Abstract
The irreversible cell cycle arrest and apoptosis induced by p53 are part of the host surveillance mechanisms for viral infection and tumor induction. Kaposi's sarcoma-associated herpesvirus (KSHV), the most recently discovered human tumor virus, is associated with the pathogenesis of Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. The K9 open reading frame of KSHV encodes a viral interferon (IFN) regulatory factor (vIRF) which functions as a repressor for cellular IFN-mediated signal transduction and as an oncoprotein to induce cell growth transformation. Here, we demonstrate that KSHV vIRF interacts with the cellular p53 tumor suppressor through the putative DNA binding region of vIRF and the central region of p53. This interaction suppresses the level of phosphorylation and acetylation of p53 and inhibits transcriptional activation of p53. As a consequence, vIRF efficiently prevents p53-mediated apoptosis. These results suggest that KSHV vIRF interacts with and inhibits the p53 tumor suppressor to circumvent host growth surveillance and to facilitate uncontrolled cell proliferation.
The p53 gene is the first tumor suppressor gene to be identified and is a common denominator in human cancer (42). Abnormalities of the p53 gene are among the most frequent molecular events in human and animal neoplasms (15, 17, 36). In about half of human tumors, p53 is directly inactivated as a result of mutations in the p53 gene. In many others, it is indirectly inactivated as a result of alterations by cellular or viral genes whose products interact with p53 (42).
The p53 tumor suppressor transmits signals arising from various forms of cellular stresses, including DNA damage, chemotherapeutic agents, and aberrant growth signal, to genes and factors that induce cell cycle arrest, cell death, and senescence. The best-characterized targets of p53-mediated cell growth control are the cell cycle inhibitor p21 and the proapoptotic protein Bax (11, 32). The p53 binds to its sequence-specific sites in the promoter regions of p21 and Bax and induces a drastic increase of their gene expression (11, 32). As a result, p21 arrests the cell cycle in the G1 phase by inhibiting cellular cyclin–cyclin-dependent kinase complex activity, and Bax induces cell death by disrupting the cellular powerhouses, the mitochondria.
The irreversible cell cycle arrest and cell death induced by p53 are considered part of host surveillance mechanisms for detecting and preventing viral infection and tumor induction (2, 3, 9, 34). To escape this host scrutiny, viral oncogene products frequently target p53 for inactivation (24). These include simian virus 40 (SV40) large T antigen (22, 27), human papillomavirus (HPV) E6 (16, 21, 38, 43), hepatitis B virus X antigen (12), adenovirus E1B (8, 20, 44), and human cytomegalovirus IE84 (40). In addition to interactions with numerous viral proteins, the covalent modifications of p53 have been shown to regulate its transcriptional activity (19). Phosphorylation at the amino-terminal transactivation domain of p53 by the DNA-activated protein kinase or ATM kinase results in increased stability (1, 23). In addition, phosphorylation in the carboxyl-terminal region by casein kinase II enhances p53 activities, including growth suppression, DNA binding activity, and transcriptional activation (31). Finally, acetylation of the carboxyl-terminal region of p53 by p300 and p300/CBP-associated factor (PCAF) leads to increased DNA binding activity (28, 37).
Interferons (IFNs) are a family of cytokines that exhibit such diverse biological effects as inhibition of cell growth and protection against viral infection. Viruses have evolved a variety of mechanisms to counteract the inhibitory effects of IFNs (41). Kaposi's sarcoma-associated herpesvirus (KSHV), the most recently discovered human tumor virus, is associated with the pathogenesis of Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease (5, 6, 35). The K9 open reading frame of KSHV exhibits significant sequence homology with cellular IFN regulatory factors (IRFs) (33). We and others have demonstrated that expression of K9 dramatically represses transcriptional activation induced by IFN-αβγ (13, 26, 45) and also leads to transformation of rodent fibroblasts, resulting in morphological change, focus formation, growth at reduced serum concentration, and tumor induction in nude mice (13, 26). Thus, the K9 gene of KSHV encodes the first viral IFN regulatory factor (vIRF) which functions as a repressor of cellular IFN-mediated signal transduction and as an oncoprotein to induce cell growth transformation.
Recent detailed studies have demonstrated that these functional activities of vIRF appear to be attributed in part to an interaction with and inhibition of p300 (4, 18, 25). Interaction of vIRF with p300 inhibits the histone acetyltransferase (HAT) activity of p300 in vitro and induces a dramatic hypoacetylation of nucleosomal histone H3 and H4 in vivo, resulting in global alteration of nucleosomal chromatin structure and inhibition of IFN-mediated gene expression (25). Thus, the modulation of p300 HAT activity is likely part of the mechanisms which vIRF employs to block cellular IFN-mediated antiviral activity (25).
Despite extensive studies of the anti-IFN activity of vIRF, little is known about the molecular mechanisms used by vIRF in cell growth transformation. In this study, we demonstrate that KSHV vIRF interacts with tumor suppressor p53 and that this interaction suppresses p53-mediated transcriptional activation of p21 and Bax, the result of which is inhibition of p53-mediated cell growth control. These results indicate that vIRF inhibits cellular tumor suppressor p53 protein to facilitate cell growth transformation.
MATERIALS AND METHODS
Cell culture and transfection.
COS-1, 293T, and Saos-2 cells were maintained in Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS). Insect (Spodoptera frugiperda) Sf9 and High 5 cell lines were maintained in Grace medium supplemented with 10% FBS. A Fugene 6 lipofection (Boehringer Mannheim) procedure was used for transfection in COS-1, 293T, and Saos-2 cells. A total of 106 cells were transfected with 1 μg of plasmid DNA. Forty-eight hours after transfection, cells were collected for luciferase reporter assays or immunoprecipitations. To establish stable cell lines expressing vIRF, Saos-2 cells were transfected with pTracer-vIRF or pTracer plasmid (Clontech, San Diego, Calif.). At 72 h posttransfection, cells were cultured with selection medium containing 1 mg of Zeocine per ml for 5 weeks.
Plasmid construction.
A DNA fragment containing the Flag-tagged vIRF sequence was subcloned into pAd-CMV-Track at HindIII and XbaI sites to generate the recombinant virus Ad-vIRF and into pAcSG at HindIII and XbaI sites to generate recombinant vIRF baculovirus. To generate glutathione S-transferase (GST) fusion protein in Escherichia coli, a DNA fragment containing each domain of vIRF or p53 was PCR amplified and cloned in frame into BamHI and XhoI sites or BamHI and EcoRI sites, respectively, of pGEX4T-3 GST fusion plasmid. DNA fragments containing each region of p53 were also subcloned in frame into plasmid pEGFP-C1 to produce enhanced green fluorescence protein (EGFP) fusion proteins. Flag-tagged wild-type (wt) and mutant vIRF were subcloned at BamHI and EcoRI sites into pTracer-A (Clontech). PG13 and p21 reporter plasmids were kindly provided by J. Alwine. All PCR-amplified product were completely sequenced to confirm the presence of authentic sequence and the absence of aberrant alteration.
Construction of recombinant viruses.
