Abstract
Distinguishing between primary (PID) and secondary (SID) immunodeficiencies, particularly in relation to hematological B-cell lymphoproliferative disorders (B-CLPD), poses a major clinical challenge. We aimed to analyze and define the clinical and laboratory variables in SID patients associated with B-CLPD, identifying overlaps with late-onset PIDs, which could potentially improve diagnostic precision and prognostic assessment. We studied 37 clinical/laboratory variables in 151 SID patients with B-CLPD. Patients were classified as “Suspected PID Group” when having recurrent-severe infections prior to the B-CLPD and/or hypogammaglobulinemia according to key ESID criteria for PID. Bivariate association analyses showed significant statistical differences between “Suspected PID”- and “SID”-groups in 10 out of 37 variables analyzed, with “Suspected PID” showing higher frequencies of childhood recurrent-severe infections, family history of B-CLPD, significantly lower serum Free Light Chain (sFLC), immunoglobulin concentrations, lower total leukocyte, and switch-memory B-cell counts at baseline. Rpart machine learning algorithm was performed to potentially create a model to differentiate both groups. The model developed a decision tree with two major variables in order of relevance: sum κ + λ and history of severe-recurrent infections in childhood, with high sensitivity 89.5%, specificity 100%, and accuracy 91.8% for PID prediction. Identifying significant clinical and immunological variables can aid in the difficult task of recognizing late-onset PIDs among SID patients, emphasizing the value of a comprehensive immunological evaluation. The differences between “Suspected PID” and SID groups, highlight the need of early, tailored diagnostic and treatment strategies for personalized patient management and follow up.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10875-024-01818-2.
Keywords: Primary immunodeficiencies; B cell chronic lymphoproliferative disorders, secondary immunodeficiency; Artificial intelligence; Clinical diagnosis; Early detection
Introduction
Immunodeficiencies are an extensively wide-ranging group of diseases characterized by qualitative and/or quantitative altered immune responses. They can be primary (PID), including more than 500 congenital monogenic germinal mutations affecting any component of the immune system (or inborn error of immunity, IEI) [1, 2] and a major proportion of patients with yet undefinable genetic basis; or secondary (SID) to diverse conditions, such as, lymphoproliferative malignant disorders or to immunosuppressive treatments, as well as other chronic disorders like malnutrition [3]. Because the prevalence of PID is much lower than that of SID, the diagnosis of PID requires exclusion of any possible cause of SID. However, the boundaries between both entities are now more debated than ever. On the one hand, the risk of hematologic malignancies is increased in most PIDs. Moreover, malignancy is the second leading cause of death among PID patients after infection, and lymphomas account for two thirds of all neoplasia found in PID patients [2, 4, 5]. On the other hand, many first-diagnosed SIDs with hematological disorders share similar clinical and analytical phenotypes to PIDs, such as Common Variable Immunodeficiency (CVID) [6]. Also, diagnosis of late-onset PIDs in adults are getting more and more prevalent. Genetic variants can influence both immunodeficiency and lymphomagenesis. Certain variants serve as drivers of neoplastic processes, like those in DNA repair genes such as PRKDC, which contribute to oncogenic cell development while also impairing B cell maturation and antibody production. Mutations in PI3KCD and PI3KR1 lead to Activated PI3K Delta Syndrome (APDS), showcasing the intersection of immunodeficiency and cancer development [7, 8]. Furthermore, variants that cause immune dysregulation, such as those in Perforin (PRF1), can disrupt anti-tumor immunosurveillance, further linking immune system dysfunction to cancer progression [9]. In contrast, other variants may simply facilitate an existing malignant process [10].
The distinction between PIDs and SIDs associated with hematological malignancies raises critical questions about their relationship—whether they represent distinct entities or varying expressions within the same clinical spectrum. Identifying biomarkers capable of distinguishing PIDs from genuine SIDs in patients with B-CLPD is crucial for enhancing diagnosis, treatment, and prognosis. This study proposes the hypothesis that there exists a “hidden” subgroup of PID patients among those classified as having SID, who may experience a unique clinical trajectory. Such patients could potentially benefit from tailored follow-up strategies and specific treatments, including immunotherapies or targeted therapies that address known altered signaling pathways, ultimately influencing their clinical outcomes and familial implications.
In this pioneering observational study, we aimed to evaluate classical and more recent clinical and immunological biomarkers for PID diagnosis in a cohort of SID with B-CLPD. We assessed the ability of these biomarkers to differentiate between late-onset PID and SID, aiming to refine the clinical understanding and management of these complex conditions.
