Abstract
The nervous system regulates perception, cognition and behavioral responses by serving as the body's primary communication system for receiving, regulating and transmitting information. Neurons are the fundamental structures and units of the nervous system. Their differentiation and maturation processes rely on the expression of specific biomarkers. Neuron-specific intracellular markers can be used to determine the degree of neuronal maturation. Neuronal cytoskeletal proteins dictate the shape and structure of neurons, while synaptic plasticity and signaling processes are intricately associated with neuronal synaptic markers. Furthermore, abnormal expression levels of biomarkers can serve as diagnostic indicators for nervous system diseases. This article reviews the markers of mature neuronal differentiation and their relationship with nervous system diseases.
Keywords: : biomarkers, cytoskeleton, nervous system diseases, neurons, synapses
Graphical Abstract
Plain language summary
Article highlights.
Mature neuron-specific intracellular markers: focuses on the most neuron-specific biomarkers so that researchers can better apply them to their experiments.
Cytoskeletal markers of mature neurons: discussion of biomarkers that maintain neuronal morphology and structure and analysis of their function.
Synaptic markers of mature neurons: understanding the development of neuronal synapses, analyzing the expression of synapse-related biomarkers and deepening the understanding of neuronal function.
Mature neuronal markers and neurological disorders: the use of neuronal biomarkers in neurological disorders and how they predict and diagnose disease.
Conclusions and future perspectives: summarizing the neuronal biomarkers discussed, discovering more novel and relevant neuronal biomarkers and looking forward to their future clinical applications.
1. Introduction
Neurons are the fundamental functional units of the nervous system, responsible for transmitting and processing information. Its structural development and maturation are crucial for the proper functioning of the brain's nervous system. During brain development and maturation, neurons undergo various changes. They send out branches, known as axons and dendrites, to the periphery and receive stimulation from specific protein molecules produced in the cell. This process allows them to form synapses with adjacent neurons, construct neural circuits and networks and maintain the normal morphology and function of neurons [1,2]. Biomarkers during differentiation of mature neurons mainly include protein molecules that are specifically expressed during neuronal development and functional maturation, and the type, localization and expression level of the biomarkers depend on the growth and function of different parts of the neuron [3–5]. Intracellular markers ensure neuronal specificity, cytoskeletal markers maintain cellular morphology during neuronal differentiation and synaptic markers ensure inter-neuronal information transfer. These markers are important for cell identification, functional observation, disease diagnosis, and prognosis [6–8]. Studying the production and expression of these biomarkers can enhance our understanding of neuronal development and maturation processes, as well as the associated molecular mechanisms.
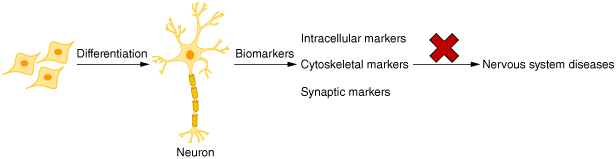
In the field of neurology, biomarkers are utilized to objectively monitor and assess normal physiology, pathological processes and therapeutic responses. Abnormal expression of certain markers has been utilized as a basis for diagnosing related diseases [9,10]. In this paper, we discuss mature neuronal biomarkers according to a roadmap (Figure 1) and analyze their localization, structure and function in mature neurons, as well as their relationship with nervous system diseases.
Figure 1.
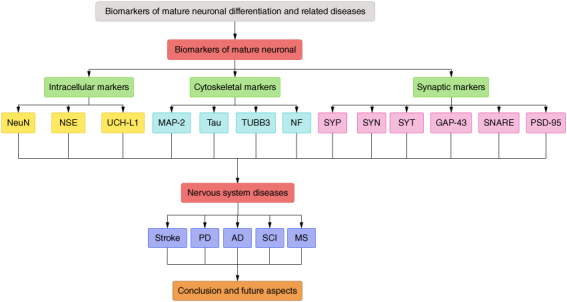
Roadmap for this review.
2. Mature neuron-specific intracellular markers
As neurons mature, the levels of neuron-specific biomarkers increase. Today, researchers use neuronal intracellular-specific markers to identify neurons and observe neuronal growth [11]. Mature neuronal cells contain a nucleus and cytoplasm. Markers for the nucleus and cytoplasm have been identified based on immunolocalization analysis [12–14]. Thus, the properties of neuron-specific intracellular markers are commonly used to identify neuronal cells. In addition, when brain injury occurs and neuronal cells are destroyed, intracellular-specific markers are released into the cerebrospinal fluid and blood, which may serve as a new tool for diagnosing disease [15,16].
2.1. Neuronal nuclei
In 1992, Mullen et al. [12] first discovered that Neuronal nuclei (NeuNs) recognizes a vertebrate nervous system and neuron-specific nuclear protein in mouse brain tissue. NeuN expression is exclusively associated with neural tissue. Unlike other biomarkers, it is primarily localized in the nucleus of mature neurons, making it the most specific marker for mature neurons (Table 1, Figure 2). A small distribution of NeuN was also found in the perinuclear cytoplasm. Immunoblotting assay revealed two bands of NeuN proteins: 46 and 48 kDa. The 46 kDa NeuN protein was predominantly located in the nucleus of neurons, while the 48 kDa NeuN protein was predominantly present in the cytoplasm, possibly due to their unique short amino acid sequences [17,18]. NeuN is encoded by the RNA binding fox-1 homolog 3 (RBFOX-3) gene, which belongs to the RNA binding fox-1 homolog (RBFOX) family of genes and regulates the differentiation of neurons in the brain and promotes their structural and functional maturation [19,20]. Additionally, there may be variations in NeuN expression among different types of neurons, as NeuN expression was not detected in certain cells such as Purkinje cells of the cerebellum, sympathetic ganglion cells, photoreceptor cells of the retina and dopaminergic neurons in the substantia nigra [18].
Table 1.
Markers of mature neurons.
| Marker | Years | Location | Function | Ref. |
|---|---|---|---|---|
| NeuN | 1992 | Cell nuclei | Recognition neurons; determine the neuronal phenotype; assess survival and death of neurons; and differential diagnosis of nerve tumors | [12,20,21] |
| NSE | 1965 | Cytoplasm | Maintain normal glucose metabolism; assess neuronal damage and neuroendocrine tumors; and monitor craniocerebral injury | [13,23–25] |
| UCH-L1 | 1983 | Cytoplasm and axons | Promote cell proliferation; protect neurons; monitor brain injury; and involvement in tumor metastasis | [14,28–30,32–34] |
| MAP-2 | 1975 | Cytoplasm and axons | Promote neuron maturation; maintain the morphological structure of neurons; modulate synaptic plasticity; and involved in signaling within neurons | [39,40,42,43] |
| Tau | 1974 | Cytoplasm and axons | Participate in the formation of microtubules and maintain the stability of neuronal cytoskeleton; affect synaptic plasticity and signal transduction; and maintain the integrity of DNA and RNA | [44,46,48–50] |
| TUBB3 | 1986 | Cytoplasm and axons | Ensure the growth and maintenance of axons; and promote the proliferation, invasion and metastasis of tumor cells | [52–54,57] |
| NF | 1957 | Axons | Maintain the growth stability of axons; monitor nerve damage; and diagnosing neurological disorders | [58–60] |
| SYP | 1985 | Synaptic vesicle | Stable membrane fusion hole; modulate vesicle circulation and synaptic plasticity; and monitor neuroendocrine tumors | [65,67–70] |
| SYN | 1977 | Synaptic vesicle | Involved in the growth and development of neurons; promote aggregation of synaptic proteins and vesicles; and maintain neurotransmitter transmission | [71,76–78] |
| SYT | 1981 | Synaptic vesicle | Controls vesicle fusion with presynaptic membranes; and maintain synaptic transmission | [79,80,82,83] |
| GAP-43 | 1981 | Presynaptic membrane | Maintain neuronal development and synaptic formation; and regulate the budding of vesicles | [85,87–89] |
| SNAP-25 | 1989 | Presynaptic membrane | Promote vesicles and presynaptic membrane fusion; and involved in axon growth | [92,93,96] |
| VAMP | 1989 | Synaptic vesicle | Participate in membrane fusion, achieve exocytosis and transmitter release; and promote elongation of axons | [91,94,96,97] |
| Syntaxin | 1992 | Presynaptic membrane | Maintain vesicle exocytosis; and involved in neural development, axon germination and growth | [91,92,95,96] |
| PSD-95 | 1981 | Postsynaptic membrane | Promote synaptic maturation; regulate intracellular signaling; and maintain synaptic plasticity. | [98,99,102,103] |
Figure 2.
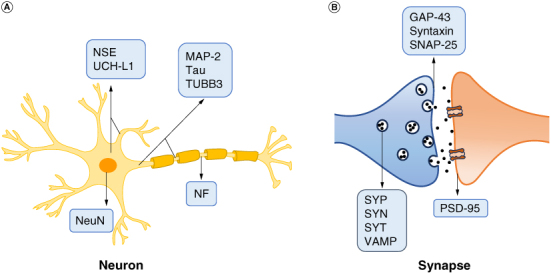
Localization of biomarkers in mature neurons. (A) In neurons, NeuN is located in the nucleus; NSE and UCH-L1 are mainly located in the cytoplasm; MAP-2, Tau and TUBB3 are mainly located in cytoplasm and axons; NF is located in the axon. (B) In neuronal synapses, SYN, SYP, SYT and VAMP are located in synaptic vesicles; GAP-43, SNAP-25 and Syntaxin are located in the presynaptic membrane; PSD-95 is localized in the postsynaptic membrane.
NeuN is involved in the development of mature neurons. The RBFOX knockout animal model constructed by Jacko et al. [20] showed defects in the development of structures such as the cytoskeleton, cytoplasmic membrane and synapses, resulting in limited electrophysiological activity. Currently, NeuN antibodies are widely used in neuroscience, stem cell biology and disease diagnosis. The anti-NeuN protein can be utilized to determine neuronal phenotype and identify neuronal differentiation. Its expression level has been used to directly assess neuronal survival or death, and NeuN-positive cells have been used as a reliable marker to quantify the efficacy of treatments in experimental therapeutic studies [20]. In addition, NeuN has potential applications in the pathological diagnosis of neuro-oncology. Zhang et al. [21] conducted immunohistochemical analysis of tumor samples from patients with neurocytoma and found that some neurocytomas had NeuN immunoreactivity in the nucleus. However, a comprehensive understanding of the role and expression pattern of NeuN in the diagnosis of different tumors is lacking, and further studies are needed to clarify the exact role of NeuN in neural development and maturation.
