Abstract
Introduction. The identification of mitotic figures is essential for the diagnosis, grading, and classification of various different tumors. Despite its importance, there is a paucity of literature reporting the consistency in interpreting mitotic figures among pathologists. This study leverages publicly accessible datasets and social media to recruit an international group of pathologists to score an image database of more than 1000 mitotic figures collectively. Materials and Methods. Pathologists were instructed to randomly select a digital slide from The Cancer Genome Atlas (TCGA) datasets and annotate 10-20 mitotic figures within a 2 mm2 area. The first 1010 submitted mitotic figures were used to create an image dataset, with each figure transformed into an individual tile at 40x magnification. The dataset was redistributed to all pathologists to review and determine whether each tile constituted a mitotic figure. Results. Overall pathologists had a median agreement rate of 80.2% (range 42.0%-95.7%). Individual mitotic figure tiles had a median agreement rate of 87.1% and a fair inter-rater agreement across all tiles (kappa = 0.284). Mitotic figures in prometaphase had lower percentage agreement rates compared to other phases of mitosis. Conclusion. This dataset stands as the largest international consensus study for mitotic figures to date and can be utilized as a training set for future studies. The agreement range reflects a spectrum of criteria that pathologists use to decide what constitutes a mitotic figure, which may have potential implications in tumor diagnostics and clinical management.
Keywords: mitotic figures, mitotic figure mimics, digital pathology, international collaboration, social media
Introduction
The identification and enumeration of mitotic figures are essential for diagnosing, classifying, and grading of various tumor types. Mitotic figures are routinely assessed in tumor types that include neuroendocrine tumors,1,2 breast carcinomas, 3 soft tissue sarcoma, 4 gastrointestinal stromal tumors (GIST),5,6 cutaneous melanoma,7,8 and thyroid tumors, 9 among others. Visual quantification of mitotic figures on hematoxylin and eosin (H&E) stained histologic sections serves as a surrogate measure for the proliferative capacity of tumors and has been shown as an independent prognostic factor of outcome in many cancer types.10–12 Mitotic counts, traditionally quantified per high-powered microscope field, have evolved to standardized area measurements in the most current classification systems in order to address the variability with different microscope lenses. 13 Therefore, mitotic counts are typically defined as the number of mitotic figures in a given unit area and are expressed in discrete ranges that are used to guide clinical decision-making and risk stratification of patients.
The identification and quantification of mitotic figures can be both subjective and time-consuming, with poor reproducibility.2,14,15 Previous studies have shown moderate agreement rates for mitotic counts, even within a standard area.16–18 While pathologists are not required to characterize mitotic figures into various phases on histologic tumor slides in routine practice, the morphologies of mitotic figures may vary widely, with distinct appearances at each phase. A deeper understanding of these variations could lead to more accurate mitotic counts. Donovan et al defined morphologies of various phases of prophase, metaphase, anaphase, and telophase that occur during mitosis. Additionally, the authors described atypical mitotic figures, and potential mitotic figure mimics. 19 Examples of mitotic figure mimics include apoptotic figures, hyperchromatic or deformed nuclei, karyorrhectic debris, inflammatory cells, and tissue artifacts.19,20 The process of distinguishing mitotic figures and atypical mitotic figures from mitotic figure mimics is challenging and is often subject to inter-observer variability.19,21 Inaccuracies in mitotic figure interpretation and quantification of mitotic counts may lead to misdiagnoses and ultimately result in inaccurate classifications of tumors in clinical practice. 14 Thus, to produce consistent and reproducible mitotic counts, it is critical for pathologists to distinguish mitotic figures and atypical mitotic figures from mitotic figure mimics, and to recognize common pitfalls encountered in making this distinction.
There is increasing interest in developing AI-assisted computational pathology tools that can automate subjective and time-consuming tasks in current pathology workflows. 22 One such potential tool would identify and count mitotic figures across a whole-slide image (WSI), resulting in a holistic measure of mitotic count, in contrast to the current method of generating a mitotic count from a limited set of areas selected by the pathologist. Given the prevalence of mitotic figure datasets from grand challenges, 23 automated systems are well positioned to solve the challenges of counting mitotic figures. However, their performance has been shown to decline in varied clinical settings due to the challenges in differentiating between mitotic figures and their mimics, which often have differing sizes, shapes, and structural elements. 24 One limitation in developing robust machine learning applications that can accurately enumerate mitotic figures across a WSI is the lack of comprehensive diverse properly labeled datasets scored by a diverse panel of expert pathologists. 25
There exists a paucity of data documenting the consistency in interpreting a large dataset of mitotic figures among pathologists from diverse backgrounds and clinical practice settings. With the availability of publicly accessible datasets and the increasing popularity of professional social media use among pathologists, we identified a unique opportunity to foster research collaborations extending beyond geographical and institutional barriers in order to generate novel insights and robust datasets. These datasets could then support the training of robust artificial intelligence-based tools within pathology workflows promising to automate tedious and error-prone tasks that can ultimately impact patient care. In this study, we leveraged publicly accessible datasets and social media to recruit an international group of 85 pathologists to collectively generate and score an image database of 1010 mitotic figures.
Design
Study Cohort
The study was announced (July 13, 2022) on Twitter (currently known as X) to recruit practicing pathologists across the world. Participants were screened based on their online professional profile and confirmed by their institutional affiliation to ensure they were at the level of a staff pathologist. No other exclusion criteria were utilized and pathologists from around the world were included.
