Abstract
The association of genotypic changes in human immunodeficiency virus (HIV) protease with reduced in vitro susceptibility to the new protease inhibitor lopinavir (previously ABT-378) was explored using a panel of viral isolates from subjects failing therapy with other protease inhibitors. Two statistical tests showed that specific mutations at 11 amino acid positions in protease (L10F/I/R/V, K20M/R, L24I, M46I/L, F53L, I54L/T/V, L63P, A71I/L/T/V, V82A/F/T, I84V, and L90M) were associated with reduced susceptibility. Mutations at positions 82, 54, 10, 63, 71, and 84 were most closely associated with relatively modest (4- and 10-fold) changes in phenotype, while the K20M/R and F53L mutations, in conjunction with multiple other mutations, were associated with >20- and >40-fold-reduced susceptibility, respectively. The median 50% inhibitory concentrations (IC50) of lopinavir against isolates with 0 to 3, 4 or 5, 6 or 7, and 8 to 10 of the above 11 mutations were 0.8-, 2.7-, 13.5-, and 44.0-fold higher, respectively, than the IC50 against wild-type HIV. On average, the IC50 of lopinavir increased by 1.74-fold per mutation in isolates containing three or more mutations. Each of the 16 viruses that displayed a >20-fold change in susceptibility contained mutations at residues 10, 54, 63, and 82 and/or 84, along with a median of three mutations at residues 20, 24, 46, 53, 71, and 90. The number of protease mutations from the 11 identified in these analyses (the lopinavir mutation score) may be useful for the interpretation of HIV genotypic resistance testing with respect to lopinavir-ritonavir (Kaletra) regimens and may provide insight into the genetic barrier to resistance to lopinavir-ritonavir in both antiretroviral therapy-naive and protease inhibitor-experienced patients.
A rebound in viral load during antiretroviral therapy is often associated with the development of phenotypic resistance to one or more of the drugs in the treatment regimen. For most antiretroviral drugs, a decline in phenotypic susceptibility has been correlated with specific mutations in the target protein for the drug of interest. Longitudinal analyses of the genetic sequences that encode human immunodeficiency virus (HIV) protease in viral isolates from patients experiencing viral rebound during protease inhibitor (PI) therapy often show the sequential accumulation of several mutations that produce changes in susceptibility (4, 15). Mutations in HIV protease both within and outside of the enzyme active site can contribute to viral resistance. The former group (primary mutations) can produce significant changes in the affinity of binding of the inhibitor to the mutant active site (8) and often occur early during rebound (11). Mutations outside of the active site have been referred to as secondary mutations and may in some cases contribute to changes in phenotypic susceptibility by upregulating the enzymatic function of the mutant protease (and thus the growth rate of the mutant virus) rather than by a direct diminution of drug binding (21). The patterns of mutations selected by different PIs have been characterized (11), although more than one genotypic pattern can emerge in different patients being treated with the same drug regimen (3). Further, although the primary mutation(s) selected by a given PI may be distinct, the accompanying secondary mutations tend to be common to the PI class. This commonality potentially limits the success of subsequent therapy following virologic failure of PI-containing regimens, since fewer new mutations may be required to produce viruses that are clinically resistant to the PI(s) in the salvage regimen.
Lopinavir (previously ABT-378) is a new PI that displays significant virologic potency in both antiretroviral therapy-naive (18) and single-PI-experienced HIV-infected subjects (S. Deeks et al., Abstr. 7th Conf. Retroviruses Opportunistic Infect., abstr. 532, 2000) when coadministered with low-dose ritonavir (RTV), which enhances and sustains plasma lopinavir levels (22). HIV strains resistant to lopinavir have been produced using in vitro passaging (2), and viral isolates from PI-experienced patients that display in vitro resistance to other PIs (particularly RTV and indinavir [IDV]) may also show reduced in vitro susceptibility to lopinavir (16). However, to date, the patterns of mutations in HIV protease associated with viral rebound on therapy with lopinavir-RTV (Kaletra) in previously antiretroviral therapy-naive individuals have not been characterized. In the absence of data from primary treatment failures, the genotypic correlates of reduced in vitro phenotypic susceptibility to lopinavir in viral isolates selected during therapy with other PIs have been examined. Definition of this relationship provides information with which to interpret the results of HIV resistance testing and insight into the genetic barrier to clinical resistance to lopinavir-RTV in either antiretroviral therapy-naive or PI-experienced individuals.
MATERIALS AND METHODS
Viral isolates.
The 112 HIV isolates used for the correlation of genotype and susceptibility to lopinavir were taken during days −6 to 1 in two lopinavir-RTV phase I/II studies: study M97-765 (56 isolates) and study M98-957 (56 isolates). Subjects entering study M97-765 had plasma HIV RNA levels between 1,000 and 100,000 copies/ml while still on their first single-PI-based treatment regimen. Subjects entering study M98-957 had plasma HIV RNA levels >1,000 copies/ml and had been treated with at least two PIs, either sequentially or simultaneously. These studies have been described elsewhere (Deeks et al., 7th Conf. Retroviruses Opportunistic Infect., abstr. 532; S. Becker et al., Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 697, 2000). Plasma HIV RNA from the baseline samples (prior to initiation of therapy with lopinavir-RTV) was amplified by PCR and incorporated into recombinant viruses for phenotypic susceptibility testing (compared to a standard wild-type [wt] recombinant virus) and genotypic sequencing of the protease gene. Samples from study M97-765 were analyzed by Virco, Inc., using the Antivirogram method (version 3.0) (10; R. Pauwels et al., Abstr. 2nd Int. Workshop HIV Drug Resist. Treatment Strategy, abstr. 51, 1998). Samples from study M98-957 were analyzed by ViroLogic, Inc., using the PhenoSense HIV assay (20). Phenotypic data were expressed as fold change in 50% inhibitory concentration (fold IC50), which was calculated by dividing the IC50 of lopinavir against the recombinant virus containing the HIV protease and reverse transcriptase genes of the baseline patient plasma sample by the IC50 against the standard wt recombinant virus. Genotype data from Virco, Inc., and ViroLogic, Inc., were determined by population sequencing and reported as sequence changes with respect to the sequences of the HXB2 and pNL4-3 laboratory wt strains, respectively. In the HIV protease gene, the pNL4-3 and HXB2 sequences differ by the identities of amino acids at position 3 (isoleucine and leucine, respectively) and position 37 (glutamine and serine, respectively). Thus, for the purposes of analyzing the combined panel of isolates, genotypic data for the M97-765 baseline samples were translated at amino acid positions 3 and 37 in HIV protease to reflect sequence changes from a common (pNL4-3) wt sequence.