2.2. Neuron-specific enolase
Neuron-specific enolase (NSE) is an enzyme with enolase activity that is specific to neurons. In 1965, Moore and McGregor [13] isolated proteins from the bovine brain and identified the soluble brain protein 14-3-2, with a molecular weight of 78 kDa, which is widely distributed in intracranial neural tissues. Subsequent studies identified the protein 14-3-2 as NSE [22]. In vertebrates, enolase isozymes consist of different subunits called α, β and γ. Among these, enolase α is ubiquitous; enolase β is muscle-specific; and enolase γ is neuron-specific. There are five known enolase isozymes, αα, ββ, γγ, αβ and αγ. The γγ and αγ dimer isozymes are found in neurons and peripheral neuroendocrine cells, respectively [22,23]. Therefore, NSE is also known as γ-enolase or enolase-2 (Eno2). The level of NSE gradually increases as neurons mature morphologically.
In vivo, NSE catalyzes the cleavage of α-phosphoglycerol to produce phosphoenolpyruvate, which maintains normal glucose metabolism and ATP production throughout glycolysis [24]. NSE is commonly used as a marker for neurons and neuroendocrine cells, making it a useful tool for assessing neuronal injury and neuroendocrine tumors. Haque et al. [23] found that the expression level of NSE was significantly elevated in rats following acute spinal cord injury (SCI), and that inhibition of NSE expression and activity could reduce secondary injury after SCI. Moreover, elevated levels of NSE, a marker for neuroendocrine cells, are associated with poorly differentiated tumors. Immunohistochemical experiments on samples from patients with adenocarcinoma tumors found that NSE expression levels increased when these sites were predisposed to malignancy [25]. This information is valuable for diagnosing, staging, and treating related tumors. Additionally, NSE also possesses neurotrophic functions. It can regulate neuronal survival, differentiation and synapse regeneration by activating the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling pathways [26]. In clinical practice, NSE is easily affected by external factors, resulting in low accuracy of results. Therefore, it is necessary to constantly improve the detection methods and technologies of NSE [27].
2.3. Ubiquitin C-terminal hydrolase L1
Ubiquitin C-terminal hydrolase L1 (UCH-L1) is a member of the ubiquitin C-terminal hydrolase family. It was originally discovered by Jackson et al. [28] in human brain extracts with a molecular weight of about 24.8 kDa, and was named ‘protein gene product 9.5’. Immunocytochemical staining showed that in the normal mouse brain, UCH-L1 showed strong and uniform cytoplasmic staining throughout the neurons [14]. The gene encoding UCH-L1, also known as the Parkinson disease 5 (PARK5) gene, is located on chromosome 4P14. Its structure consists of two lobes, one of which consists of five α-helices and the other of two α-helices and six β-folds. These α-helices and β-folds form a relatively compact α-β-α structure.UCH-L1 also contains an N-terminal domain and a C-terminal domain. Its N-terminus binds to ubiquitin and is involved in the localization and regulatory functions of UCH-L1, whereas the C-terminus regulates the stability of substrate proteins by catalyzing hydrolysis reactions and removing ubiquitin from the substrate proteins [14,29].
UCH-L1 is associated with cell proliferation, neuronal survival and synaptic plasticity [29,30]. Overexpression of UCH-L1 inhibits cell mitosis, leading to decreased cell proliferation, while underexpression of UCH-L1 results in abnormal accumulation of proteins, affecting the normal physiological functions of the brain [30,31]. Reichelt et al. [29] found that non-functional UCH-L1 impairs the proteasome and drives podocyte injury in mice with membranous nephropathy. Clinically, UCH-L1 is now widely used in the screening, diagnosis and therapeutic monitoring of patients with brain injury [32]. Recent studies have found that UCH-L1 is associated with tumor metastasis [14,33,34]. The expression of UCH-L1 in primary gastric cancer and liver metastasis of gastric cancer was detected by immunohistochemical staining in vitro. UCH-L1 promotes the metastasis of gastric cancer cells by up-regulating protein kinase B (Akt) and extracellular signal-regulated kinases (ERK1/2), and by constructing a mouse model of lung metastasis, it was found that upregulation of hypoxia-inducible factor-1α (HIF-1α) activity promoted the metastasis of breast cancer cells and lung cancer cells [33,34]. However, there is still some controversy about the mechanism of action of UCH-L1 in other tumors.
3. Cytoskeletal markers of mature neurons
The morphology and structure of neuronal cells are essential for maintaining normal function, and the cytoskeleton determines neuronal morphology [35]. As neurons differentiate and mature, the levels of cytoskeletal markers increase, regulating the polarization and stability of neurons by supporting and maintaining their morphological structure [36]. Additionally, cytoskeletal markers participate in cell transport and synaptic transmission of neurons [37]. By examining the expression of cytoskeletal markers in neurons, it can be inferred that the neurons have a normal structure and function.
3.1. Microtubule-associated protein-2
Microtubule-associated protein-2 (MAP-2) is an essential part of the cytoskeleton and influences cell structure and function by controlling microtubule stability, dynamics and localization [38]. It is a member of the MAPs family and is expressed from the mitotic stage of cell proliferation. MAP-2 is a high molecular weight protein, which was discovered by Sloboda et al. [39], using primary rat hippocampal cultures prepared from fetal rats, and it mainly exists in the axons and dendrites of neurons. It consists of multiple structural domains, including a core structural domain and multiple C-terminal structural domains, which allow MAP-2 to attach to microtubules and regulate their stability and dynamics. MAP-2 proteins have five isoforms, A, B, C, D and E. By studying the nerve tissue of rats, it was found that their distribution showed obvious specificity [40,41]. Among them, MAP-2A and MAP-2B are mainly localized in neuronal dendrites, MAP-2C is present in axons and dendrites, MAP-2D is concentrated in neuronal cytosol, and MAP-2E is exclusive to glial cells. These isoforms display varying expression during neuronal development, with MAP-2C being replaced by MAP-2A during maturation, while MAP-2B is expressed throughout development.
The level of MAP-2 expression is closely associated with neuronal maturity [42]. During the early stages of neuronal development, MAP-2 expression is low and gradually increases as neurons mature. MAP-2 regulates intracellular signaling pathways by interacting with other proteins, such as protein kinases, thereby influencing neuronal functions [43]. Synapses are key structures for communication between neurons, and MAP-2 is involved in synapse formation and information transmission. Kim et al. [40] found that MAP-2 was involved in activation-dependent synaptic plasticity in the mature hippocampal network by isolating hippocampal neurons from rat brains and inducing long-term potentiation. The absence or mutation of MAP-2 results in abnormal development and loss of function in neural synapses. However, the specific mechanisms and regulatory networks involved need to be further investigated.
3.2. Tau protein
Tau protein is another highly abundant MAP found mainly in the axons of neurons. In 1974, Iqbal et al. [44] first isolated neurofibrillary tangles and paired helical filaments proteins from the brains of Alzheimer patients. Subsequently, Weingarten et al. [45] isolated proteins essential for microtubule assembly from pig brains and named them Tau protein, revealing that paired helical filament proteins and Tau protein are the same substance. The Human Tau protein is encoded by the microtubule-associated protein tau (MAPT) gene, located on chromosome 17q21. Its structure consists of an N-terminal proline-rich region, a microtubule-binding domain, and a C-terminal, The microtubule-binding domain in Tau protein interacts with microtubules to maintain cytoskeletal stability [46]. In addition, when the gene encoding Tau protein is mutated, it reduces the affinity of Tau protein to bind microtubules and affects the formation of neuronal cytoskeleton in the brain. Tau protein is a phosphoprotein, its phosphorylation state is strictly regulated by multiple kinases and phosphatases in physiological conditions. However, when the phosphorylation process of Tau protein is imbalanced, the affinity of Tau protein for microtubules decreases, leading to an increase in Tau protein aggregation and further exacerbating microtubule instability [46,47].
Tau proteins are proteins enriched in axons and have been implicated in cytoskeleton stabilization and synaptic transmission [46]. Tau protein binds to microtubules and is translocated into axons via members of the dynamin and kinesin families, stabilizing the structure of axonal microtubules [48]. Researchers have found that induced pluripotent stem cell (iPSC)-derived neurons can secrete tau proteins, which are delivered to neighboring neurons and interact with synapses and mitochondria. When neuronal activity increases, secreted tau interacts with proteins on the outer surface of synaptic vesicles [49]. These findings reveal that Tau protein is essential for the transmission of information between nerve cells. Additionally, studies using primary neuronal cultures have shown that Tau also interacts with nucleic acids and localizes to the nucleus. Tau protein can directly bind to DNA and RNA, protecting them from oxidative damage and helping to maintain DNA and RNA integrity [50]. It provides new ideas for us to further explore the multifunctionality of Tau protein. However, the regulatory mechanism involved in Tau protein is very complex, which increases the difficulty of research.
3.3. Class III beta-tubulin
The growth and development of the nervous system is highly dependent on the cytoskeleton, the main component of which is the microtubule, which consists of heterodimers of α and β-microtubule proteins. The α/β-microtubulin heterodimer is a combination of different α and β-microtubulin isoforms. Currently, there are eight α-microtubulin isoforms and seven β-microtubulin isoforms in humans [51]. Of these, Class III beta-tubulin (TUBB3) protein expression is restricted to neurons and peaks during axon guidance and neuronal maturation [52,53]. TUBB3 was first isolated from a mouse cDNA library in 1986 [54] and has a molecular weight of approximately 50 kDa. The TUBB3 gene is located on chromosome 16q24.3 and encodes the 450 amino acid TUBB3 protein. TUBB3 consists of two identical subunits, each containing a GTP-binding site and a microtubule-binding site. The state of the GTP-binding site of the TUBB3 protein can affect the stability and dynamics of microtubules, and regulate the kinetic properties of microtubules by interacting with GTP-binding proteins. Meanwhile, the microtubule-binding site of the TUBB3 protein interacts with other microtubule proteins and participates in microtubule formation and maintenance [55,56].
TUBB3 is primarily located in axons and dendrites and is involved in neurogenesis, axon guidance, and maintenance. Latremoliere et al. [52] found that the growth rate of axons in neurons of TUBB3 knockout mice was greatly reduced compared with normal mice. Tischfield et al. [53] constructed a TUBB3 knockout mouse model revealing axon guidance defects. Downregulation or mutation of TUBB3 gene expression leads to malformations in cortical development and the development of certain related neurological disorders, such as congenital extraocular myofibrillar fibrosis type 3 (CFEOM3) [56]. Moreover, overexpression of the TUBB3 protein has been associated with various malignant tumors, including brain tumors, lung cancers, renal carcinomas and malignant melanomas. It is involved in tumor cell proliferation, invasion and metastasis [57]. Considering the close relationship between TUBB3 protein and cancer, inhibition of TUBB3 protein expression in an in vitro mouse model significantly hindered tumor growth and metastasis [51,57]. Therefore, further in-depth studies on the function and regulatory mechanism of TUBB3 protein are needed to develop new therapeutic strategies and drug targets to bring hope for the treatment of related cancers.