The AdEasy system (14) and recombinant adenoviruses Ad-p53, Ad-p53 (R273H), and Ad-GFP were kindly provided by B. Vogelstein. The recombinant adenovirus Ad-vIRF was constructed as previously described (14). All recombinant adenoviruses were amplified and titered in 293 cells. Recombinant vIRF baculovirus was constructed by using the BaculoGold system (PharMingen). High-titer recombinant baculovirus was obtained in Sf9 cells. The recombinant p53 baculovirus was kindly provided by C. Prives.
Metabolic labeling, immunoprecipitation, and immunoblotting.
Saos-2 cells were infected with recombinant adenoviruses, and High-5 cells were infected with recombinant baculoviruses at a multiplicity of infection of 5. COS-1 cells were transfected with expression plasmid by Fugene 6 transfection. After 48 h of infection and transfection, cells were collected, lysed in 1 ml of lysis buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, and protease inhibitors, and immunoprecipitated as described previously (25). For pulse-chase analysis, Saos-2 cells were rinsed three times with phosphate-buffered saline (PBS), washed once with labeling medium (RPMI 1640 minus methionine and cysteine plus 10% dialyzed fetal calf serum), then incubated with 5 ml of the same medium containing 100 μCi of [35S]methionine and [35S]cysteine (New England Nuclear, Boston, Mass.) for 3 h, and chased for 6 h. Anti-p53 antibody (DO-1) and anti-Flag antibody (M2) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.), Sigma Chemical Co. (St. Louis, Mo.), and Novagen, respectively. Anti-phospho-p53 (Ser15) and anti-phospho-p53 (Ser392) antibodies were purchased from Cell Signaling Technology (Beverly, Mass.) and New England Biolabs (Beverly, Mass.), respectively, and anti-acetylated p53 antibodies anti-p53(Ac320) and anti-p53(Ac373) were purchased from Upstate Biotechnology. Immune complexes were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting using enhanced chemiluminescence. All primary antibodies were diluted 1:2,000, and the secondary antibodies were diluted 1:10,000.
GST fusion proteins and pull-down assay.
GST fusion proteins were expressed in Escherichia coli, bound to glutathione-Sepharose beads, and eluted with buffer containing 25 mM glutathione. Purified or Sepharose-bound GST fusion proteins were mixed with lysates of Saos-2 cells infected with recombinant adenoviruses for 3 h. The precipitated protein complexes were extensively washed with lysis buffer and analyzed by SDS-PAGE.
Immunofluorescence tests.
Cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 70% ethanol for 15 min, blocked with 10% goat serum in PBS for 30 min, and reacted with 1:100-diluted primary antibody in PBS for 30 min at room temperature. After incubation, cells were washed extensively with PBS, incubated with 1:100-diluted secondary antibody (Vector Laboratories, Burlingame, Calif.) in PBS for 30 min at room temperature, and washed three times with PBS. DNA staining was performed with 1:5,000-diluted Topro-I (Molecular Probe) for 1 min. Confocal microscopy was performed with a Leica TCS SP laser-scanning microscope (Leica Microsystems, Exton, Pa.) fitted with a 100× Leica objective (PL APO, 1.4NA) and using the Leica image software. Images were collected at 512- by 512-pixel resolution. The stained cells were optically sectioned in the z axis, and the images in the different channels (photomultiplier tubes) were collected simultaneously. The step size in the z axis varied from 0.2 to 0.5 μm to obtain 30 to 50 slices per imaged file. The images were transferred to a Macintosh G3 computer (Apple Computer, Cupertino, Calif.), and NIH Image version 1.61 software was used to render the images.
Cell cycle analysis.
Cells were washed once with PBS, fixed in 70% ethanol for 15 min, and stained with staining solution (1% Triton X-100, 50 μg of propidium iodide [PI] or Hoechst 33342 per ml, 1 mg of RNase A per ml) for 30 min at room temperature. Cell cycle analysis was performed with a FACScan (Becton Dickinson, Mountain View, Calif.).
Reporter assays.
Saos-2 cells at 70% confluence in 24-well plates were transfected 1 μg of mixed plasmid DNAs (0.2 μg of reporter plasmid and various amounts of p53 and vIRF expression plasmids). All transfections included 0.2 μg of pGKβgal, which expresses β-galactosidase from a phosphoglucokinase promoter. At 48 h posttransfection, cells were washed once in PBS and lysed in 200 μl of reporter lysis buffer (Promega, Madison, Wis.). Assays for luciferase were performed with a luminometer using a luciferase assay kit (Promega, Madison, Wis.), and assays for chloramphenicol acetyltransferase (CAT) were performed with CAT assay kit (Promega). Values were normalized by β-galactosidase activity.
RESULTS
Interaction of vIRF with p53.
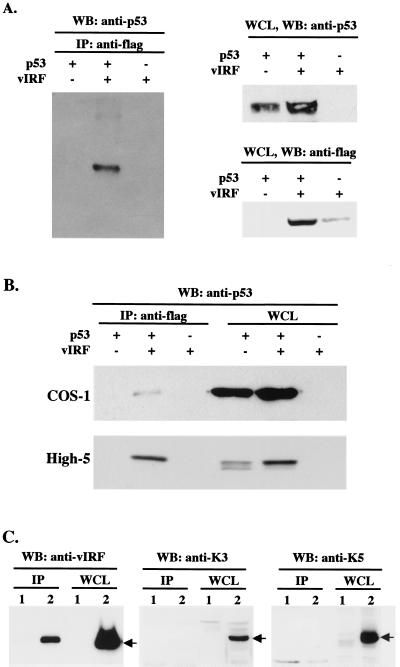
To investigate the detailed mechanisms of growth transformation used by vIRF, we examined the potential interactions of vIRF with cellular proteins that regulate cell growth control. Among numerous cellular proteins, p53 tumor suppressor was found to specifically interact with vIRF. p53-null Saos-2 cells were infected with Ad-p53 and Flag-tagged Ad-vIRF. After 48 h of infection, Saos-2 cell lysates were used for immunoprecipitation with an anti-Flag antibody, and polypeptides present in anti-Flag immune complexes were separated by SDS-PAGE, transferred to nitrocellulose, and reacted with an anti-p53 antibody. The p53 protein was readily detected in the anti-Flag immune complexes from Saos-2 cells coinfected with Ad-p53 and Ad-vIRF, whereas it was not detected from Saos-2 cells infected with Ad-p53 or Ad-vIRF alone (Fig. 1A). When Sf9 insect cells, infected with recombinant baculoviruses expressing p53 and Flag-tagged vIRF, and COS-1 cells, transfected with expression vectors for p53 and Flag-tagged vIRF, were used for coimmunoprecipitation assay, the results were essentially the same as for recombinant adenoviruses (Fig. 1B). Finally, KSHV-infected BCBL-1 cells were used to detect an interaction between vIRF and p53. Lysates of BCBL-1 cells were subjected to immunoprecipitation with an anti-p53 antibody, followed by immunoblotting with rabbit polyclonal antibodies against vIRF, K3, and K5. Approximately 5% of vIRF in KSHV-infected BCBL-1 cells interacted with cellular p53 (Fig. 1C). In contrast, K3 and K5 did not interact with p53 under the same conditions (Fig. 1C). These results demonstrate that KSHV vIRF specifically interacts with cellular p53.
FIG. 1.