Statistical methodology to identify amino acid positions in HIV protease associated with reduced in vitro susceptibility to lopinavir.
Two univariate statistical analyses were performed to determine the mutations in HIV protease associated with reduced in vitro susceptibility to lopinavir. In the first analysis, the median fold IC50 of lopinavir against isolates containing any mutation at a particular amino acid position was compared to the median fold IC50 of lopinavir against isolates that had the wt sequence at that position using the Wilcoxon rank sum test. The normal approximation (with continuity correction) was used to determine the significance level (P value) for each of the 66 out of 99 amino acid positions in HIV protease at which any variance from the pNL4-3 sequence was observed. An exact Wilcoxon rank sum test was performed for amino acid positions in which there were five or fewer isolates with a mutation. Results from the exact test were similar to those obtained using the normal approximation (with continuity correction). In the second analysis, each fold IC50 of lopinavir was converted using a Box-Cox (1) transformation as follows: (fold IC50)−0.4; this produced a distribution that was approximately symmetrical. This allowed comparison of the mean transformed values of fold IC50 of lopinavir against isolates either containing or lacking any mutation at a particular amino acid position using one-way analysis of variance (ANOVA). A P value for each of the 66 positions was determined. A modified Bonferroni adjustment (13, 23) was used to identify potentially important mutations that may not have been identified using the traditional (conservative) Bonferroni adjustment, particularly those that may occur with relatively low frequency but that contribute to high levels of phenotypic resistance to lopinavir. Therefore, a P value of 0.0062 (0.05 divided by the square root of 66) was considered significant in the above analyses.
Two multivariate linear regression analyses were performed to assess the relative association between the change in lopinavir susceptibility and individual PI mutations. The first, a (forward) stepwise linear regression model, considered all mutations with a prevalence of >4 of 112 and a positive correlation with the fold IC50 of lopinavir in the combined panel of isolates. An entry and exit P value of 0.05 was used. The second, a backward-elimination linear regression model, considered all of the mutations judged to be significantly or marginally associated with reduced susceptibility to lopinavir in the above univariate analyses. An exit P value of 0.05 was used in this model.
Assignment of mutations associated with reduced susceptibility.
Since the wt sequence at many amino acid positions can mutate to more than one amino acid, specific amino acid changes at the positions in HIV protease found to be associated with reduced in vitro susceptibility to lopinavir in the initial Wilcoxon rank sum test or ANOVA analyses were judged as either likely to contribute to reduced susceptibility or of unknown contribution based on searches of three public databases (the Los Alamos HIV Sequence Database [http://hiv-web.lanl.gov], the Antiviral Drug Resistance Online website [http://www.viral-resistance.com], and the Stanford HIV RT and Protease Sequence Database [http://hivdb.stanford.edu/hiv/index.asp]) and two review documents (7, 11). Specific amino acid changes were judged likely to contribute to reduced susceptibility if they had been previously associated with resistance to the PI class or if they were found only in the context of sequences that contained several other mutations known to confer PI resistance. Otherwise, they were judged to be of unknown contribution.
Statistical association of viral genotype with levels of reduced in vitro susceptibility to lopinavir.
Using cutoff fold changes in IC50s of 4-, 10-, 20-, and 40-fold, each of the amino acid positions judged likely to contribute to reduced susceptibility was individually evaluated using Fisher's exact test. This analysis was limited to the baseline isolates from study M98-957 since the range of IC50s in that study was much larger than that in study M97-765. Fold IC50s equal to the cutoff values were considered above the cutoff. The P value for 2-by-2 comparisons of mutation (yes, no) by the dichotomized fold change in IC50 (above cutoff, below cutoff) for each of the 11 amino acid positions was calculated for each of the four cutoff levels defined above. Based on a modification to the Bonferroni adjustment (13, 23), P values <0.0075 (0.05 divided by the square root of 44) were considered statistically significant. At amino acid positions containing more than one mutation, only the particular mutations judged likely to contribute to reduced susceptibility were scored as “yes.” Mutations judged of unknown contribution were scored as “no.”
RESULTS
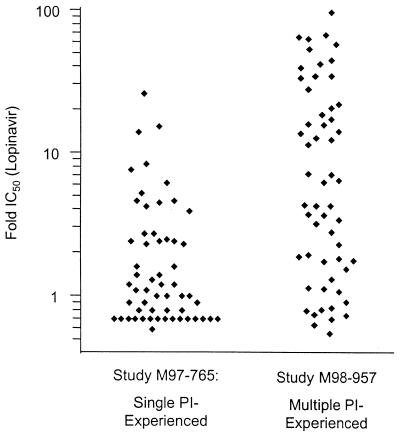
To define the genotypic correlates of reduced in vitro susceptibility to lopinavir, we examined the genotypes and phenotypes of 112 viral isolates from subjects experiencing virologic failure of therapy with one or more other PIs who entered one of two lopinavir-RTV phase I/II studies (studies M97-765 and M98-957; see Materials and Methods). The prior PI experience of subjects participating in these studies is provided in Table 1. As anticipated, the M98-957 isolates (from multiple-PI-experienced subjects) displayed markedly lower susceptibility to lopinavir (median, 16.2-fold; range, 0.5- to 96-fold) than the isolates from study M97-765 (single-PI-experienced subjects; median, 1.1-fold; range, 0.7- to 26-fold) (Fig. 1).
TABLE 1.
