of the full-length P protein (1). However, 1 μM Dɛ in trans also mediated more-efficient TP labeling than the TP cis construct, probably a reflection of its higher concentration. Semiquantitative estimates of the ratios between [35S]Met and [α-32P]deoxynucleoside monophospate labeling suggest that the priming efficiency of the separate domains approaches about 20% of that of full-length in vitro-translated DHBV P protein. The low concentrations of TP and RT-RH in the RRL system calculated from [35S]Met labeling (about 10 to 30 nM) imply high-affinity interactions within the multicomponent RNP complex that obviate the need for a covalent link, by the spacer region, of TP and RT-RH. Assuming simple two-component equilibrium conditions, a dissociation constant of 10 nM would be required to have 50% of both domains bound to each other; much lower affinities would not be compatible with the observed trans-complementation efficiency. The order of events during complex formation is presently not clear. TP and RT-RH might interact directly, or via the Dɛ RNA, or via the cellular factors associated with either of the domains, and these are not mutually exclusive. We also note that it is formally possible that TP associates only transiently with the other components of the complex, becomes labeled, and is subsequently released. However, that priming-competent RNPs were formed from bacterially expressed TP and/or RT-RH domains is clear evidence that, in an appropriate environment, both preformed proteins are able to adopt their native structures in a posttranslational fashion, in accord with a similar conclusion recently made for single-chain glutathione S-transferase-fused truncated DHBV P proteins (12).
Certainly, efforts to further improve the yields of soluble TP and RT-RH are justified, in particular for direct structural studies. However, we envisage several applications for which our system may be useful already in its present state. Apart from analytically identifying the constituent components of the active complex it should allow the complete in vitro reconstitution of an initiation complex using only purified components. A preliminary experiment using bacterially expressed TP and RT349-761 together with those proteins known to be sufficient for ligand-binding activation of steroid hormone receptors, i.e., Hsp40, Hsp70, Hsp90, Hop, and p23, plus Dɛ in trans (17) did not give any priming signals. While this negative outcome could have many reasons, a similar observation was made by others with a truncated single-chain P (J. Hu, personal communication). Thus, it is likely that there are substantial differences between hormone receptor and P protein activation, e.g., that a different set of helper proteins is required. The missing factor(s) is obviously present in RRL and therefore might be identified by supplementing the above reconstitution system with fractionated RRL. Useful information may also be obtained by using immobilized TP and RT-RH domains as an affinity matrix to define the factors in RRL that bind to each of the domains. Eventually, either of these approaches should be helpful for elucidating why HBV P protein shows no activity in RRL and thus make this medically important viral enzyme amenable to structure-function studies aimed at identifying new inhibitors.
ACKNOWLEDGMENTS
We thank Bernd Bukau for providing DnaK and GroEL antibodies.
This work was supported by the Center for Clinical Research 1 (ZKF1) of the University Hospital Freiburg and the Deutsche Forschungsgemeinschaft (DFG Na154/6-1).
REFERENCES
- 1.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck J, Bartos H, Nassal M. Experimental confirmation of a hepatitis B virus (HBV) epsilon like bulge and loop structure in avian HBV RNA encapsidation signals. Virology. 1997;227:500–504. doi: 10.1006/viro.1996.8329. [DOI] [PubMed] [Google Scholar]
- 3.Beck J, Nassal M. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol Cell Biol. 1998;18:6265–6272. doi: 10.1128/mcb.18.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck J, Nassal M. A sensitive procedure for mapping the boundaries of RNA elements binding in vitro translated proteins defines a minimal hepatitis B virus encapsidation signal. Nucleic Acids Res. 1996;24:4364–4366. doi: 10.1093/nar/24.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck J, Nassal M. Sequence- and structure-specific determinants in the interaction between the RNA encapsidation signal and reverse transcriptase of avian hepatitis B viruses. J Virol. 1997;71:4971–4980. doi: 10.1128/jvi.71.7.4971-4980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumberg B S. Hepatitis B virus, the vaccine, and the control of primary cancer of the liver. Proc Natl Acad Sci USA. 1997;94:7121–7125. doi: 10.1073/pnas.94.14.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 8.Chang L J, Hirsch R C, Ganem D, Varmus H E. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J Virol. 1990;64:5553–5558. doi: 10.1128/jvi.64.11.5553-5558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Marion P L. Amino acids essential for RNase H activity of hepadnaviruses are also required for efficient elongation of minus-strand viral DNA. J Virol. 1996;70:6151–6156. doi: 10.1128/jvi.70.9.6151-6156.