Collection of 1000 Mitotic Figures
Each pathologist who completed the survey was sent a comprehensive protocol detailing the instructions for submitting their mitotic figures. Using the online dataset from The Cancer Genome Atlas (TCGA), each participant randomly selected a diagnostic slide from the dataset of their choosing. The selection criteria was open to all available datasets on the TCGA and not limited to specific tumors where mitotic counts are routinely assessed in clinical practice. Using QuPath, an open-source bioimage analysis tool, the pathologist annotated a 2 mm2 area of tumor with the highest proliferative capacity on the tumor. 26 Within this defined area, the pathologist annotated 10-20 mitotic figures that would have been included in a mitotic count to be used for patient care in their clinical practice. The first 1000 submitted mitotic figures (with 10 extra tiles included for redundancy) were used to create an image dataset, with each figure transformed into an individual tile at 40x magnification.
Scoring of Mitotic Figures
The dataset was split into four equal parts based on the order in which the tiles were submitted and uploaded into four separate Google Forms. For each tile, participants were asked whether the tile constituted a mitotic figure (Yes/No). At the end of each of the four surveys, participants were asked to provide qualitative feedback on tiles with uncertainty. The dataset was split into parts to create a workflow that saved the rater's progress and did not need to be completed in a single setting.
Determination of Mitotic Phase and Mitotic Figure Mimics
Each submitted mitotic figure was classified as the most probable phase of mitosis by a pathologist (MJC). Identification of cells in prophase is challenging with light microscopes, given the subtle morphologic changes in this phase. Cells in mitosis that appear as dark chromosome aggregates are most likely in the final stages of prophase, commonly referred to as prometaphase. 19 The term prometaphase will be used going forward to denote mitotic figures in the later phase of prophase that can be reliably identified on histologic sections. During the review by MJC, a potential mitotic figure mimic was assigned descriptively to a subset of the tiles. Given the lack of a gold standard for what constitutes a mitotic figure from a mimic, it is not possible to definitively enumerate the exact number of tiles in these categories. This descriptive analysis provides insight into the distribution of potential mimics in each phase of the cell cycle.
Statistics
Descriptive statistics were calculated at both the pathologist and tile level. At the pathologist level, an agreement rate was calculated by the proportion of tiles classified as positive over the total number of tiles rated. At the tile level, an agreement rate was calculated by the proportion of pathologists that classified the tile as a mitotic figure over the total number of ratings. Fleiss' kappa (κ) was used as a statistical measure of inter-rater agreement, where kappa values can be interpreted as poor agreement (<0), slight agreement (0-0.2), fair agreement (0.2-0.4), moderate agreement (0.4-0.6), good agreement (0.6-0.8), and almost perfect agreement (>0.8). 27 Further, we utilized a one-way ANOVA and a subsequent Tukey's post hoc test to evaluate pathologist agreement rates based on the phases of mitosis. Additional unsupervised hierarchical clustering analysis was performed for both mitotic figures and pathologists, leading to the generation of corresponding heatmap denograms.
Results
Study Description
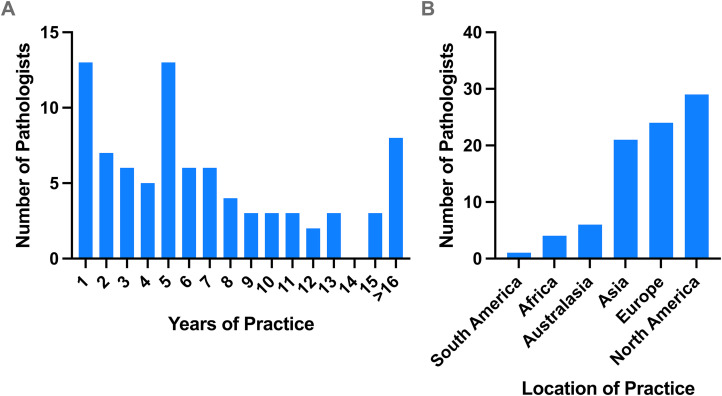
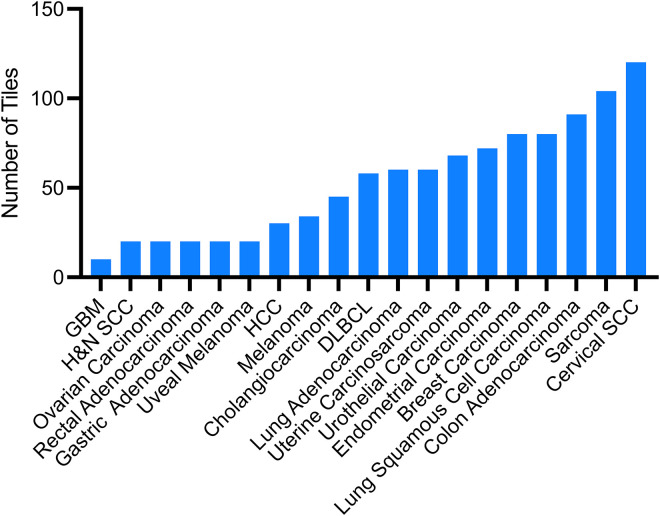
A group of 55 international staff pathologists from around the globe contributed to the mitotic figures dataset consisting of 1010 tiles, and 85 international staff pathologists scored the dataset. The group had varying years of practice and were from 24 countries (Figure 1). The dataset spans 19 distinct disease sites from the TCGA open-source database, with the largest contributors being cervical squamous carcinoma (120 tiles), sarcoma (104 tiles), and colorectal adenocarcinoma (91 tiles), followed by lung squamous cell carcinoma and breast carcinoma (both 80 tiles each) (Figure 2). The study spanned a total of 57 days from conceptualization and mitotic figure collection to scoring of the dataset.
