Abstract
Members of the antibiotic-producing bacterial genus Streptomyces undergo a complex developmental life cycle that culminates in the production of spores. Central to control of this cell differentiation process is signaling through the second messenger c-di-GMP. So far, three proteins that are directly controlled by c-di-GMP in Streptomyces have been functionally and structurally characterized: the key developmental regulators BldD and σWhiG, and the glycogen-degrading enzyme GlgX. c-di-GMP signals through BldD and σWhiG, respectively, to control the two most dramatic transitions of the Streptomyces life cycle, the formation of the reproductive aerial hyphae, and their differentiation into spore chains. Later in development, c-di-GMP activates GlgX-mediated degradation of glycogen, releasing stored carbon for spore maturation.
c-di-GMP controls Streptomyces development
3′, 5′-cyclic diguanylic acid (c-di-GMP) is synthesized from two molecules of GTP by diguanylate cyclases (DGCs) characterized by GGDEF domains and degraded by c-di-GMP-specific phosphodiesterases (PDEs) carrying EAL or HD-GYP domains (Figure 1B). These domains are named after conserved residues found in their active sites [1]. Homologs of DGCs and PDEs are found in all major bacterial phyla [2], making c-di-GMP a nearly universal signaling molecule through which bacteria sense and respond to the environment. Despite its ubiquity, studies on c-di-GMP signaling have primarily been limited to Gram-negative bacteria, where it controls processes such as motility, biofilm formation and virulence [1,3-5].
Figure 1. c-di-GMP levels control Streptomyces development.
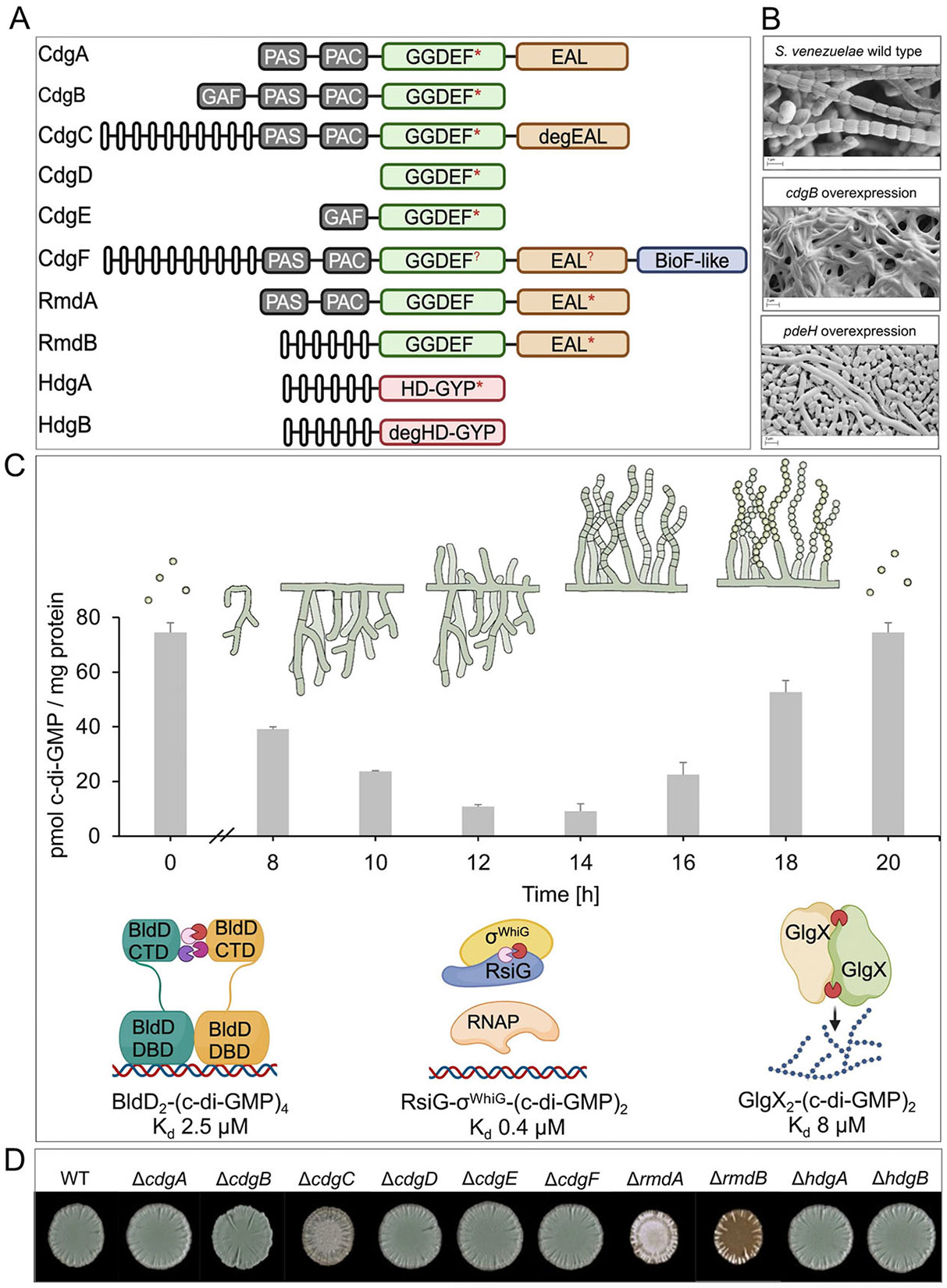
(A) Scanning electron micrographs showing that overexpression of a c-di-GMP-synthesizing enzyme (CdgB) prevents the initiation of development, trapping Streptomyces in vegetative growth (the classic ‘Bald’ phenotype), whereas overexpression of a c-di-GMP-degrading enzyme (PdeH) causes precocious hypersporulation. Wild-type S. venezuelae is shown for comparison. Cells were grown on maltose-yeast extract-malt extract (MYM) agar for four days at 30 °C prior to imaging. (B) Domain organization of the 10 c-di-GMP-metabolizing enzymes in S. venezuelae. GGDEF domains are shown in green, EAL domains in brown, HD-GYP domains in pink and the BioF-like domain of CdgF in purple. Predicted transmembrane helices are shown as open bars and N-terminal GAF, PAS and PAC signaling domains are shown in grey. WT, wild type; deg, degenerate; *, enzymatically active domain; ?, enzymatic activity unknown. (C) Colony phenotypes of mutants for each of the 10 genes encoding c-di-GMP-metabolizing enzymes in S. venezuelae. (D) Representation of the Streptomyces life cycle showing the three main developmental stages: vegetative growth, aerial growth, and sporulation. c-di-GMP levels (grey bars) vary across the life cycle [16]. During the vegetative phase, high c-di-GMP levels mediate the oligomerization of the developmental master repressor BldD into an active dimer, blocking the expression of a large regulon of sporulation genes. High c-di-GMP levels also directly mediate the sequestration of the sporulation-specific sigma factor σWhiG by the antisigma factor RsiG. As c-di-GMP levels begin to drop at the start of the transition to sporulation, the BldD dimer dissociates, resulting in the loss of DNA binding, and thereby allowing the transcriptional cascade leading to sporulation to become activated [9,22]. Subsequently, as c-di-GMP levels drop further, σWhiG is released from the antisigma factor RsiG, allowing the activation of late sporulation gene expression [29]. As sporulation proceeds, c-di-GMP levels begin to rise, and the binding of c-di-GMP to GlgX leads to a long-range conformational change in its active site, activating the enzyme and causing the degradation of glycogen in maturing spores [16]. The Kds of BldD, RsiG-σWhiG and GlgX for c-di-GMP shown are consistent with this order of events. Note that GlgX is abundant only during sporulation. CTD, C-terminal domain; DBD, DNA-binding domain; RNAP, RNA polymerase.