3.4. Neurofilament
Neurofilament (NF) proteins, which are intermediate filament proteins, are highly conserved and are mainly found in neuronal axons. They were discovered by Maxfield et al. [58] in 1957, when they were isolated from squid axons. NF proteins are arranged in parallel along the long axis of the axon and consist of neurofilament light chain (NFL), neurofilament medium chain (NFM), neurofilament heavy chain, alpha-linked proteins, and peripheral proteins. The first three proteins are often referred to as the “neurofilament triplet”. NFL is widely expressed and can self-assemble to form functional fibers, but neurofilament medium chain or neurofilament heavy chain alone cannot form functional fibers [59]. In addition, these NFs share the same structural domains, including an N-terminal head domain, a helical core rod domain of 1a, 1b, and 2a/b, and a variable C-terminal tail domain [60]. The process of NF proteins assembly begins with the dimerization of NFL or α-internexin with other NFs, which then aggregate in an antiparallel manner, resulting in the formation of tetramers. Eight transversely associated tetramers then arrange themselves to form a cylindrical structure. Subsequent annealing processes facilitate the longitudinal and lateral extension of the cylindrical NF, ultimately leading to the formation of NFs with a diameter of 10 nm [61] (Figure 3). As neuron matures, nestin and waveform proteins are replaced by NF proteins. Axons gradually elongate, and connect, and protein transportation, assembly and modification processes commence.
Figure 3.
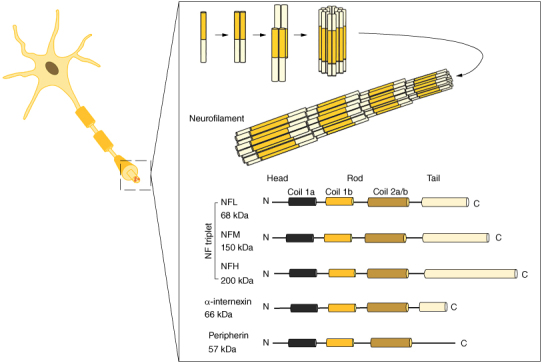
Assembly and structure of NF proteins. NF is formed by the formation of NFL or α-internexin by binding to the conserved rod domain to form a coiled spiral dimer with other NFs, two dimers assembled in a contralateral parallel fashion to form a tetramer, and eight laterally bound tetramers to form a cylindrical structure, which is lengthened longitudinally and compressed to form a slender filamentous NF. The domains of all neurofilament proteins consist of an N-terminal domain, a helical core containing 1a, 1b and 2a/b, and a variable C-terminal tail domain.
NF proteins act as part of the cytoskeleton and are localized in neuronal axons. The exact function of NF is unknown, but it can stabilize the morphology of axons and regulate the growth and positioning of axons and dendrites to achieve efficient and high-speed nerve conduction [60]. Also, NF interacts with other proteins and organelles, such as mitochondria and microtubules. This suggests that NF proteins may have additional undiscovered functions beyond axon stability [62]. The expression of NF proteins is limited to mature neurons and is one of the earliest visible markers in the mature nervous system. NFL, the most significant component of NF, is particularly important, and its expression level can be used to assess axon functionality. Gaetani et al. [59] observed in patients with amyotrophic spinal sclerosis an abnormally elevated level of cerebrospinal fluid NFL expression leading to axonal dysfunction and neurodegeneration. Thus, NF proteins are highly specific for neuronal cell injury and death and are widely used as valuable markers in the diagnosis of neurological disorders. In addition to axons, a large number of studies can be used to explore the function of NF in the future.
4. Synaptic markers of mature neurons
Neurons in the CNS form interconnected networks through synapses, which are the functional connections between neurons and the primary sites of information transmission [63]. Neuronal synaptic markers are specific proteins or molecules used to label neuronal synapses. By observing the expression levels of synaptic markers, we can understand the development of neuronal synapses and study the connections and communication modes between neurons [64]. This deepens our understanding of the function of the nervous system.
4.1. Synaptophysin (SYP)
SYP is a protein located on the membrane of synaptic vesicles in neurons. It was discovered by Jahn et al. [65] in 1985 from rat brain synaptic vesicles and is also known as p38. SYP has a relative molecular mass of 38 KDa and accounts for approximately 7–10% of total synaptic vesicle proteins. In humans and mammals, the SYP gene is located at Xp11.2-p11.23 on chromosome X and is highly conserved. The structure of SYP consists of four transmembrane structural domains, a short N-terminal signal peptide, and a long C-terminal hydrophobic peptide, the latter two of which are exposed on the cytoplasmic surface of the vesicle membrane. The presence of a hydrophilic region between the first and second transmembrane structural domains of SYP allows it to be encapsulated in the vesicle membranes of synaptic vesicles of neurons and to interact with other synaptic proteins [66]. Furthermore, the N-terminus of SYP contains many highly conserved amino acid sequences, while the C-terminus interacts with various proteins to regulate vesicle cycling [67]. SYP is expressed early in neurogenesis and is significantly upregulated during synaptogenesis [65]. Therefore, SYP can be used as a marker of synaptogenesis and neuronal maturation.
SYP regulates membrane fusion, cytokinesis, vesicle recycling, and synaptic plasticity by interacting with a variety of proteins. Hsiao et al. [68] discovered that SYP controls membrane fusion pore dynamics during Ca2+-triggered exocytosis and soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins are crucial in the formation of membrane fusion pores at the onset of cytotoxicity. Specifically, the transmembrane structural domain III of SYP interacts with the SNARE complex to stabilize the membrane fusion pores. Chang et al. [67] studied mouse synapses and found that SYP interacts with dynamin to regulate vesicle fusion, maintain synaptic plasticity and ensure normal neurotransmitter release. Under normal physiological conditions, SYP is involved in the recycling of vesicle-associated membrane protein (VAMP) in the vesicular cycle. Gordon et al. [69] found that SYP in mouse hippocampal neurons can transport synaptic VAMP to vesicles, making them capable of fusion, while SYP deficiency causes VAMP to disperse along axons, become trapped on the plasma membrane and limit vesicle endocytosis. Furthermore, Konukiewitz et al. [70] conducted immunohistochemical staining of tumor tissues and found that SYP could be used as a new clinical diagnostic marker for neuroendocrine tumors, especially for the effective diagnosis of pancreatic neuroendocrine tumors. However, its accuracy and specificity need to be analyzed for clinical application
4.2. Synapsin
Synapsin (SYN) is mainly situated on the surface of synaptic vesicles and is a phosphorylated protein closely linked to neuronal development and neurotransmitter release. The SYN protein family consists of three isoforms: SYN I, SYN II and SYN III. SYN I was originally discovered by Paul Greengard in 1977 from bovine brains [71], followed by SYN II in slices of rat cerebral cortex [72]. SYN III was discovered in 1998 during the early work study of the Human Genome Project [73]. In humans, SYN I, SYN II and SYN III are produced by selective splicing and located on chromosomes X, 3 and 22. The N-terminal domain of SYN is highly conserved, and the C-terminal domain is relatively unstable. This allows SYN proteins to interact with other proteins to form a complex regulatory network that regulates synaptic development and messaging [74]. SYN is mainly localized at synapses, whereas SYN III is mainly found in extrasynaptic regions of the adult brain [75]. This suggests that SYN III, unlike SYN I and II, does not play a major role in synaptic activity, but rather in early neural development.
Interneuronal communication relies on exocytosis from synaptic vesicles. SYN, a presynaptic vesicle membrane protein, can regulate synaptic vesicle dynamics. Recent studies have shown that SYN aggregates vesicles through liquid-liquid phase separation, providing a new organizational framework for synapses [76]. Additionally, SYN binds to related proteins and facilitates the aggregation of synaptic proteins and vesicles to uphold neurotransmitter transmission. Yu et al. [77] conducted electrophysiological and electron microscopic examinations on synaptic vesicles of Hidradenitis elegans and found that SYN regulates neurotransmitter release by binding vesicles to cytoskeletal proteins in the axon. In the absence of SYN, vesicles were unable to be captured at the release site and thus could not fuse with the membrane, failing neurotransmitter release. SYN has also been implicated in synaptic plasticity. Song et al. [78] showed in hippocampal neurons cultured from SYN subtypes knockout mice that synaptic growth was delayed and axon branching was absent, which is not conducive to axon growth and development, and that different isoforms of the SYN family had different roles at the presynaptic terminals, which provided a new way of thinking about the development, and different subtypes of SYN family had different effects on presynaptic terminals. However, the specific mechanism of action and the relationship between subtypes are still unclear.
4.3. Synaptotagmin
The Synaptotagmin (SYT) family is a crucial regulator of calcium-dependent membrane fusion events, located on synaptic vesicles in brain neurons. The SYT protein, identified by Matthew et al. [79] in 1981, has a molecular weight of 65 KDa, consists of 220 amino acids, and mainly affects neurotransmitter release and membrane translocation. Structurally, SYT has an N-terminal site in the vesicle and a C-terminal site in the cytoplasm and also includes a transmembrane domain, a parafollicular domain and a calcium-binding C2 domain consisting of C2A and C2B. These domains are linked to transmembrane domains on vesicles via parafollicular junctional domains to form the SYT core framework. In the presence of Ca2+, the C2 domain inserts into the vesicle membrane, mediating vesicle fusion and regulating transmitter release [80]. Humans have seventeen SYT isoforms, with SYT1, SYT2 and SYT7 regulating synaptic vesicle release, SYT3 involved in endocytosis of synaptic proteins, SYT4 and SYT11 responsible for vesicle transport, SYT6 involved in BDNF release, SYT10 as a neuroprotective effector protein and SYT11 involved in membrane repair, among others [81]. The functional diversity of these isoforms contributes to understanding nervous system functioning.
