Abstract
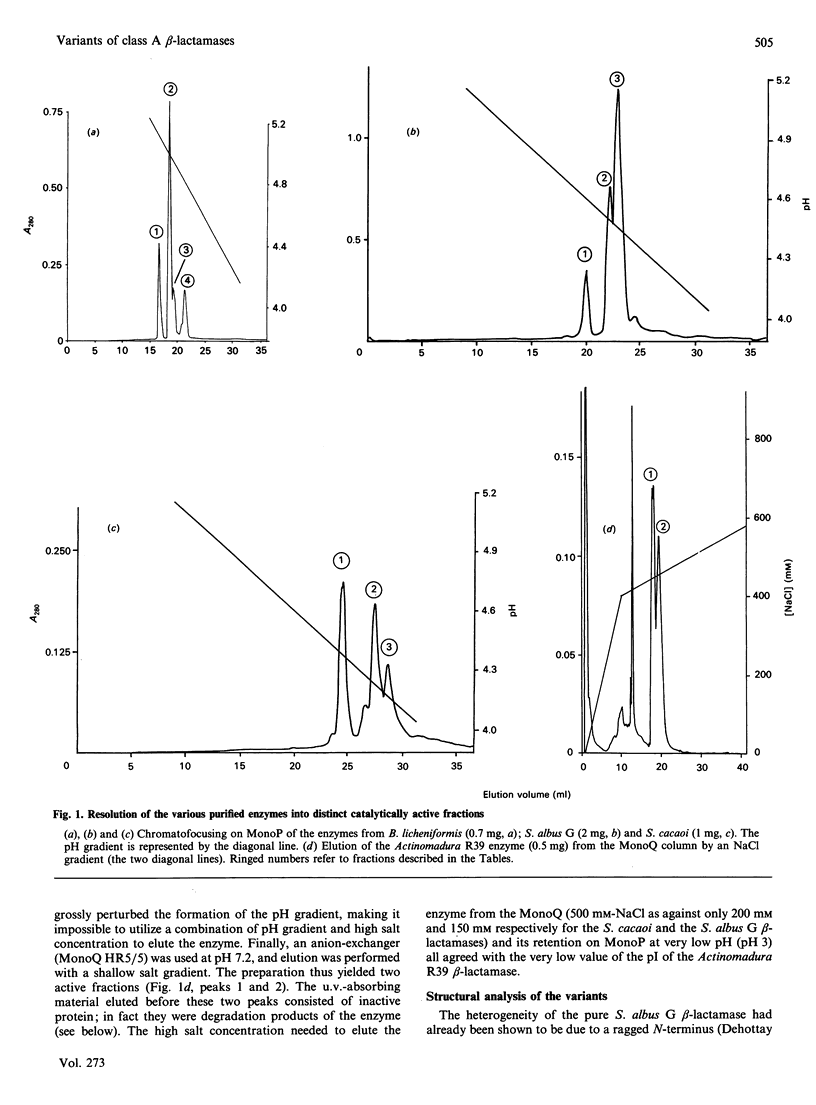
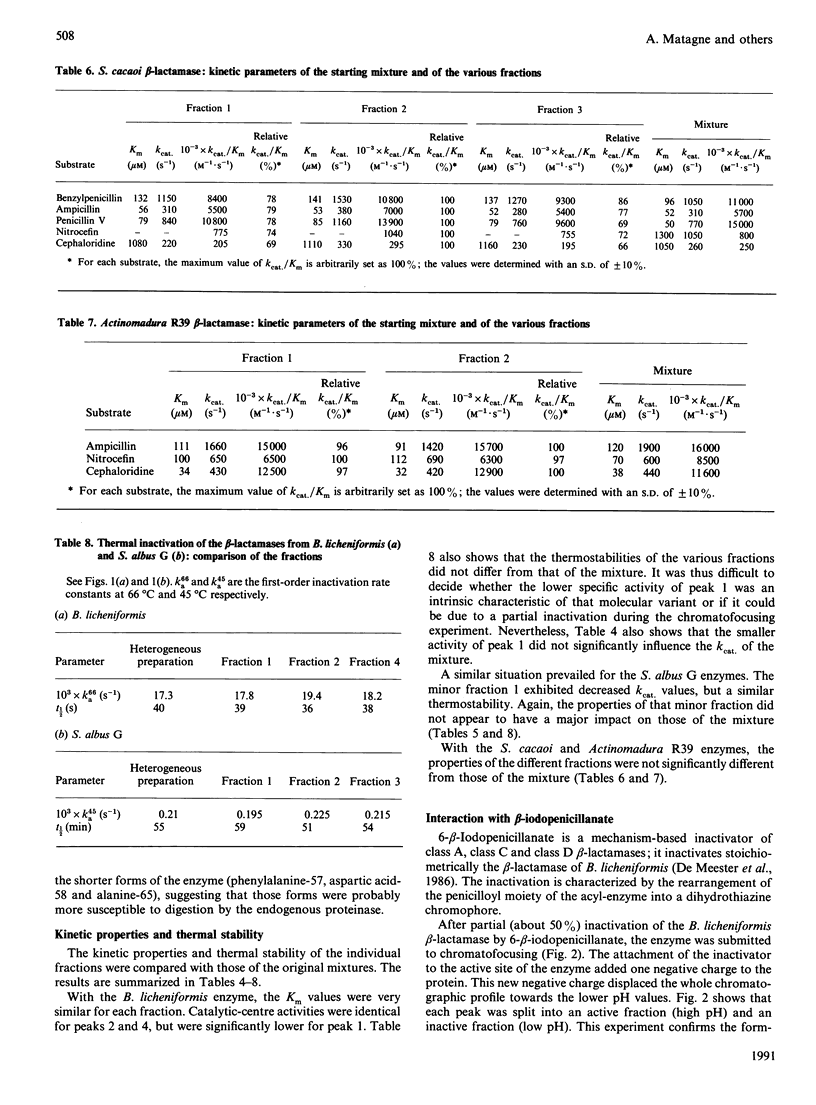
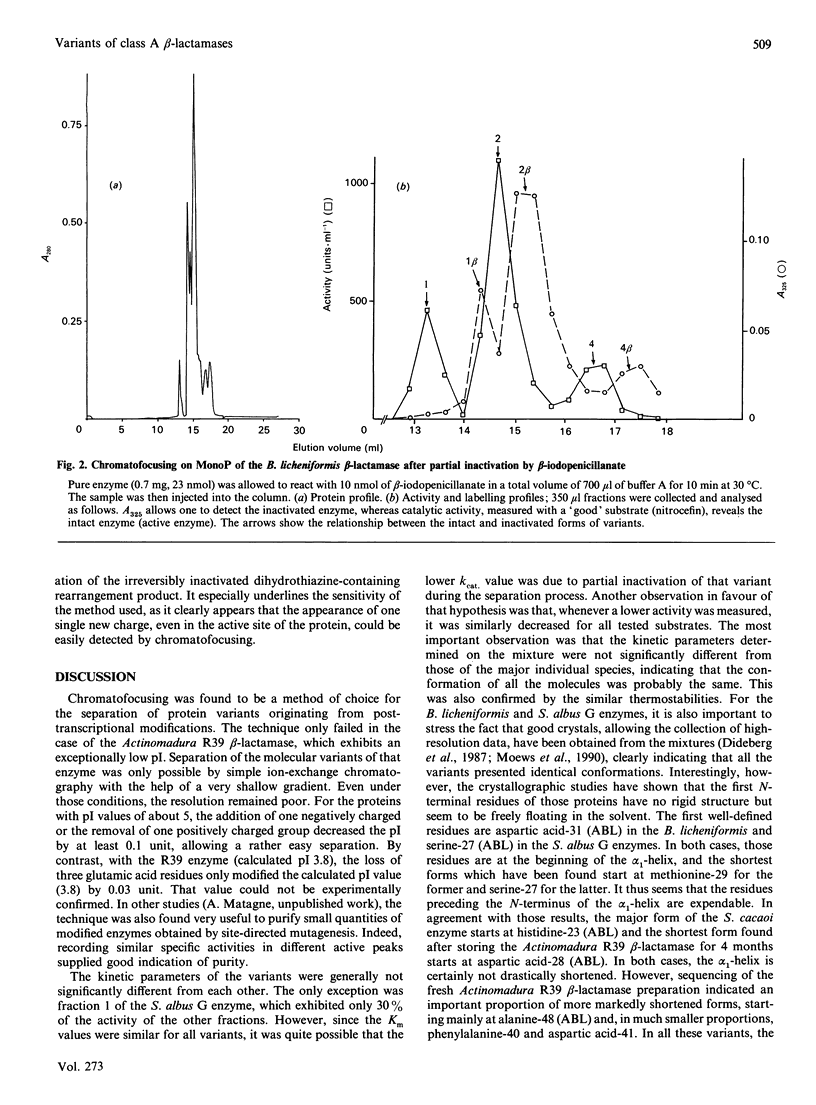
Four beta-lactamases excreted by Gram-positive bacteria exhibited microheterogeneity when analysed by chromatofocusing or ion-exchange chromatography. Ragged N-termini were in part responsible for the charge variants, but deamidation of an asparagine residue was also involved, at least for the Bacillus licheniformis enzyme. The activity of a contaminating proteinase could also be demonstrated in the case of Actinomadura R39 beta-lactamase. With that enzyme, proteolysis resulted in partial inactivation, but the inactivated fragments were easily separated from the active forms. With these, as with the other enzymes, the kinetic parameters of the major variants were identical with those of the mixture within the limits of experimental error, so that the catalytic properties of these enzymes can be determined with the 'heterogeneous' preparations.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Meadway R. J. Chemical structure of bacterial penicillinases. Nature. 1969 Apr 5;222(5188):24–26. doi: 10.1038/222024a0. [DOI] [PubMed] [Google Scholar]
- Brive C., Barthelemy M., Bouanchaud D. H., Labia R. Microhétérogénéité en électrofocalisation analytique de beta-lactamases d'origine plasmidique. Ann Microbiol (Paris) 1977 Oct;128(3):309–317. [PubMed] [Google Scholar]
- De Meester F., Frère J. M., Waley S. G., Cartwright S. J., Virden R., Lindberg F. 6-beta-Iodopenicillanate as a probe for the classification of beta-lactamases. Biochem J. 1986 Nov 1;239(3):575–580. doi: 10.1042/bj2390575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester F., Joris B., Reckinger G., Bellefroid-Bourguignon C., Frère J. M., Waley S. G. Automated analysis of enzyme inactivation phenomena. Application to beta-lactamases and DD-peptidases. Biochem Pharmacol. 