Figure 1.
Demographics of pathologists scoring the mitotic figures dataset. Years of practice among the staff pathologists (A) and location of practice categorized by continent (B).
Figure 2.
Distribution of tumor types used in the 1010 mitotic figures dataset. Abbreviations: GBM, glioblastoma multiforme; H&N SCC, head and neck squamous cell carcinoma; HCC, hepatocellular carcinoma; DLBC, diffuse large B-cell lymphoma; Cervical SCC, cervical squamous cell carcinoma.
Assessment of Interobserver Variability
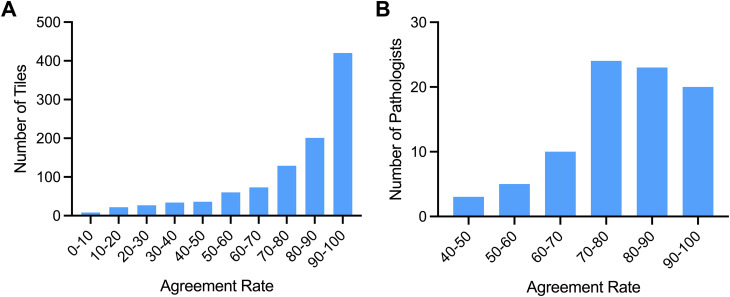
At the pathologist level, there was a median agreement rate of 80.2% of the mitotic figure tiles (mean 78.5 ± SD 12.4), with an agreement range of 42.0% to 95.7% (Figure 3A). At the tile level, there was a median agreement rate of 87.0% (mean 78.5 ± SD 22.2) with an agreement range of 1.2% to 100%, reflecting a subset of tiles with mixed or low agreement. Of all tiles submitted, 41.6% had a high agreement score in the 90%-100% category (Figure 4B). The average kappa value of 0.284 indicates fair inter-rater agreement across all tiles.
Figure 3.
Agreement rates among pathologists across the overall dataset (A). Mitotic figure tiles categorized based on agreement rate from 85 pathologists (B).
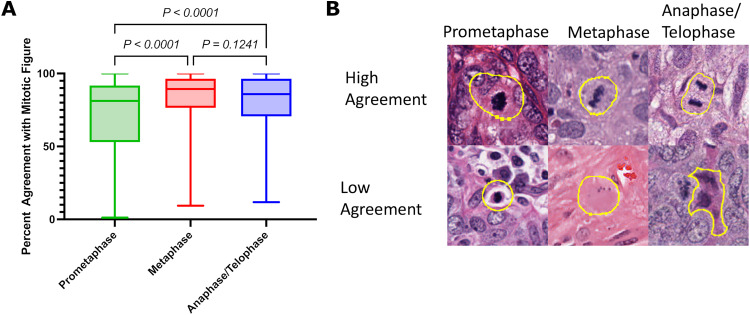
Figure 4.
Agreement rates with mitotic figures stratified based on phase of mitosis (A). Representative images of different mitosis phases of high and low agreement (B, 40x magnification).
Agreement Rate and Potential Mimics Based on Phase of Mitosis
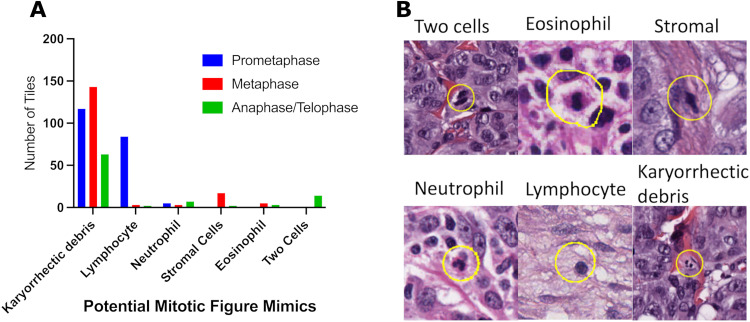
Mitotic figures in prometaphase showed lower agreement rates (median 81.2%) compared to mitotic figures in metaphase (median 89.4, p < 0.0001), and anaphase/telophase (median 85.9%, p < 0.0001), with no differences between other groups (Figure 4). In order to better understand the variability in mitotic figure enumeration, each mitotic figure was assessed for potential mimics. A potential mimic was assigned in 467 out of 1010 (46.2%) tiles within the dataset. Mitotic figures with low agreement rates were most likely to resemble mitotic figure mimic, with the most common mitotic figure mimic being karyorrhectic debris, immune cells, stromal cells, and two cells in close proximity. All phases of mitosis could mimic karyorrhectic debris, whereas lymphocytes were most likely to resemble mitosis in prometaphase, and stromal cells were likely to mimic mitosis in metaphase (Figure 5).
Figure 5.
Mitotic figures within the dataset characterized by potential mitotic figure mimics (A). Representative microscopy images of mitotic figure mimics observed in the dataset (B).