Viral isolates used in the correlation of genotypic and phenotypic susceptibility to lopinavir
| Study | No. of isolates | Median no. of previous PIs | % Identity of previous PIa:
|
Fold change in IC50 of lopinavir
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| IDV | NFV | RTV | SQV | Mean | Median | Range | |||
| M97-765 | 56 | 1 | 41 | 38 | 7 | 16 | 2.8 | 1.1 | 0.7–26.0 |
| M98-957 | 56 | 3 | 86 | 57 | 77 | 71 | 16.2 | 5.2 | 0.5–96.0 |
For study M98-957, previous PIs were tallied irrespective of dose and simultaneous or sequential PI treatment.
FIG. 1.
Phenotypic susceptibilities of baseline isolates from studies M97-765 and M98-957 to lopinavir.
Identification of amino acid positions associated with reduced susceptibility to lopinavir.
To define the genotypic changes correlated with reduced susceptibility to lopinavir, the entire amino acid sequences of HIV proteases in the panel of 112 isolates were analyzed. Mutations, compared to the pNL4-3 wt sequence, were found at a total of 66 out of 99 amino acid positions. Mixtures of mutant and wt amino acids were scored as mutant. Since the average trough plasma lopinavir concentrations in HIV-infected subjects (produced by a dose of 400 mg of lopinavir and 100 mg of RTV twice daily) are ≥75-fold above the serum-adjusted IC50 of lopinavir against wt HIV (18), we required the identification of sets of mutations associated with large as well as small phenotypic changes. Thus, rather than treating phenotypic susceptibility as a categorical response variable, we investigated methods of analysis that used a continuous response variable to define phenotype. Because the phenotype did not follow a normal distribution, the Wilcoxon rank sum test, which compared the median fold IC50 against those isolates scored as mutant to the median fold IC50 against those with the wt amino acid at that position, was initially employed. In this initial analysis, mutations at 11 amino acid positions in HIV protease (positions 54, 82, 10, 71, 46, 20, 90, 84, 24, 53, and 63, in order of greatest to least significance) were found to be statistically significantly associated (P < 0.0062 [0.05/{661/2}]) with the loss of phenotypic susceptibility to lopinavir (Table 2). The association with six additional amino acid positions (73, 43, 93, 58, 33, and 12) was found to be marginally significant (0.0062 < P < 0.05). The association of genotype with the fold IC50 of IDV, nelfinavir (NFV), RTV, and saquinavir (SQV) was analyzed in the same manner (complete data were not available for amprenavir [APV]). With one exception (position 93), this method of analysis identified amino acid positions at which mutations have previously been shown to be selected during therapy with PIs and to produce cross-resistance to the PI class (rather than specifically associated with resistance to one PI, such as the D30N mutation). This high correlation lends credence to the relevance of this method of identifying mutations that are associated with reduced in vitro susceptibility (and thus potential in vivo cross-resistance) to lopinavir-RTV.
TABLE 2.
Amino acid positions associated with reduced in vitro susceptibility to PIs in combined panel of viral isolates
| Inhibitor | Amino acid positions associated with reduced susceptibilitya at:
|
|
|---|---|---|
| P < 0.0062 | 0.0062 < P < 0.05 | |
| Lopinavir | 54, 82, 10, 71, 46, 20, 90, 84, 24, 53, 63 | 73, 43, 93, 58, 33, 12 |
| IDV | 10, 54, 82, 46, 90, 93, 71, 84, 20, 73, 63 | 24, 53, 58, 62, 60 |
| NFV | 10, 90, 54, 71, 84, 20, 73, 88, 93, 46, 33 | 82, 19, 77, 63 |
| RTV | 54, 82, 10, 90, 84, 63, 71, 20, 24, 53, 46, 73 | 43, 33, 93, 58, 14 |
| SQV | 90, 10, 84, 71, 73, 54, 63, 20 | 53, 19, 33, 93, 48, 67 |
Ordered from greatest to least degree of significance (Wilcoxon rank sum test).
To confirm the associations of genotype and phenotype identified using the Wilcoxon rank sum test, we performed a second, independent statistical analysis using transformed values of fold IC50. Because the distribution of IC50s was highly asymmetric, each value of fold IC50 was converted to (fold IC50)−0.4 using a Box-Cox transformation (1) to achieve a distribution that was approximately symmetric. For each amino acid position, the mean transformed values of fold IC50 of lopinavir against isolates either containing or lacking each particular mutation were compared using ANOVA. The amino acid positions at which mutations were found to be statistically associated (P < 0.0062) with a loss of susceptibility to lopinavir were exactly the same as those identified using the Wilcoxon rank sum test, although in slightly different order (positions 54, 82, 10, 71, 46, 20, 90, 84, 24, 53, and 63, in order of greatest to least degree of statistical significance). Additionally, the association with changes at positions 73, 43, 33, 58, 93, and 12 was marginally significant, as observed in the Wilcoxon analysis.
Identity of specific amino acid changes associated with reduced susceptibility.
Since multiple amino acid substitutions have been observed at many of the amino acid positions in HIV protease, we sought to identify which particular amino acid changes are likely to contribute to reduced in vitro susceptibility to lopinavir. Each individual mutation in the combined panel of viral isolates at the 17 amino acid positions found to be either statistically or marginally significantly associated with phenotype was assigned as either likely to contribute to reduced susceptibility or of unknown contribution based on several considerations. A particular amino acid was deemed likely to contribute to reduced susceptibility if it previously had been associated with PI resistance through either virologic or clinical studies. In cases where a link with PI resistance was not established, database searches were performed (see Materials and Methods). If the particular mutation was found only in the context of sequences that contained several mutations known to confer PI resistance, it was deemed likely to contribute to reduced susceptibility. In contrast, if the mutation was a common polymorphism or appeared in multiple sequences that did not contain multiple other mutations known to confer PI resistance, it was deemed of unknown contribution.