Normal brain function relies on precise control of membrane fusion events. SYT serves as both a promoter and inhibitor of vesicle fusion. The C2A and C2B domains of SYT spontaneously oligomerize and inhibit fusion by binding to Ca2+. Until an excess of Ca2+ breaks down the SYT ring, membrane fusion, and release are triggered [80]. SYT is essential for maintaining reliable synaptic transmission. Lebowitz et al. [82] discovered that neurotransmitter transmission depends on SYT1 and synchronized release in response to initial stimulation in murine neuronal cells. Loss of SYT blocks rapid synchronized neurotransmitter release, leading to reduced synaptic transmission and short-term depression, as well as insensitivity to presynaptic Ca2+. Ullah et al. [83] found that vesicles need to undergo processes such as docking and initiation before fusion can occur and that SYT interacts with receptors on the plasma membrane (e.g., Syntaxin1, SNAP-25) to promote vesicle docking and initiate vesicle fusion via Ca2+ influx. In addition, Tawfik et al. [84] found that stimulation of SYT7 in the mouse brain increased the number of vesicles fused through SYT1 but negatively affected their fusion rate, suggesting both synergistic and competitive interactions between synaptic receptors. It can be concluded that the mechanism of SYT action is complex and the study is limited by biological complexity.
4.4. Growth associated protein-43
Growth associated protein-43 (GAP-43), also known as B-50 or neuromodulin, is a low molecular weight phosphoprotein primarily found in the growth cone terminals and presynaptic membranes of neurons. It was discovered in 1981 by Skene and Willard [85] through intraocular methionine injection in rabbits. The human GAP-43 gene is located on chromosome 3 and contains two promoters: P1 and P2. GAP-43 expression is mediated by a helix-loop-helix mechanism acting on the active P2 promoter, from which the majority of GAP-43 mRNA is also derived [86]. Although GAP-43 is classified as a neuron-specific protein, there is growing evidence that it is not restricted to neurons. Caprara et al. [87] found that GAP-43 was expressed in adult mouse myoblasts, myotubes and adult skeletal muscle fibers, and was located between the calcium-releasing unit and mitochondria of mammalian skeletal muscle, where it was involved in maintaining intracellular calcium homeostasis.
During neuronal development, there is a significant increase in the expression level of GAP-43 protein. Subsequently, GAP-43 influences ion channels and signaling by phosphorylating and releasing calmodulin, which promotes the polymerization of microtubulin and actin at the growth cone of the presynaptic membrane and regulates vesicle outgrowth [87,88]. Overexpression of GAP-43 protein promotes synaptic plasticity and enhances connections and communication between neurons. However, reduced GAP-43 expression hinders neurotransmitter release, leading to disease [86]. Wang et al. [89] established an optic nerve injury model in rats and found that inhibition of GAP-43 could promote retinal cell apoptosis, promote the expression of GAP-43, and increase the number of axons in the optic nerve. Additionally, Cheng et al. [90] provided optogenetic stimulation to stroke mice and found that the upregulation of nerve growth factor and brain-derived neurotrophic factor (BDNF) was accompanied by an increase in GAP-43 levels. The activation of nerve growth factor and BDNF signaling promotes neuroprotection, synaptogenesis, and neural regeneration, making GAP-43 an important effector driven by BDNF.
4.5. Soluble N-ethylmaleimide-SNARE
SNARE complex proteins are the core components connecting vesicles to the plasma membrane and are mainly composed of synaptosomal-associated protein 25 (SNAP-25), VAMP/Synaptobrevin, and syntaxin [91,92]. SNAP-25 was discovered by Oyler et al. [93] in 1989 in the mouse brain, with a molecular weight of 25KDa, and is expressed in the presynaptic membrane of neurons. VAMP was extracted from synaptic vesicles in rat brains by Baumert et al. [94] in 1989, with a molecular weight of 18KDa, and it consists of a SNARE structural domain and a C-terminal transmembrane structural domain. Syntaxin, which is the largest family of proteins in the SNARE complex, was discovered in 1992 by Bennett et al. [95] using monoclonal antibody-immunoprecipitation techniques and is located in the presynaptic membrane. It consists of one SNARE domain, a C-terminal transmembrane region, and an N-terminal regulatory domain. SNAP-25 has two SNARE motifs, and VAMP and Syntaxin each contain one SNARE motif, which assembles sequentially from the N-terminus to the C-terminus to form a four-helix bundle SNARE complex that releases energy for inducing membrane fusion [91] (Figure 4).
Figure 4.
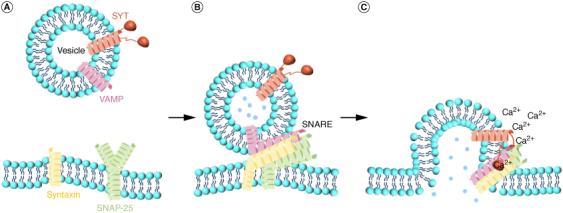
SNARE complex formation and neurotransmitter release. (A) The release of neurotransmitters requires the involvement of the SNARE complex as well as the regulatory protein SYT (red). Components of the SNARE complex include SNAP-25 (green), Syntaxin (yellow), and VAMP (purple). (B) Assembly of SNARE complexes. Syntaxin and VAMP bind to the SNARE domain of SNAP-25, respectively, to form a parallel four-helix bundle. (C) When calcium flows in, through Ca2+ binding to SYT, the vesicular membrane and plasma membrane fuse and open the fusion pore, allowing neurotransmitter release.
The SNARE complex is a core protein for membrane fusion during neurotransmitter release. Upon activation of SYT by Ca2+, it interacts with t-SNARE on the cell membrane, leading to the fusion of the cell membrane and plasma membrane for cytosolic and transmitter release [92]. However, the loss of proteins in the SNARE complex results in reduced synaptic vesicular exocytosis and decreased neurotransmitter release [96]. SNARE complex is critical for the growth and development of organisms [91]. They are involved in major membrane fusion events during mouse fertilization, such as acrosomal vesicle cell division and female and male gamete fusion. Syntaxin deletion causes physiological abnormalities and survival difficulties in embryonic mice. In nervous system development, SNAP-25, VAMP-2 and Syntaxin-1A are involved in the sprouting and growth of axons in nerve cells [91,96]. The SNARE complex is essential for cognitive function and long-term memory consolidation. Vanguilder et al. [97] observed that the protein expression levels of the hippocampal region in rats of different ages showed a significant decrease in proteins related to neurotransmitter release, including SNAP-25, VAMP-2, Syntaxin-1, SYT1 and SYP as age increased. In summary, the SNARE complex is well known for its role in neurotransmitter release, but it is also present throughout the life stages of an organism, and the functions of the SNARE complex involved in this stage are yet to be discovered.
4.6. Postsynaptic density protein-95 (PSD-95)
The postsynaptic density (PSD) is a complex of postsynaptic membrane signaling molecules, and PSD-95 is the most abundant and important scaffolding protein in the PSD [98]. In 1981 Sampedro et al. [99] found PSD-95 in the PSD through immunohistochemical study in the brains of mice and rats. It has also been known as synapse-associated protein 90, with a molecular weight of 95 KDa, and is a member of the related guanylate kinase family (MAGUK). PSD-95 comprises three PSD-95/Discs large/Zonula occludens-1 (PDZ) structural domains at the N-terminal end (PDZ1, PDZ2 and PDZ3), a Src homology 3 (SH3) structural domain in the middle, and a guanosine kinase (GUK) structural domain at the C-terminal end, in which the three structural domains, PDZ3, SH3 and GUK, are tightly associated to form a conserved supermodule PSG [100]. Hamilton et al. [101] combined discrete molecular dynamics and single-molecule forster resonance energy transfer to characterize PSG supermodules revealed that domains in PSG supermodules can interact with each other as well as recognize and bind key synaptic ligands. Meanwhile, the PDZ structural domain regulates function by binding to specific protein sequences, the SH3 structural domain interacts with proteins to regulate signaling and the GUK structural domain has protein kinase activity [98,100]. Thus, the different structural domains in PSD-95 bind to relevant protein receptors and signaling molecules and are involved in synapse formation, regulation of intracellular signaling, and maintenance of synaptic plasticity.
PSD-95 is a key protein involved in synapse development and maturation. Husseini et al. [102] discovered that overexpression of PSD-95 in hippocampal neurons drove synapse maturation and increased the number of dendritic spines. Conversely, in PSD-95 knockout mice, synaptic aggregation was disrupted, accompanied by abnormal dendritic spine development and transmitter delivery. Furthermore, PSD-95 heavily influences the localization of postsynaptic AMPA-type glutamate receptor (AMPAR) and NMDA-type glutamate receptor, which are crucial scaffolding proteins for cell signaling molecules. This enables rapid and efficient synaptic transmission by precisely juxtaposing AMPAR with presynaptic release sites. PSD-95 also binds directly to subunits of NMDA-type glutamate receptor and stabilizes its expression on the synaptic surface. Downregulation of PSD-95 leads to the loss of AMPAR-containing synapses, thus affecting the normal signaling function [98]. Additionally, Zhang et al. [103] found that PSD-95 is associated with synaptic plasticity. Inhibiting PSD-95 expression disrupts synaptic plasticity and hinders the cAMP-responsive element-binding protein/BDNF pathway, resulting in cognitive dysfunction in mice. Conversely, elevating PSD-95 expression enhances cognitive function in mice. Nevertheless, the applicability of these findings may be limited by differences between animal models and the human nervous system.
5. Mature neuronal markers & neurological disorders
Neurological diseases are characterized by complex pathophysiological mechanisms. Our understanding of the pathogenesis, diagnosis and treatment of neurological diseases still has significant gaps. Studies have found that abnormalities in biomarker levels are associated with neurological diseases and change with disease onset and progression [16,104,105]. Additionally, biomarkers serve to predict and diagnose diseases and provide directions for exploring the mechanisms of diseases and their potential therapeutic options [106,107].
5.1. Stroke
Stroke is a major cause of disability and neurological disorders globally. Markers are now being used to diagnose, assess and predict stroke severity due to limitations in neuroimaging techniques. Onatsu et al. [108] found that serum NSE and Tau levels were positively correlated with infarct size in stroke patients, indicating potential as predictors of severe clinical manifestations. Bi et al. [109] discovered that Tau promotes neuronal damage after stroke, while Tau-deficient mice are protected from brain damage and neurological deficits. Pekny et al. [110] found that high levels of NFL in the blood were associated with poor clinical prognosis after stroke. Sandelius et al. [111] observed a transient increase in cerebrospinal fluid GAP-43 in ischemic stroke patients, which was correlated with stroke severity, cerebral white matter lesions, and degree of atrophy. Haifeng Lu et al. [112] found that SYT3 was upregulated in ischemic stroke mice, and its knockdown prevented ischemic injury and promoted recovery. However, the association between the above stroke biomarkers and the pathological mechanisms involved is not clear. The association between stroke biomarkers and clinical manifestations needs to be further analyzed and applied in clinical trials, which is expected to be an effective tool for stroke prevention and diagnosis.
