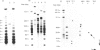Abstract
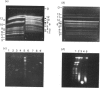
Long DNA molecules of greater than 10(5) bp (0.1 Mbp) are easily broken by pipetting. Therefore, chromosomal DNA is generally isolated after embedding cells in a protective coat of agarose. The embedded DNA can then be cut into long pieces and fractionated on gels using pulsed fields, but these pieces are again easily broken if the resolved DNA molecules are recovered from the gels. We now describe a novel gel matrix, a 'cellular' gel, that permits the recovery of resolved fragments from gels in a form that enables facile manipulation without shear. This facilitates purification and restriction mapping of fragments of 0.1-1.0 Mbp. We illustrate the utility of the method by mapping chromosome III of baker's yeast, which has a length of approximately 0.36 Mbp. This method should facilitate purification and restriction mapping of yeast artificial chromosomes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L., Yee C. N. Restriction site mapping is in separation theory. Comput Appl Biosci. 1988 Mar;4(1):97–101. doi: 10.1093/bioinformatics/4.1.97. [DOI] [PubMed] [Google Scholar]
- Bellon B. Construction of restriction maps. Comput Appl Biosci. 1988 Mar;4(1):111–115. doi: 10.1093/bioinformatics/4.1.111. [DOI] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Cook P. R. A general method for preparing intact nuclear DNA. EMBO J. 1984 Aug;3(8):1837–1842. doi: 10.1002/j.1460-2075.1984.tb02056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson A., Waterston R., Kiff J., Sulston J., Kohara Y. Genome linking with yeast artificial chromosomes. Nature. 1988 Sep 8;335(6186):184–186. doi: 10.1038/335184a0. [DOI] [PubMed] [Google Scholar]
- Durand R., Bregegere F. An efficient program to construct restriction maps from experimental data with realistic error levels. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):703–716. doi: 10.1093/nar/12.1part2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., Smith T. F., Ralph W. W. Mapping the order of DNA restriction fragments. Gene. 1983 Apr;22(1):19–29. doi: 10.1016/0378-1119(83)90060-4. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Laas W., Patterson D. Fractionation of large mammalian DNA restriction fragments using vertical pulsed-field gradient gel electrophoresis. Somat Cell Mol Genet. 1986 Mar;12(2):185–195. doi: 10.1007/BF01560665. [DOI] [PubMed] [Google Scholar]
- Garza D., Ajioka J. W., Burke D. T., Hartl D. L. Mapping the Drosophila genome with yeast artificial chromosomes. Science. 1989 Nov 3;246(4930):641–646. doi: 10.1126/science.2510296. [DOI] [PubMed] [Google Scholar]
- Krawczak M. Algorithms for the restriction-site mapping of DNA molecules. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7298–7301. doi: 10.1073/pnas.85.19.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan G. P., Maina C. V., Szalay A. A. Plasmid mapping computer program. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):717–729. doi: 10.1093/nar/12.1part2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R. Automatic construction of restriction site maps. Nucleic Acids Res. 1982 Jan 11;10(1):217–227. doi: 10.1093/nar/10.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polner G., Dorgai L., Orosz L. PMAP, PMAPS: DNA physical map constructing programs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):227–236. doi: 10.1093/nar/12.1part1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Smith S. B., Aldridge P. K., Callis J. B. Observation of individual DNA molecules undergoing gel electrophoresis. Science. 1989 Jan 13;243(4888):203–206. doi: 10.1126/science.2911733. [DOI] [PubMed] [Google Scholar]
- Tuffery P., Dessen P., Mugnier C., Hazout S. Restriction map construction using a 'complete sentences compatibility' algorithm. Comput Appl Biosci. 1988 Mar;4(1):103–110. [PubMed] [Google Scholar]