Following the above assignments, those positions (20, 24, 33, 43, 63, and 82) at which more than one mutant amino acid was present were reanalyzed using both the Wilcoxon rank sum test and ANOVA, considering only those mutations judged likely to contribute to reduced susceptibility to lopinavir. By limiting the analysis to the specific subset of mutations at the above sites, it was found that the P values for both tests did not change substantially (i.e., positions 20, 24, 63, and 82 and positions 33 and 43 remained statistically and marginally associated, respectively, with reduced susceptibility to lopinavir). Thus, 11 specific mutations were found to be consistently associated with reduced in vitro susceptibility to lopinavir (Table 3). This set of mutations is very similar to those reported to be selected during therapy with IDV (5) or RTV (16) and contains many of the mutations associated with resistance to the PI class (7, 11). The exception is the F53L mutation, which has only previously been reported in one isolate from a subject receiving RTV (24) but appeared in 13 of 112 isolates examined in this analysis.
TABLE 3.
Median fold changes in IC50 against viral isolates containing mutations associated with reduced in vitro susceptibility to lopinavir
| Mutation | Isolates containing mutation
|
Isolates lacking mutation
|
Pa by:
|
|||||
|---|---|---|---|---|---|---|---|---|
| n | Fold IC50
|
n | Fold IC50
|
|||||
| Median | Mean | Median | Mean | Wilcoxon | ANOVA | |||
| L10F/I/R/V | 69 | 4.6 | 14.5 | 43 | 0.9 | 1.4 | <0.0001 | <0.0001 |
| K20M/R | 14 | 30.5 | 29.3 | 98 | 1.6 | 6.6 | 0.0001 | <0.0001 |
| L24I | 10 | 14.0 | 26.7 | 102 | 1.7 | 7.8 | 0.0011 | 0.0008 |
| M46I/L | 41 | 4.2 | 15.8 | 71 | 1.1 | 5.8 | 0.0003 | 0.0002 |
| F53L | 13 | 4.6 | 24.6 | 99 | 1.6 | 7.5 | 0.0019 | 0.0014 |
| I54L/T/V | 38 | 16.0 | 23.7 | 74 | 1.1 | 2.1 | <0.0001 | <0.0001 |
| L63P | 88 | 2.5 | 11.5 | 24 | 1.1 | 2.1 | 0.0026 | 0.0023 |
| A71I/L/V/RT | 65 | 3.6 | 11.2 | 47 | 1.1 | 7.1 | 0.0002 | 0.0001 |
| V82A/F/T | 40 | 14.0 | 21.5 | 72 | 1.1 | 2.8 | <0.0001 | <0.0001 |
| I84V | 19 | 6.1 | 14.8 | 93 | 1.4 | 8.4 | 0.0007 | 0.0004 |
| L90M | 48 | 3.4 | 12.4 | 64 | 1.1 | 7.3 | 0.0004 | 0.0003 |
Since the M97-765 (Virco) and M98-957 (ViroLogic) baseline isolates were analyzed using different single-cycle phenotypic assays, a sensitivity analysis that introduced a twofold discordance between the assays was performed. A twofold discordance in one direction did not affect which mutations were statistically significant using the Wilcoxon rank sum test and produced only a minor change in the outcome of the confirmatory analysis (ANOVA), wherein the L63P mutation changed from statistically to marginally significant. A twofold discordance in the other direction did not affect the statistical significance of any of the 11 mutations but produced an apparent statistically significant association of the G73A/S/T mutation.
Analysis of in vitro susceptibility to lopinavir with respect to the number of mutations.
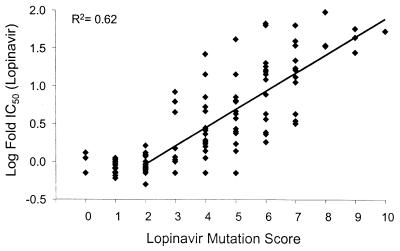
The susceptibility (log fold lopinavir IC50) of the combined panel of isolates as a function of the number of the above 11 mutations identified as likely to contribute to reduced susceptibility to lopinavir (designated the lopinavir mutation score) is shown in Fig. 2. In general, isolates with mutation scores of 2 or less displayed wt susceptibility (only 2 of 34 of these isolates contained primary mutations at position 82, 84, or 90). In contrast, isolates with a mutation score of 3 or more displayed correspondingly greater degrees of reduced susceptibility to lopinavir, as illustrated by the log-linear regression line (R2 = 0.62). Virtually all (71 of 73) of these isolates had changes at position 82, 84, and/or 90, consistent with the role of primary mutations, in combination with secondary mutations, in producing changes in susceptibility to PIs. The estimated slope of the regression line was 0.24 log fold IC50/mutation (95% confidence interval: 0.20 to 0.28 log fold IC50/mutation). Transforming the regression to the original (linear) scale of measurement provided the following expression for estimation of the lopinavir fold IC50 as a function of the mutation score: 0.303 × 1.74(lopinavir mutation score).
FIG. 2.
Log fold IC50 of lopinavir with respect to the lopinavir mutation score.
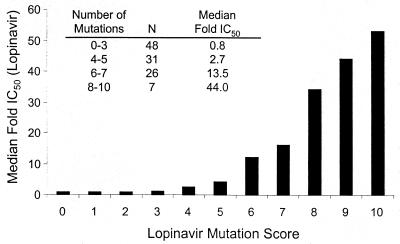
The correlation between phenotype and genotype, as defined by the lopinavir mutation score, was much higher than that found by simply modeling the log fold IC50 of lopinavir as a function of every variation from the pNL4-3 wt sequence (R2 = 0.38; data not shown). This difference indicates that the changes in lopinavir IC50 are primarily a consequence of the set of 11 mutations identified as being associated with reduced in vitro susceptibility and that the remaining variations from the wt sequence are present at low prevalence and/or contribute little to changes in susceptibility in vitro to lopinavir. The median IC50s of lopinavir against isolates within the combined panel with mutation scores of 0 to 3, 4 or 5, 6 or 7, and 8 to 10 mutations were 0.8-, 2.7-, 13.5-, and 44.0-fold higher than the IC50 against wt HIV, respectively (Fig. 3).
FIG. 3.
Median fold IC50 of lopinavir with respect to the number of mutations associated with reduced in vitro susceptibility to lopinavir.