Interaction of vIRF with p53. (A) Interaction of vIRF with p53 in recombinant adenovirus-infected Saos-2 cells. p53-null Saos-2 cells were infected with Ad-p53 and/or Ad-vIRF as indicated at the top. After 48 h, cell extracts were used for immunoprecipitations (IP) with an anti-Flag antibody, followed by Western blot assay with an anti-p53 antibody. p53 and vIRF expression in whole-cell lysates (WCL) of infected Saos-2 cells were determined by Western blotting with anti-p53 and anti-Flag antibodies. (B) Interaction of vIRF with p53 in COS-1 cells transfected with p53 and vIRF expression vectors and in High-5 cells infected with recombinant p53 and vIRF baculoviruses. As indicated at the top, COS-1 cells were transfected with p53 and/or Flag-tagged vIRF expression vector, and High-5 insect cells were infected with recombinant p53 and/or vIRF baculoviruses. After 48 h, cell extracts were used for immunoprecipitations with an anti-Flag antibody, followed by Western blot assay (WB) with an anti-p53 antibody (left). Whole-cell lysates (WCL) of transfected COS-1 cells and infected High-5 cells were used to show p53 expression (right) and vIRF expression (data not shown). In addition, vIRF expression did not alter the level of SV40 large T antigen interaction of p53 in COS-1 cells (data not shown). (C) Interaction of vIRF with p53 in KSHV-infected BCBL-1 cells. Lysates of KSHV-negative BJAB (lane 1) and KSHV-infected BCBL-1 (lane 2) cells were used for immunoprecipitation (IP) with an anti-p53 antibody, followed by Western blot assay (WB) with anti-vIRF, anti-K3, and anti-K5 antibodies. A whole-cell lysate (WCL) was used to show vIRF, K3, and K5 expression.
Subcellular colocalization of vIRF with p53.
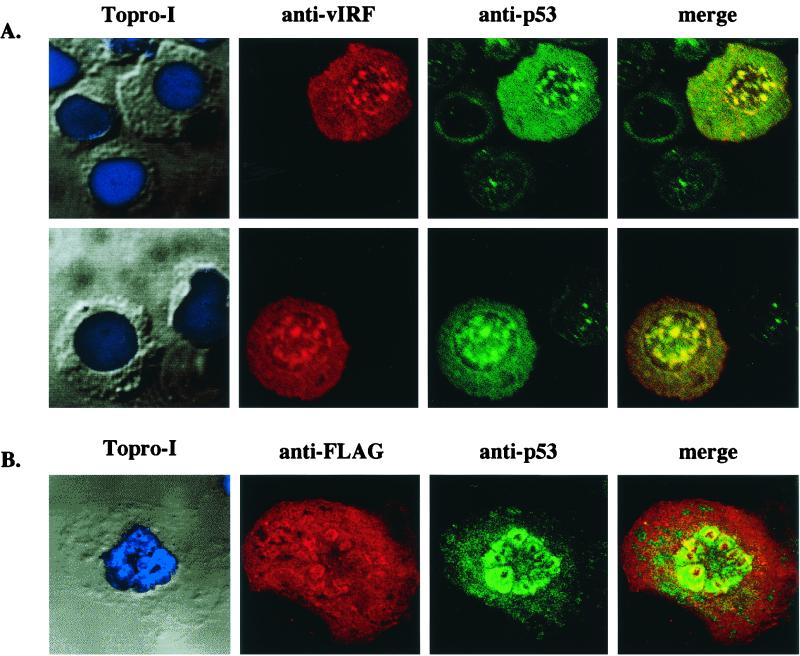
To further investigate an interaction of vIRF with p53, we examined their subcellular localization by indirect immunofluorescence tests. KSHV-infected BCBL-1 cells were fixed, reacted with anti-vIRF and anti-p53 antibodies, and examined under a confocal immunofluorescence microscope. Both vIRF and p53 proteins were present throughout the cytoplasm and nucleus, with a high degree of overlapping staining between them (Fig. 2A). In addition, COS-1 cells transfected with expression vector containing the Flag-tagged vIRF were used for the confocal immunofluorescence assay. vIRF was also colocalized with p53 in the nucleus of COS-1 cells (Fig. 2B). Thus, confocal immunofluorescence tests demonstrate that a considerable amount of vIRF is colocalized with cellular p53, further suggesting a specific interaction of vIRF with p53.
FIG. 2.
Colocalization of vIRF with p53. (A) Colocalization of vIRF with p53 in KSHV-infected BCBL-1 cells. KSHV-infected BCBL-1 cells were fixed and reacted with rabbit polyclonal anti-vIRF and mouse monoclonal anti-p53 antibodies. vIRF protein was detected with an anti-rabbit secondary antibody conjugated with Alexa 568 (red), and p53 protein was detected with an anti-mouse secondary antibody conjugated with Alexa 488 (green). Cells were visualized with Nomarski optics after Topro-I nuclear staining (blue). These two panels are representatives of 10 different fields. (B) Colocalization of vIRF with p53 in COS-1 cells. COS-1 cells were transfected with expression vector containing the Flag-tagged vIRF gene. At 48 h posttransfection, cells were fixed and reacted with mouse monoclonal anti-Flag and rabbit polyclonal anti-p53 antibodies. p53 protein was detected with an anti-rabbit secondary antibody conjugated with Alexa 488 (green), and the Flag-tagged vIRF protein was detected with an anti-mouse secondary antibody conjugated with Alexa 568 (red). Cells were visualized with Nomarski optics after Topro-I nuclear staining (blue). The immunofluorescence test was performed with a Leica confocal immunofluorescence microscope. The yellow color in merged panels indicates colocalization of the red and green labels. The data were reproduced in three independent experiments.
A drastic increase of p53 staining was detected in BCBL-1 cells with vIRF expression compared to that in BCBL-1 cells without vIRF expression (Fig. 2A). However, expression of vIRF in 293T, COS-1, U2OS, and BJAB cells altered neither the level of p53 protein staining nor the level of p53 protein expression (Fig. 2B and data not shown). In addition, vIRF expression did not alter the stability of p53 protein at detectable levels (data not shown). These results indicate that factors other than vIRF likely affect p53 expression in KSHV-infected BCBL-1 cells.
The putative DNA binding region of vIRF is necessary for p53 interaction.