Streptomyces, which belong to the phylum Actinobacteria (synonym Actinomycetota) are filamentous bacteria with a fascinating developmental life cycle involving progression from vegetative growth to the production of reproductive aerial hyphae, which differentiate into long chains of exospores (Figure 1A, D) [6-8]. In a key study, the endogenous c-di-GMP metabolism of Streptomyces venezuelae was overwhelmed by engineering the overexpression of either a DGC or a PDE, aiming to raise or deplete, respectively, c-di-GMP levels [9]. Both had dramatic consequences. Overexpressing the DGC blocked the initiation of development, giving a classic bald phenotype, while overexpressing the PDE caused precocious hypersporulation (Figure 1A). These results suggested that high c-di-GMP levels block differentiation, trapping Streptomyces in vegetative growth, whereas decreased levels of c-di-GMP accelerate entry into development, promoting sporulation [9]. Consistent with this conclusion, the loss of individual enzymes involved in c-di-GMP metabolism affects development [10-15]. S. venezuelae has 10 such enzymes (Figure 1B), and deletion studies show that loss of either of the DGCs, CdgB or CdgC, enhances sporulation whereas deletion of the PDEs RmdA or RmdB delays development (Figure 1C) [13]. The DGCs CdgA, CdgB and CdgC, and the PDE RmdB are the most highly conserved c-di-GMP metabolizing enzymes in the genus Streptomyces [13,14].
Consistent with c-di-GMP playing a central role in development, data showed that c-di-GMP levels are high in early vegetative growth and drop progressively to reach a minimum at around 14 h of growth in liquid sporulation medium, coinciding with the initiation of differentiation (Figure 1D) [16]. c-di-GMP levels then rise during spore formation, reaching a maximum when development is complete (Figure 1D). How these c-di-GMP levels are controlled across development is certain to be complex. As shown in Figure 1B, five of the 10 c-di-GMP metabolizing enzymes in S. venezuelae are composite GGDEF-EAL proteins carrying both the synthetic and degradative domains, and most of the enzymes carry multiple regulatory domains (e.g. GAF, PAS, PAC) that are likely to control enzymatic activity in response to unknown regulatory inputs [13,14,17]. In addition, four genes encoding DGCs – cdgA, cdgB, cdgC and cdgE – are direct targets of BldD regulation (see below), creating the possibility of negative regulatory feedback loops [9-11,13,17].
c-di-GMP binds BldD to control the onset of development
The striking phenotypic consequences of changing c-di-GMP levels in Streptomyces suggested that this nucleotide second messenger must interact directly with the regulatory network that controls the life cycle. The first direct target of c-di-GMP to be discovered in Streptomyces was BldD, identified as a c-di-GMP binding protein in affinity pull-down assays using a c-di-GMP capture compound [9,18]. BldD is the master repressor of Streptomyces development. It sits at the apex of the regulatory network, repressing a large set of sporulation genes, including many genes of the core transcriptional regulatory cascade itself, in addition to genes encoding proteins critical for sporulation septation and the segregation of chromosomes into spores [7,9,10,19]. Critically, the ability of BldD to bind DNA and repress its target genes requires complex formation with c-di-GMP, which acts as a to act as a “brake”, prolonging vegetative growth and blocking entry into development [9].
The crystal structure of BldD bound to c-di-GMP together with biochemical studies revealed a unique molecular mechanism of c-di-GMP signaling (Figure 2). BldD has two domains, an N-terminal DNA-binding domain (DBD) and a C-terminal domain (CTD) that was of unknown function [9,20,21]. Biochemical experiments demonstrated that the CTD is a c-di-GMP-binding domain, and that binding c-di-GMP causes the CTD to dimerize [9]. Remarkably, the crystal structure showed that the subunits of the CTD dimer are separated by ~10 Å, with no protein-protein contacts. Instead, a tetrameric cage of c-di-GMP bridges between the two subunits (Figure 2) [9]. Consequently, high levels of c-di-GMP drive dimerization of BldD (Figures 1D and 2), leading to repression of the BldD regulon of sporulation genes during the vegetative growth stage, thereby acting as a checkpoint to control the initiation of development. As c-di-GMP levels begin to drop at the start of the transition to sporulation, the BldD dimer dissociates into monomers, causing it to dissociate from DNA, thus allowing the transcriptional cascade leading to sporulation to become activated (Figure 1D) [9,22]. c-di-GMP can assume different oligomeric states to execute different functions [1,3,5], but to date the tetrameric form of c-di-GMP seen in BldD is unique [22].
Figure 2. Dimerization of the developmental master repressor BldD is mediated by the formation of tetrameric c-di-GMP bound between the two CTDs.