5.2. Parkinson's disease
Parkinson's disease (PD) is a neurodegenerative disorder associated with abnormal accumulation of associated proteins. The disease is typically identified at an advanced stage of complete neuronal degeneration and lacks timely and effective treatment. Therefore, markers are essential to detect the disease in its early stages for prevention [22,113]. Research has shown that PD pathogenesis involves NSE, UCH-L1, Tau and SNARE complex. Papuć et al. [15] found elevated levels of NSE in the cerebrospinal fluid of patients with PD patients, which is considered an important marker of axonal degeneration in PD. UCH-L1 has been linked to the formation of Lewy bodies, a key pathological feature of PD [104]. Reduced levels of UCH-L1 in the fluid are associated with cognitive dysfunction in PD and can serve as a potential marker for diagnosing cognitive dysfunction in PD patients [104,114]. Meanwhile, α-synuclein is involved in the pathogenesis of PD, and Tau promotes the aggregation and proliferation of α-synuclein [115,116]. Knocking out the Tau protein in PD mice resulted in reduced symptoms [116]. Additionally, the aggregation of α-synuclein and its binding to the SNARE complex lead to synaptic dysfunction and could serve as new easily accessible markers [10]. Further studies on the link between biomarkers and clinical manifestations of PD are needed to determine the specificity and accuracy between them. Meanwhile, the expression of PD biomarkers can be synthesized and more PD biomarkers can be explored to provide a basis for early intervention and treatment of PD in the future.
5.3. Alzheimer's disease
Alzheimer's disease (AD) is a neurodegenerative disease, and the pathological deposition of Tau is a hallmark of AD. Currently, the detection of phosphorylated Tau in plasma and cerebrospinal fluid can be used as a criterion for the diagnosis of AD. Anti-Tau regimens have become a focus for the treatment of AD in the clinic, and related Tau marker technology has been widely used in clinical trials and practice [9]. In addition to Tau protein deposition, synaptic dysfunction is also a key feature of AD. Lanxia Meng et al. [117] found that the overexpression of SYN in the hippocampus resulted in synaptic dysfunction and cognitive impairment in AD mice. Meanwhile, Jia et al. [106] concluded that exosomes GAP-43, SNAP-25 and SYT can be used as effective biomarkers for predicting AD 5–7 years prior to cognitive impairment, which provides new evidence for clinical AD screening. Sandelius et al. [118] found that GAP-43 was significantly expressed in AD patients and correlated with the degree of neuro progenitor fiber tangles and β-amyloid in the hippocampus, amygdala and cortex. Recently Kivisäkk et al. [105] observed elevated levels of PSD-95 and SNAP-25 proteins in the cerebrospinal fluid of AD patients and used them as markers associated with synaptic pathology in AD. In summary, AD biomarkers are closely related to the pathomechanisms and symptoms of the disease. Monitoring the specificity between AD and these markers can improve the diagnostic rate of AD. Currently, the combination of AD biomarkers and animal model studies is insufficient. Therefore, there is a need to widely apply biomarkers to animal experiments and develop therapeutic programs based on the pathological processes involved.
5.4. Spinal cord injury
SCI is a complex and serious neurological disorder for which no effective treatment has been found. The use of markers to prevent and monitor the occurrence of SCI is currently of interest to scholars. McCoy et al. [119] showed that SCI leads to upregulation of NSE, and NSE inhibitors attenuate the activation of related kinases and promote functional recovery after SCI. Therefore, inhibition of NSE could be a potential therapeutic strategy to prevent neurodegeneration and promote nerve cell regeneration and repair after SCI. NSE and NF were found to be significantly elevated in the blood of patients with acute SCI, and they were used as markers of acute SCI [23,59]. In addition, SCI is associated with hyperphosphorylation of Tau, and it was found that cerebrospinal fluid and serum Tau phosphorylation levels were significantly elevated in SCI rats, and there was a positive correlation between Tau protein levels and SCI severity [120]. Recently, Stukas et al. [16] found that cerebrospinal fluid and serum levels of UCH-L1 were significantly elevated in patients with acute SCI, ranging from 10 to 100-times higher than normal. After the condition of SCI patients improved, UCH-L1 levels decreased significantly. This suggests that UCH-L1 can be used as a marker to reflect the severity of SCI injury and prognosis. Using the expression characteristics of these biomarkers in SCI for clinical observation is a promising avenue for future research. Constructing SCI animal models to observe the relationship between biomarkers and pathology will provide new targets and directions for future SCI treatment and intervention.
5.5. Multiple sclerosis
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system caused by autoimmunity, which leads to tissue damage and disability. Neuroaxonal damage is a critical factor in the development of permanent disability in MS, resulting in abnormal expression of markers [121]. According to Disanto et al. [122] observed that NFL levels were significantly higher in MS patients than in normal subjects, and the correlation between serum NFL levels, disease activity and severity can be used to monitor tissue damage and treatment outcomes. Also, abnormal Tau levels have been associated with chronic axonal damage. Virgilio et al. [107] concluded that cerebrospinal fluid Tau levels in MS patients predicted the accumulation of early disability in MS patients predicted the accumulation of early disability and may have predictive value in diagnosing MS, leading to early treatment. Furthermore, disability in MS is primarily caused by axonal loss, and Petrova et al. [123] found a notable decrease in spinal synapses in the spinal cord of MS patients post-mortem, using immunohistochemistry with SYP and SYN. Although some progress has been made in the research of MS biomarkers, the specific mechanism of MS biomarkers is still unclear. Therefore, we need to conduct animal experiments and clinical diagnostic analyses of MS patients and explore more MS-related biomarkers to provide a more reliable basis for the prevention and treatment of MS patients.
6. Conclusion
Mature neuronal biomarkers are valuable tools in neuroscience research as they aid in understanding neuronal development. In nerve regeneration research, biomarkers are commonly used to identify cells and observe cell growth status. Different biomarkers are differentially expressed at different stages of neuronal differentiation. For example, in the early stages of neuronal development, NeuN can be used for labeling; however, as neuronal development matures, more options are available for labeling with NSE and MAP-2. In addition, neuronal cytoskeletal and synaptic markers are also widely used for neuronal identification labeling and functional assessment. TUBB3 and MAP-2 and Tau maintain the normal morphology of neurons and promote axonal and dendritic extension and branching formation.NF stabilizes axonal and dendritic growth and localization, enabling efficient message transmission. Synaptic vesicle biomarkers (SYP, SYN, SYT) are involved in vesicle transport and cycling to ensure normal neurotransmitter release. Presynaptic membrane biomarkers (GAP-43) are involved in the regulation of vesicle outgrowth and work in synergy with vesicle biomarkers to promote neurotransmitter release. SNARE complex acts as a bridge between the vesicle and plasma membrane, initiating membrane fusion and the release of neurotransmitters from the vesicle into the synaptic gap. The postsynaptic membrane biomarker (PSD-95) receives presynaptic release signals, integrates released neurotransmitters and maintains synaptic plasticity.
Recent studies have implicated markers such as presynaptic proteins (Bassoon and Piccolo), neurotoxins (Neurexin) and postsynaptic proteins (SHANK, Homer and SAPAP) in the regulation of neuronal growth, development and function [124–126]. Bassoon and Piccolo are components of the presynaptic region that maintain synaptic integrity and regulate synaptic vesicle release. Neurexin is central to transsynaptic cell adhesion and signaling during synapse specification and maintenance, and Shank, Homer and SAPAP establish the PSD of glutamatergic synapses through a dense network of molecular interactions. However, few studies have been conducted on these biomarkers, and their specific mechanisms of action and functional relationships are unclear.
7. Future perspective
As neuroscience research continues, it is believed that biomarkers of mature neurons will play an even more important role in future research. Researchers should search for new neuronal biomarkers to support the identification and classification of neurons. In addition, in neurological diseases, nerve cells are damaged and the associated biomarkers are released into body fluids. Observation of biomarkers from cerebrospinal fluid and blood have become a diagnostic criterion for related diseases as an easy way to screen for diseases and is in the stage of clinical application. However, the research between biomarkers and neurological diseases is still in the stage of animal experiments, and in the future, biomarkers can be tested with human tissues and then further applied to clinical patients. In addition, the developmental and functional regulatory mechanisms of the nervous system are very complex, and more biomarkers need to be combined to observe neuronal development. Currently, single-cell technology is developing rapidly, and utilizing this technology to detect the expression of biomarkers in neuronal cells and explore their association with neurological diseases will bring broader therapeutic prospects.
Acknowledgments
We thank the Department of Neurology, Kailuan General Hospital, North China University of Science and Technology, and the Key Laboratory of Neurobiology and Function of Hebei Province for providing the research platform.
Funding Statement
This work was supported by the Hebei Province Innovation Capacity Improvement Program in 2020 - Special Project for the Construction of Science and Technology R&D Platform and New R&D Institution (grant number: 20567622H). Hebei Province Medical applicable technology tracking project in 2023 (grant number: GZ2023049).
Author contributions
X Yuan: Writing – original draft, Writing – review & editing. W Li: Writing – original draft, Writing – review & editing. Q Yan: Writing – review. Y Ou: Writing – review & editing. Q Long: Writing – review. P Zhang: Writing – original draft, Writing – review & editing, Funding acquisition.
Financial disclosure
This work was supported by the Hebei Province Innovation Capacity Improvement Program in 2020 - Special Project for the Construction of Science and Technology R&D Platform and New R&D Institution (grant number: 20567622H). Hebei Province Medical applicable technology tracking project in 2023 (grant number: GZ2023049).
Competing interests disclosure
The authors have no other competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Scott-Solomon E, Boehm E, Kuruvilla R. The sympathetic nervous system in development and disease. Nat Rev Neurosci. 2021;22(11):685–702. doi: 10.1038/s41583-021-00523-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batool S, Raza H, Zaidi J, Riaz S, Hasan S, Syed NI. Synapse formation: from cellular and molecular mechanisms to neurodevelopmental and neurodegenerative disorders. J Neurophysiol. 2019;121(4):1381–1397. doi: 10.1152/jn.00833.2018 [DOI] [PubMed] [Google Scholar]
- 3.Subbarayan R, Girija DM, Rao SR. Human umbilical cord tissue stem cells and neuronal lineages in an injectable caffeic acid-bioconjugated gelatin hydrogel for transplantation. J Cell Physiol. 2019;234(3):1967–1977. doi: 10.1002/jcp.26834 [DOI] [PubMed] [Google Scholar]
- 4.Bufalo MC, Almeida MES, Jensen JR, et al. Human sensory neuron-like cells and glycated collagen matrix as a model for the screening of analgesic compounds. Cells. 2022;11(2):247. doi: 10.3390/cells11020247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sultan N, Amin LE, Zaher AR, Grawish ME, Scheven BA. Neurotrophic effects of dental pulp stem cells on trigeminal neuronal cells. Sci Rep. 2020;10(1):19694. doi: 10.1038/s41598-020-76684-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartoli F, Misiak B, Crocamo C, Carrà G. Glial and neuronal markers in bipolar disorder: a meta-analysis testing S100B and NSE peripheral blood levels. Prog Neuropsychopharmacol Biol Psychiatry. 2020;101:109922. doi: 10.1016/j.pnpbp.2020.109922 [DOI] [PubMed] [Google Scholar]
- 7.Bomont P. The dazzling rise of neurofilaments: physiological functions and roles as biomarkers. Curr Opin Cell Biol. 2021;68:181–191. doi: 10.1016/j.ceb.2020.10.011 [DOI] [PubMed] [Google Scholar]
- 8.Aronson JK, Ferner RE. Biomarkers-A General Review. Curr Protocols Pharmacol. 2017;76:9.23.21–29.23.17. doi: 10.1002/cpph.19 [DOI] [PubMed] [Google Scholar]; • Reviews the value and clinical applications of biomarkers.
