Abstract
Aim
Knowledge of how circadian rhythm influences brain health remains limited. We aimed to investigate the associations of accelerometer‐measured circadian rest‐activity rhythm (CRAR) with incident dementia, cognitive dysfunction, and structural brain abnormalities in the general population and underlying biological mechanisms.
Methods
Fifty‐seven thousand five hundred and two participants aged over 60 years with accelerometer data were included to investigate the association of CRAR with incidental dementia. Non‐parametric CRAR parameters were utilized, including activity level during active periods of the day (M10), activity level during rest periods of the day (L5), and the relative difference between the M10 and L5 (relative amplitude, RA). Associations of CRAR with cognitive dysfunction and brain structure were studied in a subset of participants. Neuroimaging‐transcriptomics analysis was utilized to identify the underlying molecular mechanisms.
Results
Over 6.86 (4.94–8.78) years of follow‐up, 494 participants developed dementia. The risk of incident dementia was associated with decreasing M10 (hazard ratio [HR] 1.45; 95% conference interval [CI], 1.28–1.64) and RA (HR 1.37; 95% CI, 1.28–1.64), increasing L5 (HR 1.14, 95% CI 1.07–1.21) and advanced L5 onset time (HR 1.12; 95% CI, 1.02–1.23). The detrimental associations were exacerbated by APOE ε4 status and age (>65 years). Decreased RA was associated with lower processing speed (Beta −0.04; SE 0.011), predominantly mediated by abnormalities in subcortical regions and white matter microstructure. The genes underlying CRAR‐related brain regional structure variation were enriched for synaptic function.
Conclusions
Our study underscores the potential of intervention targeting at maintaining a healthy CRAR pattern to prevent dementia risk.
Keywords: brain structure, circadian rest‐activity rhythm, cognition, dementia, UK biobank
Introduction
Circadian rhythm disruption often co‐occurs with neurodegenerative diseases, including Alzheimer's disease and related dementias. 1 The question of whether circadian rhythm disruption is a cause or a consequence of dementia is warmly debated. As a pivotal behavioral indicator of circadian rhythm, circadian rest‐activity rhythm (CRAR) has been associated with the risk of multiple health outcomes. 2 , 3 , 4 Emerging evidence has suggested that disrupted CRAR is associated with the acceleration of progression to dementia in individuals with potentially existing early‐stage dementia‐related pathologies, 5 , 6 wherein reverse causality may have occurred. Investigation of the association between CRAR and dementia risk among a more cognitively intact population could help tackle the above puzzle. However, it remains unknown whether CRAR disruption predicts the incidence of dementia in the healthy elderly population. Additionally, growing evidence has suggested that dementia is determined by gene–environmental interactions, therefore, it is intriguing to explore the joint effects of stratified genetic risk for dementia, APOE ε4, with CRAR conferring risk for dementia.
More importantly, the mechanisms underlying the association between CRAR and dementia remain incompletely elucidated. Brain structure is unusually considered as the intermediate phenotype explaining dementia risk, 7 which is affected by genetic variations and underlying neurological pathologies. To date, only a few previous studies with limited sample size have reported associations of CRAR disruption with atrophy in the medial temporal lobe or impaired white matter microstructures, but did not comprehensively assess structural changes across the entire brain. 8 , 9 , 10 It remains unknown whether the brain structural abnormalities mediate the associations between CRAR and dementia or cognitive dysfunction. Furthermore, animal studies have implied that circadian rhythm disruption potentially promoted oxidative stress and cell death, impaired amyloid clearance, or disturbed synaptic plasticity, then possibly has led to brain atrophy. 11 Nevertheless, the biological process underlying the impact of CRAR on brain changes remains elusive, which is critical in identifying potential targets for preventing brain dysfunction. Benefitting from the large‐scale brain imaging and genetic data of UK Biobank, it is feasible to testify whether these neurobiological mechanisms underlie associations between CRAR disruption and brain structural abnormalities using neuroimaging‐transcriptomic analysis.
Utilizing individual data from a large‐scale population‐based cohort study of UK Biobank, we sought to answer three main aims. Our primary aim was to determine the associations of accelerometer‐measured CRAR in various nonparametric measures (including amplitude, phase and stability) with incident dementia and cognitive function in healthy elderly population, and to investigate whether APOE ε4 allele modified such associations. Second, we aimed to examine whether changes in brain structure mediate the association between CRAR and cognition. Lastly, we sought to explore the potential neurobiological mechanisms underlying associations between CRAR disruption and changes in brain structure.
Materials and Methods
Study design and participants
The study was based on data from the UK Biobank study, a large‐scale prospective cohort study including approximately half a million participants aged 37 to 73 years and recruited from 22 centers across the United Kingdom between 2006 and 2010. 12 All participants provided consent for researchers to access their national health‐related hospital and death records. The National Health Service (NHS) North West Centre for Research Ethics Committee approved the UKB study (reference 11/NW/0382), which conforms to the provisions of the Declaration of Helsinki. The participants provided information on sociodemographic characteristics, lifestyle behaviors, environmental exposures, and health‐related factors collected through a combination of a touchscreen questionnaire and nurse‐led verbal interviews. Between February 2013 and December 2015, A total of 106,053 (44.8%) participants agreed to take part and were provided with a wrist‐worn Axivity AX3 accelerometer (Axivity, Newcastle upon Tyne, UK). 13 Finally, 103,712 raw accelerometer datasets (field ID 90001) were collected for data analysis. Further details of the UKB study are available online (https://www.ukbiobank.ac.uk). The workflow of the study and the exclusion criteria for the participants are summarized in the Supplementary materials (Supplementary Fig. S1; Supplementary Methods).
Accelerometry data
Over 100,000 participants wore an Axivity AX3 triaxial accelerometer on their dominant wrist for 7 consecutive days in the years 2013 to 2015. The accelerometers were configured to collect data at 100 Hz with a dynamic range of ±8 g for the current study. The raw acceleration signals were calibrated to gravity to ensure data accuracy. Nonwear time was defined as consecutive stationary episodes lasting for at least 1 h, during which the standard deviation of all three axes of the accelerometer was less than 13.0 m. More detailed information on data processing and analysis has been published elsewhere. 14
We employed nonparametric analyses with the physical activity intensity data into 5‐s epochs (UK Biobank field ID 90004) to derive seven CRAR parameters: (i) L5: the average activity level during the least active 5‐h period, reflecting activity during periods of rest; (ii) M10: the average activity level during the most active 10‐h period, indicating the intensity of activity during the awake period; (iii) relative amplitude (RA): a measure that quantifies the relative difference between the M10 and L5 over a 24‐h period. It is calculated using the formula RA = (M10‐L5)/(M10 + L5). A higher RA value signifies a greater distinction between activity levels during the most or least active periods of the day; 15 (iv) L5 onset time: indicating whether inactivity (likely sleep) occurs earlier or later in the day; (v) M10 onset time: indicating whether a person is more active earlier or later in the day; (vi) interdaily stability (IS): the day‐to‐day stability of the rhythm (higher values reflect higher robustness of circadian rhythm); and (vii) intradaily variability (IV): the fragmentation of rest‐activity rhythm (higher values reflect higher circadian fragmentation). These measures have been defined and reviewed in detail elsewhere. 16
Outcome
Dementia diagnoses in the UK Biobank dataset were ascertained based on records from hospital inpatient and death registries. Hospital inpatient records were obtained from the Hospital Episode Statistics for England, the Scottish Morbidity Record for Scotland, and the Patient Episode Database for Wales. Death registry records were acquired from the National Health Service Digital for England and Wales as well as the Information and Statistics Division for Scotland. Furthermore, dementia diagnoses were also identified from primary care data utilizing disease codes. The diagnoses of dementia were recorded using the tenth edition of the International Classification of Diseases (ICD) coding system. The complete list of codes utilized in this study can be found in Supplementary Table S1. Incident dementia events were defined as diagnoses made subsequent to the accelerometer measurements. At the time of analysis, hospital admission data were available for participants until 15 January 2021. Therefore, participants were censored at the date associated with the incidence of dementia, date of death, or last known follow‐up (15 January 2021), whichever occurred first. The distribution of time‐to‐event was illustrated in Supplementary Fig. S2.