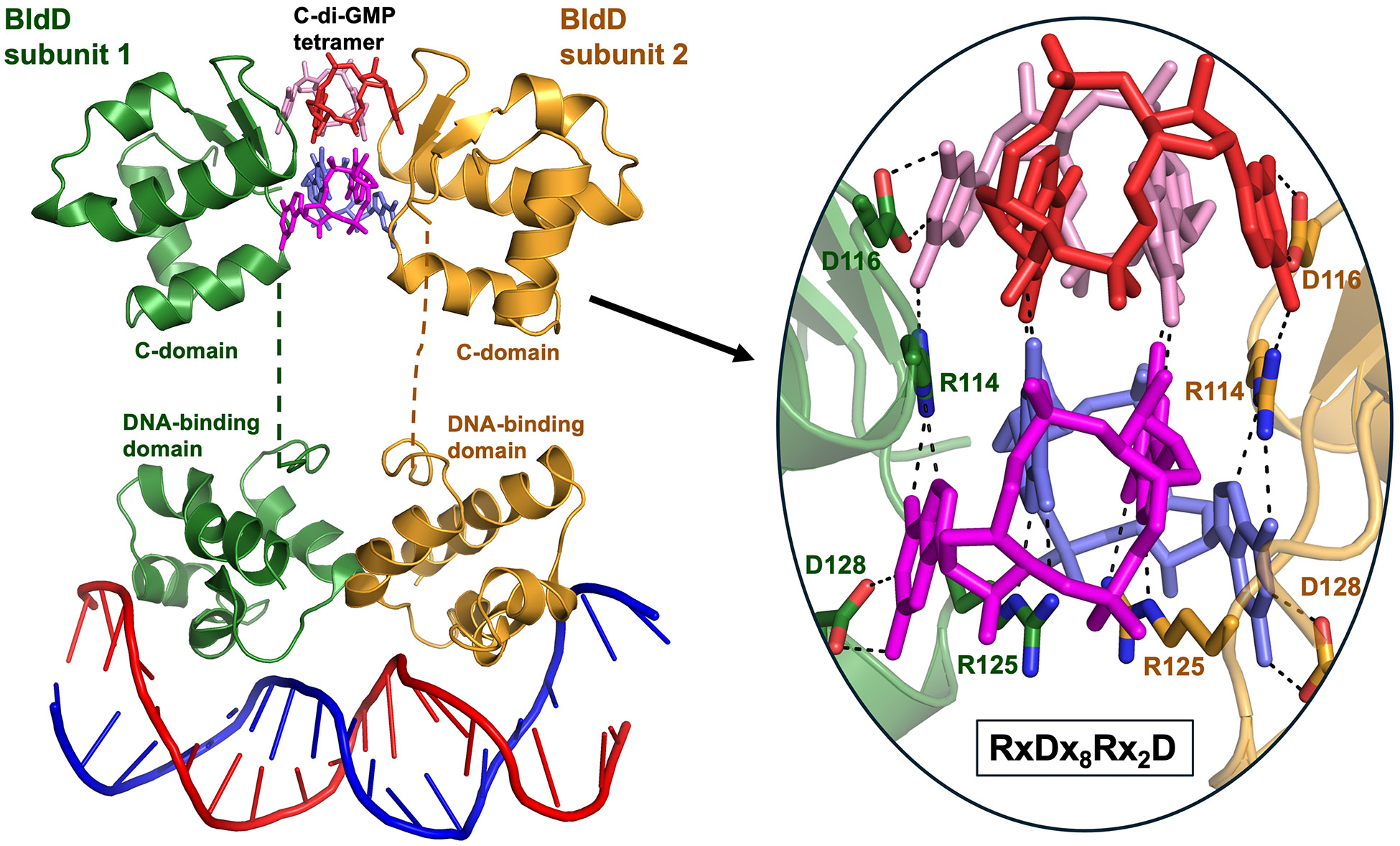
Model of the BldD2-(c-di-GMP)4-DNA complex, based on the ~2 Å resolution structure of the CTD2-(c-di-GMP)4 complex combined with a ~4.5 Å resolution structure of the DBD-DNA region [9]. One BldD subunit is colored wheat and the other green. For clarity, each of the four c-di-GMP molecules is colored differently. The c-di-GMP molecules are shown as sticks and the DNA as a cartoon. The linker region between the CTD and the DBD (shown as dashed lines in wheat or green) is disordered in both subunits, indicating their conformational flexibility. The inset shows the mechanism of selective binding of the c-di-GMP tetramer by the RxDx8Rx2D BldD signature motif. Dashes indicate polar interactions.
c-di-GMP is monomeric in solution at physiological concentrations [23], and the pathway that leads to the fully assembled BldD2-(c-di-GMP)4 complex is not completely understood. It is clear, however, that BldD binds c-di-GMP through an ordered sequential mechanism [22]. The structure of a BldD assembly intermediate bound to one c-di-GMP dimer was captured, showing that BldD still forms a dimer in this intermediate. Guided by these structures, it was possible to prevent the binding of the second c-di-GMP dimer by mutating Asp116 to Ala, trapping the half-loaded assembly intermediate for functional assessment. Both in vitro and in vivo, the BldD(D116A) retained some weak DNA-binding activity [22]. However, the bldD D116A mutant has a null phenotype, showing that the ability to regulate entry into development requires assembly of the full BldD2-(c-di-GMP)4 complex [22].
BldD is present throughout most filamentous Actinobacteria, and its RxDx8Rx2D c-di-GMP-binding signature motif (Figure 2) is conserved in all homologs [9]. Further, all these bacteria have DGCs, suggesting that BldD-(c-di-GMP) is likely to control key developmental processes in filamentous species throughout the phylum. Consistent with this suggestion, BldD-(c-di-GMP) has been shown to control sporangium formation in Actinoplanes missouriensis, an intriguing filamentous actinobacterial species that produces flagellated, motile spores [24,25].
In addition to their fascinating developmental life cycles, Streptomyces and their filamentous actinobacterial relatives are also of importance because they are the most abundant source of antibiotics and other natural products used in medicine [26]. Although BldD does not directly control antibiotic biosynthesis in the model species S. venezuelae, in other filamentous Actinobacteria it has been shown to directly control the expression of the biosynthetic gene clusters (BGCs) for several clinically important antibiotics. These include erythromycin in Saccharopolyspora erythraea [27], avermectin in Streptomyces avermitilis [28], and daptomycin in Streptomyces roseosporus [28], showing that the BldD-(c-di-GMP)-mediated regulation of developmental processes in Actinobacteria extends to their medically and commercially important specialized metabolism.
c-di-GMP binds RsiG-σWhiG to control the differentiation of aerial hyphae into spores
While BldD2-(c-di-GMP)4 regulates the onset of development, a 2020 study revealed that c-di-GMP also directly intervenes later in the Streptomyces life cycle by regulating the activity of the sporulation-specific sigma factor σWhiG [29]. This alternative sigma factor is highly conserved in the genus Streptomyces, where it is required for the differentiation of aerial hyphae into spores [29,30]. The specific role of σWhiG in Streptomyces was revealed using ChIP-seq (chromatin immunoprecipitation followed by sequencing) combined with global transcriptional profiling, which showed that the main function of σWhiG is to activate transcription of just two genes, whiH and whiI. WhiH and WhiI are both transcription factors, each responsible for regulating a large set of late-stage sporulation genes [31,32], explaining how σWhiG controls the differentiation of aerial hyphae into spores.