Cluster Analysis of Pathologist Agreement in Mitotic Figure Classification
To better understand the clustering of mitotic figures and pathologists, heatmap denograms following unsupervised hierarchical clustering were generated (Supplemental Figures 1 and 3). Representative mitotic figures from the clusters are included in Supplemental Figures 2 and 4. Additional analysis of the results from the hierarchical clustering showed no association between the clusters of pathologists and their subspecialty, region of practice, or years of practice that was previously collected in the study.
Discussion
This study leveraged a social media platform to recruit a large cohort of pathologists to generate and score the largest dataset of mitotic figures to date, 1010 proposed mitotic figures. This approach allows for rapidly generating robust datasets generated by an international cohort of pathologists. In this study, we were able to move from conceptualization through recruitment, onboarding, and complete primary data collection in 57 days. The use of public datasets minimized barriers for data sharing, such as data transfer agreements and multi-institutional ethics approval. To our knowledge, this is the largest international consensus study for mitotic figures to date, with the overall goal of examining the consistency in interpreting mitotic figures among pathologists.
We found a spectrum of opinions on what constitutes a mitotic figure, which is consistent with existing literature.15,16,19 A key finding was the lower agreement for prometaphase mitotic figures compared to other phases of mitosis. As all of the mitotic figures within the dataset were entities pathologists would classify as mitotic figures in their clinical practice, it is unsurprising that a vast majority of the dataset is in the upper agreement range (>80%). A subset of mitotic figures in the intermediate agreement range highlights the subjectivity of the task of identifying mitotic figures (Supplemental Figure 1). Individual pathologists also have a spectrum of agreement rates, with clusters of pathologists having variable thresholds for what constitutes a mitotic figure (Supplemental Figure 3). Despite having 85 pathologists that served as raters, the study was insufficiently powered to determine if this varying threshold was influenced by subspecialty, region of practice, or years of practice.
Pathologists’ interpretation of mitotic figures is inherently variable and previous studies have documented intermediate agreement rates for mitotic counts (κ = 0.34-0.64).17,28,29 Our study presented a lower kappa value (κ = 0.284), indicating fair agreement. One possible explanation for this lower agreement rate is the vast diversity of our sample of pathologists compared to previous studies. A second explanation, highlighted by participant feedback, is the unique challenge of interpreting mitotic figures on WSI due to limitations of scan quality and the inability to refocus the specimen—a functionality that is possible on analog microscopes. Studies have previously documented the decrease in accuracy in detecting mitoses using WSI rather than microscopy.30,31 Distinguishing mitotic figures and atypical mitotic figures from mitotic figure mimics, which include apoptotic bodies, hyperchromatic nuclei, karyorrhectic debris, inflammatory cells, two cells in close proximity, and tissue artifacts,2,19 is one of the most error-prone steps in determining mitotic counts.
Due to the pitfalls surrounding the identification and enumeration of mitotic figures, other means of obtaining proliferation markers have been variably utilized. Immunohistochemistry with phosphorylated histone H3 (pHH3), a marker highly specific to mitotic figures, can yield improved mitotic figure counting accuracy and efficiency. 32 However, implementation is not practical in routine practice as this added step lends itself to diagnostic pitfalls with immunohistochemistry, increases in operational costs, and further delays in slide preparation. Further, studies defining the use of mitotic figures in grading and diagnosing cancers may need to be repeated to account for changes in the threshold for classification. The Ki67 proliferation index is routinely used in tumors, such as non-pulmonary neuroendocrine tumors, and is a more general marker of proliferation in which expression is not limited to cells undergoing mitosis.33,34 However, Ki67 indices are not predictive for all cancer types, and there are similar challenges with the lack of standardization and variation in quantification and staining methodologies.34,35
With major advancements in computational tools, automated mitotic figure detection and enumeration have garnered significant research interest. However, proposed models for automated mitotic figure classification have room for improvement and tend to lack generalizability in different disease sites and clinical settings.25,36–38 A majority of these models are trained by limited publicly available mitosis detection datasets from grand challenges such as MITOS, 39 AMIDA, 40 MITOS-ATYPIA, 41 TUPAC, 42 and MIDOG, 23 that provides standardized WSIs for researchers to train and test automated mitoses detection models, with recent models adopting increasingly complex neural networks using a combination of these datasets. Models are only as good as the data they are trained on, and datasets are often limited in the breadth of disease types and in selection of mitotic figures by a small number of observers. Ideally, automated mitotic figure models should be developed with datasets that account for mitotic figures within different disease sites, include mitotic figures in different phases of cell division, account for variations in tissue processing that may lead to staining variability, and allow variations in slide scanning quality. Previous datasets present binary information: each cell is classified as either a mitotic figure or not. Our dataset is unique as we have 85 observers, allowing for an agreement or consensus rate for each cell. Instead of training models with a simple binary system, our dataset affords a way for weighted confidence to be integrated into the training set, with high agreement mitotic figures given proportionally more weight.
Our study has several limitations. We recognize the challenge with assigning binary values to mitotic figures where uncertainty is present. In order to limit the number of repetitive inputs from staff pathologists who volunteered their time to contribute to this study, we did not capture confidence levels from pathologists when scoring the mitotic figures. Moreover, as there is no gold standard for what definitively does and does not constitute a mitotic figure, we were unable to include true negatives in our dataset which would have increased the robustness of the collected data and reduced bias when scoring the dataset. Further, it is possible that providing a dataset of images that other colleagues have denoted as mitotic figures might influence scorers to agree when evaluating tiles containing mitotic figures with nonspecific features. During the collection of mitotic figures, pathologists were instructed to choose a histologic slide from a disease site of their choosing. The use of TCGA slides was both a limitation and advantage for this study. Slides were prepared and scanned at various institutions, resulting in heterogeneous H&E appearance and scanning preparation. 43 This heterogeneity may have made the identification of mitotic figures more challenging for participants, but it also helped provide a more diverse data set that can be utilized in subsequent training systems.