Participants completed a battery of cognitive tests organized by the UK Biobank. We chose to analyze tests done by at least 10% of participants. Seven cognitive tests were grouped into three cognitive domains: Processing speed (Reaction time test, Trail making test A and Symbol digit substitution test), Executive function (Fluid intelligence test and Trail making test B) and Memory (Numeric memory test and Pairs matching test). Full details of the computation of cognitive test scores and generation of cognitive domains can be found in the Supplementary Methods.
Quality‐controlled T1‐weighted neuroimaging data processed with FreeSurfer were used in the present study. FreeSurfer was used to extract the cortical measures, including surface area, volume, and mean cortical thickness of atlas regions and volumes of subcortical regions. For this study, the FreeSurfer aparc atlas (field ID = 192), consisting of 66 cortical regions, and the ASEG atlas (field ID = 190), consisting of 33 subcortical regions, were utilized. Two quality‐controlled imaging‐derived phenotypes obtained from diffusion tensor imaging (DTI) provided by the UKB were finally used in this study, namely, fractional anisotropy (FA) and mean diffusivity (MD). DTIFIT (part of the FMRIB Software Library's Diffusion Toolbox) was used to generate FA and MD maps. Next, probabilistic tractography using the AutoPtx package from FSL was conducted to extract 27 tracts throughout the entire brain in FA maps. Masks of these tracts, derived from the FA maps, were then applied to locate the corresponding tracts on the MD maps. Finally, weighted tract‐averaged FA and MD values for each tract were generated, providing a summary measure of FA and MD along the tracts of interest. These measures allowed us to assess the microstructural integrity of specific white matter tracts in the brain.
Statistical analyses
The events of all outcomes we investigated were sufficient according to the rule‐of‐thumb estimation, which requires at least 10 events per variable in the model. 17 In addition, to minimize potential inferential bias, multiple imputations were conducted to assign any missing covariate values using the “mice” R package. 18 According to Sterne et al.'s recommendations for multiple imputation, 19 we included the exposures, covariates, disease and mortality outcomes and the time to these outcomes in the imputation procedure.
Cox proportional hazard regression analysis, with time since wearing the accelerometer set as the start of follow‐up, was used to examine the associations of CRAR parameters with dementia outcomes. Hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) were calculated. The proportionality of the hazards assumption was assessed using the Schoenfeld residuals technique 20 and no violation of the assumption was found. Hazard ratios (HRs) and corresponding 95% confidence intervals (95% CIs) were calculated. CRAR parameters were analyzed as continuous values with the HR expressed per 1‐SD decrease. The L5 and M10 onset time were converted from time data (hh: mm) to decimal data (##0.00). Specifically, for L5 onset time, we added 24 h to participants whose L5 onset started the following day. 21 Additionally, quartiles were utilized to categorize the participants. The highest quartiles of M10, RA, and IS and the lowest quartiles of L5 and IV served as references. We identified three categories of participants regarding L5 onset time and M10 onset time based on having a peak time at the population mean for the study population, more than 1 SD above the mean (delayed phase), and 1 SD below the mean (advanced).
We conducted comprehensive adjustments in various Cox proportional hazard models. Model 1 adjusted for age, sex and education level, and Model 2 additionally adjusted for ethnicity, the Townsend index of deprivation, recruitment center and season of accelerometer wear. Model 3 further adjusted for dementia‐related common risk factors, including smoking status, alcohol intake; healthy diet score, obesity, history of diabetes, hypertension, depression, and APOE ε4 carrier status. The codes of covariates utilized in our study can be found in Supplementary Table S2. By applying these preprocessing steps, we aimed to ensure that the data used in the regression analyses were appropriately adjusted for confounding factors and transformed to facilitate meaningful comparisons and interpretations. In addition, we used restricted cubic spline models fitted for Cox proportional hazards models with three knots to flexibly model and visualize the dose–response relationships of each exposure with incident dementia. 2 Interaction analyses on both multiplicative and additive scales and joint associations of CRAR parameters with age, sex and APOE ε4 carrier status were performed. We also conducted subgroup analyses stratified by age, sex and APOE ε4 carrier status categories.
Linear regression analysis was performed on a smaller subset of the data. Prior to the analyses, we regressed out the effects of brain size from all T1 measures using linear regression. In linear regression analysis investigating the association of CRAR parameters with structural brain changes and cognitive performance, we additionally adjusted the time interval between the accelerometer assessment and cognitive test/brain scans. By applying these preprocessing steps, we aimed to ensure that the data used in the regression analyses were appropriately adjusted for confounding factors and transformed to facilitate meaningful comparisons and interpretations. Exploratory mediation analyses were carried out to couple the imaging findings with the cognitive findings using the mediation package version 4.5.0 in R (http://CRAN.R-project.org/package=mediation). We reported the proportion of the total effect of the CRAR parameters on cognitive performance mediated by the MRI measures with P‐values calculated through quasi‐Bayesian approximation using 5000 simulations.
We used the Allen Human Brain Atlas (AHBA) transcriptomic dataset collected from six human donor brains through ENIGMA TOOLBOX (https://enigma-toolbox.readthedocs.io/en/latest/pages/10.genemaps/index.html). The microarray expression data were initially generated using abagen following established recommendations. 22 , 23 Preprocessing steps included intensity‐based filtering and selecting representative microarray probes for each gene in both hemispheres, matching microarray samples to brain parcels from the Desikan‐Killiany and aggregating within parcels and across donors. Subsequently, genes across donors with similarity below a defined threshold (r < 0.2) across donors were removed, resulting in a total of 12,668 genes for further analysis (using the Desikan‐Killiany atlas). Regional expression levels of each gene were then compiled to create a 65 × 12,668 (cortical regions) and 14 × 12,668 (subcortical regions) regional transcription matrix.
Partial least squares (PLS) regression was utilized to relate the regional brain transcriptome to the regional brain morphology associated with CRAR parameters. 23 PLS combines dimension reduction and linear regression, identifying components from X (the 65 × 12,668 (cortical regions) or 14 × 12,688 (subcortical regions) predictor matrix of regional mRNA measurements) that have maximum covariance with Y (the 65 × 1 (cortical regions) and 14 × 1 (subcortical regions) regional brain morphology). The first PLS component (PLS1) was used to weigh and rank gene predictor variables. The statistical significance of the variance explained by PLS1 was tested by permuting the response variables 1000 times to test the null hypothesis that PLS1 did not explain more variance in Y than chance. The error in estimating each gene's PLS1 weight was assessed by bootstrapping, and the ratio of the weight of each gene to its bootstrapped standard error was used to rank its contribution to PLS1. All PLS and bootstrapping analyses were conducted in MATLAB. We tested the null hypothesis of zero weight for each gene using a false‐discovery‐rate inverse‐quantile‐transformation correction to account for winner's curse bias using R version 4.2.2. 24 Only genes that survived this correction at Q < 0.05 were included in the enrichment analyses. Finally, the g:Profiler toolset 25 was applied to perform a gene ontological (GO) enrichment analysis of the weighted genes defined by PLS1. We filtered the resulting list of GO terms, retaining only those that were significantly enriched at Q < 0.05.