During vegetative growth, σWhiG is present in Streptomyces cells, yet the expression of its target genes does not increase until later in development. This suggested σWhiG is post-translationally regulated.. It was initially unclear how σWhiG activity is regulated until a bacterial two-hybrid genomic library screen was used to identify its cognate antisigma factor, RsiG [29]. Unlike many antisigma factors, which are often encoded adjacent to their cognate sigma partners [33], rsiG lies distant from whiG in Streptomyces genomes (in S. venezuelae the two genes are found ~1.5 Mbp apart). The structure of the RsiG-σWhiG complex revealed that binding to RsiG results in compaction of σWhiG, which prevents its interaction with RNA polymerase. Crucially, the structure also revealed a c-di-GMP dimer at the RsiG-σWhiG interface (Figure 3), even though no c-di-GMP was added prior to crystallization, meaning it copurified with the complex following overexpression in E. coli. This represents the first, and to date only example of a sigma-antisigma pair whose association requires c-di-GMP. Most of the direct contacts to the c-di-GMP dimer are mediated by the antisigma factor RsiG, and RsiG alone is able to bind c-di-GMP (with a Kd of 6.5 μM). This binding affinity increases to a Kd of 0.4 μM with the addition of σWhiG, most likely because σWhiG provides additional direct contacts to c-di-GMP.
Figure 3. c-di-GMP arms RsiG as an antisigma, allowing it to bind and sequester the sporulation-specific σ factor, σWhiG.
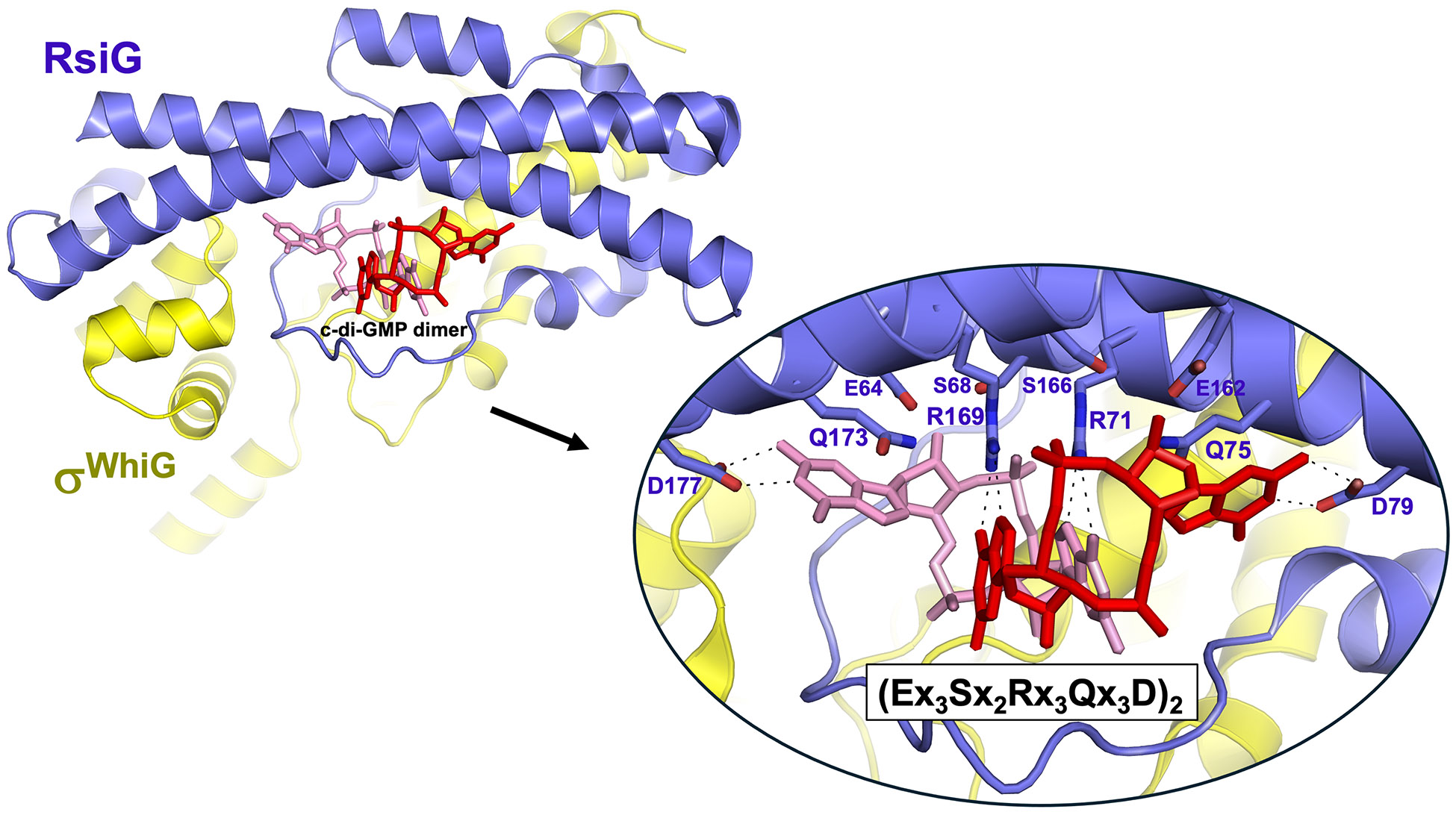
Ribbon diagram of the overall structure of the S. venezuelae RsiG-(c-di-GMP)2-σWhiG complex [29]. The antisigma RsiG is colored blue and σWhiG is colored yellow. The two partially intercalated c-di-GMP molecules are shown as pink or red sticks. The inset shows the selective binding mechanism of the c-di-GMPs by residues of the two copies of Ex3Sx2Rx3Qx3D RsiG signature motif, one present on each of the two helices of the RsiG antiparallel coiled coil, selectively bind the c-di-GMP nucleotides. Dashes indicate polar interactions.