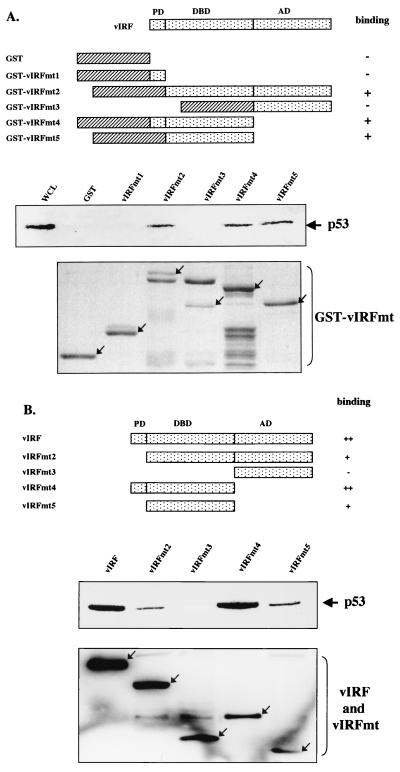
Cellular IRFs contain a conserved DNA binding domain at the amino terminus and a divergent activation domain at the carboxyl terminus (7). The amino terminus of vIRF shows significant homology to the amino-terminal DNA binding domain of IRF, while the carboxyl terminus of vIRF is divergent from the carboxyl activation domain of IRF (33). In addition, KSHV vIRF contains 80 amino acids at the amino terminus that are not homologous with cellular IRFs (33). This region contains six repeats of a proline-rich PX2–3P motif. To map the regions of vIRF required for p53 interaction, GST-vIRF fusion proteins containing the individual domains of vIRF were used for in vitro pull-down assays (Fig. 3A). Lysates of Saos-2 cells infected with Ad-p53 were precleared with 5 μg of GST and then incubated with 5 μg of GST or GST-vIRF fusion proteins. Polypeptides associated with GST-vIRF fusion proteins after pull-down assays were immunoblotted with an anti-p53 antibody. This demonstrated that GST-vIRF fusion proteins containing the potential DNA binding domain of vIRF could bind to p53 in vitro (Fig. 3A). In contrast, GST and GST-vIRF fusion proteins containing the amino-terminal proline-rich region or the carboxyl activation region did not bind to p53 under the same conditions (Fig. 3A).
FIG. 3.
Mutational analysis of vIRF for p53 interaction. (A) Summary of GST-vIRF fusion constructs and in vitro GST pull-down assay. Individual domains of vIRF were cloned into the pGEX4T-1 vector to generate GST-vIRF fusion proteins. Lysates of Saos-2 cells infected with Ad-p53 were precleared with 5 μg of GST, followed by incubation with 5 μg of GST or GST-vIRF fusion proteins. Polypeptides associated with the GST-vIRF fusion proteins were subject to Western blotting with an anti-p53 antibody. A whole-cell lysate (WCL) which represents 5% of cellular p53 was used for a positive control. Arrows in the bottom indicate GST and GST-vIRF mutant fusion proteins (GST-vIRFmt) stained with Coomassie blue solution. Boxes with slashed lines indicate GST, and boxes with dots indicate a domain of vIRF, PD, proline-rich domain; DBD, DNA binding domain; AD, activation domain. + and − indicate positive and negative binding of GST-vIRF fusion proteins to p53. (B) Summary of vIRF mutants and in vivo interaction of vIRF mutants with p53. COS-1 cells were transfected with the Flag-tagged wt vIRF and mutants vIRFmt2 to -5. Cell lysates were used for immunoprecipitation with an anti-Flag antibody, followed by Western blotting with an anti-p53 antibody to detect p53. PD, proline-rich domain; DBD, DNA binding domain; AD, activation domain. Western blotting of whole-cell lysates with an anti-Flag antibody showed equivalent expression of wt and mutant vIRF: arrows indicate the Flag-tagged wt and mutant vIRF (bottom). +, ++, and − indicate weak, strong, and no binding of vIRF mutants to p53.
The interaction between vIRF and p53 was further assessed by in vivo coimmunoprecipitation assay. Four deletion mutations were generated as follows: vIRFmt2 has a deletion of the first 80 amino acids, which contain the amino-terminal proline-rich sequence; vIRFmt3 has a deletion of amino acid residues 1 to 255, which contain the amino-terminal proline-rich region and the putative central DNA binding region; vIRFmt4 has a deletion of amino acid residues 255 to 449, which contain the carboxyl-terminal putative activation region; and vIRFmt5 has deletions of both the amino-terminal proline-rich region and the carboxyl activation region (Fig. 3B). To demonstrate expression of these deletion mutants, the Flag-tagged vIRF mutants were cloned into the pcDNA3.1 vector and expressed in COS-1 cells. After transfection, whole-cell lysates were used for immunoblotting with an anti-Flag antibody. Wild-type and mutant vIRF were expressed at somewhat variable but still comparable levels in COS-1 cells (Fig. 3B, bottom). The same cell lysates were used for immunoprecipitation with an anti-Flag antibody, followed by immunoblotting with an anti-p53 antibody to detect the presence of p53 in anti-Flag immune complexes (Fig. 3B, middle). The results showed that vIRFmt2, vIRFmt4, and vIRFmt5 interacted with p53, whereas vIRFmt3 did not. In addition, repeated experiments showed that wt vIRF and vIRFmt4, containing both the amino-terminal proline-rich region and the central DNA binding region, exhibited stronger interaction with p53 than vIRFmt2 and vIRFmt5 in the in vivo binding assay but not the in vitro GST pull-down assay (Fig. 3). Thus, these results demonstrate that the central putative DNA binding region of vIRF is necessary for binding to p53 and that the amino-terminal proline-rich region of vIRF likely enhances its p53 binding activity in vivo.
Multiple regions of p53 interact with vIRF.
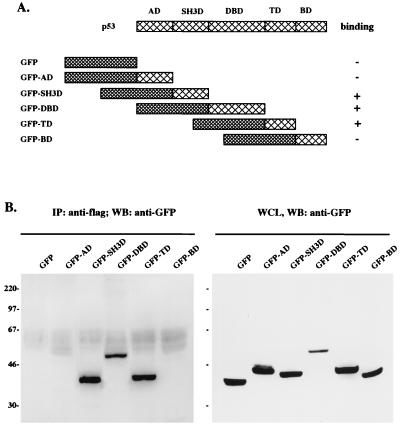
To further delineate an interaction between vIRF and p53, we attempted to define the regions of p53 required for this interaction. The p53 protein consists of five distinctive domains: an activation domain, a proline-rich SH3 binding (SH3B) domain, a DNA binding domain, a tetramerization domain, and a basic domain (Fig. 4A) (34, 42). Because of a high level of nonspecific binding of the GST-p53 fusion protein, we used a GFP-p53 fusion protein for in vivo coimmunoprecipitation assays. A series of GFP-p53 fusion proteins containing individual domains of p53 were constructed as described in Fig. 4A. The Flag-tagged vIRF expression vector and the various GFP-p53 fusion expression vectors were cotransfected into 293T cells. The GFP expression vector without p53 sequence was also included as a control. At 48 h posttransfection, whole-cell lysates were used for immunoblot analysis with an anti-GFP antibody. GFP-p53 fusion proteins were expressed at comparable levels in COS-1 cells (Fig. 4B, right). The same cell lysates were used for immunoprecipitation with an anti-Flag antibody, followed by immunoblotting with an anti-GFP antibody. These experiments demonstrated that the amino-terminal SH3B domain, the central DNA binding domain, and the carboxyl tetramerization domain of p53 were capable of binding to vIRF in vivo, whereas the amino-terminal transactivation domain and the carboxyl-terminal basic domain of p53 were not (Fig. 4B, left). Furthermore, GFP did not interact with vIRF under the same conditions (Fig. 4B, left). These results suggest that multiple regions of p53, including the amino-terminal SH3B domain, the central DNA binding domain, and the carboxyl tetramerization domain, are involved in an interaction with vIRF.
FIG. 4.