The field of pathology is becoming increasingly digitized, allowing pathologists novel ways to rapidly share data and contribute to international research collaborations with the promise of furthering patient care. 44 In this study, we leverage an extensive international professional network of pathologists on social media to develop and score the largest dataset of mitotic figures to date and to capture the subjectivity in interpreting mitotic figures. This project stands as an example of providing equitable and inclusive opportunities for research among a group of scholars connected through social media. We anticipate the increasing adoption of technology-driven approaches to generate robust datasets that can fuel artificial-intelligence-based applications in pathology and drastically reduce the number of subjective and time-consuming tasks for pathologists. Initiatives, such as these, can assist in implementing well-defined guidelines to standardize mitotic counts and assist pathologists in tumor assessment.
Supplemental Material
Supplemental material, sj-pptx-1-ijs-10.1177_10668969241234321 for The 1000 Mitoses Project: A Consensus-Based International Collaborative Study on Mitotic Figures Classification by Sherman Lin, Christopher Tran, Ela Bandari, Tommaso Romagnoli, Yueyang Li, Michael Chu, Abinaya S. Amirthakatesan, Adam Dallmann, Andrii Kostiukov, Angel Panizo, Anjelica Hodgson, Anna R. Laury, Antonio Polonia, Ashley E. Stueck, Aswathy A. Menon, Aurélien Morini, Birsen Özamrak, Caroline Cooper, Celestine Marie G. Trinidad, Christian Eisenlöffel, Dauda E. Suleiman, David Suster, David A. Dorward, Eman A. Aljufairi, Fiona Maclean, Gulen Gul, Irene Sansano, Irma E. Erana-Rojas, Isidro Machado, Ivana Kholova, Jayanthi Karunanithi, Jean-Baptiste Gibier, Jefree J. Schulte, Joshua J.X. Li, Jyoti R. Kini, Katrina Collins, Laurence A. Galea, Louis Muller, Luca Cima, Luiz M. Nova-Camacho, Marcus Dabner, Matthew J. Muscara, Matthew G. Hanna, Mehdi Agoumi, Nicholas J. P. Wiebe, Nicola K. Oswald, Nusrat Zahra, Olaleke O. Folaranmi, Oleksandr Kravtsov, Orhan Semerci, Namrata N. Patil, Preethi Muthusamy Sundar, Prem Charles, Priyadarshini Kumaraswamy Rajeswaran, Qi Zhang, Rachael van der Griend, Raghavendra Pillappa, Raul Perret, Raul S. Gonzalez, Robyn C. Reed, Sachin Patil, Xiaoyin “Sara” Jiang, Sumaira Qayoom, Susan Prendeville, Swikrity U. Baskota, Thanh-Truc Tran, Thar-Htet San, Tiia-Maria Kukkonen, Timothy J. Kendall, Toros Taskin, Tristan Rutland, Varsha Manucha, Vincent Cockenpot, Yale Rosen, Yessica P. Rodriguez-Velandia, Zehra Ordulu and Matthew J. Cecchini in International Journal of Surgical Pathology
Acknowledgments
We extend our sincere gratitude to the global network of pathologists whose invaluable insights and dedication significantly contributed to this study. Their willingness to invest time and expertise was instrumental in achieving this project. We would not be able to accomplish this feat without their belief in innovative approaches in advancing patient care within the field of pathology.
Footnotes
Author Contributions: We would especially like to thank the following individuals for their outstanding contributions: Dr. Amandeep Aneja, Cooper Medical School of Rowan University Hospital, Department of Pathology and Laboratory Medicine; Dr. Alp Usubutun, Hacettepe University School of Medicine, Department of Pathology; Dr. Cacey W. Peters, Pathology Watch; Dr. Camila Tomikawa; Dr. David J. Escobar, Northwestern University Feinberg School of Medicine, Department of Pathology; Dr. Jesus Cienfuegos, National Institute of Neurology and Neurosurgery Manuel Velasco Suarez, Mexico City, Neuropathology Department; Dr. Lilia M. Bernal, Hospital General con Especialidades Juan Ma. de Salvatierra, Laboratorio de Patología; Dr. Naomi Hardy, University of Maryland School of Medicine, Department of Pathology; Dr. Peter Braubach, Hannover Medical School, Institute for Pathology; Dr. Renan Ribeiro e Ribeiro, Instituto do Câncer do Estado de São Paulo (ICESP), Anatomia Patológica; Dr. Shingi Bopoto, University of Zimbabwe Faculty of Medicine, Department of Pathology; Dr. Vinh H. Nguyen, Ho Chi Minh City University Medical Center, Department of Pathology and Cytology.
Authors' Note: Christopher Tran currently affiliated with Department of Pathology, InterHospital Laboratory Partnership, Stratford, ON, Canada, and Swikrity U. Baskota is affiliated with Department of Pathology and Laboratory Medicine, University of California Davis Health System, Sacramento, CA, USA.