The manner in which RsiG interacts with the c-di-GMP dimer is unprecedented: it binds the second messenger via two copies of a novel Ex3Sx2Rx3Qx3D motif, one on each of two helices that form an antiparallel coiled coil (Figure 3) [29,34]. Typically, in other effector proteins that bind c-di-GMP dimers, the four guanine bases make stacking interactions to adopt a fully intercalated conformation [35-39]. In contrast, when bound to RsiG, the two molecules of c-di-GMP are partially intercalated, with only two of the guanine bases stacked, and the other two rotated out to stack against conserved glutamine residues and interact with conserved aspartic acid residues found in the repeated signature motif (Figure 3). The c-di-GMP dimer functions to bind the sigma-antisigma pair together through direct contacts and by promoting the folding of a loop in RsiG that forms part of the interface with σWhiG, thereby stabilizing the complex. A mutant RsiG protein that cannot bind c-di-GMP (having alanines substituting for arginine and aspartic acid residues in both signature motifs) was unable to impede σWhiG activity, both in vitro and in vivo, confirming that c-di-GMP is required for stable complex formation [29]. Consequently, formation of the RsiG-(c-di-GMP)2-σWhiG complex is favored during vegetative growth when c-di-GMP levels are high, resulting in sequestration of σWhiG from RNAP. When c-di-GMP levels drop at the onset of reproductive growth, the complex disassociates, releasing σWhiG to direct RNAP to transcribe the genes encoding the late-stage sporulation regulators WhiH and WhiI (Figure 1D). The Kd values of BldD and RsiG-σWhiG for c-di-GMP provide a potentially simple explanation for why BldD dissociates from DNA before σWhiG is released from RsiG. BldD binds c-di-GMP with a Kd of 2.5 μM [9], whereas the Kd for [RsiG + σWhiG] is 0.4 μM [29]. Therefore, as c-di-GMP levels decline, BldD would become inactive as a repressor before σWhiG is released from its antisigma (Figure 1D).
Distribution and evolution of the antisigma factor RsiG
RsiG is unique among both antisigma factors and c-di-GMP receptors, sharing no homology with characterized examples of either. A search of representative bacterial genomes found that distribution of this novel c-di-GMP-binding antisigma factor is restricted to the phylum Actinobacteria [40]. The majority of the actinobacterial homologs identified resemble S. venezuelae RsiG with twin c-di-GMP-binding motifs. However, five of the representative actinobacterial genomes were found to encode RsiG homologs that have only one c-di-GMP-binding motif. The structure of one of these, from Rubrobacter radiotolerans, revealed that the single-motif RsiG homologs dimerize to form an antiparallel coiled coil such that they bind c-di-GMP and cognate σWhiG partners in a similar manner to the monomeric twin-motif form of RsiG. Based on the distribution of the two forms of RsiG within the Actinobacteria, it seems likely that the single-motif protein is the ancestral form of RsiG and that an intragenic duplication event gave rise to the twin-motif form of the protein inherited by many Actinobacteria [40].
Members of the genus Rubrobacter are not filamentous and do not sporulate. This means that in this genus, RsiG2-(c-di-GMP)2-σWhiG must regulate a different biological process from the homologs first characterized in Streptomyces. Indeed, it was shown that in Rubrobacter, RsiG2-(c-di-GMP)2-σWhiG regulates the biosynthesis of type IV pili [40], surface appendages that mediate diverse functions in bacteria including motility, biofilm formation, surface sensing, and DNA uptake [41]. Additionally, in Rubrobacter multiple DGCs and PDEs were predicted to be under the control of RsiG2-(c-di-GMP)2-σWhiG, suggesting the presence of regulatory feedback loops evocative of those observed for Streptomyces BldD-(c-di-GMP). This example thus illustrates how homologous transcriptional regulators can undergo major structural and functional shifts over the course of their evolution.
c-di-GMP binds GlgX to stimulate enzymatic degradation of glycogen stores during spore maturation
In addition to BldD- and σWhiG -mediated regulation of gene expression, recent studies revealed that c-di-GMP controls the timing of glycogen degradation by acting as an allosteric activator of the glycogen debranching enzyme GlgX [16]. Like BldD, GlgX was identified as a c-di-GMP binding protein by affinity pull-down assay using a c-di-GMP capture compound. Glycogen is a highly branched homopolysaccharide consisting of α-1,4-linked glucose subunits in the linear oligosaccharide chains, and α-1,6-linked glucose at branching points [42]. Many bacterial species accumulate glycogen for carbon and energy storage, especially under conditions of carbon excess while limited for another nutrient (e.g. nitrogen) [43,44].
The storage and degradation of glycogen in time and space is linked to developmental functions and regulation in Streptomyces [45]. In Streptomyces coelicolor, deposition of the biopolymer occurs in two discrete ‘tissues’ of the developing colony. Phase I deposition takes place at the interface of the vegetative and aerial hyphae, likely serving as an energy source for aerial growth. Phase II storage occurs in immature spore chains within aerial hyphae. Notably, S. coelicolor possesses two isoforms of glycogen branching enzyme, GlgBI, associated with Phase I deposition, and GlgBII, associated with Phase II deposition [46.47]. In S. venezuelae, uncontrolled degradation of glycogen caused by overexpressing the glycogen debranching enzyme GlgX results in delayed sporulation and a reduced number of viable spores [16]. Interestingly, the glycogen content of spores decreases as they mature. Spores harvested from S. venezuelae plates three days after inoculation contain about 160 μg glycogen per mg total protein, whereas those harvested after 14 days after inoculation contain only 5 μg glycogen per mg total protein (Katrin Wrede and Natalia Tschowri, unpublished), suggesting that glycogen degradation is part of the spore maturation process, and that carbon released from the biopolymer supports spore formation and vitality.
Together with the glycogen phosphorylase GlpP, GlgX constitutes the core of glycogen catabolism. While GlpP catalyzes the depolymerization of α-(1→4)-glucosidic linkages, GlgX removes α-(1→6)-linked glucose residues in glycogen [42]. Two mechanisms account for the correct timing of GlgX-mediated glycogen degradation during spore maturation. First, glgX expression is developmentally regulated so that the enzyme accumulates during sporulation. Second, activation of GlgX requires binding of c-di-GMP [16]. A GlgX-c-di-GMP structure showed that the enzyme utilizes a novel ExRx6R signature motif to bind the second messenger (Figure 4). In complex with c-di-GMP, GlgX forms an antiparallel, head-to-tail dimer in which monomeric c-di-GMP binds at each end of the protein dimer to stabilize an active conformation (Figures 1D and 4). Dimerization of GlgX is stimulated by c-di-GMP binding but is not strictly dependent on it, and activation of the enzyme occurs through c-di-GMP-induced long-range conformational changes resulting in structural rearrangements at the active site of the enzyme. Importantly, GlgX has a relatively moderate affinity for c-di-GMP (~ 8 μM) explaining why enzyme activity depends on relatively high levels of the second messenger, which reach their peak during sporulation [16] (Figure 1D). Thus, the accumulation of GlgX and of c-di-GMP in the late stages of development ensures the controlled release of stored carbon for spore maturation. Notably, glycogen debranching enzymes are widespread in eukaryotes, archaea and bacteria [42], but the ExRx6R c-di-GMP binding motif is only conserved in glycogen debranching enzymes from streptomycetes and a few other Actinobacteria. This suggests that the link between c-di-GMP signaling and GlgX-mediated glycogen degradation evolved as a specific feature of Streptomyces development.