All statistical tests were two‐sided, and a P‐value of less than 0.05 was regarded as statistically significant. To account for multiple comparisons, all P‐values in fully adjusted models were corrected via the false‐discovery rate (FDR). 26 We conducted all statistical analyses by using R version 4.2.2 and MATLAB 2022.
Results
Association of CRAR with incident dementia
Baseline demographics and health characteristics of participants stratified by incident dementia status are provided in Table 1. The risk of incident dementia was significantly associated with decreased M10 and RA, increased L5 and advanced L5 onset time (Fig. 1; Supplementary Table S3). The results remained robust after additionally adjusting for sleep duration, sleep efficiency and moderate to vigorous physical activity (Supplementary Table S3) and in a series of sensitivity analysis by conducting COX regression analysis in complete data restricted to participants without any missing covariate data (Supplementary Table S4 and S5 ), or by excluding events that occurred within the first year of follow‐up (Supplementary Table S6) or by censoring up to December 31, 2019 (Supplementary Table S7 ), or by using competing risk regression (Supplementary Table S8 ). Furthermore, RCS models showed that the risk of incident dementia decreased rapidly until M10 at approximately 50 g/min and then became relatively flat (P for nonlinearity <0.001) (Fig. 1).
Table 1.
Baseline characteristics of participants by incident dementia status
| Characteristics | No incident dementia (n = 57,008) | Incident dementia (n = 494) | Total (n = 57,502) |
|---|---|---|---|
| Age at accelerometry (years) | 62.37 ± 7.83 | 69.95 ± 5.05 | 62.42 ± 7.83 |
| Female (Yes/No) | 51,392 (56.67) | 246 (47.22) | 51,638 (56.62) |
| Non‐White ethnicity (Yes/No) | 2715 (2.99) | 14 (2.69) | 2729 (2.99) |
| Townsend deprivation index | −1.75 ± 2.81 | −1.85 ± 2.78 | −1.75 ± 2.81 |
| Recruitment regions (Yes/No) | |||
| England | 81,411 (89.78) | 486 (93.28) | 81,897 (89.8) |
| Wales | 3397 (3.75) | 9 (1.73) | 3406 (3.73) |
| Scotland | 5871 (6.47) | 26 (4.99) | 5897 (6.47) |
| Education level | |||
| Degree or above | 39,723 (43.81) | 205 (39.35) | 39,928 (43.78) |
| Any other qualification | 43,446 (47.91) | 224 (42.99) | 43,670 (47.88) |
| No qualification | 7510 (8.28) | 92 (17.66) | 7602 (8.34) |
| Season of accelerometer wear (Yes/No) | |||
| Spring | 20,524 (22.63) | 112 (21.5) | 20,636 (22.63) |
| Summer | 23,643 (26.07) | 153 (29.37) | 23,796 (26.09) |
| Autumn | 27,129 (29.92) | 150 (28.79) | 27,279 (29.91) |
| Winter | 19,383 (21.38) | 106 (20.35) | 19,489 (21.37) |
| Healthy diet score | 2.69 ± 1.17 | 2.86 ± 1.20 | 2.69 ± 1.17 |
| Smoking status | |||
| Never | 52,177 (57.54) | 244 (46.83) | 52,421 (57.48) |
| Previous | 32,833 (36.21) | 249 (47.79) | 33,082 (36.27) |
| Current | 5669 (6.25) | 28 (5.37) | 5697 (6.25) |
| Alcohol consumption | |||
| Not current | 5365 (5.92) | 57 (10.94) | 5422 (5.95) |
| Two or less times a week | 41,985 (46.3) | 203 (38.96) | 42,188 (46.26) |
| Three or more times a week | 43,329 (47.78) | 261 (50.1) | 43,590 (47.8) |
| Health status (Yes/No) | |||
| Obesity | 17,616 (19.43) | 113 (21.69) | 17,729 (19.44) |
| Diabetes history | 4319 (4.76) | 64 (12.28) | 4383 (4.81) |
| Hypertension history | 24,571 (27.1) | 219 (42.03) | 24,790 (27.18) |
| Cardiovascular diseases history | 22,143 (24.42) | 237 (45.49) | 22,380 (24.54) |
| Depression history | 8018 (8.84) | 49 (9.4) | 8067 (8.85) |
| APOE ε4 carrier (Yes/No) | 25,548 (28.17) | 229 (43.95) | 25,777 (28.26) |
| Circadian markers | |||
| L5 (counts/min) | 3.15 ± 1.18 | 3.27 ± 1.29 | 3.15 ± 1.18 |
| M10 (counts/min) | 47.42 ± 15.00 | 39.74 ± 13.80 | 47.37 ± 15.01 |
| RA (arbitrary units) | 0.87 ± 0.06 | 0.83 ± 0.08 | 0.87 ± 0.06 |
| L5 onset time (h) | 24.87 ± 1.09 | 24.75 ± 1.16 | 24.87 ± 1.09 |
| M10 onset time (h) | 8.53 ± 1.34 | 8.31 ± 1.16 | 8.53 ± 1.34 |
| Interdaily stability (arbitrary units) | 0.54 ± 0.13 | 0.58 ± 0.12 | 0.54 ± 0.13 |
| Intradaily variability (arbitrary units) | 0.91 ± 0.24 | 0.91 ± 0.22 | 0.91 ± 0.24 |
| Sleep duration (hours) | 7.28 ± 0.89 | 7.20 ± 1.04 | 7.28 ± 0.89 |
| Sleep efficiency (%) | 76.17 ± 7.24 | 75.02 ± 8.74 | 76.16 ± 7.25 |
| Total MVPA volume (minutes/week) | 156.61 ± 148.85 | 97.58 ± 115.38 | 156.27 ± 148.74 |
Note: Values are expressed as No. (%) for categorical variables and mean ± SD for continuous variables, unless specified otherwise. Includes imputed values for covariates.
Abbreviations: APOE, apolipoprotein E; L5, the least active 5‐h period; M10, the most active 10‐h period; RA, relative amplitude; MVPA, moderate to vigorous physical activity; PRS, polygenic risk score; SD, standard deviation.
Fig. 1.
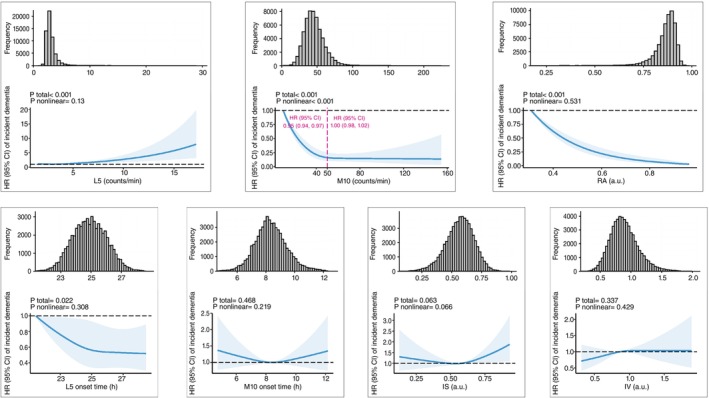
Associations between CRAR parameters and incident dementia. Restricted cubic spline models fitted for Cox proportional hazards models with three knots. Dashed vertical red lines represent the turning point for the association between M10 and the incidence of dementia. The HRs were adjusted for age, sex, education levels, ethnicity, the Townsend index of deprivation, recruitment center, season of accelerometer wear, smoking status, alcohol intake, healthy diet score, obesity, history of diabetes, hypertension, depression and APOE ε4 status. APOE, apolipoprotein E; CI, confidence interval; CRAR, circadian rest‐activity rhythm; HR, hazard ratio; L5, the least active 5‐h period; M10, the most active 10‐h period; RA, relative amplitude; IS, interdaily stability; IV, intradaily variability; a.u., arbitrary unit.