Data Availability: The dataset that was accumulated and information related to the scoring generated as part of this study is available upon request.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Not applicable, because this article does not contain any studies with human or animal subjects. All slides were from the publicly available TCGA database.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Ongoing funding support is gratefully acknowledged from the Megan J. Davey Opportunity Fund and the Cancer Pathology Translational Research Grant from the Ontario Molecular Pathology Research Network and Ontario Institute for Cancer Research.
Informed Consent: Not applicable, because this article does not contain any studies with human or animal subjects.
Trial Registration: Not applicable, because this article does not contain any clinical trials. All slides were from the publicly available TCGA database.
ORCID iDs: Sherman Lin https://orcid.org/0009-0009-1005-062X
Angel Panizo https://orcid.org/0000-0001-8405-7378
Anjelica Hodgson https://orcid.org/0000-0002-4069-2000
Antonio Polonia https://orcid.org/0000-0001-8312-1681
Irma E. Erana-Rojas https://orcid.org/0000-0002-9022-3739
Isidro Machado https://orcid.org/0000-0001-7318-1330
Jefree J. Schulte https://orcid.org/0000-0003-3920-0301
Joshua J.X. Li https://orcid.org/0000-0003-2425-0314
Katrina Collins https://orcid.org/0000-0002-9603-6731
Laurence A. Galea https://orcid.org/0000-0001-7968-1846
Luca Cima https://orcid.org/0000-0002-3445-1622
Luiz M. Nova-Camacho https://orcid.org/0000-0002-7625-7895
Nicola K. Oswald https://orcid.org/0000-0002-8347-3385
Olaleke O. Folaranmi https://orcid.org/0000-0002-0237-8934
Orhan Semerci https://orcid.org/0000-0001-7863-3228
Rachael van der Griend https://orcid.org/0000-0002-8605-9841
Robyn C. Reed https://orcid.org/0000-0002-8523-2139
Sumaira Qayoom https://orcid.org/0000-0003-1139-2444
Toros Taskin https://orcid.org/0000-0003-2909-2933
Yale Rosen https://orcid.org/0000-0001-6987-4134
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Travis WD, Rush W, Flieder DB, et al. Survival analysis of 200 pulmonary neuroendocrine tumors with clarification of criteria for atypical carcinoid and its separation from typical carcinoid. Am J Surg Pathol. 1998;22(8):934-944. doi: 10.1097/00000478-199808000-00003 [DOI] [PubMed] [Google Scholar]
- 2.Baak JPA. Mitosis counting in tumors. Hum Pathol. 1990;21(7):683-685. doi: 10.1016/0046-8177(90)90026-2 [DOI] [PubMed] [Google Scholar]
- 3.Ibrahim A, Lashen AG, Katayama A, et al. Defining the area of mitoses counting in invasive breast cancer using whole slide image. Mod Pathol Off J U S Can Acad Pathol Inc. 2022;35(6):739-748. doi: 10.1038/s41379-021-00981-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillou L, Coindre JM, Bonichon F, et al. Comparative study of the national cancer institute and French federation of cancer centers sarcoma group grading systems in a population of 410 adult patients with soft tissue sarcoma. J Clin Oncol Off J Am Soc Clin Oncol. 1997;15(1):350-362. doi: 10.1200/JCO.1997.15.1.350 [DOI] [PubMed] [Google Scholar]
- 5.Parab TM, DeRogatis MJ, Boaz AM, et al. Gastrointestinal stromal tumors: a comprehensive review. J Gastrointest Oncol. 2019;10(1):144-154. doi: 10.21037/jgo.2018.08.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer S, Rubin BP, Lux ML, et al. Prognostic value of KIT mutation type, mitotic activity, and histologic subtype in gastrointestinal stromal tumors. J Clin Oncol Off J Am Soc Clin Oncol. 2002;20(18):3898-3905. doi: 10.1200/JCO.2002.03.095 [DOI] [PubMed] [Google Scholar]
- 7.Marsch AF, McKee RM, Hinds BR. Morphologic forms and classification of dermal mitotic figure density in primary cutaneous melanoma: a retrospective study. Am J Dermatopathol. 2020;42(1):35-40. doi: 10.1097/DAD.0000000000001453 [DOI] [PubMed] [Google Scholar]
- 8.Keung EZ, Gershenwald JE. The eighth edition American joint committee on cancer (AJCC) melanoma staging system: implications for melanoma treatment and care. Expert Rev Anticancer Ther. 2018;18(8):775-784. doi: 10.1080/14737140.2018.1489246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baloch ZW, Asa SL, Barletta JA, et al. Overview of the 2022 WHO classification of thyroid neoplasms. Endocr Pathol. 2022;33(1):27-63. doi: 10.1007/s12022-022-09707-3 [DOI] [PubMed] [Google Scholar]