- 9.Ossenkoppele R, Van Der Kant R, Hansson O. Tau biomarkers in Alzheimer's disease: towards implementation in clinical practice and trials. Lancet Neurol. 2022;21(8):726–734. doi: 10.1016/S1474-4422(22)00168-5 [DOI] [PubMed] [Google Scholar]
- 10.Agliardi C, Meloni M, Guerini FR, et al. Oligomeric α-Syn and SNARE complex proteins in peripheral extracellular vesicles of neural origin are biomarkers for Parkinson's disease. Neurobiol Dis. 2021;148:105185. doi: 10.1016/j.nbd.2020.105185 [DOI] [PubMed] [Google Scholar]
- 11.Church KA, Rodriguez D, Vanegas D, et al. Models of microglia depletion and replenishment elicit protective effects to alleviate vascular and neuronal damage in the diabetic murine retina. J Neuroinflamm. 2022;19(1):300. doi: 10.1186/s12974-022-02659-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116(1):201–211. doi: 10.1242/dev.116.1.201 [DOI] [PubMed] [Google Scholar]
- 13.Moore BW, Mcgregor D. Chromatographic and electrophoretic fractionation of soluble proteins of brain and liver. J Biol Chem. 1965;240:1647–1653. doi: 10.1016/S0021-9258(18)97483-1 [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Zhang N, Li M, Hong T, Meng W, Ouyang T. Ubiquitin C-terminal hydrolase-L1: a new cancer marker and therapeutic target with dual effects (Review). Oncol Lett. 2023;25(3):123. doi: 10.3892/ol.2023.13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papuć E, Rejdak K. Increased cerebrospinal fluid S100B and NSE reflect neuronal and glial damage in Parkinson's disease. Front Aging Neurosci. 2020;12:156. doi: 10.3389/fnagi.2020.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Biomarkers associated with axonal degeneration in the cerebrospinal fluid of Parkinson's patients revealed.
- 16.Stukas S, Gill J, Cooper J, et al. Characterization of cerebrospinal fluid ubiquitin C-terminal hydrolase L1 as a biomarker of human acute traumatic spinal cord injury. J Neurotrauma. 2021;38(15):2055–2064. doi: 10.1089/neu.2020.7352 [DOI] [PubMed] [Google Scholar]
- 17.Gusel'nikova VV, Korzhevskiy DE. NeuN as a neuronal nuclear antigen and neuron differentiation marker. Acta Naturae. 2015;7(2):42–47. doi: 10.32607/20758251-2015-7-2-42-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan W, Zhang YP, Hou Z, et al. Novel insights into neun: from neuronal marker to splicing regulator. Mol Neurobiol. 2016;53(3):1637–1647. doi: 10.1007/s12035-015-9122-5 [DOI] [PubMed] [Google Scholar]
- 19.Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284(45):31052–31061. doi: 10.1074/jbc.M109.052969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacko M, Weyn-Vanhentenryck SM, Smerdon JW, et al. Rbfox splicing factors promote neuronal maturation and axon initial segment assembly. Neuron. 2018;97(4):853–868.e856. doi: 10.1016/j.neuron.2018.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Fu W, Zheng L, et al. A clinicopathological and molecular analysis of sellar/suprasellar neurocytoma mimicking pituitary adenoma. Front Endocrinol. 2022;13:861540. doi: 10.3389/fendo.2022.861540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isgrò MA, Bottoni P, Scatena R. Neuron-specific enolase as a biomarker: biochemical and clinical aspects. Adv Exp Med Biol. 2015;867:125–143. doi: 10.1007/978-94-017-7215-0_9 [DOI] [PubMed] [Google Scholar]
- 23.Haque A, Polcyn R, Matzelle D, Banik NL. New insights into the role of neuron-specific enolase in neuro-inflammation, neurodegeneration, and neuroprotection. Brain Sci. 2018;8(2):33. doi: 10.3390/brainsci8020033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernández R, Blanco S, Peragón J, Pedrosa J, Peinado M. Hypobaric hypoxia and reoxygenation induce proteomic profile changes in the rat brain cortex. Neuromol Med. 2013;15(1):82–94. doi: 10.1007/s12017-012-8197-7 [DOI] [PubMed] [Google Scholar]
- 25.Mao X, Liu J, Hu F, et al. Serum NSE is early marker of transformed neuroendocrine tumor after EGFR-TKI treatment of lung adenocarcinoma. Cancer Management Res. 2022;14:1293–1302. doi: 10.2147/CMAR.S349082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner A, Obermajer N, Kos J. Gamma-Enolase C-terminal peptide promotes cell survival and neurite outgrowth by activation of the PI3K/Akt and MAPK/ERK signalling pathways. Biochem J. 2012;443(2):439–450. doi: 10.1042/BJ20111351 [DOI] [PubMed] [Google Scholar]
- 27.Luijerink L, Waters KA, Machaalani R. Immunostaining for NeuN does not show all mature and healthy neurons in the human and pig brain: focus on the hippocampus. Appl Immunohistochem Mol Morphol: AIMM. 2021;29(6):e46–e56. doi: 10.1097/PAI.0000000000000925 [DOI] [PubMed] [Google Scholar]
- 28.Jackson P, Thompson RJ. The demonstration of new human brain-specific proteins by high-resolution two-dimensional polyacrylamide gel electrophoresis. J Neurol Sci. 1981;49(3):429–438. doi: 10.1016/0022-510X(81)90032-0 [DOI] [PubMed] [Google Scholar]
- 29.Reichelt J, Sachs W, Frömbling S, et al. Non-functional ubiquitin C-terminal hydrolase L1 drives podocyte injury through impairing proteasomes in autoimmune glomerulonephritis. Nat Commun. 2023;14(1):2114. doi: 10.1038/s41467-023-37836-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Q, Zhang H, Li Y, Liu C, Wang S, Liao X. UCH-L1 inhibition suppresses tau aggresome formation during proteasomal impairment. Mol Neurobiol. 2018;55(5):3812–3821. doi: 10.1007/s12035-017-0558-7 [DOI] [PubMed] [Google Scholar]
- 31.Wang M. Dysfunctional UCH-L1 inhibits proteostasis. Nature Rev Nephrol. 2023;19(7):424. doi: 10.1038/s41581-023-00730-z [DOI] [PubMed] [Google Scholar]
- 32.Korley FK, Jain S, Sun X, et al. Prognostic value of day-of-injury plasma GFAP and UCH-L1 concentrations for predicting functional recovery after traumatic brain injury in patients from the US TRACK-TBI cohort: an observational cohort study. Lancet Neurol. 2022;21(9):803–813. doi: 10.1016/S1474-4422(22)00256-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goto Y, Zeng L, Yeom CJ, et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1α. Nat Commun. 2015;6:6153. doi: 10.1038/ncomms7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu YY, Yang M, Zhao M, et al. The de-ubiquitinase UCHL1 promotes gastric cancer metastasis via the Akt and Erk1/2 pathways. Tumour Biol: J Int Soc Oncodevelopment Biol Med. 2015;36(11):8379–8387. doi: 10.1007/s13277-015-3566-0 [DOI] [PubMed] [Google Scholar]
- 35.Di Giaimo R, Penna E, Pizzella A, Cirillo R, Perrone-Capano C, Crispino M. Cross talk at the cytoskeleton-plasma membrane interface: impact on neuronal morphology and functions. Int J Mol Sci. 2020;21(23):9133. doi: 10.3390/ijms21239133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pisciotta A, Lunghi A, Bertani G, et al. PEDOT: PSS promotes neurogenic commitment of neural crest-derived stem cells. Front Physiol. 2022;13:930804. doi: 10.3389/fphys.2022.930804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nag TC, Kathpalia P, Wadhwa S. Microtubule alterations may destabilize photoreceptor integrity: age-related microtubule changes and pattern of expression of MAP-2, Tau and hyperphosphorylated Tau in aging human photoreceptor cells. Exp Eye Res. 2020;198:108153. doi: 10.1016/j.exer.2020.108153 [DOI] [PubMed] [Google Scholar]
- 38.Bodakuntla S, Jijumon AS, Villablanca C, Gonzalez-Billault C, Janke C. Microtubule-associated proteins: structuring the cytoskeleton. Trends Cell Biol. 2019;29(10):804–819. doi: 10.1016/j.tcb.2019.07.004 [DOI] [PubMed] [Google Scholar]
- 39.Sloboda RD, Rudolph SA, Rosenbaum JL, Greengard P. Cyclic AMP-dependent endogenous phosphorylation of a microtubule-associated protein. Proc Natl Acad Sci U S A. 1975;72(1):177–181. doi: 10.1073/pnas.72.1.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y, Jang YN, Kim JY, et al. Microtubule-associated protein 2 mediates induction of long-term potentiation in hippocampal neurons. FASEB J. 2020;34(5):6965–6983. doi: 10.1096/fj.201902122RR [DOI] [PubMed] [Google Scholar]; • Suggests that the mature neuronal cytoskeletal marker MAP-2 is involved in synaptic plasticity.