Mutational analysis of p53 for vIRF interaction. (A) Summary of p53 mutants. Individual domains of p53 were cloned in frame into the GFP vector to generate GFP-p53 fusion proteins. Boxes with large cross lines indicate individual domains of p53; boxes with small cross lines indicate GFP. AD, activation domain; SH3D, SH3B domain; DBD, DNA binding domain; TD, tetramerization domain; BD, basic domain. + and − indicate positive and negative binding of GFP-p53 fusion proteins to vIRF. (B) Identification of the vIRF binding domains of p53. 293T cells were cotransfected with Flag-tagged wt vIRF and GFP-p53 fusion constructs. Cell lysates were used for immunoprecipitation (IP) with an anti-Flag antibody, followed by Western blotting (WB) with an anti-GFP antibody to detect GFP-p53 fusion proteins (left). Western blotting of whole-cell lysates with an anti-GFP antibody showed GFP-p53 expression in transfected 293T cells (right). Sizes are indicated in kilodaltons.
vIRF suppresses the serine phosphorylation of p53.
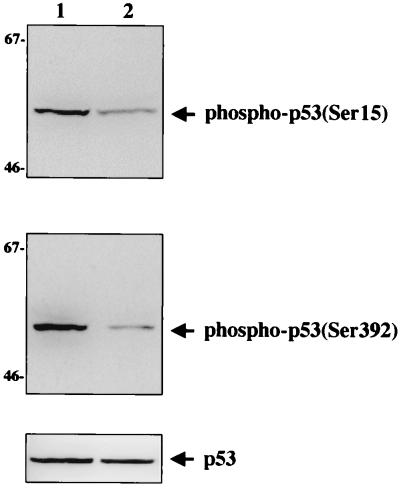
Upon transmission of stress signals, the rapid activation of p53 is often achieved through modifications, i.e., phosphorylation and acetylation (19). Various stress-activated kinases phosphorylate p53 at multiple serine sites (1, 23, 31). DNA-dependent protein kinase and ATM phosphorylate serine residue 15 (1, 23), and casein kinase II phosphorylates serine residue 392; these phosphorylations enhance p53 activities (31). To examine the effects of vIRF interaction on the phosphorylation of p53, we constructed Saos-2 cells stably expressing vIRF (Saos-2/vIRF cells). At 48 h after infection with Ad-p53, lysates of Saos-2 and Saos-2/vIRF cells were used for immunoblotting with antibodies specific for p53 phosphorylated at serine residues 15 and 392. The levels of phosphorylation at both serine residues of p53 were significantly lower in Saos-2/vIRF cells than in Saos-2 cells (Fig. 5). A similar level of p53 was expressed in both Saos-2 and Saos-2/vIRF cells after infection with Ad-p53 (Fig. 5, bottom). These results demonstrate that vIRF suppresses the level of serine phosphorylation of p53.
FIG. 5.
Suppression of in vivo p53 phosphorylation by vIRF expression. Identical amounts of proteins from Saos-2 cells (lane 1) and Saos-2/vIRF (lane 2) cells were used for Western blot analysis with antibodies specific for p53 phosphorylated at serine residue 15 (top) and at serine residue 392 (middle). Western blot assay of whole-cell lysate with horseradish peroxidase-conjugated anti-pan-p53 antibody showed equivalent expression of p53 in both cells (bottom). Sizes are indicated in kilodaltons.
vIRF suppresses the acetylation of p53.
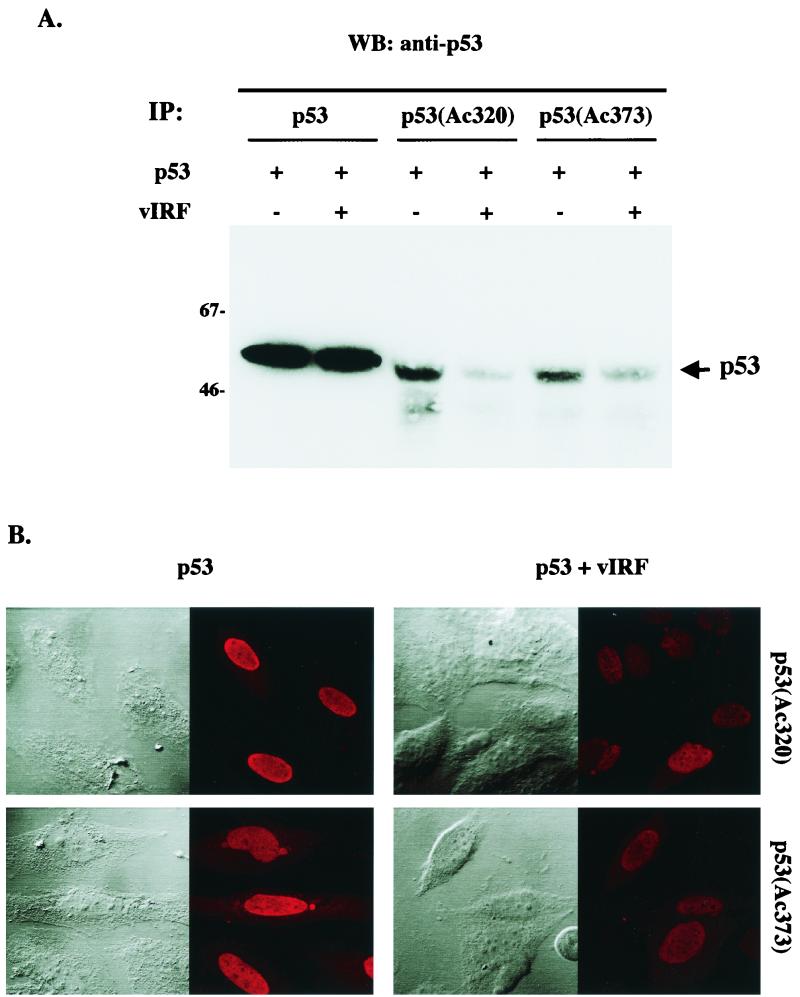
Besides phosphorylation, p53 is also acetylated at multiple lysine residues at its carboxyl terminus by the cellular transcriptional coactivator p300/CBP and PCAF and this modification has also been shown to increase its DNA binding and transcription activities (28, 37). To further investigate the effect of vIRF interaction on the modification of p53, we examined the level of p53 acetylation in the presence of vIRF expression. At 48 h after infection with Ad-p53 and Ad-vIRF, Saos-2 cell lysates were used for immunoprecipitations with an anti-pan-p53, anti-p53(Ac320), or anti-p53(Ac373) antibody. The anti-p53(Ac320) antibody specifically reacts with the acetylated form of p53 at lysine residue 320, and the anti-p53(Ac373) antibody specifically reacts with the acetylated form of p53 at lysine residue 373. The amount of p53 protein after immunoprecipitation was assessed by immunoblotting with an anti-pan-p53 antibody that reacted with all forms of p53. While p53 was expressed at equivalent levels in Ad-p53-infected and Ad-p53/Ad-vIRF coinfected Saos-2 cells (Fig. 6A, first two lanes), the levels of acetylation at both lysine residue 320 and lysine residue 373 of p53 were significantly reduced in Saos-2 cells with vIRF expression compared to Saos-2 cells without vIRF expression (Fig. 6A, last four lanes). This result was further confirmed by immunofluorescence tests using the same antibodies (Fig. 6B). These results demonstrate that vIRF expression leads to hypoacetylation of the p53 protein.