In the subsequent interaction analysis, CRAR parameters were reclassified into two groups according to their associations with incident dementia (Fig. 1; Supplementary Table S3): L5 (quartiles 1–3, quartile 4), M10 (<50 g/min, ≥50 g/min), RA (quartiles 2–4, quartile 1), and L5 onset time (intermediate to delayed, advanced), to facilitate the interpretation of interaction effects. Subgroup analyses revealed that the detrimental effect of lower M10 on incident dementia was more pronounced at younger ages, while the effect of advanced L5 onset time on incident dementia was higher in females (Supplementary Fig. S3). Through joint analysis and additive interaction analyses, the detrimental effects of higher L5, lower M10 and RA on incident dementia were exacerbated by APOE ε4 carrier status (Table 2; Supplementary Table S9). In addition, the detrimental effects of lower M10 and RA on incident dementia were amplified by older age (≥65 years).
Table 2.
Interaction effects of CRAR parameters and APOE ε4 status on incident dementia
| Multiplicative interaction | Additive interaction | |||||
|---|---|---|---|---|---|---|
| CRARs | APOE ε4 non‐carrier | APOE ε4 carrier | HR (95% CI) † ; P | RERI (95% CI) † | AP (95% CI) † | SI (95% CI) † |
| L5 | ||||||
| Quarter 1–3 | 1.00 [Reference] | 3.43 (2.78, 4.25); <0.001 | 1.32 (0.90, 1.93); 0.161 | 1.48 (0.27, 2.69) | 0.29 (0.10, 0.49) | 1.58 (1.11, 2.26) |
| Quarter 4 | 1.11 (0.84, 1.48); 0.467 | 5.03 (3.88, 6.51); <0.001 | ||||
| M10 | ||||||
| >50 | 1.00 [Reference] | 4.32 (2.86, 6.54); <0.001 | 0.84 (0.53, 1.33); 0.467 | 1.59 (0.05, 3.13) | 0.23 (0.02, 0.45) | 1.38 (0.98, 1.95) |
| ≤50 | 1.85 (1.31, 2.63); <0.001 | 6.77 (4.79, 9.57); <0.001 | ||||
| Relative amplitude | ||||||
| Quarter 2–4 | 1.00 [Reference] | 4.12 (3.26, 5.22); <0.001 | 0.80 (0.56, 1.15); 0.231 | 1.86 (0.25, 3.46) | 0.26 (0.08, 0.44) | 1.43 (1.07, 1.92) |
| Quarter 1 | 2.15 (1.65, 2.80); <0.001 | 7.13 (5.46, 9.31); <0.001 | ||||
| L5 onset | ||||||
| Intermediate to delayed | 1.00 [Reference] | 3.84 (3.14, 4.68); <0.001 | 0.87 (0.57, 1.35); 0.539 | 0.19 (−1.13, 1.50) | 0.04 (−0.25, 0.34) | 1.06 (0.71, 1.59) |
| Advanced | 1.29 (0.94, 1.76); 0.116 | 4.31 (3.19, 5.83); <0.001 | ||||
Abbreviations: AP, attributable proportion due to interaction; APOE, apolipoprotein E; CI, confidence interval; CRAR, circadian rest‐activity rhythm; HR, hazard ratio; L5, the least active 5‐h period; M10, the most active 10‐h period; RERI, relative excess risk due to interaction; SI, the synergy index.
Multivariable Cox model was adjusted for age, sex, education level, ethnicity, Townsend index of deprivation, recruitment center, season of accelerometer wear, smoking status, alcohol intake, healthy diet score, obesity, history of diabetes, hypertension, depression, and APOE ε4 carrier.
Associations between CRAR and cognition
Higher L5 and lower RA and M10 had a significant relationship with lower performance in processing speed, while the association between M10 and processing speed was no longer significant after FDR correction. Higher L5 and advanced L5 and M10 onset time were positively associated with lower performance in executive function. Furthermore, decreased IS and increased IV were associated with better performance in all cognitive domains (Supplementary Table S10). Additionally, RCS models did not show any significant nonlinear associations (Fig. 2), except for the relationship between M10 and cognitive performance, which displayed a turning point at approximately 50 g/min (P for nonlinearity <0.05).
Fig. 2.
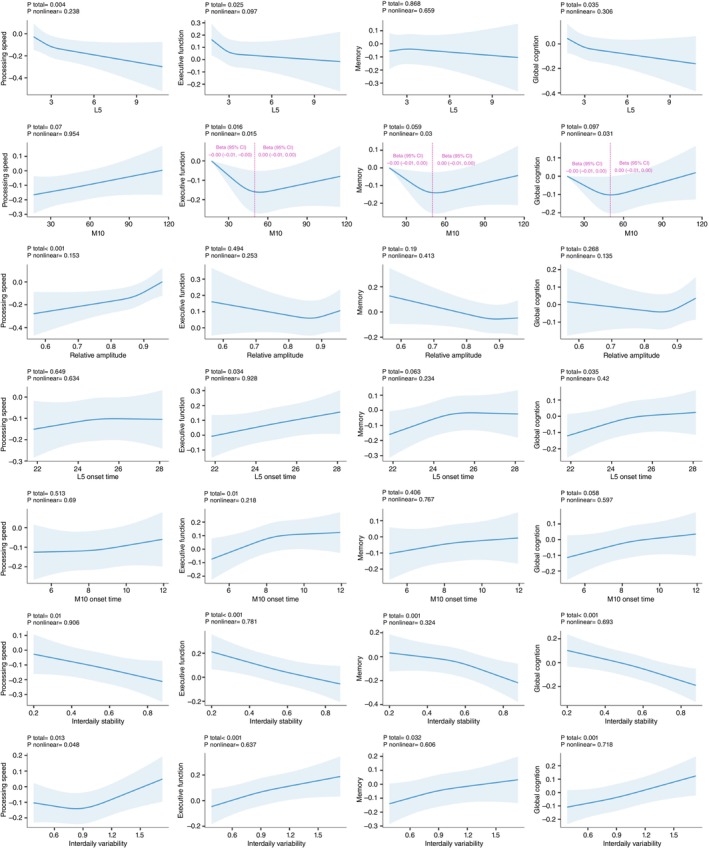
Associations between CRAR parameters and cognitive function. Restricted cubic spline models fitted for linear regression models with three knots. Dashed vertical red lines represent the turning point for the association between M10 and cognition. The Betas were adjusted for age, sex, education levels, ethnicity, the Townsend index of deprivation, recruitment center, season of accelerometer wear, smoking status, alcohol intake, healthy diet score, obesity, history of diabetes, hypertension, depression and APOE ε4 status. APOE, apolipoprotein E; CRAR, circadian rest‐activity rhythm; L5, the least active 5‐h period; M10, the most active 10‐h period.
Associations between CRAR and changes in brain structure
Both higher M10 and RA were significantly associated with larger volumes of cerebral white matter and greater global surface areas in both the left and right hemispheres (Fig. 3, Supplementary Table S11). Furthermore. higher M10 was associated with a larger volume of cerebrospinal fluid (Supplementary Table S11). Additionally, higher IS was associated with lower brain thickness and a larger volume of cerebrospinal fluid. The associations were still significant after FDR correction. No significant association was observed between other CRAR parameters and global brain structure.
Fig. 3.