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Robinson W S, Marion P L. Selected mutations of the duck hepatitis B virus P gene RNase H domain affect both RNA packaging and priming of minus-strand DNA synthesis. J Virol. 1994;68:5232–5238. doi: 10.1128/jvi.68.8.5232-5238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies J F, Hostomska Z, Hostomsky Z, Jordan S R, Matthews D A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 12.Hu J, Anselmo D. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J Virol. 2000;74:11447–11455. doi: 10.1128/jvi.74.24.11447-11455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci USA. 1996;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J, Toft D O, Seeger C. Hepadnavirus assembly and reverse transcription require a multi component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane J F. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 16.Knaus T, Nassal M. The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem loop structure that is critical for its function. Nucleic Acids Res. 1993;21:3967–3975. doi: 10.1093/nar/21.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosano H, Stensgard B, Charlesworth M C, McMahon N, Toft D. The assembly of progesterone receptor hsp90 complexes using purified proteins. J Biol Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- 18.Lanford R E, Kim Y H, Lee H, Notvall L, Beames B. Mapping of the hepatitis B virus reverse transcriptase TP and RT domains by transcomplementation for nucleotide priming and by protein-protein interaction. J Virol. 1999;73:1885–1893. doi: 10.1128/jvi.73.3.1885-1893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanford R E, Notvall L, Beames B. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J Virol. 1995;69:4431–4439. doi: 10.1128/jvi.69.7.4431-4439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanford R E, Notvall L, Lee H, Beames B. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J Virol. 1997;71:2996–3004. doi: 10.1128/jvi.71.4.2996-3004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer M P, Rüdiger S, Bukau B. Molecular basis for interactions of the DnaK chaperone with substrates. Biol Chem. 2000;381:877–885. doi: 10.1515/BC.2000.109. [DOI] [PubMed] [Google Scholar]
- 22.Nassal M. Hepatitis B virus replication: novel roles for virus-host interactions. Intervirology. 1999;42:100–116. doi: 10.1159/000024970. [DOI] [PubMed] [Google Scholar]
- 23.Nassal M. Macromolecular interactions in hepatitis B virus replication and particle formation. In: Cann A J, editor. Frontiers in molecular biology: DNA virus replication. Vol. 26. Oxford, United Kingdom: Oxford University Press; 2000. pp. 1–40. [Google Scholar]
- 24.Nassal M, Rieger A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J Virol. 1996;70:2764–2773. doi: 10.1128/jvi.70.5.2764-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollack J R, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieger A, Nassal M. Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J Virol. 1996;70:585–589. doi: 10.1128/jvi.70.1.585-589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaaf S G. Dissertation. Heidelberg, Germany: University of Heidelberg; 1999. [Google Scholar]
- 30.Schaaf S G, Beck J, Nassal M. A small 2′ OH and base dependent recognition element downstream of the initiation site in the RNA encapsidation signal is essential for hepatitis B virus replication initiation. J Biol Chem. 1999;274:37787–37794. doi: 10.1074/jbc.274.53.37787. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher R J, Hurst R, Sullivan W P, McMahon N J, Toft D O, Matts R L. ATP-dependent chaperoning activity of reticulocyte lysate. J Biol Chem. 1994;269:9493–9499. [PubMed] [Google Scholar]
- 32.Seeger C, Leber E H, Wiens L K, Hu J. Mutagenesis of a hepatitis B virus reverse transcriptase yields temperature sensitive virus. Virology. 1996;222:430–439. doi: 10.1006/viro.1996.0440. [DOI] [PubMed] [Google Scholar]
- 33.Seeger C, Mason W S. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith D F. Sequence motifs shared between chaperone components participating in the assembly of progesterone receptor complexes. Biol Chem. 1998;379:283–288. doi: 10.1515/bchm.1998.379.3.283. [DOI] [PubMed] [Google Scholar]
- 35.Tavis J E, Ganem D. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavis J E, Massey B, Gong Y. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, ɛ. J Virol. 1998;72:5789–5796. doi: 10.1128/jvi.72.7.5789-5796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavis J E, Perri S, Ganem D. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G H, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 40.Wang G H, Zoulim F, Leber E H, Kitson J, Seeger C. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J Virol. 1994;68:8437–8442. doi: 10.1128/jvi.68.12.8437-8442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber M, Bronsema V, Bartos H, Bosserhoff A, Bartenschlager R, Schaller H. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J Virol. 1994;68:2994–2999. doi: 10.1128/jvi.68.5.2994-2999.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W, Hendrickson W A, Crouch R J, Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 44.Zoulim F, Seeger C. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J Virol. 1994;68:6–13. doi: 10.1128/jvi.68.1.6-13.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]