FIG. 6.
Suppression of in vivo p53 acetylation by vIRF expression. (A) Western blot assay of in vivo p53 acetylation. Saos-2 cells were infected with recombinant Ad-p53 and/or Ad-vIRF as indicated at the top. At 48 h postinfection, cell lysates were used for immunoprecipitation (IP) with anti-p53, anti-p53(Ac320), and anti-p53(Ac373) antibodies, followed by Western blot (WB) analysis with the horseradish peroxidase-conjugated anti-pan-p53 antibody. The data were reproduced in two independent experiments. (B) Immunofluorescence test of in vivo p53 acetylation. The Saos-2 cells described above were stained with anti-p53(Ac320) and anti-p53(Ac373) antibodies. Cells were visualized with Nomarski optics. Equivalent levels of p53 were detected in both cells with an anti-p53 antibody (see above). The immunofluorescence test was performed with a Leica confocal immunofluorescence microscope.
Downregulation of p53-mediated transcriptional activation by vIRF.
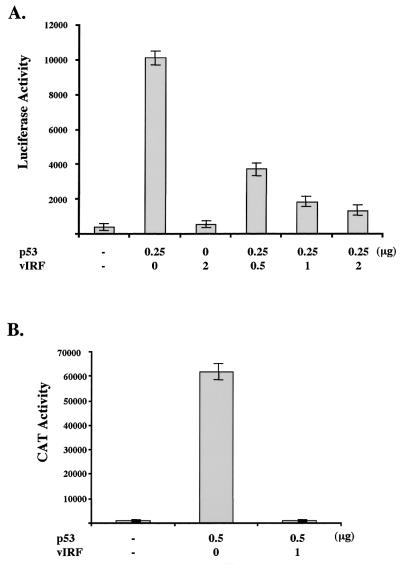
p53 is a transcriptional activator that binds to sequence-specific binding sites at the promoter region of numerous cellular genes and activates their transcription (10, 42). To determine the effect of vIRF expression on p53-mediated transcriptional activation, a PG13-luciferase reporter that contains a synthetic promoter of 13 tandem copies of an endogenous p53 DNA binding site (11) was transfected into p53-null Saos-2 cells together with p53 and/or vIRF expression vectors. While p53 dramatically induced PG13 promoter activity, vIRF expression significantly inhibited p53-mediated activation of PG13 promoter activity, and this inhibition was dependent on the dose of vIRF (Fig. 7A).
FIG. 7.
Inhibition of p53 transcriptional activation by vIRF. (A) Inhibition of p53-mediated activation of PG13 promoter activity by vIRF. PG13-luciferase (0.25 μg) and pGKβgal (0.25 μg) reporter plasmids were transfected into p53-null Saos-2 cells together with 0.25 μg of p53 expression vector and different amounts of vIRF expression vector as indicated at the bottom. Luciferase activity was measured 48 h posttransfection, and luciferase values were normalized by β-galactosidase activity. Luciferase activity is represented as the average of three independent experiments. (B) Inhibition of p53-mediated activation of p21 promoter activity by vIRF. p21-CAT (0.25 μg) and pGKβgal (0.25 μg) reporter plasmids were transfected into p53-null Saos-2 cells together with 0.5 μg of p53 expression vector and 1 μg of vIRF expression vector as indicated at the bottom. CAT activity was measured 48 h posttransfection by a Fuji phosphoimager, and values were normalized by β-galactosidase activity. CAT activity is represented as the average of two independent experiments.
One of the well-characterized cellular targets of p53-mediated transcriptional activation is the cyclin-dependent kinase inhibitor p21 gene (11). p53 binds to the sequence-specific binding sites in the promoter region of p21, leading to a drastic increase of p21 transcription (11). To examine the effect of vIRF expression on p21 promoter activity, Saos-2 cells were transfected with a p21 promoter CAT reporter construct together with p53 and vIRF expression constructs. p21 promoter activity was dramatically activated by p53 expression, whereas this activation was almost abolished by vIRF expression (Fig. 7B). These results demonstrate that vIRF expression strongly inhibits p53-mediated transcriptional activation.
Inhibition of p53-mediated upregulation of p21 and Bax protein by vIRF.
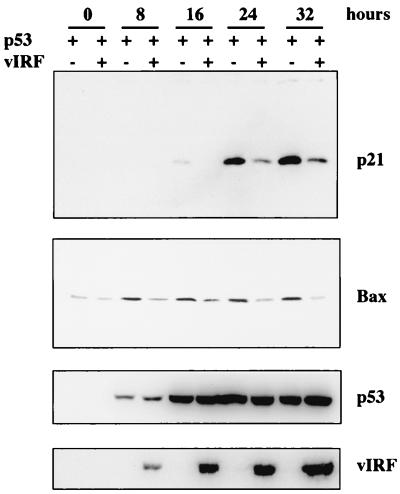
Cell cycle inhibitor p21 and proapoptotic Bax have been identified as targets for p53-mediated transcriptional activation (11, 32). To further demonstrate an effect of vIRF on p53-mediated transcriptional activation, we examined the level of p21 and Bax protein in the presence and absence of vIRF expression. Lysates of p53 null Saos-2 cells were collected at different time points after infection with a mixture of Ad-p53 plus Ad-GFP or Ad-p53 plus Ad-vIRF and immunoblotted with anti-p21 and anti-Bax antibodies. An increase of p21 protein in Saos-2 cells infected with recombinant Ad-p53 plus Ad-GFP was detected 16 h postinfection, further enhanced 24 h postinfection, and prolonged until 32 h postinfection (Fig. 8). In contrast, p21 protein in Saos-2 cells coinfected with Ad-p53 plus Ad-vIRF was not detected until 24 h postinfection (Fig. 8). In addition, the level of p21 protein was significantly lower in Saos-2 cells coinfected with Ad-p53 plus Ad-vIRF than in Saos-2 cells infected with Ad-p53 plus Ad-GFP (Fig. 8). Bax protein was expressed at a low but detectable level in Saos-2 cells before infection with recombinant adenoviruses and was induced at an appreciable level after infection with Ad-p53 plus Ad-GFP (Fig. 8). However, the increase of Bax protein by p53 expression was almost abolished by vIRF expression (Fig. 8). Similar levels of p53 were expressed in the presence and absence of vIRF expression (Fig. 9). These results further demonstrate that vIRF suppresses p53-mediated upregulation of p21 and Bax expression.
FIG. 8.
Inhibition of p53-mediated upregulation of p21 and Bax protein by vIRF. Lysates of p53-null Saos-2 cells were collected at different time points after infection with a mixture of Ad-p53 plus Ad-GFP or Ad-p53 plus Ad-vIRF as indicated at the top and subject to Western blotting with anti-p21, anti-Bax, anti-p53, and anti-Flag (vIRF) antibodies, as indicated at the right. Equivalent titers of recombinant adenoviruses were used for infection.
FIG. 9.