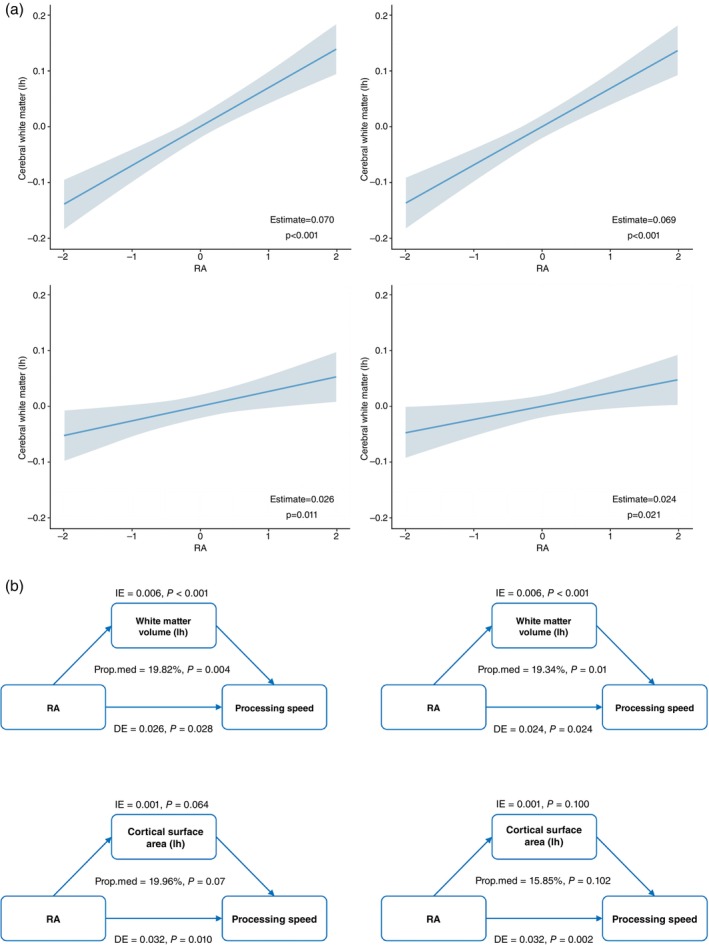
The association between RA and global brain structure, mediation analysis of the association between RA, global brain structures, and processing speed. (a) shows the significant association of RA with global brain structure (white matter volumes and cortical surface areas). (b) shows the mediating effect of global brain structure in the association of RA and processing speed. DE, direct effect; IE, indirect effect; lh, left hemisphere; Prop.med, proportion of mediation; rh, right hemisphere; RA, relative amplitude.
Among CRAR parameters, M10 and RA showed the primary significant relationships with regional brain structure (Fig. 4, Supplementary Table S12). Higher M10 was predominantly associated with smaller surface areas of subregions of the cingulate and larger surface areas in subregions of the occipital lobe, as well as in the precentral and superior parietal cortex. Higher RA was predominantly associated with a larger surface area in regions of the occipital lobe and a smaller surface area in the left insula. The relationships were mostly reversed in terms of thickness. Regarding subcortical structure, higher M10 was significantly associated with larger volumes of the anterior corpus callosum, as well as smaller volumes of the amygdala, accumbens area and caudate. Higher RA was associated with larger volumes of the ventral DC, left thalamus proper, most regions of the corpus callosum, brainstem, right pallidum, and left cerebellum cortex as well as smaller volumes of the lateral ventricle. In terms of white matter microstructure measured in DTI, higher M10 was associated with higher FA and lower MD in the posterior thalamic radiation and was additionally associated with lower MD in the anterior thalamic radiation. Higher RA was predominantly associated with higher FA and lower MD in the posterior and anterior thalamic radiation, superior and inferior longitudinal fasciculus, inferior fronto‐occipital fasciculus and cingulate gyrus (Fig. 3, Supplementary Table S13).
Fig. 4.
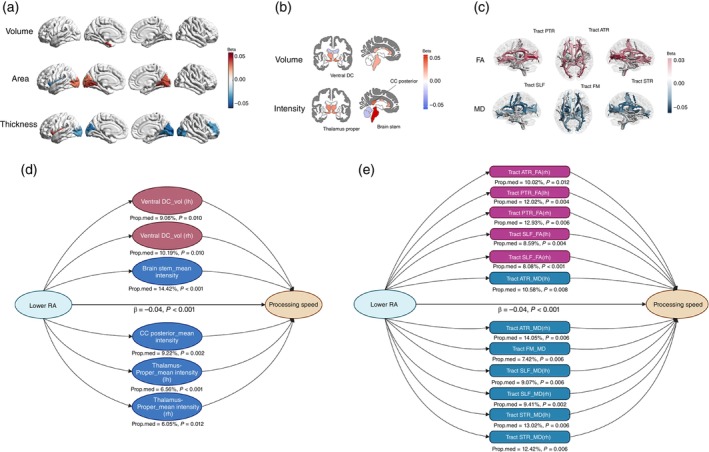
The association of RA with regional brain structures and white matter microstructures. (a–c) show the association of RA with regional brain structures (a) cortical volume, cortical surface area, cortical thickness; (b) subcortical volume and subcortical intensity; (c) FA and MD. (d) shows the significant mediating effect of regional brain structures in the association of RA and processing speed. (e) shows the significant mediating effect of white matter microstructures in the association of RA and processing speed. ATR, anterior thalamic radiation; CC, corpus callosum; DE, direct effect; FA, fractional anisotropy; FM, forceps minor; IE, indirect effect; lh, left hemisphere; MD, mean diffusivity; Prop.med, proportion of mediation; PTR, posterior thalamic radiation; RA, relative amplitude; rh, right hemisphere; SLF, superior longitudinal fasciculus; STR, superior thalamic radiation; Ventral DC, Ventral Diencephalon; vol, volume.
Mediating effects of brain structures on the associations between CRAR parameters and cognitive function
The global white matter volume significantly mediated the association between RA and processing speed (Fig. 3 ). Regarding the regional brain structure, only subcortical structures and white matter microstructures exhibited mediating effects (Fig. 4, Supplementary Table S14 ). The volume of the ventral DC, mean intensities of the brainstem and posterior corpus callosum, and white matter microstructures in the anterior and superior thalamic radiation and superior longitudinal fasciculus mediated the association between RA and processing speed. Additionally, the mean thickness of the left precentral cortex mediated the association between IS and executive function and memory as well as processing speed (nominally significant).
The association of gene expression profiles with regional brain structure variations related to CRAR
We utilized PLS regression analysis to identify patterns of gene expression that correlated with the anatomical distribution of altered brain structure associated with CRAR parameters. The beta coefficients indicated the strength of the association between circadian amplitude (L5, M10, RA) and brain regional morphologies (cortical area, cortical thickness, subcortical volume, and subcortical intensity). The first PLS component (PLS1) explained the highest percentage of variance (>20%) in the relationship of brain structure changes and circadian amplitudes, and the gene expression weights derived from PLS1 exhibited a strong positive correlation with the beta values (Supplementary Fig. S4), exceeding the association expected by chance (permutation test, P < 0.05). Through (Gene Ontology) GO enrichment analyses, we discovered sets of biological processes most strongly linked to the identified genes (Fig. 5), most of which were shared by different CRAR parameters. Genes expressed more prominently in regions with higher beta values were predominantly enriched for GO terms related to neuron projection development, transsynaptic signaling, synaptic transmission, and synaptic plasticity. Specifically, cortical structural changes related to RA were associated with alcohol and sterol metabolism. Subcortical volume changes related to L5 were particularly associated with dendrite function. Additionally, when exploring biological mechanisms underlying the association of subcortical intensities and CRAR parameters, we observed that subcortical intensities were linked to ribosome‐related processes under the influence of the L5 circadian marker. Under the influence of the RA circadian marker, subcortical intensities were associated with visual function (Fig. 5).
Fig. 5.