Figure 4. c-di-GMP controls the enzymatic activity of the glycogen debranching enzyme GlgX.
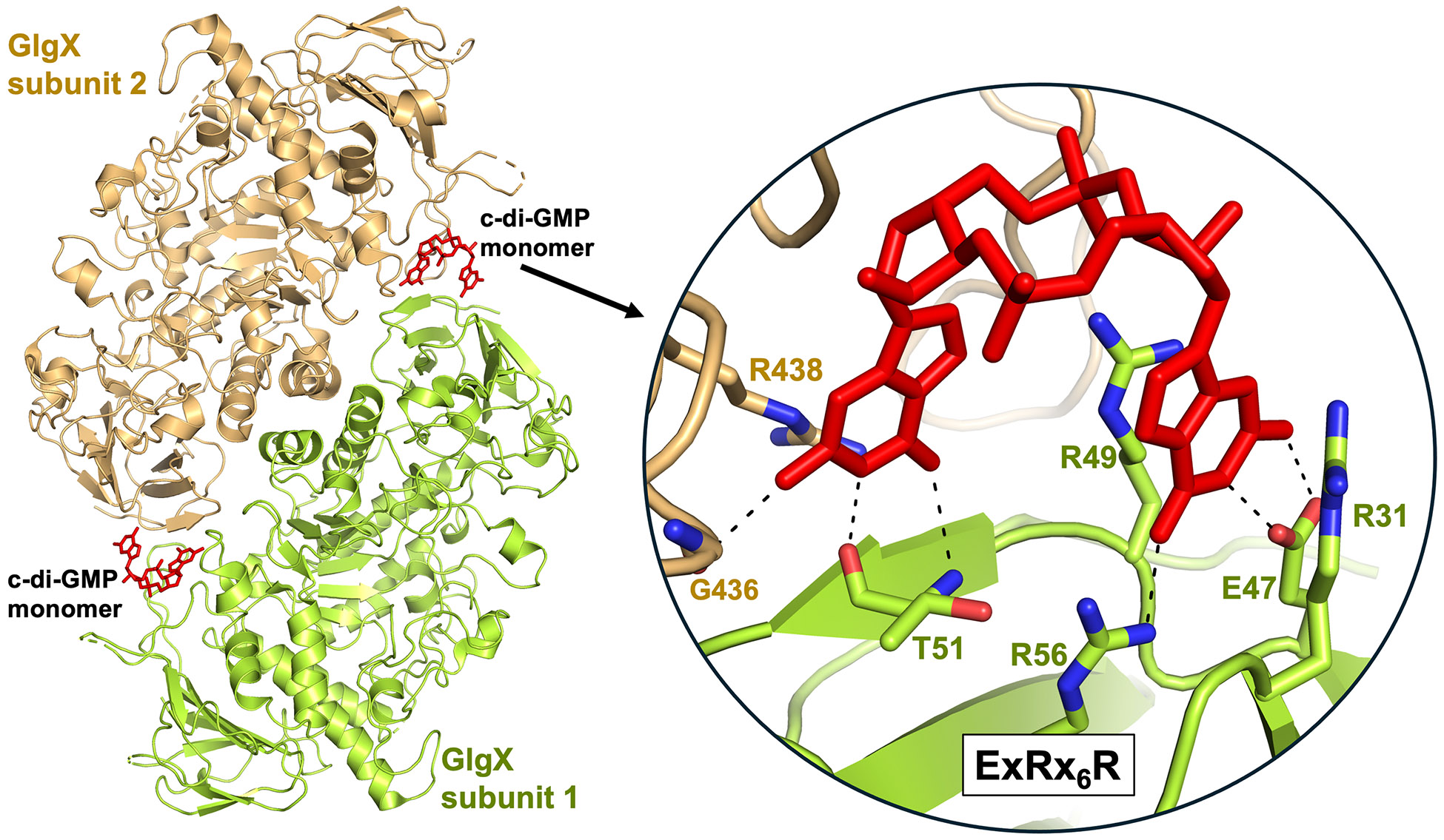
Ribbon diagram of the dimeric S. venezuelae GlgX2-c-di-GMP2 complex [16], with one GlgX subunit colored wheat and the other green. Shown as red sticks are the c-di-GMP molecules bound at the subunit interfaces at each end of the antiparallel GlgX dimer. The inset shows how the residues of the GlgX ExRx6R signature motif selectively bind c-di-GMP. Dashes indicate polar interactions.
Conclusions
Research from the last decade on c-di-GMP signaling in Streptomyces has revolutionized our view of the regulatory mechanisms that control progression through the life cycle of these biotechnologically important bacteria. The rise and fall of c-di-GMP levels, in concert with the Kds of the three effectors BldD, RsiG-σWhiG and GlgX, determine the timing and order of events during differentiation. Across development, the measured c-di-GMP levels range from 10-75 pmol c-di-GMP per mg total protein (Figure 1D) [16]. If the protein concentration of the S. venezuelae cytoplasm were ~100 μg μl−1, this would equate to c-di-GMP concentrations of 1-7.5 μM, and the Kd values of the three known Streptomyces c-di-GMP binding proteins sit in this approximate range (Figure 1D). To allow c-di-GMP dynamics to be monitored at the single-cell level – especially important in a differentiating, multicellular organism like Streptomyces – it will be important in the future to use c-di-GMP biosensors to visualize signaling heterogeneity by live cell microscopy [48,49].