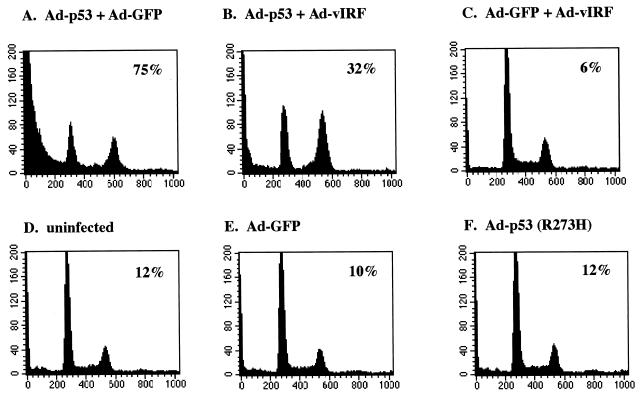
Inhibition of p53-mediated apoptosis by vIRF. Exponentially growing Saos-2 cells were infected with equivalent titers of recombinant adenoviruses as indicated. At 48 h postinfection, cells were stained for the chromosomal DNA with PI and analyzed on a FACScan flow cytometer. Numbers inside boxes indicate percentages of cells in the subdiploid phase of cell cycle, representing apoptotic cells. Results are representative of three individual experiments.
Inhibition of p53-mediated apoptosis by vIRF.
To examine the consequences of vIRF inhibition of p53 transcriptional activation, we examined the effect of vIRF on p53-mediated apoptosis. p53-null Saos-2 cells were infected with equivalent titers of recombinant Ad-p53 together with Ad-GFP or Ad-vIRF. In addition, Ad-p53 (R273H), carrying the p53 R273H mutant which is not able to induce apoptosis, was included as a control. At 48 h postinfection, cells were stained with PI and analyzed by flow cytometry to determine the level of apoptosis. Expression of wt p53 in Saos-2 cells induced extensive apoptosis, as indicated by the subdiploid cell population, whereas coexpression of vIRF significantly blocked p53-induced apoptosis: 75% apoptosis of Ad-p53- and Ad-GFP-infected Saos-2 cells, versus 32% apoptosis of Ad-p53- and Ad-vIRF-infected Saos-2 cells (Fig. 9A and B). Interestingly, a large portion of cells which were protected from p53-mediated apoptosis by vIRF expression appeared to be accumulated at the G2/M phase of cell cycle (Fig. 9B). In addition, infection of Saos-2 cells with Ad-GFP, Ad-p53 (R273H), or Ad-vIRF did not induce apoptosis under the same conditions (Fig. 9C to F). These results demonstrate that vIRF expression significantly blocks p53-mediated apoptosis.
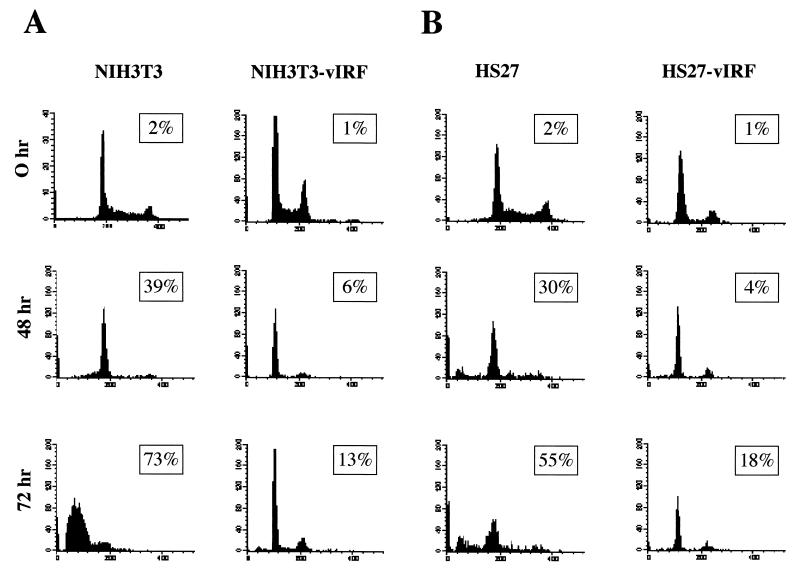
The p53 tumor suppressor transmits signals arising from various forms of cellular stresses, including growth factor depletion, to induce apoptosis (42). We have previously shown that expression of vIRF in rodent NIH 3T3 fibroblasts and human HS27 fibroblasts induces transformation, resulting in morphological change and/or focus formation (25, 26). These cells were used to further examine effects of vIRF on apoptosis induced by growth factor depletion. NIH 3T3, NIH 3T3-vIRF, HS27, and HS27-vIRF cells were incubated in medium containing 0.5% serum condition for 48 and 72 h, stained with PI, and analyzed by flow cytometry. These experiments showed that vIRF expression markedly protected these cells from growth factor depletion-induced apoptosis: 73% of NIH 3T3 cells underwent apoptosis after 72 h of serum starvation, whereas 13% of NIH 3T3-vIRF cells underwent apoptosis; 55% of HS27 cells underwent apoptosis after 72 h of serum starvation, whereas 18% of HS27-vIRF cells underwent apoptosis (Fig. 10). These results further indicate that vIRF expression markedly inhibits p53-mediated apoptosis.
FIG. 10.
vIRF expression confers resistance to apoptosis induced by growth factor depletion. A total of 5 × 106 NIH 3T3 and NIH 3T3-vIRF mouse fibroblasts and HS27 and HS27-vIRF human fibroblasts were collected at 0, 48, and 72 h after incubation with medium containing 0.5% FBS, stained for chromosomal DNA with PI, and analyzed on a FACScan flow cytometer. Numbers inside boxes indicate percentages of subdiploid cells of the cell cycle, representing apoptotic cells. Results are representative of three individual experiments.
DISCUSSION
The irreversible cell cycle arrest and apoptosis induced by p53 are considered part of host surveillance mechanisms for viral infection and tumor induction (2, 3, 9, 34). In this study, we demonstrate that KSHV vIRF interacts with the cellular p53 tumor suppressor and that this interaction inhibits modification and transcriptional activation of p53. As a consequence, vIRF expression significantly inhibits p53-mediated apoptosis. These results indicate that an inhibition of p53 function by KSHV vIRF is likely important to maintain persistent infection and develop virus-associated malignancies.
p53 consists of five distinct domains that serve different functions: amino-terminal transactivation domain, growth suppression SH3B domain, central core DNA binding domain, tetramerization domain, and carboxyl basic domain (34, 42). SV40 large T antigen binds to the core DNA binding domain (22, 27), adenovirus E1B binds to the amino-terminal transactivation domain (8, 20, 44), and HPV E6 binds to both the core DNA binding and carboxyl-terminal basic region (16, 38, 43). Like HPV E6, KSHV vIRF targets multiple regions of p53, including the growth suppression SH3B domain, the central core DNA binding domain, and the tetramerization domain. However, unlike HPV E6, which alters p53 protein stability (39), vIRF does not appear to alter its stability. Thus, KSHV vIRF targets p53 tumor suppressor similarly but not identically to other viral oncoproteins. In addition, our preliminary study indicates that BCBL-1 and BC-1 cells contain the wt p53 amino acid sequence, indicating that p53 in KSHV-infected cells can function to carry out cell growth control (unpublished results).