Materials and Methods
Patients and Study Design
We performed a single center ambispective observational cohort study in SID patients with B-CLPD evaluated at our institution from 2015 to 2023. The diagnosis of B-CLPD was established based on the WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue [11]. SID in these patients was diagnosed based on the criteria outlined in the latest clinical guidelines [12], which include recurrent or severe infections, a reduction in serum IgG, IgA, and/or IgM by at least two standard deviations below the normal mean, and a failure to mount an antibody response to polysaccharide and/or protein antigens, in the presence of diagnosed malignancy. As a comparison group, 17 CVID diagnosed patients with clinical exome analyses were studied. Latest clinical criteria of CVID [13] were met and secondary causes had been excluded in all of them.
Infection categories - recurrent, severe and persistent -, were defined according to consensus criteria [14]. Follow-up duration time was calculated from the date of referral to the Immunology Department to the last contact or the date of death. Comprehensive baseline data was collected, that included clinical manifestations with specific and timely history of infectious complications, tumor staging, laboratory findings. For the ‘history of childhood infections’ criterion, we relied on patients’ recollections and available medical records. To ensure accuracy, we only considered a history of recurrent and/or severe infections for patients who reported hospital referrals, frequent antibiotic treatments (multiple cycles per year), or significant disruptions to their daily life (primarily school life) due to recurring illnesses.
All procedures followed were in accordance with the ethical standards of the institutional research committee and with the Helsinki Declaration of 1975, as revised in 2000. The study received approval from the hospital’s Institutional Research Ethics Committee (20/072-E and 19/219-E).
Laboratory Assessment
We performed an extensive baseline immunological evaluation that included: differential white blood cell count, serum IgG, IgA, IgM and IgG subclasses (IgG1, IgG2); complement factors C3 and C4; specific antibody responses to the 13-valent pneumococcal (PPV), Typhim Vi (TV) polysaccharide, and tetanus-toxoid (TT) vaccines before vaccination and 30 days post-vaccination. Serum IgG, IgA, and IgM, IgG1, IgG2, C3 and C4 concentrations were quantified by turbidimetry using commercial kits on an Optilite analyzer (The Binding Site Group Ltd, Birmingham, UK). Reference values were provided for each measured component. Peripheral blood samples were analyzed using the lyse-no-wash method on a FACSCanto II flow cytometer. The BD Multitest TBNK kit (Becton-Dickinson, San Jose, CA) was utilized to measure CD4+ and CD8+ T-lymphocytes, CD19+ B lymphocytes, and CD56+ NK cells. Absolute counts and percentages were expressed relative to total lymphocytes. The reference ranges were adapted from a study of a similarly aged Spanish healthy population. Adequate specific Ab responses were defined as individuals obtaining a fold increase (FI) ≥ 3 for PPV and TT and ≥ 5 for TV according to published data in PID and SID, respectively [15, 16]. Further assessment such as microbiology serological tests and peripheral blood PCR for EBV and CMV was performed as per routine use.
Statistical Analysis
Data were collected using the Excel spreadsheet (Microsoft, Inc., Redmond, WA, USA). Descriptive data and continuous variables are presented as mean ± standard deviation (SD) or median values (interquartile range, Q1 - Q3), according to the normal or non-normal distribution of the data. Categorical variables were described as counts and percentages of subjects. Bivariate association analyses included the chi-squared test or the Fisher’s exact test for categorical variables, and the Student’s t test or the Wilcoxon’s ranksum test, as appropriate. The variables with statistically significant differences in the bivariate association analyses were employed to construct a decision tree aimed to distinguish between PID and SID. The decision tree was fitted using recursive partitioning and validated with the bootstrap method. Statistical analyses were conducted using R software (version 4.3.1) and the following packages: caret (6.0–94), Rpart (4.1.23) and boot (1.3–30).
Results
Demographic and Clinical Features of SID Patients with BCLPD
During the year 2023, 151 SID patients with B-CLPD (mean age 67, with SD of 13.53 years, and a 3:2 female: male sex ratio) were enrolled. Most of the patients were referred to our department in a range of 0 to 10 years after BCLPD diagnosis, except for those with more recent diagnosis that were studied at cancer diagnosis. All participants underwent thorough clinical assessment during their routine clinical examination, where they provided information on 19 different clinical variables related to their medical history, including timelines of infections, autoimmune diseases, enteropathies, and malignancies. Additionally, each participant was tested for 18 immunological and analytical variables (Supplementary Table 1).