- 10.Olar A, Wani KM, Sulman EP, et al. Mitotic index is an independent predictor of recurrence-free survival in meningioma. Brain Pathol Zurich Switz. 2015;25(3):266-275. doi: 10.1111/bpa.12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson JF, Soong SJ, Balch CM, et al. Prognostic significance of mitotic rate in localized primary cutaneous melanoma: an analysis of patients in the multi-institutional American joint committee on cancer melanoma staging database. J Clin Oncol Off J Am Soc Clin Oncol. 2011;29(16):2199-2205. doi: 10.1200/JCO.2010.31.5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medri L, Volpi A, Nanni O, et al. Prognostic relevance of mitotic activity in patients with node-negative breast cancer. Mod Pathol. 2003;16(11):1067-1075. doi: 10.1097/01.MP.0000093625.20366.9D [DOI] [PubMed] [Google Scholar]
- 13.Cree IA, Tan PH, Travis WD, et al. Counting mitoses: sI(ze) matters!. Mod Pathol. 2021;34(9):1651-1657. doi: 10.1038/s41379-021-00825-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yigit N, Gunal A, Kucukodaci Z, Karslioglu Y, Onguru O, Ozcan A. Are we counting mitoses correctly? Ann Diagn Pathol. 2013;17(6):536-539. doi: 10.1016/j.anndiagpath.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 15.Zhang R, jiao Chen H, et al. Reproducibility of the Nottingham modification of the scarff-bloom-richardson histological grading system and the complementary value of Ki-67 to this system. Chin Med J (Engl). 2010;123(15):1976-1982. [PubMed] [Google Scholar]
- 16.Coindre JM, Trojani M, Contesso G, et al. Reproducibility of a histopathologic grading system for adult soft tissue sarcoma. Cancer. 1986;58(2):306-309. doi:10.1002/1097-0142(19860715)58:2 < 306::AID-CNCR2820580216 > 3.0.CO;2-7 [DOI] [PubMed] [Google Scholar]
- 17.Frierson HF, Wolber RA, Berean KW, et al. Interobserver reproducibility of the Nottingham modification of the bloom and richardson histologic grading scheme for infiltrating ductal carcinoma. Am J Clin Pathol. 1995;103(2):195-198. doi: 10.1093/ajcp/103.2.195 [DOI] [PubMed] [Google Scholar]
- 18.Garbe C, Eigentler TK, Bauer J, et al. Mitotic rate in primary melanoma: interobserver and intraobserver reliability, analyzed using H&E sections and immunohistochemistry: mitotic rate in primary melanoma - reliability. JDDG J Dtsch Dermatol Ges. 2016;14(9):910-915. doi: 10.1111/ddg.12797 [DOI] [PubMed] [Google Scholar]
- 19.Donovan TA, Moore FM, Bertram CA, et al. Mitotic figures—normal, atypical, and imposters: a guide to identification. Vet Pathol. 2021;58(2):243-257. doi: 10.1177/0300985820980049 [DOI] [PubMed] [Google Scholar]
- 20.Al-Janabi S, van Slooten HJ, Visser M, van der Ploeg T, van Diest PJ, Jiwa M. Evaluation of mitotic activity index in breast cancer using whole slide digital images. PLoS One. 2013;8(12):e82576. doi: 10.1371/journal.pone.0082576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saygin I, Cakir E, Ercin ME. Interobserver variability in mitotic count for meningioma grading: how can we reduce it? Turk Neurosurg. 2020;30(5):643-650. doi: 10.5137/1019-5149.JTN.26252-19.2 [DOI] [PubMed] [Google Scholar]
- 22.Cui M, Zhang DY. Artificial intelligence and computational pathology. Lab Invest. 2021;101(4):412-422. doi: 10.1038/s41374-020-00514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aubreville M, Wilm F, Stathonikos N, et al. A comprehensive multi-domain dataset for mitotic figure detection. Sci Data. 2023;10(1):484. doi: 10.1038/s41597-023-02327-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathew T, Kini JR, Rajan J. Computational methods for automated mitosis detection in histopathology images: a review. Biocybern Biomed Eng. 2021;41(1):64-82. doi: 10.1016/j.bbe.2020.11.005 [DOI] [Google Scholar]
- 25.Mathew T, Ajith B, Kini JR, Rajan J. Deep learning-based automated mitosis detection in histopathology images for breast cancer grading. Int J Imaging Syst Technol. 2022;32(4):1192-1208. doi: 10.1002/ima.22703 [DOI] [Google Scholar]
- 26.Bankhead P, Loughrey MB, Fernández JA, et al. Qupath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleiss JL, Levin B, Paik MC. Statistical methods for rates and proportions. 1st ed. New Jersey: Wiley; 2003. doi: 10.1002/0471445428 [DOI] [Google Scholar]
- 28.Meyer JS, Alvarez C, Milikowski C, et al. Breast carcinoma malignancy grading by bloom-richardson system vs proliferation index: reproducibility of grade and advantages of proliferation index. Mod Pathol Off J U S Can Acad Pathol Inc. 2005;18(8):1067-1078. doi: 10.1038/modpathol.3800388 [DOI] [PubMed] [Google Scholar]
- 29.Rakha EA, Bennett RL, Coleman D, Pinder SE, Ellis IO. Review of the national external quality assessment (EQA) scheme for breast pathology in the UK. J Clin Pathol. 2017;70(1):51-57. doi: 10.1136/jclinpath-2016-203800 [DOI] [PubMed] [Google Scholar]