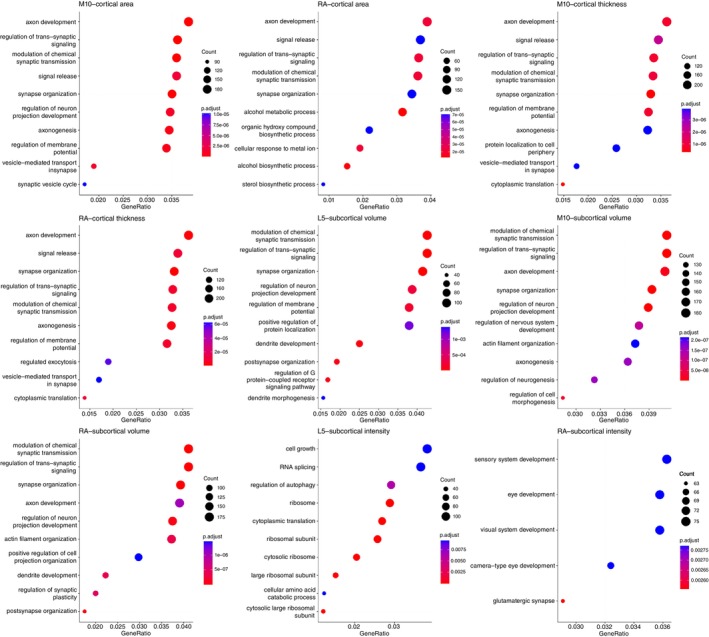
GO enrichment analysis. CI, confidence interval; GO, Gene Ontology; HR, hazard ratio; L5, the least active 5‐h period; M10, the most active 10‐h period; RA, relative amplitude.
Discussion
In this large‐scale cohort study, we found that lower amplitude and advanced phase of CRAR were associated with incident dementia and cognitive decline in the healthy elderly population. These associations were particularly pronounced in individuals carrying the APOE ε4 allele and those aged over 65 years. Strikingly, the association between the amplitude of CRAR and cognition was predominantly mediated by alterations in subcortical structure and white matter microstructures. Neuroimaging‐transcriptomic analysis further indicated that synaptic pathways play a crucial role in the association between the amplitude of CRAR and alterations in brain structure. This study has elucidated mechanisms underlying the association between circadian rhythm dysregulation and incident dementia, which may be related to changes in subcortical structure, white matter microstructures, and synaptic dysfunction.
Our findings are in line with evidence from recent observational studies suggesting that decreased amplitude of CRAR is strongly associated with incident dementia and cognitive decline. 8 , 9 , 27 , 28 Findings from previous studies on the relationship between circadian phases and the incidence of dementia have been mixed. 1 It has been suggested that individuals with mild cognitive impairment present a phase advance of melatonin 29 (the most accurate circadian marker in humans) and core body temperature. 30 Furthermore, the Rotterdam Study indicated earlier L5 onset with increased dementia risk in the next 2 years, which supports our findings. In contrast, several other studies have indicated that delayed acrophase is associated with cognitive decline or dementia. 31 , 32 Our study has the advantages of a large sample size and inclusion of a healthy elderly population excluding early‐onset dementia cases, providing sufficient statistical power for our analyses. However, further systematic reviews and meta‐analyses are needed to thoroughly investigate the association between circadian phase and incident dementia. Our primary previous findings indicate that lower day‐to‐day stability and higher fragmentation of CRAR as risk factors for dementia and cognitive decline, 8 , 9 , 27 , 28 we did not observe a significant association between these factors and the incidence of dementia in healthy elderly participants. However, previous findings were primarily based on older participants (>80 years), who exhibited a greater vulnerability to developing dementia. In addition, our study demonstrated that older age and APOE ε4 allele exaggerated the association between aberrant CRAR and incident dementia. Therefore, we propose that lower day‐to‐day stability and higher fragmentation of CRAR are specifically associated with the incidence of dementia in highly vulnerable populations rather than the healthy elderly. In conclusion, we suggest that the amplitude and phase of CRAR can serve as more accurate indicators of dementia compared to IS and IV.
To the best of our knowledge, no prior study has examined the effects of interactions of CRAR with APOE ε4 carrier status with the incidence of dementia. A similar effect of the additive interaction between low late‐life physical activity and the APOE ε4 allele on AD risk was reported in a sample of general practice patients, 33 which supported our findings to some extent. APOE ε4 has been shown to modulate various biological processes involved in the pathogenesis of dementia, 34 including synaptic plasticity, neurogenesis, and neuroinflammatory processes. Disruptions in CRAR may disrupt these pathways in a manner synergistic with the effects of APOE ε4, potentially resulting in a greater risk of dementia. Further animal research is needed to elucidate the underlying mechanisms and clarify the nature of this interaction.
No previous studies have systematically investigated the relationship between CRAR and regional cortical structure. A negative association of cortical surface area with RA and M10 was observed in most cortical regions except the cingulate cortex. Brain surface area is thought to reflect the structural integrity of gray matter and to be associated with the number and sizes of cells within radial columns perpendicular to the pial surface. 35 The contraction of the brain area has been considered affecting functional networks in schizophrenia. 36 In contrast, cortical thickness showed an opposite association with the amplitude of CRAR. Cortical thickness refers to the depth of the outer layer of neural tissues in the cerebrum. 37 Cortical thinning has been hypothesized to reflect the intracortical myelination, 38 which can displace the boundary observed in MR images between the gray and white matter toward the brain surface, resulting in an apparent thinning of the reconstructed cortex in MR images. Furthermore, the cortical thickness presents a consistent spatial contrast pattern with functional connectivity, even though these measurements represent different aspects of the brain's structure and function. In conclusion, cortical surface area and thickness could reflect brain connectivity, although they were not observed to mediate the relationship between the amplitude of CRAR and cognitive function in our study.
Our findings underscore the crucial involvement of altered subcortical structure (predominantly of the thalamus and corpus callosum) in the relationship between the amplitude of CRAR and cognitive performance. The thalamus, serving as the primary relay center of the brain, plays a crucial role in processing and transmitting sensory information to the cortex, as well as receiving inputs from limbic structures. 39 Accumulating evidence indicates that the thalamus is involved in cognitive function. 40 , 41 , 42 In addition, the thalamus was recently found to contribute to the risk of dementia in a large population‐based cohort of 5522 cognitively normal participants from the Rotterdam Study. 43 The corpus callosum is the primary interhemispheric fiber connection, enabling communication and information exchange between the cerebral hemispheres. 44 Furthermore, atrophy of the corpus callosum has been observed in individuals in the early stages of dementia. 45 In conclusion, brain connections are strongly related to the amplitude of CRAR. Our study and previous studies with small sample sizes investigating white matter microstructures have also confirmed the important role of brain connections in the relationship between the amplitude of CRAR and cognition. 10 , 46 , 47 Longitudinal studies including imaging data collected before and after dementia diagnosis are needed to replicate and confirm our findings.
It is important to draw attention to the potential biological mechanisms underlying the effect of CRAR. Previous animal studies have indicated that circadian modulation in both neuronal and glial cells is involved, specifically in relation to synaptic plasticity. 48 , 49 , 50 Synaptic degeneration represents a pivotal pathophysiological process in dementia and is more robustly associated with the extent of cognitive decline in Alzheimer's disease than the accumulation of amyloid plaques. 51 Our neuroimaging‐transcriptomics study suggests that synaptic function, which is fundamental to neural communication, plays a critical role in the association between CRAR and changes in brain structure. Strikingly, the separate neuroimaging‐transcriptomics analyses for cortical and subcortical structure indicated that the identified genes were consistently enriched in molecular pathways related to synaptic function, suggesting that cortical surface area and thickness may represent the connection between brain regions. Therefore, these findings provide a convincing argument that synaptic dysfunction may be the central mechanism underlying the effect of circadian rhythm disruption on cognitive dysfunction and dementia development. Future mediation analyses of synaptic pathways underlying the association between CRAR and cognition/incident dementia are needed.