1987 Jul 15;36(14):2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- Dehottay P., Dusart J., De Meester F., Joris B., Van Beeumen J., Erpicum T., Frère J. M., Ghuysen J. M. Nucleotide sequence of the gene encoding the Streptomyces albus G beta-lactamase precursor. Eur J Biochem. 1987 Jul 15;166(2):345–350. doi: 10.1111/j.1432-1033.1987.tb13521.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Wéry J. P., Dehottay P., Dusart J., Erpicum T., Frère J. M., Ghuysen J. M. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem J. 1987 Aug 1;245(3):911–913. doi: 10.1042/bj2450911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houba S., Willem S., Duez C., Molitor C., Dusart J., Frère J. M., Ghuysen J. M. Nucleotide sequence of the gene encoding the active-site serine beta-lactamase from Actinomadura R39. FEMS Microbiol Lett. 1989 Dec;53(3):241–246. doi: 10.1016/0378-1097(89)90224-3. [DOI] [PubMed] [Google Scholar]
- Izui K., Nielsen J. B., Caulfield M. P., Lampen J. O. Large exopenicillinase, initial extracellular form detected in cultures of Bacillus licheniformis. Biochemistry. 1980 Apr 29;19(9):1882–1886. doi: 10.1021/bi00550a023. [DOI] [PubMed] [Google Scholar]
- Joris B., De Meester F., Galleni M., Reckinger G., Coyette J., Frere J. M., Van Beeumen J. The beta-lactamase of Enterobacter cloacae P99. Chemical properties, N-terminal sequence and interaction with 6 beta-halogenopenicillanates. Biochem J. 1985 May 15;228(1):241–248. doi: 10.1042/bj2280241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzini M. V., Nojima S., Dusart J., Ogawara H., Dehottay P., Frere J. M., Ghuysen J. M. Cloning and amplified expression in Streptomyces lividans of the gene encoding the extracellular beta-lactamase from Streptomyces cacaoi. J Gen Microbiol. 1987 Oct;133(10):2915–2920. doi: 10.1099/00221287-133-10-2915. [DOI] [PubMed] [Google Scholar]