- 41.Ferhat L, Bernard A, Ribas De Pouplana L, Ben-Ari Y, Khrestchatisky M. Structure, regional and developmental expression of rat MAP2d, a MAP2 splice variant encoding four microtubule-binding domains. Neurochem Int. 1994;25(4):327–338. doi: 10.1016/0197-0186(94)90139-2 [DOI] [PubMed] [Google Scholar]
- 42.Sánchez C, Díaz-Nido J, Avila J. Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog Neurobiol. 2000;61(2):133–168. doi: 10.1016/S0301-0082(99)00046-5 [DOI] [PubMed] [Google Scholar]
- 43.Plucarová J, Jansen S, Narasimhan S, et al. Specific phosphorylation of microtubule-associated protein 2c by extracellular signal-regulated kinase reduces interactions at its Pro-rich regions. J Biol Chem. 2022;298(10):102384. doi: 10.1016/j.jbc.2022.102384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iqbal K, Wisniewski HM, Grundke-Iqbal I, Korthals JK, Terry RD. Chemical pathology of neurofibrils. Neurofibrillary tangles of Alzheimer's presenile-senile dementia. J Histochem Cytochem. 1975;23(7):563–569. doi: 10.1177/23.7.1141687 [DOI] [PubMed] [Google Scholar]
- 45.Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci U S A. 1975;72(5):1858–1862. doi: 10.1073/pnas.72.5.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye H, Han Y, Li P, Su Z, Huang Y. The role of post-translational modifications on the structure and function of tau protein. J Mol Neurosci. 2022;72(8):1557–1571. doi: 10.1007/s12031-022-02002-0 [DOI] [PubMed] [Google Scholar]
- 47.Tapia-Rojas C, Cabezas-Opazo F, Deaton CA, Vergara EH, Johnson GVW, Quintanilla RA. It's all about tau. Prog Neurobiol. 2019;175:54–76. doi: 10.1016/j.pneurobio.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scholz T, Mandelkow E. Transport and diffusion of Tau protein in neurons. Cell Mol Life Sci. 2014;71(16):3139–3150. doi: 10.1007/s00018-014-1610-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tracy TE, Madero-Pérez J, Swaney DL, et al. Tau interactome maps synaptic and mitochondrial processes associated with neurodegeneration. Cell. 2022;185(4):712–728.e714. doi: 10.1016/j.cell.2021.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sultan A, Nesslany F, Violet M, et al. Nuclear tau, a key player in neuronal DNA protection. J Biol Chem. 2011;286(6):4566–4575. doi: 10.1074/jbc.M110.199976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duly AMP, Kao FCL, Teo WS, Kavallaris M. βIII-tubulin gene regulation in health and disease. Front Cell Dev Biol. 2022;10:851542. doi: 10.3389/fcell.2022.851542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Latremoliere A, Cheng L, Delisle M, et al. Neuronal-specific TUBB3 is not required for normal neuronal function but is essential for timely axon regeneration. Cell Rep. 2018;24(7):1865–1879.e1869. doi: 10.1016/j.celrep.2018.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tischfield MA, Baris HN, Wu C, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140(1):74–87. doi: 10.1016/j.cell.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan KF, Cleveland DW. Identification of conserved isotype-defining variable region sequences for four vertebrate beta tubulin polypeptide classes. Proc Natl Acad Sci U S A. 1986;83(12):4327–4331. doi: 10.1073/pnas.83.12.4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radwitz J, Hausrat TJ, Heisler FF, et al. Tubb3 expression levels are sensitive to neuronal activity changes and determine microtubule growth and kinesin-mediated transport. Cell Mol Life Sci. 2022;79(11):575. doi: 10.1007/s00018-022-04607-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puri D, Barry BJ, Engle EC. TUBB3 and KIF21A in neurodevelopment and disease. Front Neurosci. 2023;17:1226181. doi: 10.3389/fnins.2023.1226181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Person F, Wilczak W, Hube-Magg C, et al. Prevalence of βIII-tubulin (TUBB3) expression in human normal tissues and cancers. Tumour Biol. 2017;39(10):1010428317712166. doi: 10.1177/1010428317712166 [DOI] [PubMed] [Google Scholar]
- 58.Maxfield M, Hartley RWJr. Dissociation of the fibrous protein of nerve. Biochim Biophys Acta. 1957;24(1):83–87. doi: 10.1016/0006-3002(57)90149-X [DOI] [PubMed] [Google Scholar]
- 59.Gaetani L, Blennow K, Calabresi P, Di Filippo M, Parnetti L, Zetterberg H. Neurofilament light chain as a biomarker in neurological disorders. J Neurol Neurosurg Psychiatry. 2019;90(8):870–881. doi: 10.1136/jnnp-2018-320106 [DOI] [PubMed] [Google Scholar]; • NfL can serve as a diagnostic, prognostic, and monitoring biomarker for neurological disorders.
- 60.Khalil M, Teunissen CE, Otto M, et al. Neurofilaments as biomarkers in neurological disorders. Nature Rev Neurol. 2018;14(10):577–589. doi: 10.1038/s41582-018-0058-z [DOI] [PubMed] [Google Scholar]
- 61.Herrmann H, Aebi U. Intermediate filaments: structure and assembly. Cold Spring Harb Perspect Biol. 2016;8(11):a018242. doi: 10.1101/cshperspect.a018242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan A, Rao MV, Veeranna, Nixon RA. Neurofilaments and neurofilament proteins in health and disease. Cold Spring Harb Perspect Biol. 2017;9(4):a018309. doi: 10.1101/cshperspect.a018309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Südhof TC. The cell biology of synapse formation. J Cell Biol. 2021;220(7):e202103052. doi: 10.1083/jcb.202103052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maiti P, Bowers Z, Bourcier-Schultz A, Morse J, Dunbar GL. Preservation of dendritic spine morphology and postsynaptic signaling markers after treatment with solid lipid curcumin particles in the 5xFAD mouse model of Alzheimer's amyloidosis. Alzheimer's Res Ther. 2021;13(1):37. doi: 10.1186/s13195-021-00769-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Jahn R, Schiebler W, Ouimet C, Greengard P. A 38,000-dalton membrane protein (p38) present in synaptic vesicles. Proc Natl Acad Sci U S A. 1985;82(12):4137–4141. doi: 10.1073/pnas.82.12.4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kwon SE, Chapman ER. Synaptophysin regulates the kinetics of synaptic vesicle endocytosis in central neurons. Neuron. 2011;70(5):847–854. doi: 10.1016/j.neuron.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang CW, Hsiao YT, Jackson MB. Synaptophysin regulates fusion pores and exocytosis mode in chromaffin cells. J Neurosci. 2021;41(16):3563–3578. doi: 10.1523/JNEUROSCI.2833-20.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsiao YT, Jackson MB. Synaptophysin transmembrane domain III controls fusion pore dynamics in Ca(2+)-triggered exocytosis. Biophys. J. 2023;122(11):1962–1973. doi: 10.1016/j.bpj.2022.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gordon SL, Harper CB, Smillie KJ, Cousin MA. A fine balance of synaptophysin levels underlies efficient retrieval of synaptobrevin II to synaptic vesicles. PLOS ONE. 2016;11(2):e0149457. doi: 10.1371/journal.pone.0149457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Konukiewitz B, Jesinghaus M, Kasajima A, Klöppel G. Neuroendocrine neoplasms of the pancreas: diagnosis and pitfalls. Virchows Arch. 2022;480(2):247–257. doi: 10.1007/s00428-021-03211-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ueda T, Greengard P. Adenosine 3′:5′-monophosphate-regulated phosphoprotein system of neuronal membranes. I. Solubilization, purification, and some properties of an endogenous phosphoprotein. J Biol Chem. 1977;252(14):5155–5163. doi: 10.1016/S0021-9258(17)40170-0 [DOI] [PubMed] [Google Scholar]
- 72.Forn J, Greengard P. Depolarizing agents and cyclic nucleotides regulate the phosphorylation of specific neuronal proteins in rat cerebral cortex slices. Proc Natl Acad Sci U S A. 1978;75(10):5195–5199. doi: 10.1073/pnas.75.10.5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosaka M, Südhof TC. Synapsin III, a novel synapsin with an unusual regulation by Ca2+. J Biol Chem. 1998;273(22):13371–13374. doi: 10.1074/jbc.273.22.13371 [DOI] [PubMed] [Google Scholar]
- 74.Longhena F, Faustini G, Brembati V, Pizzi M, Benfenati F, Bellucci A. An updated reappraisal of synapsins: structure, function and role in neurological and psychiatric disorders. Neurosci Biobehav Rev. 2021;130:33–60. doi: 10.1016/j.neubiorev.2021.08.011 [DOI] [PubMed] [Google Scholar]
- 75.Porton B, Wetsel WC, Kao HT. Synapsin III: role in neuronal plasticity and disease. Semin Cell Develop Biol. 2011;22(4):416–424. doi: 10.1016/j.semcdb.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sansevrino R, Hoffmann C, Milovanovic D. Condensate biology of synaptic vesicle clusters. Trends Neurosci. 2023;46(4):293–306. doi: 10.1016/j.tins.2023.01.001 [DOI] [PubMed] [Google Scholar]
- 77.Yu SC, Liewald JF, Shao J, Steuer Costa W, Gottschalk A. Synapsin is required for dense core vesicle capture and cAMP-dependent neuropeptide release. J Neurosci. 2021;41(19):4187–4201. doi: 10.1523/JNEUROSCI.2631-20.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song SH, Augustine GJ. Synapsin isoforms and synaptic vesicle trafficking. Mol Cells. 2015;38(11):936–940. doi: 10.14348/molcells.2015.0233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matthew WD, Tsavaler L, Reichardt LF. Identification of a synaptic vesicle-specific membrane protein with a wide distribution in neuronal and neurosecretory tissue. J Cell Biol. 1981;91(1):257–269. doi: 10.1083/jcb.91.1.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu J, Mcdargh ZA, Li F, Krishnakumar SS, Rothman JE, O'shaughnessy B. Synaptotagmin rings as high-sensitivity regulators of synaptic vesicle docking and fusion. Proc Natl Acad Sci U S A. 2022;119(38):e2208337119. doi: 10.1073/pnas.2208337119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolfes AC, Dean C. The diversity of synaptotagmin isoforms. Curr Opin Neurobiol. 2020;63:198–209. doi: 10.1016/j.conb.2020.04.006 [DOI] [PubMed] [Google Scholar]
- 82.Lebowitz JJ, Banerjee A, Qiao C, Bunzow JR, Williams JT, Kaeser PS. Synaptotagmin-1 is a Ca(2+) sensor for somatodendritic dopamine release. Cell Rep. 2023;42(1):111915. doi: 10.1016/j.celrep.2022.111915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ullah N, Maaiden EE, Uddin MS, Ashraf GM. Synaptotagmin-1: a multi-functional protein that mediates vesicle docking, priming, and fusion. Curr Protein Pept Sci. 2021;22(6):470–478. doi: 10.2174/1389203722666210325110231 [DOI] [PubMed] [Google Scholar]