The strengths of this study include the large sample size, healthy elderly population, prospective design, and extensive measurement of covariates, which empower the study to robustly investigate the association of CRAR and the incidence of late‐onset dementia. Furthermore, the study's assessment of cognition, region‐specific brain structure, mediation analysis, and the incorporation of brain regional gene expression profiles enrich the exploration of underlying mechanisms. Our study has several limitations that should be acknowledged. First, the UK Biobank sample consists of individuals who are relatively healthier and reside in less socioeconomically deprived areas than the general UK population. Our study population may not be suitable for estimating the prevalence and incidence of dementia on a global scale. However, the valid estimates and comparisons of exposure‐outcome associations derived from our study are broadly applicable and generalizable. 52 , 53 Second, approximately 97% of participants in UK Biobank belong to white ethnicity. The lack of ethnic diversity can impede the generalization of our findings, as health disparities can exist among different ethnic groups. Third, we did not conduct longitudinal studies to investigate the associations of CRAR parameters with cognitive performance and neuroimaging due to the relatively small sample size of repeated measures. Fourth, our neuroimaging‐transcriptomics analysis utilized gene expression data derived from donors unaffected by neurological or psychiatric diseases. Therefore, the spatial expression profiles we examined are based solely on unaffected individuals, and our conclusions are limited to the intrinsic variability within this group. To fully understand the changes in gene expression that occur in dementia and their relationship with brain structure changes related to CRAR parameters, it will be important to incorporate gene expression data from individuals with dementia and obtain widespread cortical coverage.
In conclusion, our findings suggest that reduced amplitude of CRAR is predominantly associated with an increased risk of dementia in the healthy elderly population, especially among APOE ε4 carriers and individuals aged over 65. The association between the amplitude of CRAR and cognition was predominantly mediated by alterations in the subcortical structure and white matter microstructures. Furthermore, our analysis demonstrated that higher levels of intrinsic gene expression related to synaptic function contributed to brain structural changes associated with the amplitude of CRAR. These findings extend current understanding of the potential role of disrupted CRAR in the onset of dementia and highlight the need for future studies to identify the underlying mechanisms.
Disclosure statement
The authors declare no competing interests.
Funding information
This work was supported by the National Natural Science Foundation of China (grant 82101555). YL was supported by the Startup Funding for National Natural Science Foundation of China of Guangdong Provincial People's Hospital (grant 8210051457). HF and YL were supported by the International Postdoctoral Exchange Fellowship Program of China. This research was also supported by Guangzhou Municipal Key Discipline in Medicine (2021–2023), Guangzhou High‐level Clinical Key Specialty, and Guangzhou Research‐oriented Hospital.
Author contributions
Y.Y.L., Y.P.L. and J.Z. conceived and designed the research; Y.L. and H.F. made the statistical analysis with the help of J.D., L.Y. and H.X. In addition, Y.L. and H.F. interpreted the results and drafted the manuscript. J.Z. had full access to all the data in the study. All authors gave their final approval and agreed to be accountable for all aspects of the work. Y.P.L. and Y.Y.L. had final responsibility for the decision to submit for publication.
Supporting information
Appendix S1. Supplementary Information.
Acknowledgments
This research has been conducted using the UK Biobank resource under application number 58082. We thank the participants of the UK Biobank.
[Correction added on 22 October 2024, after first online publication: The copyright line was changed.]
Contributor Information
Yannis Yan Liang, Email: liangyan@link.cuhk.edu.hk.
Yaping Liu, Email: yaping.liu@cuhk.edu.hk.
Data availability statement
The accelerometer, demographics, health, neuroimaging, and genotype data from the UK Biobank dataset are available at (www.ukbiobank.ac.uk/) by application. The Allen Human Brain regional gene expression data were available from ENIGMA TOOLBOX (https://enigma-toolbox.readthedocs.io/en/latest/).
References
- 1. Leng Y, Musiek ES, Hu K, Cappuccio FP, Yaffe K. Association between circadian rhythms and neurodegenerative diseases. Lancet Neurol. 2019; 18: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feng H, Yang L, Ai S et al. Association between accelerometer‐measured amplitude of rest‐activity rhythm and future health risk: A prospective cohort study of the UK biobank. Lancet Healthy Longev. 2023; 4: e200–e210. [DOI] [PubMed] [Google Scholar]
- 3. Yang L, Feng H, Ai S et al. Association of accelerometer‐derived circadian abnormalities and genetic risk with incidence of atrial fibrillation. NPJ Digit. Med. 2023; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng H, Yang L, Liang YY et al. Associations of timing of physical activity with all‐cause and cause‐specific mortality in a prospective cohort study. Nat. Commun. 2023; 14: 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li P, Gao L, Gaba A et al. Circadian disturbances in Alzheimer's disease progression: A prospective observational cohort study of community‐based older adults. Lancet Healthy Longev. 2020; 1: e96–e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao L, Li P, Gaykova N et al. Circadian rest‐activity rhythms, delirium risk, and progression to dementia. Ann. Neurol. 2023; 93: 1145–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kendall KM, Rees E, Escott‐Price V et al. Cognitive performance among carriers of pathogenic copy number variants: Analysis of 152,000 UK biobank subjects. Biol. Psychiatry 2017; 82: 103–110. [DOI] [PubMed] [Google Scholar]
- 8. Roh HW, Choi JG, Kim NR et al. Associations of rest‐activity patterns with amyloid burden, medial temporal lobe atrophy, and cognitive impairment. EBioMedicine 2020; 58: 102881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiao Q, Shadyab AH, Rapp SR et al. Rest‐activity rhythms and cognitive impairment and dementia in older women: Results from the Women's Health Initiative. J. Am. Geriatr. Soc. 2022; 70: 2925–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McMahon M, Malneedi Y, Worthy DA, Schnyer DM. Rest‐activity rhythms and white matter microstructure across the lifespan. Sleep 2021; 44: zsaa266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoyt KR, Obrietan K. Circadian clocks, cognition, and Alzheimer's disease: Synaptic mechanisms, signaling effectors, and chronotherapeutics. Mol. Neurodegener. 2022; 17: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sudlow C, Gallacher J, Allen N et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015; 12: e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doherty A, Jackson D, Hammerla N et al. Large scale population assessment of physical activity using wrist worn accelerometers: The UK biobank study. PLoS One 2017; 12: e0169649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Someren EJ, Hagebeuk EE, Lijzenga C et al. Circadian rest‐activity rhythm disturbances in Alzheimer's disease. Biol. Psychiatry 1996; 40: 259–270. [DOI] [PubMed] [Google Scholar]
- 15. Lyall LM, Wyse CA, Graham N et al. Association of disrupted circadian rhythmicity with mood disorders, subjective wellbeing, and cognitive function: A cross‐sectional study of 91 105 participants from the UK biobank. Lancet Psychiatry 2018; 5: 507–514. [DOI] [PubMed] [Google Scholar]
- 16. Blume C, Santhi N, Schabus M. 'nparACT' package for R: A free software tool for the non‐parametric analysis of actigraphy data. MethodsX 2016; 3: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Riley RD, Ensor J, Snell KIE et al. Calculating the sample size required for developing a clinical prediction model. BMJ 2020; 368: m441. [DOI] [PubMed] [Google Scholar]