Association of mutations at different amino acid positions with different degrees of reduced susceptibility.
The accumulation of mutations during rebound in plasma HIV RNA on PI therapy is sequential, with one or more primary mutations appearing early followed by several secondary mutations that are generally correlated with a higher degree of reduced susceptibility (4, 5, 15). Since the plasma lopinavir levels in HIV-infected subjects are sustained at many multiples above the IC50 against wt HIV, it was of interest to probe the correlation of particular mutations with a high degree of reduced susceptibility to lopinavir. For this purpose, we chose the subset of isolates from study M98-957, since the range of phenotype was much greater than that for the M97-765 baseline isolates (Fig. 1). For each of the above 11 mutations, two-by-two tables were constructed; these tables consisted of the number of isolates either containing or lacking a mutation and the number of isolates displaying susceptibility greater or less than arbitrary cutoff susceptibility values. To explore a broad range of phenotypes, we chose cutoff values of 4-, 10-, 20-, and 40-fold changes in IC50. The P values from Fisher's exact test of each comparison are shown in Table 4. Not all amino acid positions were found to be statistically associated (P < 0.0075 [0.05/{441/2}]) with phenotype by using any of the cutoff values, illustrating the strength of using a continuous (Wilcoxon or ANOVA) rather than categorical response variable to describe the relationship of genotype to phenotype. Nonetheless, the majority of mutations were either statistically or marginally (0.0075 < P < 0.05) associated with a change in phenotype to above one or more of the cutoff levels. Thus, a change in susceptibility to lopinavir by >4-fold was uncommon in the absence of a mutation at position 82 or 54 (7 of 30 and 6 of 29 isolates, respectively), and only 2 isolates and 1 isolate lacking those mutations, respectively, displayed a >10-fold change in phenotype. Similarly, mutations at positions 10, 63, 71, and 84 were most closely associated with 4- and/or 10-fold changes in phenotype. In contrast, the F53L and K20M/R mutations, which were present in isolates with a median lopinavir mutation score of 8, were most closely associated with high levels of reduced susceptibility (20- and 40-fold, respectively).
TABLE 4.
Association of viral genotype with levels of reduced susceptibility to lopinavir
| Mutation | No. of occurrences |
Pa for cutoff of fold lopinavir IC50
|
|||
|---|---|---|---|---|---|
| 4 | 10 | 20 | 40 | ||
| L10F/I/R/V | 45 | 0.0013 | 0.0013 | 0.0261 | ns |
| K20M/R | 13 | ns | ns | 0.0028 | ns |
| L24I | 8 | ns | ns | ns | ns |
| M46I/L | 27 | ns | ns | ns | ns |
| F53L | 9 | ns | ns | 0.0479 | 0.0173 |
| I54L/T/V | 27 | <0.0001 | <0.0001 | <0.0001 | 0.0016 |
| L63P | 49 | 0.0371 | 0.0160 | ns | ns |
| A71I/L/V/T | 35 | 0.0137 | 0.0301 | ns | ns |
| V82A/F/T | 26 | <0.0001 | <0.0001 | 0.0005 | 0.0011 |
| I84V | 14 | 0.0121 | ns | ns | ns |
| L90M | 32 | ns | ns | ns | ns |
P values <0.0075 were considered statistically significant; P values between 0.05 and 0.0075 were considered marginally significant; P values >0.05 were considered not significant (ns).
The mean trough levels of lopinavir in plasma achieved by lopinavir doses of 400 mg and RTV doses of 100 mg twice daily exceed the serum protein-adjusted IC50 of lopinavir against wt HIV by >75-fold (18). Consequently, clinical resistance manifested as in vivo virologic failure is expected to require a high level of reduced phenotypic susceptibility to lopinavir. Although a single consensus pattern of mutations producing large changes in phenotype was not evident, each of the 16 viral isolates in the panel that displayed >20-fold-reduced susceptibility to lopinavir contained mutations at amino acid positions 10, 54, 63, and either 82 or 84. In addition, the median number of the remaining mutations (at positions 20, 24, 46, 53, 71, and 90) was three (range, zero to five). Although the mutations at positions 20 and 53, in the context of multiple other mutations, were associated with high-level reduced susceptibility, only 8 of the above 16 isolates had one or both of these mutations, suggesting that additional genotypic patterns can also produce marked phenotypic changes.
Relative association of mutations with reduced susceptibility to lopinavir.
Mutations in HIV protease occur together in complex patterns that produce changes in susceptibility to PIs (5, 15). Since the M97-765 and M98-957 baseline isolates were selected during therapy with other PIs rather than with lopinavir-RTV, the relative association of individual mutations with changes in lopinavir susceptibility was assessed using multivariate analyses (see Materials and Methods). A (forward) stepwise linear regression model that considered a total of 42 amino acid positions (all positions with a mutation prevalence of >4 of 112 and a positive correlation with fold IC50 of lopinavir) showed that 6 of the above 11 mutations (positions 54, 46, 10, 82, 84, and 20) were independently associated (P < 0.05) with reduced susceptibility to lopinavir (Table 5). In a separate analysis, the set of 17 mutations either statistically significantly or marginally associated with reduced susceptibility to lopinavir in the two univariate analyses (Table 2) were considered in a backward-elimination stepwise linear regression model. The same set of six mutations was found in this analysis to be independently associated with reduced susceptibility (Table 5).
TABLE 5.
Multivariate analyses evaluating the association between lopinavir susceptibility and individual protease mutations
| Amino acid position |
Pa |
|---|---|
| 10 | 0.0001 |
| 20 | 0.0181 |
| 46 | 0.0048 |
| 54 | 0.0001 |
| 82 | 0.0002 |
| 84 | 0.0016 |
Final models were identical for the (forward) stepwise and backward-elimination logistic regression analyses.
Phenotypic comparison of lopinavir and other PIs.