The combination of physiological and structural analyses of c-di-GMP signaling in Streptomyces has proven to be extremely powerful for the discovery of unprecedented and unpredictable mechanisms through which a nucleotide second messenger can affect protein function, as exemplified by the BldD2-(c-di-GMP)4 and RsiG-(c-di-GMP)2-σWhiG complexes. BldD, RsiG-σWhiG, and GlgX all bind c-di-GMP via distinct and novel motifs, and none of the three proteins share homology with any previously identified c-di-GMP binding proteins or with each other. These examples suggest that the true diversity of c-di-GMP effectors is likely vastly underappreciated. As c-di-GMP is a nearly universal second messenger in bacteria, further characterization of the molecular mechanisms that underpin c-di-GMP signaling in diverse phyla will be an important focus of future research efforts aimed at understanding how bacteria coordinate responses to dynamic environmental conditions.
Acknowledgements
We thank Elijah Jones for helpful discussion. Research in the Gallagher laboratory is funded by startup provided through Cornell University’s College of Agriculture and Life Sciences. Research in the Tschowri laboratory is funded by the German Research Foundation (DFG) and by the European Research Council (ERC, SecMessFunctions, 101039556). Research in the Brennan laboratory is funded by a James B. Duke Distinguished Biochemistry Professorship. Research in the Schumacher laboratory is funded by the US National Institutes of Health (R35GM130290) and a Nanaline H. Duke Endowed Chair. Research in the Buttner lab was funded by the UK Biotechnology and Biological Sciences Research Council.
References
- 1.Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 2017, 15:271–284. [DOI] [PubMed] [Google Scholar]
- 2.Römling U, Galperin MY, Gomelsky M: Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 2013, 77:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hengge R, Pruteanu M, Stülke J, Tschowri N, Turgay K: Recent advances and perspectives in nucleotide second messenger signaling in bacteria. microLife 2023, 4:uqad015. **An extremely readable and comprehensive account of recent developments in the field of bacterial nucleotide second messenger signaling.
- 4.Burroughs AM, Zhang D, Schäffer DE, Iyer LM, Aravind L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res 2015, 43:10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou S-H, Galperin MY. Diversity of cyclic di-GMP-binding proteins and mechanisms. J Bacteriol 2016, 198:32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flärdh K, Buttner MJ: Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 2009, 7: 36–49. [DOI] [PubMed] [Google Scholar]
- 7.Bush MJ, Tschowri N, Schlimpert S, Flärdh K, Buttner MJ: c-di-GMP signaling and the regulation of developmental transitions in Streptomyces. Nat Rev Microbiol 2015, 13:749–760. [DOI] [PubMed] [Google Scholar]
- 8.McCormick JR, Flärdh K: Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev 2012, 36:206–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ: Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 2014, 158:1136–1147. ** Shows that c-di-GMP controls entry into development by driving dimerization of BldD, the master repressor of the sporulation regulatory cascade. The structure shows the two C-terminal domains of BldD have no protein-protein contacts and are instead bridged by a unique tetrameric cage of c-di-GMP.
- 10.den Hengst CD, Tran NT, Bibb MJ, Chandra C, Leskiw BK, Buttner MJ. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol 2010, 78:361–379. [DOI] [PubMed] [Google Scholar]
- 11.Tran NT, den Hengst CD, Gomez-Escribano J-P, Buttner MJ. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J Bacteriol 2011, 193:3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hull TD, Ryu MH, Sullivan MJ, Johnson RC, Klena NT, Geiger RM, Gomelsky M, Bennett JA. Cyclic Di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J Bacteriol 2012, 194:4642–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Bassam MM, Haist J, Neumann SA, Lindenberg S, Tschowri N: Expression patterns, genomic conservation and input into developmental regulation of the GGDEF/EAL/HD-GYP domain proteins in Streptomyces. Front Microbiol 2018, 9:2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Latoscha A, Wörmann ME, Tschowri N: Nucleotide second messengers in Streptomyces. Microbiology 2019, 165:1153–1165. [DOI] [PubMed] [Google Scholar]
- 15. Haist J, Neumann SA, Al-Bassam MM, Lindenberg S, Elliot MA, Tschowri N: Specialized and shared functions of diguanylate cyclases and phosphodiesterases in Streptomyces development. Mol Microbiol 2020, 114:808–822. ** Determines the regulon of the two key DGCs CdgC and CdgB and of the two central PDEs RmdA and RmdB in S. venezuelae. The study shows that c-di-GMP affects expression of cell division genes and of chaplin and rodlin genes that are needed for the formation of the hydrophobic sheath of aerial hyphae and spores.
- 16. Schumacher MA, Wörmann ME, Henderson M, Salinas R, Latoscha A, Al-Bassam MM, Singh KS, Barclay E, Gunka K, Tschowri N: Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP. Nat Commun 2022, 13:5834. ** Demonstrates in biochemical and structural detail how, late in development, rising c-di-GMP levels directly activate GlgX-mediated degradation of glycogen, releasing stored carbon for spore development.
- 17.Tschowri N: Cyclic dinucleotide-controlled regulatory pathways in Streptomyces species. J Bacteriol 2016, 198:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteomics 2012, 75:4874–4878. [DOI] [PubMed] [Google Scholar]
- 19.Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol Microbiol 2001, 40:257–269. [DOI] [PubMed] [Google Scholar]
- 20.Kim IK, Lee CJ, Kim MK, Kim JM, Yim HS, Cha SS, Kang SO. Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2). Mol Microbiol 2006, 60:1179–1193. [DOI] [PubMed] [Google Scholar]
- 21.Kim JM, Won HS, Kang SO. The C-terminal domain of the transcriptional regulator BldD from Streptomyces coelicolor A3(2) constitutes a novel fold of winged-helix domains. Proteins: Structure, function and bioinformatics 2014, 82:1093–1098. [DOI] [PubMed] [Google Scholar]
- 22.Schumacher MA, Zeng W, Findlay KC, Buttner MJ, Brennan RG, Tschowri N: The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex. Nucleic Acids Res 2017, 45:6923–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentner M, Allan MG, Zaehringer F, Schirmer T, Grzesiek S. Oligomer formation of the bacterial second messenger c-di-GMP: reaction rates and equilibrium constants indicate a monomeric state at physiological concentrations. J Am Chem Soc 2012, 134:1019–29. [DOI] [PubMed] [Google Scholar]
- 24.Mouri Y, Konishi K, Fujita A, Tezuka T, Ohnishi Y. Regulation of sporangium formation by BldD in the rare actinomycete Actinoplanes missouriensis. J Bacteriol 2017, 199:e00840–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buttner MJ. Actinoplanes swims into the molecular age. J Bacteriol 2017, 199:e00070–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopwood DA: Streptomyces in nature and medicine: the antibiotic makers. Oxford University Press, New York, NY. 2007. [Google Scholar]
- 27.Chng C, Lum AM, Vroom JA, Kao CM. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc Natl Acad Sci 2008, 105:11346–11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan H, Lu X, Sun D, Zhuang S, Chen Q, Chen Z, Li J, Wen Y. BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Mol Microbiol 2020, 113:123–142. **Shows that in some Streptomyces species, the BldD-(c-di-GMP)-mediated regulation of developmental processes extends to the direct control of the production of medically and commercially important antibiotics.