- Matagne A., Misselyn-Bauduin A. M., Joris B., Erpicum T., Granier B., Frère J. M. The diversity of the catalytic properties of class A beta-lactamases. Biochem J. 1990 Jan 1;265(1):131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moews P. C., Knox J. R., Dideberg O., Charlier P., Frère J. M. Beta-lactamase of Bacillus licheniformis 749/C at 2 A resolution. Proteins. 1990;7(2):156–171. doi: 10.1002/prot.340070205. [DOI] [PubMed] [Google Scholar]
- Mézes P. S., Blacher R. W., Lampen J. O. Processing of Bacillus cereus 569/H beta-lactamase I in Escherichia coli and Bacillus subtilis. J Biol Chem. 1985 Jan 25;260(2):1218–1223. [PubMed] [Google Scholar]
- Piron-Fraipont C., Duez C., Matagne A., Molitor C., Dusart J., Frère J. M., Ghuysen J. M. Cloning and amplified expression in Streptomyces lividans of the gene encoding the extracellular beta-lactamase of Actinomadura R39. Biochem J. 1989 Sep 15;262(3):849–854. doi: 10.1042/bj2620849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Sarvas M., Garoff H., Helenius A. Membrane-bound and secreted forms of penicillinase from Bacillus licheniformis. J Mol Biol. 1978 Dec 25;126(4):673–690. doi: 10.1016/0022-2836(78)90015-3. [DOI] [PubMed] [Google Scholar]
- Thatcher D. R. Beta-lactamase (Bacillus licheniformis). Methods Enzymol. 1975;43:653–664. doi: 10.1016/0076-6879(75)43130-5. [DOI] [PubMed] [Google Scholar]
- Thatcher D. R. The partial amino acid sequence of the extracellular beta-lactamase I of Bacillus cereus 569/H. Biochem J. 1975 May;147(2):313–326. doi: 10.1042/bj1470313. [DOI] [PMC free article] [PubMed] [Google Scholar]