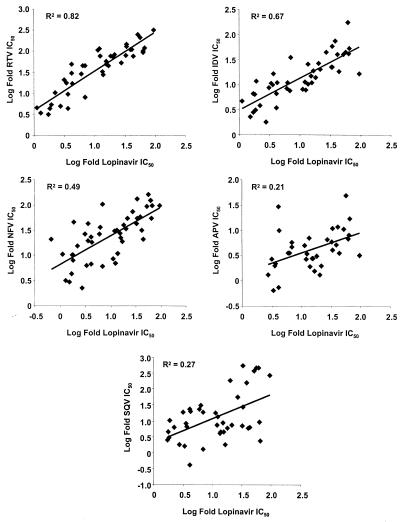
To provide more information regarding viral isolates with reduced susceptibility to lopinavir, the relative susceptibilities (log fold IC50) of the M98-957 baseline isolates to lopinavir were compared to the susceptibilities of the panel to other PIs (each comparison was restricted to those isolates displaying >2.5-fold-reduced susceptibility to one of the two drugs being compared). The correlation was highest between lopinavir and RTV (R2 = 0.82) and intermediate between lopinavir and either IDV (R2 = 0.67) or NFV (R2 = 0.49). In contrast, the correlation between susceptibility to lopinavir and either SQV (R2 = 0.27) or APV (R2 = 0.21) was relatively low (Fig. 4). The median fold changes in IC50 against the set of 16 isolates in the combined panel with >20-fold-reduced susceptibility to lopinavir for the PIs were as follows: lopinavir, 40-fold; RTV, 92-fold; IDV, 40-fold; NFV, 56-fold; SQV, 18-fold; and APV, 6.5-fold (data for APV were missing for one viral isolate from study M98-957 with a lopinavir fold IC50 of >20). The numbers of viral isolates in this subset with >20-fold-reduced susceptibility to the other PIs were the following: RTV, 16 of 16; IDV, 14 of 16; NFV, 16 of 16; SQV, 9 of 16; and APV, 1 of 15.
FIG. 4.
Relative susceptibility of M98-957 baseline viruses to lopinavir and other PIs.
DISCUSSION
In this study, statistical methods were used to identify 11 mutations in HIV protease that correlate with reduced in vitro susceptibility to lopinavir within a panel of viruses selected in vivo by other PIs. Two separate univariate analyses, both using a continuous response variable to describe phenotype, gave essentially identical results. Furthermore, one of these methods (the Wilcoxon rank sum test), applied to the other PIs for which the complete data set was available, identified sets of mutations associated with reduced susceptibility similar to those previously reported to be selected by and/or to produce cross-resistance to those PIs (11). Since the panel of isolates used for this analysis was selected during therapy with PIs other than lopinavir-RTV, the set of 11 mutations does not necessarily describe the development of de novo resistance to lopinavir-RTV (i.e., this analysis would not identify mutations that might be uniquely selected by lopinavir but not by other PIs). This limitation is illustrated by the fact that the D30N mutation was not found to be associated with reduced susceptibility to NFV. That lack of association is presumably due to the fact that other genotypes, selected either by other PIs (4, 15) or by NFV itself (3), also confer resistance to NFV. The analysis is further limited by the prior PI treatment experience of subjects entering studies M97-765 and M98-957 (predominantly IDV, NFV, RTV, and/or SQV). Thus, the potential effects of unique mutations selected by other PIs on the susceptibility to lopinavir are not addressed with this analysis. In spite of these limitations, the mutations identified in this analysis provide a set of common mutations that might be expected to occur during virologic rebound on therapy with lopinavir-RTV. In this context, mutations at 9 of the 11 positions defined by the lopinavir mutation score (positions 10, 20, 24, 46, 53, 54, 63, 71, and 82) have been observed at least once following viral rebound on lopinavir-RTV therapy in subjects previously treated with one or more of the other PIs (A. Molla et al., unpublished results). These observations provide additional insight into the possible relevance of this set of mutations as determinants of changes in susceptibility that may be clinically relevant.
Six of the 11 mutations determined by univariate analyses to be associated with reduced susceptibility (positions 10, 20, 46, 54, 82, and 84) were confirmed to be independently associated in both multivariate analyses. The independent association with phenotype strongly suggests that these mutations, in the context of different combinations of other mutations, can contribute directly to incrementally reduced susceptibility to lopinavir. However, the precise role of each mutation will be best examined by in vitro site-directed mutagenesis. The role of the remaining five mutations (positions 24, 53, 63, 71, and 90) as determinants or simply markers of reduced susceptibility by virtue of association with other mutations will also require experimental assessment. Nonetheless, the associations found in this analysis may be useful for estimating the potential for reduced susceptibility to lopinavir in patients failing therapy with other PIs.
The 11 mutations associated with reduced in vitro susceptibility to lopinavir were identified using a conservative approach (P < 0.0062), as dictated by a modified Bonferroni adjustment (13, 23). With larger sample sizes, other mutations might be found to be associated with reduced susceptibility (e.g., those marginally associated with reduced phenotypic susceptibility from the present analysis [amino acid positions 73, 43, 93, 58, and 33]). In particular, it is likely that mutations in addition to those at positions 20 and 53 will be found to contribute incrementally, in combination with multiple other mutations, to high-level in vitro resistance (e.g., the K20M/R and F53L mutations are only present in 8 of 16 and 5 of 16 viral isolates, respectively, in the panel with >20-fold-reduced susceptibility). For example, the L33F mutation occurred in only 3 of 112 isolates examined, but 2 of those displayed >20-fold-reduced susceptibility to lopinavir. Likewise, the K43T mutation was present in three isolates with >10-fold-reduced (range, 18- to 53-fold-reduced) susceptibility to lopinavir. While not statistically associated with phenotype because of small numbers, these mutations may still contribute to reduced susceptibility to lopinavir. In this regard, the appearance of L33F as a new mutation following plasma HIV RNA rebound in a previously PI-experienced subject who began lopinavir-RTV therapy with multiple mutations in protease (17) lends credence to this hypothesis.