- 18. Van Buuren S, Groothuis‐Oudshoorn K. Mice: Multivariate imputation by chained equations in R. J. Stat. Softw. 2011; 45: 1–67. [Google Scholar]
- 19. Sterne JA, White IR, Carlin JB et al. Multiple imputation for missing data in epidemiological and clinical research: Potential and pitfalls. BMJ 2009; 338: b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nguyen DV, Rocke DM. Partial least squares proportional hazard regression for application to DNA microarray survival data. Bioinformatics 2002; 18: 1625–1632. [DOI] [PubMed] [Google Scholar]
- 21. Thorburn‐Winsor EA, Neufeld SAS, Rowthorn H et al. Device‐measured sleep onset and duration in the development of depressive symptoms in adolescence. J. Affect. Disord. 2022; 310: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arnatkeviciute A, Fulcher BD, Fornito A. A practical guide to linking brain‐wide gene expression and neuroimaging data. NeuroImage 2019; 189: 353–367. [DOI] [PubMed] [Google Scholar]
- 23. Afanador NL, Tran TN, Buydens LM. Use of the bootstrap and permutation methods for a more robust variable importance in the projection metric for partial least squares regression. Anal. Chim. Acta 2013; 768: 49–56. [DOI] [PubMed] [Google Scholar]
- 24. Bigdeli TB, Lee D, Webb BT et al. A simple yet accurate correction for winner's curse can predict signals discovered in much larger genome scans. Bioinformatics 2016; 32: 2598–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raudvere U, Kolberg L, Kuzmin I et al. G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019; 47: W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat Methodol. 1995; 57: 289–300. [Google Scholar]
- 27. Lananna BV, McKee CA, King MW et al. Chi3l1/YKL‐40 is controlled by the astrocyte circadian clock and regulates neuroinflammation and Alzheimer's disease pathogenesis. Sci. Transl. Med. 2020; 12: eaax3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tranah GJ, Blackwell T, Stone KL et al. Circadian activity rhythms and risk of incident dementia and mild cognitive impairment in older women. Ann. Neurol. 2011; 70: 722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naismith SL, Hickie IB, Terpening Z et al. Circadian misalignment and sleep disruption in mild cognitive impairment. J. Alzheimers Dis. 2014; 38: 857–866. [DOI] [PubMed] [Google Scholar]
- 30. Ortiz‐Tudela E, Martinez‐Nicolas A, Diaz‐Mardomingo C et al. The characterization of biological rhythms in mild cognitive impairment. Biomed. Res. Int. 2014; 2014: 524971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jeon SY, Byun MS, Yi D et al. Circadian rest‐activity rhythm and longitudinal brain changes underlying late‐life cognitive decline. Psychiatry Clin. Neurosci. 2023; 77: 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bokenberger K, Strom P, Dahl Aslan AK et al. Association between sleep characteristics and incident dementia accounting for baseline cognitive status: A prospective population‐based study. J. Gerontol. A Biol. Sci. Med. Sci. 2017; 72: 134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luck T, Riedel‐Heller SG, Luppa M et al. Apolipoprotein E epsilon 4 genotype and a physically active lifestyle in late life: Analysis of gene‐environment interaction for the risk of dementia and Alzheimer's disease dementia. Psychol. Med. 2014; 44: 1319–1329. [DOI] [PubMed] [Google Scholar]
- 34. Lin YT, Seo J, Gao F et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer's disease phenotypes in human iPSC‐derived brain cell types. Neuron 2018; 98: 1141–1154.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koelkebeck K, Miyata J, Kubota M et al. The contribution of cortical thickness and surface area to gray matter asymmetries in the healthy human brain. Hum. Brain Mapp. 2014; 35: 6011–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palaniyappan L, Mallikarjun P, Joseph V, White TP, Liddle PF. Regional contraction of brain surface area involves three large‐scale networks in schizophrenia. Schizophr. Res. 2011; 129: 163–168. [DOI] [PubMed] [Google Scholar]
- 37. Han F, Gu Y, Brown GL, Zhang X, Liu X. Neuroimaging contrast across the cortical hierarchy is the feature maximally linked to behavior and demographics. NeuroImage 2020; 215: 116853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fjell AM, Grydeland H, Krogsrud SK et al. Development and aging of cortical thickness correspond to genetic organization patterns. Proc. Natl. Acad. Sci. U. S. A. 2015; 112: 15462–15467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. El‐Boustani S, Sermet BS, Foustoukos G, Oram TB, Yizhar O, Petersen CCH. Anatomically and functionally distinct thalamocortical inputs to primary and secondary mouse whisker somatosensory cortices. Nat. Commun. 2020; 11: 3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weaver NA, Kuijf HJ, Aben HP et al. Strategic infarct locations for post‐stroke cognitive impairment: A pooled analysis of individual patient data from 12 acute ischaemic stroke cohorts. Lancet Neurol. 2021; 20: 448–459. [DOI] [PubMed] [Google Scholar]
- 41. Cremers LG, de Groot M, Hofman A et al. Altered tract‐specific white matter microstructure is related to poorer cognitive performance: The Rotterdam study. Neurobiol. Aging 2016; 39: 108–117. [DOI] [PubMed] [Google Scholar]
- 42. Cassel JC, Ferraris M, Quilichini P et al. The reuniens and rhomboid nuclei of the thalamus: A crossroads for cognition‐relevant information processing? Neurosci. Biobehav. Rev. 2021; 126: 338–360. [DOI] [PubMed] [Google Scholar]
- 43. van der Velpen IF, Vlasov V, Evans TE et al. Subcortical brain structures and the risk of dementia in the Rotterdam study. Alzheimers Dement. 2023; 19: 646–657. [DOI] [PubMed] [Google Scholar]
- 44. Bloom JS, Hynd GW. The role of the corpus callosum in interhemispheric transfer of information: Excitation or inhibition? Neuropsychol. Rev. 2005; 15: 59–71. [DOI] [PubMed] [Google Scholar]
- 45. Hensel A, Wolf H, Kruggel F et al. Morphometry of the corpus callosum in patients with questionable and mild dementia. J. Neurol. Neurosurg. Psychiatry 2002; 73: 59–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palmer JR, Duffy SL, Meares S et al. Rest‐activity functioning is related to white matter microarchitecture and modifiable risk factors in older adults at‐risk for dementia. Sleep 2021; 44: zsab007. [DOI] [PubMed] [Google Scholar]
- 47. Smagula SF, Chahine L, Metti A et al. Regional gray matter volume links rest‐activity rhythm fragmentation with past cognitive decline. Am. J. Geriatr. Psychiatry 2020; 28: 248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Du XF, Li FN, Peng XL et al. Circadian regulation of developmental synaptogenesis via the hypocretinergic system. Nat. Commun. 2023; 14: 3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hayashi Y, Koyanagi S, Kusunose N et al. The intrinsic microglial molecular clock controls synaptic strength via the circadian expression of cathepsin S. Sci. Rep. 2013; 3: 2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McCauley JP, Petroccione MA, D'Brant LY et al. Circadian modulation of neurons and astrocytes controls synaptic plasticity in hippocampal area CA1. Cell Rep. 2020; 33: 108255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nilsson J, Cousins KAQ, Gobom J et al. Cerebrospinal fluid biomarker panel of synaptic dysfunction in Alzheimer's disease and other neurodegenerative disorders. Alzheimers Dement. 2023; 19: 1775–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collins R. What makes UK biobank special? Lancet 2012; 379: 1173–1174. [DOI] [PubMed] [Google Scholar]
- 53. Fry A, Littlejohns TJ, Sudlow C et al. Comparison of sociodemographic and health‐related characteristics of UK biobank participants with those of the general population. Am. J. Epidemiol. 2017; 186: 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supplementary Information.
Data Availability Statement
The accelerometer, demographics, health, neuroimaging, and genotype data from the UK Biobank dataset are available at (www.ukbiobank.ac.uk/) by application. The Allen Human Brain regional gene expression data were available from ENIGMA TOOLBOX (https://enigma-toolbox.readthedocs.io/en/latest/).