- 30.Tabata K, Uraoka N, Benhamida J, et al. Validation of mitotic cell quantification via microscopy and multiple whole-slide scanners. Diagn Pathol. 2019;14(1):65. doi: 10.1186/s13000-019-0839-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lashen A, Ibrahim A, Katayama A, et al. Visual assessment of mitotic figures in breast cancer: a comparative study between light microscopy and whole slide images. Histopathology. 2021;79(6):913-925. doi: 10.1111/his.14543 [DOI] [PubMed] [Google Scholar]
- 32.Ibrahim A, Toss MS, Makhlouf S, Miligy IM, Minhas F, Rakha EA. Improving mitotic cell counting accuracy and efficiency using phosphohistone-H3 (PHH3) antibody counterstained with haematoxylin and eosin as part of breast cancer grading. Histopathology. 2023;82(3):393-406. doi: 10.1111/his.14837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed ST, Ahmed AM, Musa DH, Sulayvani FK, Al-Khyatt M, Pity IS. Proliferative Index (Ki67) for prediction in breast duct carcinomas. Asian Pac J Cancer Prev APJCP. 2018;19(4):955-959. doi: 10.22034/APJCP.2018.19.4.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang LH, Gonen M, Hedvat C, Modlin IM, Klimstra DS. Objective quantification of the Ki67 proliferative Index in neuroendocrine tumors of the gastroenteropancreatic system: a comparison of digital image analysis with manual methods. Am J Surg Pathol. 2012;36(12):1761-1770. doi: 10.1097/PAS.0b013e318263207c [DOI] [PubMed] [Google Scholar]
- 35.Young HTM, Carr NJ, Green B, Tilley C, Bhargava V, Pearce N. Accuracy of visual assessments of proliferation indices in gastroenteropancreatic neuroendocrine tumours. J Clin Pathol. 2013;66(8):700-704. doi: 10.1136/jclinpath-2012-201217 [DOI] [PubMed] [Google Scholar]
- 36.Malon C, Brachtel E, Cosatto E, et al. Mitotic figure recognition: agreement among pathologists and computerized detector. Anal Cell Pathol Amst. 2012;35(2):97-100. doi: 10.3233/ACP-2011-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. Aggnet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging. 2016;35(5):1313-1321. doi: 10.1109/TMI.2016.2528120 [DOI] [PubMed] [Google Scholar]
- 38.Mahmood T, Arsalan M, Owais M, Lee MB, Park KR. Artificial intelligence-based mitosis detection in breast cancer histopathology images using faster R-CNN and deep CNNs. J Clin Med. 2020;9(3):749. doi: 10.3390/jcm9030749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MITOS Dataset. MITOS dataset. Accessed July 24, 2023. http://ludo17.free.fr/mitos_2012/dataset.html
- 40.Veta M, Van Diest PJ, Willems SM, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20(1):237-248. doi: 10.1016/j.media.2014.11.010 [DOI] [PubMed] [Google Scholar]
- 41.MITOS-ATYPIA-14 - Grand Challenge. grand-challenge.org. Accessed July 24, 2023. https://mitos-atypia-14.grand-challenge.org/Dataset/
- 42.Veta M, Heng YJ, Stathonikos N, et al. Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge. Med Image Anal. 2019;54:111-121. doi: 10.1016/j.media.2019.02.012 [DOI] [PubMed] [Google Scholar]
- 43.Jahn SW, Plass M, Moinfar F. Digital pathology: advantages, limitations and emerging perspectives. J Clin Med. 2020;9(11):3697. doi: 10.3390/jcm9113697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022;35(1):23-32. doi: 10.1038/s41379-021-00919-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pptx-1-ijs-10.1177_10668969241234321 for The 1000 Mitoses Project: A Consensus-Based International Collaborative Study on Mitotic Figures Classification by Sherman Lin, Christopher Tran, Ela Bandari, Tommaso Romagnoli, Yueyang Li, Michael Chu, Abinaya S. Amirthakatesan, Adam Dallmann, Andrii Kostiukov, Angel Panizo, Anjelica Hodgson, Anna R. Laury, Antonio Polonia, Ashley E. Stueck, Aswathy A. Menon, Aurélien Morini, Birsen Özamrak, Caroline Cooper, Celestine Marie G. Trinidad, Christian Eisenlöffel, Dauda E. Suleiman, David Suster, David A. Dorward, Eman A. Aljufairi, Fiona Maclean, Gulen Gul, Irene Sansano, Irma E. Erana-Rojas, Isidro Machado, Ivana Kholova, Jayanthi Karunanithi, Jean-Baptiste Gibier, Jefree J. Schulte, Joshua J.X. Li, Jyoti R. Kini, Katrina Collins, Laurence A. Galea, Louis Muller, Luca Cima, Luiz M. Nova-Camacho, Marcus Dabner, Matthew J. Muscara, Matthew G. Hanna, Mehdi Agoumi, Nicholas J. P. Wiebe, Nicola K. Oswald, Nusrat Zahra, Olaleke O. Folaranmi, Oleksandr Kravtsov, Orhan Semerci, Namrata N. Patil, Preethi Muthusamy Sundar, Prem Charles, Priyadarshini Kumaraswamy Rajeswaran, Qi Zhang, Rachael van der Griend, Raghavendra Pillappa, Raul Perret, Raul S. Gonzalez, Robyn C. Reed, Sachin Patil, Xiaoyin “Sara” Jiang, Sumaira Qayoom, Susan Prendeville, Swikrity U. Baskota, Thanh-Truc Tran, Thar-Htet San, Tiia-Maria Kukkonen, Timothy J. Kendall, Toros Taskin, Tristan Rutland, Varsha Manucha, Vincent Cockenpot, Yale Rosen, Yessica P. Rodriguez-Velandia, Zehra Ordulu and Matthew J. Cecchini in International Journal of Surgical Pathology