The additional analysis of phenotype as a categorical response variable also provides insight into particular mutations that are likely to appear in isolates over a broad phenotypic range (i.e., those likely to be selected early) as opposed to those that appear predominantly in isolates that display high-level changes in susceptibility (i.e., those likely to be accumulated later). Several of the mutations closely associated with resistance to other PIs (e.g., those at positions 82, 54, 10, 63, 71, and 84) were found to be most closely associated with relatively modest (4-fold and/or 10-fold) changes in susceptibility. Reduced baseline susceptibility (fourfold or less) or the presence of one or two mutations or both have been found to be predictive of diminished virologic response to PI regimens (7, 14), including those containing RTV-SQV (6, 9, 25), IDV (19), and NFV (A. K. Patick et al., Abstr. 2nd Int. Workshop HIV Drug Resist. Treatment Strategy, abstr. 57, 1998). In contrast, neither a fourfold change in baseline susceptibility nor the presence of three or more mutations at positions 10, 54, 71, and 82 at baseline was associated with diminished response to lopinavir-RTV in single-PI-experienced patients (Kempf et al., 7th Conf. Retroviruses Opportunistic Infect., abstr. 731). This difference is likely to be a consequence of the high, sustained plasma lopinavir levels provided by the lopinavir-RTV regimen. Consequently, the identification of secondary mutations that accumulate on top of a platform of other mutations resulting in a greater degree of reduced in vitro susceptibility to lopinavir is important for the optimal interpretation of HIV genotypic resistance tests. In this context, the association of the K20R/M and F53L mutations (along with multiple other mutations) with increases in lopinavir IC50 of >20- and >40-fold, respectively, may provide useful information for interpreting in vivo cross-resistance to lopinavir-RTV.
Viruses with one or two (nearly exclusively secondary) mutations displayed wt susceptibility to lopinavir. In isolates with 3 or more mutations, virtually all of which contained at least 1 primary mutation, the average change in phenotypic susceptibility per mutation (out of the above set of 11 mutations) was 0.241-log-fold (1.74-fold) per mutation. The median fold IC50 values for isolates containing 6 or 7 and 8 to 10 mutations were 13.5- and 44-fold, respectively. Although the relative contributions of the above 11 mutations to the incremental change in susceptibility to lopinavir are not expected to be equal, a greater number of mutations is usually associated with more profoundly reduced susceptibility (5, 15), and the average change in phenotype over a range of mutations may be useful in assessing the likelihood of substantial activity against a given isolate in vivo. Since trough plasma lopinavir levels with the 400-mg lopinavir dose in combination with the 100-mg RTV dose average more than 75-fold above the serum-adjusted IC50 of lopinavir against wt HIV (18), the results of this analysis suggest that lopinavir-RTV is likely to exert significant antiviral activity in subjects whose viruses contain 6 or 7, and possibly more, of the 11 mutations identified with reduced in vitro susceptibility to lopinavir. Although this analysis is unlikely to completely describe the de novo development of resistance to lopinavir-RTV in previously antiretrovirus treatment-naive patients, the incremental loss of susceptibility over many mutations suggests that the in vivo genetic barrier to resistance to lopinavir-RTV may be high, particularly in subjects who are PI naive and who have not received a PI-resistant virus through transmission. This conclusion is supported by the observation that, in subjects experiencing a rebound in plasma HIV RNA to >1,000 copies while on lopinavir-RTV, evidence of viral evolution in HIV protease and changes in susceptibility to lopinavir (compared to the corresponding baseline isolates) were only evident in subjects whose baseline viruses contained at least 4 of the 11 mutations identified in this analysis (17).
In addition to providing insight into the genetic barrier to phenotypic resistance to lopinavir-RTV, the set of mutations identified in these analyses is useful for the analysis of virologic response to lopinavir-RTV therapy with respect to baseline genotype. Previous studies examining this relationship with other PIs have focused on a small number of key mutations (19, 25). However, that approach is likely to provide incomplete information with respect to lopinavir-RTV regimens because of the strong association of those mutations with only modest levels of reduced susceptibility to lopinavir (well below the sustained plasma lopinavir levels). The Data Analysis Plan of the HIV Resistance Collaborative Working Group contains a provision for treating the number of mutations associated with PI resistance as a covariate (7). However, that list is “generic” for the PI class and contains mutations that are clearly not associated with reduced susceptibility to lopinavir. The number of mutations from the list of 11 identified in this analysis (lopinavir mutation score) provides an alternate covariate with which to investigate the genotypic susceptibility breakpoint(s) for lopinavir-RTV (12). Details of those studies will be published elsewhere.
Finally, the set of mutations found by this analysis to be associated with reduced susceptibility is almost identical to those selected with IDV (5) and RTV (15). Moreover, the phenotypic susceptibility of the combined panel of isolates to lopinavir strongly correlated with susceptibility to RTV or IDV. These results suggest a high potential for cross-resistance of resistant isolates selected in vivo with lopinavir-RTV to either RTV or IDV. The phenotypic correlations between lopinavir and the other PIs tested, particularly SQV and APV, were lower; nonetheless, the lopinavir mutation score includes multiple secondary protease mutations associated with resistance to the PI class. Thus, the potential for cross-resistance of isolates selected by lopinavir-RTV to other PIs will require careful assessment.
ACKNOWLEDGMENTS
We gratefully acknowledge the assistance of Yolanda Lie and Nick Hellman (ViroLogic) and Kurt Hertogs and Brendan Larder (Virco) in obtaining the phenotypes and genotypes of the M98-957 and M97-765 baseline isolates, respectively.