Upon transmission of stress signals, the rapid activation of p53 is often achieved through modifications, including phosphorylation and acetylation (34, 42). Various stress-activated kinases phosphorylate p53 at multiple serine sites. DNA-dependent protein kinase and ATM phosphorylate serine residue 15 (1, 23), and casein kinase II phosphorylates serine residue 392; these phosphorylations enhance p53 activities (31). Many tumor viruses have been shown to alter the level of p53 modifications, which is part of their defense against host growth surveillance (19, 34, 42). We demonstrate that KSHV vIRF also significantly suppresses the level of serine phosphorylation at amino acids 15 and 392 of p53. Such aberrant levels of p53 phosphorylation are likely associated with the cell growth transformation induced by vIRF.
Recent studies have also shown that p53 is acetylated at multiple lysine residues in the carboxyl region by p300/CBP and PCAF and that this acetylation increases its DNA binding affinity and transcriptional activation (28, 37). p300/CBP and PCAF exhibit specificity toward different lysine residues of p53; lysine 320 is the preferential target for PCAF, whereas lysine 373 is the target for p300/CBP but not for PCAF (28, 37). Adenovirus has been shown to alter the level of p53 acetylation at two different levels: E1B interacts with p53 and inhibits its acetylation induced by PCAF (29); E1A interacts with p300/CBP and PCAF and represses their acetylation activity toward p53 (30). Previously, we have also shown that vIRF interacts with cellular p300 HAT protein and that this interaction not only inhibits p300 HAT activity but also displaces PCAF from p300 complexes (25). Thus, similar to adenovirus E1A and E1B, KSHV vIRF affects p53 acetylation at multiple levels: the interaction of p53, the inhibition of p300 acetylation activity, and the displacement of PCAF from p300. These results raise several issues. Does vIRF directly interact with p53, or do other cellular proteins, such as p300, bridge these two protein? Is the reduced level of p53 acetylation due to an inhibition of p300 HAT activity by vIRF or by physical hindrance by vIRF interaction? Future studies should be directed to answering these questions.
p53 is a transcriptional activator that binds to sequence-specific binding sites in the promoter regions of numerous cellular genes and activates their transcription (10, 42). In addition, phosphorylation and acetylation have been shown to activate p53-mediated transcriptional activation, which leads ultimately to apoptosis or the inhibition of cell division (19). In this report, we demonstrate that vIRF interacts with p53 and that this interaction suppresses its modification and transcriptional activation, resulting in an inhibition of p53-mediated apoptosis. Our studies suggest that vIRF contributes to KSHV-associated pathogenesis at two different levels: inhibition of the p53 tumor suppressor, which is likely involved in facilitating cell growth transformation, and alteration of p300 cotranscriptional factor activity, which is likely involved in deregulating host IFN-mediated anti-viral activity. Our studies also elucidate how a virus-captured cellular gene has evolved to circumvent host immune surveillance and cell growth control mechanisms to achieve persistent infection and pathogenesis. Finally, the continuously growing list of viral proteins which apparently interact and interfere with p53 tumor suppressor further emphasizes that p53 is an important cellular regulator to control viral infection and tumor induction.
ACKNOWLEDGMENTS
H. Nakamura and M. Li contributed equally to this work.
We especially thank R. Means for critical discussion and reading of the manuscript, B. Damania, J. Alwine, C. Prives, and B. Vogelstein for providing reagents, and X. Alvarez for confocal analysis.
This work was partly support by Public Health Service grants CA82057 and RR00168. J. Jung is a Leukemia & Lymphoma Society Scholar.
REFERENCES
- 1.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 2.Bates S, Hickman E S, Vousden K H. Reversal of p53-induced cell-cycle arrest. Mol Carcinog. 1999;24:7–14. doi: 10.1002/(sici)1098-2744(199901)24:1<7::aid-mc2>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Bates S, Vousden K H. Mechanisms of p53-mediated apoptosis. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burysek L, Yeow W S, Lubyova B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 8.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 9.el-Deiry W S. p21/p53, cellular growth control and genomic integrity. Curr Top Microbiol Immunol. 1998;227:121–137. doi: 10.1007/978-3-642-71941-7_6. [DOI] [PubMed] [Google Scholar]
- 10.el-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 11.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 12.Feitelson M A, Zhu M, Duan L X, London W T. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene. 1993;8:1109–1117. [PubMed] [Google Scholar]
- 13.Gao S-J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 14.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holden R J, Mooney P A. The p53 paradox in the pathogenesis of tumor progression. Med Hypotheses. 1999;52:483–485. doi: 10.1054/mehy.1998.0741. [DOI] [PubMed] [Google Scholar]
- 16.Howley P M, Munger K, Romanczuk H, Scheffner M, Huibregtse J M. Cellular targets of the oncoproteins encoded by the cancer associated human papillomaviruses. Princess Takamatsu Symp. 1991;22:239–248. [PubMed] [Google Scholar]
- 17.Hussain S P, Harris C C. p53 mutation spectrum and load: the generation of hypotheses linking the exposure of endogenous or exogenous carcinogens to human cancer. Mutat Res. 1999;428:23–32. doi: 10.1016/s1383-5742(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 18.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Jayaraman L, Prives C. Covalent and noncovalent modifiers of the p53 protein. Cell Mol Life Sci. 1999;55:76–87. doi: 10.1007/s000180050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 21.Kubbutat M H, Vousden K H. New HPV E6 binding proteins: dangerous liaisons? Trends Microbiol. 1998;6:173–175. doi: 10.1016/s0966-842x(98)01267-0. [DOI] [PubMed] [Google Scholar]
- 22.Lane D P, Crawford L V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 23.Lees-Miller S P, Chen Y R, Anderson C W. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine A J. The p53 tumor suppressor gene and gene product. Princess Takamatsu Symp. 1989;20:221–230. [PubMed] [Google Scholar]
- 25.Li M, Damania B, Alvarez X, Ogryzko V, Ozato K, Jung J U. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol Cell Biol. 2000;20:8254–8263. doi: 10.1128/mcb.20.21.8254-8263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linzer D I, Levine A J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Colosimo A L, Yang X J, Liao D. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol Cell Biol. 2000;20:5540–5553. doi: 10.1128/mcb.20.15.5540-5553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 31.Meek D W, Simon S, Kikkawa U, Eckhart W. The p53 tumour suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 1990;9:3253–3260. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 33.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 34.Prives C, Hall P A. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Ganem D. Lytic growth of Kaposi's sarcoma-associate herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 36.Roemer K. Mutant p53: gain-of-function oncoproteins and wild-type p53 inactivators. Biol Chem. 1999;380:879–887. doi: 10.1515/BC.1999.108. [DOI] [PubMed] [Google Scholar]
- 37.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffner M, Takahashi T, Huibregtse J M, Minna J D, Howley P M. Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J Virol. 1992;66:5100–5105. doi: 10.1128/jvi.66.8.5100-5105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 40.Speir E, Modali R, Huang E S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 41.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 375–399. [Google Scholar]
- 42.Vogelstein B, Lane D, Levine A J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 43.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 44.White E. Regulation of apoptosis by the transforming genes of the DNA tumor virus adenovirus. Proc Soc Exp Biol Med. 1993;204:30–39. doi: 10.3181/00379727-204-43631. [DOI] [PubMed] [Google Scholar]
- 45.Zimring J C, Goodbourn S, Offermann M K. Human herpesvirus 8 encodes an interferon regultory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]