- 84.Tawfik B, Martins JS, Houy S, et al. Synaptotagmin-7 places dense-core vesicles at the cell membrane to promote Munc13-2- and Ca(2+)-dependent priming. Elife. 2021;10:e64527. doi: 10.7554/eLife.64527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Skene JH, Willard M. Axonally transported proteins associated with axon growth in rabbit central and peripheral nervous systems. J Cell Biol. 1981;89(1):96–103. doi: 10.1083/jcb.89.1.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chung D, Shum A, Caraveo G. GAP-43 and BASP1 in axon regeneration: implications for the treatment of neurodegenerative diseases. Front Cell Dev Biol. 2020;8:567537. doi: 10.3389/fcell.2020.567537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Caprara GA, Perni S, Morabito C, Mariggiò MA, Guarnieri S. Specific association of growth-associated protein 43 with calcium release units in skeletal muscles of lower vertebrates. Eur J Histochem. 2014;58(4):2453. doi: 10.4081/ejh.2014.2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holahan MR. A shift from a pivotal to supporting role for the growth-associated protein (GAP-43) in the coordination of axonal structural and functional plasticity. Front Cell Neurosci. 2017;11:266. doi: 10.3389/fncel.2017.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang N, Yang W, Xiao T, et al. Possible role of miR-204 in optic nerve injury through the regulation of GAP-43. Mol Med Rep. 2018;17(3):3891–3897. doi: 10.3892/mmr.2017.8341 [DOI] [PubMed] [Google Scholar]
- 90.Cheng MY, Wang EH, Woodson WJ, et al. Optogenetic neuronal stimulation promotes functional recovery after stroke. Proc Natl Acad Sci U S A. 2014;111(35):12913–12918. doi: 10.1073/pnas.1404109111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cupertino RB, Kappel DB, Bandeira CE, et al. SNARE complex in developmental psychiatry: neurotransmitter exocytosis and beyond. J Neural Transm. 2016;123(8):867–883. doi: 10.1007/s00702-016-1514-9 [DOI] [PubMed] [Google Scholar]
- 92.Han J, Pluhackova K, Böckmann RA. The multifaceted role of SNARE proteins in membrane fusion. Front Physiol. 2017;8:5. doi: 10.3389/fphys.2017.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oyler GA, Higgins GA, Hart RA, et al. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol. 1989;109(6 Pt 1):3039–3052. doi: 10.1083/jcb.109.6.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baumert M, Maycox PR, Navone F, De Camilli P, Jahn R. Synaptobrevin: an integral membrane protein of 18,000 daltons present in small synaptic vesicles of rat brain. EMBO J. 1989;8(2):379–384. doi: 10.1002/j.1460-2075.1989.tb03388.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257(5067):255–259. doi: 10.1126/science.1321498 [DOI] [PubMed] [Google Scholar]
- 96.Tang BL. SNAREs and developmental disorders. J Cell Physiol. 2021;236(4):2482–2504. doi: 10.1002/jcp.30067 [DOI] [PubMed] [Google Scholar]; •• An overview of the composition, function, and association of the SNARE complex with pathogenic potential
- 97.Vanguilder HD, Yan H, Farley JA, Sonntag WE, Freeman WM. Aging alters the expression of neurotransmission-regulating proteins in the hippocampal synaptoproteome. J Neurochem. 2010;113(6):1577–1588. doi: 10.1111/j.1471-4159.2010.06719.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vallejo D, Codocedo JF, Inestrosa NC. Posttranslational modifications regulate the postsynaptic localization of PSD-95. Mol Neurobiol. 2017;54(3):1759–1776. doi: 10.1007/s12035-016-9745-1 [DOI] [PubMed] [Google Scholar]
- 99.Sampedro MN, Bussineau CM, Cotman CW. Postsynaptic density antigens: preparation and characterization of an antiserum against postsynaptic densities. J Cell Biol. 1981;90(3):675–686. doi: 10.1083/jcb.90.3.675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maslov I, Hendrix J. Unmasking a two-faced protein. Elife. 2022;11:e83482. doi: 10.7554/eLife.83482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hamilton GL, Saikia N, Basak S, et al. Fuzzy supertertiary interactions within PSD-95 enable ligand binding. Elife. 2022;11:e77242. doi: 10.7554/eLife.77242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290(5495):1364–1368. doi: 10.1126/science.290.5495.1364 [DOI] [PubMed] [Google Scholar]
- 103.Zhang Y, Zhou Q, Lu L, et al. Copper induces cognitive impairment in mice via modulation of cuproptosis and CREB Signaling. Nutrients. 2023;15(4):972. doi: 10.3390/nu15040972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dong L, Chang Q, Ma J, et al. Associations of blood UCH-L1 and NfL levels with cognitive dysfunction in Parkinson's disease patients. Neurosci Lett. 2023;804:137219. doi: 10.1016/j.neulet.2023.137219 [DOI] [PubMed] [Google Scholar]
- 105.Kivisäkk P, Carlyle BC, Sweeney T, et al. Increased levels of the synaptic proteins PSD-95, SNAP-25, and neurogranin in the cerebrospinal fluid of patients with Alzheimer's disease. Alzheimer's Res Ther. 2022;14(1):58. doi: 10.1186/s13195-022-01002-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jia L, Zhu M, Kong C, et al. Blood neuro-exosomal synaptic proteins predict Alzheimer's disease at the asymptomatic stage. Alzheimer's Dement. 2021;17(1):49–60. doi: 10.1002/alz.12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Virgilio E, Vecchio D, Crespi I, et al. Cerebrospinal Tau levels as a predictor of early disability in multiple sclerosis. Mult Scler Relat Disord. 2021;56:103231. doi: 10.1016/j.msard.2021.103231 [DOI] [PubMed] [Google Scholar]
- 108.Onatsu J, Vanninen R, Jäkälä P, et al. Tau, S100B and NSE as blood biomarkers in acute cerebrovascular events. In Vivo. 2020;34(5):2577–2586. doi: 10.21873/invivo.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bi M, Gladbach A, Van Eersel J, et al. Tau exacerbates excitotoxic brain damage in an animal model of stroke. Nat Commun. 2017;8(1):473. doi: 10.1038/s41467-017-00618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pekny M, Wilhelmsson U, Stokowska A, Tatlisumak T, Jood K, Pekna M. Neurofilament light chain (NfL) in blood-A biomarker predicting unfavourable outcome in the acute phase and improvement in the late phase after stroke. Cells. 2021;10(6):1537. doi: 10.3390/cells10061537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sandelius Å, Cullen NC, Källén Å, et al. Transient increase in CSF GAP-43 concentration after ischemic stroke. BMC Neurol. 2018;18(1):202. doi: 10.1186/s12883-018-1210-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu H, Chen S, Nie Q, et al. Synaptotagmin-3 interactions with GluA2 mediate brain damage and impair functional recovery in stroke. Cell Rep. 2023;42(3):112233. doi: 10.1016/j.celrep.2023.112233 [DOI] [PubMed] [Google Scholar]
- 113.Vandendriessche C, Kapogiannis D, Vandenbroucke RE. Biomarker and therapeutic potential of peripheral extracellular vesicles in Alzheimer's disease. Adv Drug Deliv Rev. 2022;190:114486. doi: 10.1016/j.addr.2022.114486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang KK, Yang Z, Sarkis G, Torres I, Raghavan V. Ubiquitin C-terminal hydrolase-L1 (UCH-L1) as a therapeutic and diagnostic target in neurodegeneration, neurotrauma and neuro-injuries. Expert Opin Ther Targets. 2017;21(6):627–638. doi: 10.1080/14728222.2017.1321635 [DOI] [PubMed] [Google Scholar]
- 115.Bassil F, Brown HJ, Pattabhiraman S, et al. Amyloid-beta (Aβ) plaques promote seeding and spreading of alpha-synuclein and tau in a mouse model of lewy body disorders with Aβ pathology. Neuron. 2023;111(22):3699. doi: 10.1016/j.neuron.2023.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pan L, Li C, Meng L, et al. Tau accelerates α-synuclein aggregation and spreading in Parkinson's disease. Brain. 2022;145(10):3454–3471. doi: 10.1093/brain/awac171 [DOI] [PubMed] [Google Scholar]
- 117.Meng L, Zou L, Xiong M, et al. A synapsin I cleavage fragment contributes to synaptic dysfunction in Alzheimer's disease. Aging Cell. 2022;21(5):e13619. doi: 10.1111/acel.13619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sandelius Å, Portelius E, Källén Å, et al. Elevated CSF GAP-43 is Alzheimer's disease specific and associated with tau and amyloid pathology. Alzheimer's Dement. 2019;15(1):55–64. doi: 10.1016/j.jalz.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mccoy HM, Polcyn R, Banik NL, Haque A. Regulation of enolase activation to promote neural protection and regeneration in spinal cord injury. Neural Regen Res. 2023;18(7):1457–1462. doi: 10.4103/1673-5374.361539 [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Suggests that neuronal biomarkers can be used as potential therapeutic targets to promote neuronal survival and regeneration.
- 120.Caprelli MT, Mothe AJ, Tator CH. Hyperphosphorylated tau as a novel biomarker for traumatic axonal injury in the spinal cord. J Neurotrauma. 2018;35(16):1929–1941. doi: 10.1089/neu.2017.5495 [DOI] [PubMed] [Google Scholar]
- 121.Barro C, Benkert P, Disanto G, et al. Serum neurofilament as a predictor of disease worsening and brain and spinal cord atrophy in multiple sclerosis. Brain. 2018;141(8):2382–2391. doi: 10.1093/brain/awy154 [DOI] [PubMed] [Google Scholar]
- 122.Disanto G, Barro C, Benkert P, et al. Serum Neurofilament light: a biomarker of neuronal damage in multiple sclerosis. Ann Neurol. 2017;81(6):857–870. doi: 10.1002/ana.24954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Petrova N, Nutma E, Carassiti D, et al. Synaptic loss in multiple sclerosis spinal cord. Ann Neurol. 2020;88(3):619–625. doi: 10.1002/ana.25835 [DOI] [PubMed] [Google Scholar]
- 124.Krohs C, Körber C, Ebbers L, et al. Loss of miR-183/96 alters synaptic strength via presynaptic and postsynaptic mechanisms at a central synapse. J Neurosci. 2021;41(32):6796–6811. doi: 10.1523/JNEUROSCI.0139-20.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fuccillo MV, Pak C. Copy number variants in neurexin genes: phenotypes and mechanisms. Curr Opin Genet Dev. 2021;68:64–70. doi: 10.1016/j.gde.2021.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hassani Nia F, Woike D, Bento I, et al. Structural deficits in key domains of Shank2 lead to alterations in postsynaptic nanoclusters and to a neurodevelopmental disorder in humans. Mol Psychiatry. 2024;29(6):1683–1697. doi: 10.1038/s41380-022-01882-3 [DOI] [PMC free article] [PubMed] [Google Scholar]