- 29. Gallagher KA, Schumacher MA, Bush MJ, Bibb MJ, Chandra G, Holmes NA, Zeng W, Henderson M, Zhang H, Findlay KC, Brennan RG, Buttner MJ: c-di-GMP arms an anti-σ to control progression of multicellular differentiation in Streptomyces. Mol Cell 2020, 77:586–599. **c-di-GMP controls the differentiation of reproductive hyphae into spores by arming a novel anti-σ (RsiG) to bind and sequester the sporulation-specific σ factor, σWhiG.
- 30.Flärdh K, Findlay KC, Chater KF: Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 1999, 145:2229–2243. [DOI] [PubMed] [Google Scholar]
- 31.Al-Bassam MM, Bibb MJ, Bush MJ, Chandra G, Buttner MJ: Response regulator heterodimer formation controls a key stage in Streptomyces development. PLoS Genetics 2014, 10: e1004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Becher PG, Verschut V, Bibb MJ, Bush MJ, Molnár BP, Barane E, Persson J, Al-Bassam MM, Chandra C, Song L, Challis GL, Buttner MJ, Flärdh K: Developmentally regulated volatiles geosmin and 2-methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal. Nature Microbiol 2020, 5:821–829. **Shows that the volatiles geosmin and 2-methylisoborneol that give Streptomyces their characteristic earthy odor are produced under developmental control as an integral part of the sporulation process, completing the Streptomyces life cycle by attracting soil arthropods that serve as vectors for spore dispersal.
- 33.Paget MS: Bacterial sigma factors and anti-sigma factors; Structure, function and distribution. Biomolecules 2015, 5:1245–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou S-H, Galperin MY. Cyclic di-GMP in streptomycetes: A new conformation, new binding mode, new receptor, and a new mechanism to control cell development. Mol Cell 2020, 77:443–445. [DOI] [PubMed] [Google Scholar]
- 35.Krasteva PJ, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 2010, 327:866–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuyama BY, Krasteva PV, Baraquet C, Harwood CS, Sondermann H, Navarro MVAS. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc Natl Acad Sci USA 2016, 113:E209–E218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher MA, Zeng W. Structures of the activator of K. pneumonia biofilm formation, MrkH, indicates PilZ domains involved in c-di-GMP and DNA binding. Proc Natl Acad Sci USA 2016, 113:10067–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F, He Q, Su K, Gao F, Huang Y, Lin Z, Zhu D, Gu L. The PilZ domain of MrkH represents a novel DNA binding motif. Protein Cell 2016, 7:766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raju H, Sharma R. Crystal structure of BrlR with c-di-GMP. Biochem Biophys Res Commun 2017, 490:260–264. [DOI] [PubMed] [Google Scholar]
- 40. Schumacher MA, Gallagher KA, Holmes NA, Chandra C, Henderson M, Kysela DT, Brennan RG, Buttner MJ: Evolution of a σ-(c-di-GMP)-anti-σ switch. Proc Natl Acad Sci USA 2021, 118:e2105447118. ** Shows that an antisigma-(c-di-GMP)-sigma switch has been co-opted during evolution to regulate distinct biological functions in unicellular and filamentous bacteria, controlling type IV pilus production in Rubrobacter and the differentiation of reproductive hyphae into spores in Streptomyces. It also shows that the antisigma originated as a dimer and evolved to become a monomer through an intragenic duplication event.
- 41.Craig L, Forest KT, Maier B: Type IV pili: dynamics, biophysics and functional consequences. Nat Rev Microbiol 2019, 17:429–440. [DOI] [PubMed] [Google Scholar]
- 42.Cifuente JO, Comino N, Trastoy B, D'Angelo C, Guerin ME. Structural basis of glycogen metabolism in bacteria. Biochem J 2019, 476:2059–2092. [DOI] [PubMed] [Google Scholar]
- 43.Preiss J. Glycogen: Biosynthesis and Regulation. EcoSal Plus 2014, 6. 10.1128/ecosalplus.ESP-0015-2014. [DOI] [PubMed] [Google Scholar]
- 44.Schniete JK, Brüser T, Horn MA, Tschowri N: Specialized biopolymers: versatile tools for microbial resilience. Curr Opin Microbiol 2024, 77:102405. [DOI] [PubMed] [Google Scholar]
- 45.Yeo M, and Chater K The interplay of glycogen metabolism and differentiation provides an insight into the developmental biology of Streptomyces coelicolor. Microbiology 2005, 151:855–861. [DOI] [PubMed] [Google Scholar]
- 46.Bruton CJ, Plaskitt KA, Chater KF. Tissue-specific glycogen branching isoenzymes in a multicellular prokaryote, Streptomyces coelicolor A3(2). Mol Microbiol 1995, 18:89–99. [DOI] [PubMed] [Google Scholar]
- 47.Plaskitt KA, Chater KF. Influences of developmental genes on localized glycogen deposition in colonies of a mycelial prokaryote, Streptomyces coelicolor A3(2): a possible interface between metabolism and morphogenesis. Philos Trans Roy Soc B 1995, 347:105–121. [Google Scholar]
- 48. Kaczmarczyk A, van Vliet S, Jakob RP, Teixeira RD, Scheidat I, Reinders A, Klotz A, Maier T, Jenal U. A genetically encoded biosensor to monitor dynamic changes of c-di-GMP with high temporal resolution. Nature Commun 2024, 15:3920. ** Describes the construction of a fluorescence reporter with a large dynamic range that allows in vivo c-di-GMP levels to be monitored at the single cell level, and its application in Caulobacter and Pseudomonas. The reporter fuses two copies of the c-di-GMP-binding domain of BldD to the N- and C- termini of circularly permutated EGFP. The BldD domain homodimerizes in the presence of the ligand, resulting in a dramatic change in fluorescence.
- 49.Halte M, Wörmann ME, Bogisch M, Erhardt M, Tschowri N: BldD-based bimolecular fluorescence complementation for in vivo detection of the second messenger cyclic di-GMP. 2022, Mol Microbiol 117:705–713. [DOI] [PubMed] [Google Scholar]