REFERENCES
- 1.Box G E P, Cox D R. An analysis of transformations. J R Stat Soc Ser B. 1964;26:211–243. [Google Scholar]
- 2.Carrillo A, Stewart K, Sham H L, Norbeck D W, Kohlbrenner W E, Leonard J M, Kempf D J, Molla A. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J Virol. 1998;72:7532–7541. doi: 10.1128/jvi.72.9.7532-7541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condra J, Holder J J, Schleif W A, Bakshi K, Danovich R M, Graham D J, Shivaprakash M, Holmes K, Saah A J, Leavitt R Y, Chodakewitz J A, Emini E A. Genetic correlates of virological response to an indinavir-containing salvage regimen in patients with nelfinavir failure. Antiviral Ther. 1999;4(Suppl. 1):44. [Google Scholar]
- 4.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 5.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deeks S G, Hellmann N S, Grant R M, Parkin N T, Petropoulos C J, Becker M, Symonds W, Chesney M, Volberding P A. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J Infect Dis. 1999;179:1375–1381. doi: 10.1086/314775. [DOI] [PubMed] [Google Scholar]
- 7.DeGruttola V, Dix L, D'Aquila R, Holder D, Phillips A, Ait-Khaled M, Baxter J, Clevenbergh P, Hammer S, Harrigan R, Katzenstein D, Lanier R, Miller M, Para M, Yerly S, Zolopa A, Murray J, Patick A, Miller V, Castillo S, Pedneault L, Mellors J. The relation between baseline HIV drug resistance and response to antiretroviral therapy: reanalysis of retrospective and prospective studies using a standardized data analysis plan. Antiviral Ther. 2000;5:41–48. doi: 10.1177/135965350000500112. [DOI] [PubMed] [Google Scholar]
- 8.Gulnik S V, Suvorov L I, Liu B S, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 9.Harrigan P R, Hertogs K, Verbiest W, Pauwels R, Larder B, Kemp S, Bloor S, Yip B, Hogg R S, Alexander C, Montaner J S. Baseline HIV drug resistance profile predicts response to ritonavir-SQV protease inhibitor therapy in a community setting. AIDS. 1998;13:1863–1871. doi: 10.1097/00002030-199910010-00008. [DOI] [PubMed] [Google Scholar]
- 10.Hertogs K, Debethune M P, Miller V, Ivens T, Schel P, Vancauwenberge A, Vandeneynde C, Vangerwen V, Azijn H, Vanhoutte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. International AIDS Society—USA Panel. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 12.Kempf D, Brun S, Rode R, Isaacson J, King M, Xu Y, Real K, Hsu A, Granneman R, Lie Y, Hellmann N, Bernstein B, Sun E. Identification of clinically relevant phenotypic and genotypic breakpoints for ABT-378/r in multiple PI-experienced, NNRTI-naive patients. Antiviral Ther. 2000;5(Suppl. 3):70. [Google Scholar]
- 13.Mantel N. Assessing laboratory evidence for neoplastic activity. Biometrics. 1980;36:381–399. [PubMed] [Google Scholar]
- 14.Miller V, Cozzi-Lepri A, Hertogs K, Gute P, Larder B, Bloor S, Klauke S, Rabenau H, Phillips A, Staszewski S. HIV drug susceptibility and treatment response to mega-HAART regimen in patients from the Frankfurt HIV cohort. Antiviral Ther. 2000;5:49–55. doi: 10.1177/135965350000500113. [DOI] [PubMed] [Google Scholar]
- 15.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H-M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A B, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 16.Molla A, Vasavanonda S, Kumar G, Sham H L, Johnson M, Grabowski B, Denissen J F, Kohlbrenner W, Plattner J J, Leonard J M, Norbeck D W, Kempf D J. Human serum attenuates the activity of protease inhibitors toward wild-type and mutant human immunodeficiency virus. Virology. 1998;250:255–262. doi: 10.1006/viro.1998.9383. [DOI] [PubMed] [Google Scholar]
- 17.Molla A, Brun S, Mo H, Real K, Poddig J, Bernstein B, Hertogs K, Larder B, Lie Y, Hellmann N, Vasavanonda S, Chernyavskiy T, Freimuth W, Japour A, Sun E, Kempf D. Genotypic and phenotypic analysis of viral isolates from subjects with detectable viral load on therapy with ABT-378/ritonavir (ABT-378/r) Antiviral Ther. 2000;5(Suppl. 3):30. [Google Scholar]
- 18.Murphy R L, Brun S, Hicks C, Eron J J, Gulick R, King M, White A C, Jr, Benson C, Thompson M, Kessler H A, Hammer S, Bertz R, Hsu A, Japour A, Sun E. ABT-378/ritonavir plus stavudine and lamivudine for the treatment of antiretroviral-naïve adults with HIV-1 infection: 48-week results. AIDS. 2001;15:F1–F9. doi: 10.1097/00002030-200101050-00002. [DOI] [PubMed] [Google Scholar]
- 19.Para M F, Glidden D V, Coombs R W, Collier A C, Condra J H, Craig C, Bassett R, Leavitt R, Snyder S, McAuliffe V, Boucher C. Baseline human immunodeficiency virus type 1 phenotype, genotype, and RNA response after switching from long-term hard-capsule saquinavir to indinavir or soft-gel-capsule saquinavir in AIDS Clinical Trials Group protocol 333. J Infect Dis. 2000;182:733–743. doi: 10.1086/315769. [DOI] [PubMed] [Google Scholar]
- 20.Petropoulos C J, Parkin N T, Limoli K L, Lie Y S, Wrin T, Huang W, Tian H, Smith D, Winslow G A, Capon D J, Whitcomb J M. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schock H B, Garsky V M, Kuo L C. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials—compensatory modulations of binding and activity. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 22.Sham H L, Kempf D J, Molla A, Marsh K C, Kumar G N, Chen C-M, Kati W, Stewart K, Lal R, Hsu A, Betebenner D, Korneyeva M, Vasavanonda S, McDonald E, Saldivar A, Wideburg N, Chen X, Niu P, Park C, Jayanti V, Grabowski B, Granneman G R, Sun E, Japour A J, Leonard J M, Plattner J J, Norbeck D W. ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob Agents Chemother. 1998;42:3218–3224. doi: 10.1128/aac.42.12.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tukey J W, Ciminera J L, Heyse J F. Testing the statistical certainty of a response to increasing doses of a drug. Biometrics. 1985;41:295–301. [PubMed] [Google Scholar]
- 24.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zolopa A R, Shafer R W, Warford A, Montoya J G, Hsu P, Katzenstein D, Merigan T C, Efron B. HIV-1 genotypic resistance patterns predict response to SQV-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann Intern Med. 1999;131:813–821. doi: 10.7326/0003-4819-131-11-199912070-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]