Our hypothesis was not simply to confirm the resemblance of SID to PID, which can be expected due to immune deficiencies caused by therapy or underlying cancer. Instead, we aimed to identify which SID cases may actually be undiagnosed PID cases. The critical distinction we made was to assess patients’ clinical features prior to the initiation of therapy. To this purpose, our cohort was initially split into two groups (“Suspected-PID Group” and “SID Group”), based on two key criteria for CVID diagnostic framework: increased susceptibility to infection - before the diagnosis of the B-CLPD; and marked decrease of IgG and marked decrease of IgA with or without low IgM levels - IgG levels were considered as ‘low’ when below 500 mg/dL [13]. The “Suspected-PID Group” comprised 110 patients (55 NHL, 22 CLL, 7 LH, 24 MGUS, 8 MM and Wäldestrom, and 3 other B-cell neoplasia; 14 of them having more than one B-CLPD). The “SID Group” comprised 41 patients (16 NHL, 12 CLL, 1 LH, 4 MM and Wäldestrom, 11 MGUS and 1 other B-cell neoplasia; 7 of them having more than one B-CLPD).
When assessed, 21 patients (13.91%) of the cohort were on active treatment. Of these, 8 (5.3%) were on anti-CD20 therapy, representing 38.1% of the patients on active therapy.
In the ‘Suspected-PID’ group, 14.55% were on treatment at the time of the study, compared to 12.19% in the ‘SID’ group.
Differences between “Suspected-PID” and “SID” Groups
The bivariate analysis of the 37 variables considered in this study showed statistically significant differences in 10 variables between the two groups—six clinical and four analytical—excluding the initial grouping criteria (‘Recurrent respiratory tract infections before malignancy diagnosis’ and ‘Immunoglobulin G levels’). (Table 1).
Table 1.
Bivariate analysis of demographic and immunological characteristics of the main cohort groups. Statistically significance was considered when p < 0.05. Statistically significant variables are shown in bold
| “SUSPECTED PID GROUP” N = 110 |
“SID GROUP” N = 41 |
P | |
|---|---|---|---|
| Age (years) | 67.50 (57.25–75.75) | 71.00 (65.00–77.00) | 0.046 |
| Age at B-CLPD diagnosis | 53.57 (15.35) | 59.54 (11.91) | 0.037 |
| Rituximab treatment | 63 (61.76) | 13 (38.24) | 0.028 |
| Complete remission | 56 (61.54) | 15 (44.12) | 0.121 |
| Childhood recurrent/severe infections | 57 (53.77) | 0 (0) | < 0.001 |
| Recurrent/severe infections pre-BCLPD | 53 (51.46) | 1 (2.56) | < 0.001 |
| Zoster infection | 26 (24.07) | 4 (10.53) | 0.122 |
| Autoimmune disease | 33 (30.84) | 7 (17.95) | 0.181 |
| Malabsorption | 32 (29.63) | 6 (15.38) | 0.126 |
| Family history of B-CLPD | 41 (37.96) | 5 (12.82) | 0.006 |
| IgRT | 72 (67.29) | 15 (39.47) | 0.004 |
| IgG at B-CLPD diagnosis | 574.00 (387.50–928.00) | 712.00 (494.00–1325.00) | 0.027 |
| sFLC kappa | 10.40 (6.50–17.40) | 16.90 (10.80–23.40) | 0.002 |
| sFLC lambda | 9.10 (5.70–16.40) | 17.00 (11.10–24.10) | < 0.001 |
| Sum kappa + lambda | 19.10 (11.90–36.80) | 36.00 (26.70–73.20) | < 0.001 |
| Switched memory B cells (smB) | 0.00 (0.00–6.50) | 7.80 (0.00–23.65) | 0.010 |
| NK cells | 10.00 (6.00–16.00) | 6.50 (2.75–10.75) | 0.069 |
The data are presented as the number of patients (%), except for age: median (IQR); and age at BCLPD diagnosis: mean (SD). For laboratory values, the data are expressed as median (IQR). Statistical significance is shown as p < 0.01
The analysis highlighted significant distinctions particularly in the rates of recurrent and severe respiratory infections during childhood (p < 0.001) and the presence of a family history of lymphoid malignancies (p = 0.006). These differences suggest a potential genetic basis for PIDs and emphasize their importance in early diagnosis and familial counseling.
In addition, the analysis revealed that the “Suspected-PID Group” showed a significantly greater requirement of rituximab administration for cancer control and of Intravenous Immunoglobulin Replacement Therapy (IgRT) compared to the “SID Group” (61.76% vs. 38.24%, p = 0.028; and 67.29% vs. 39.47%, p = 0.004, respectively). Furthermore, the mean age at B-CLPD diagnosis was significantly lower in the “Suspected-PID Group” (53.57 years) compared to the “SID Group” (59.54 years, p = 0.037). These findings suggest distinct clinical profiles that may assist in differentiating between SID and suspected PID within the context of B-CLPD. Also, interestingly, 85.71% of the “Suspected-PID” group who received anti-CD20 therapy had completed their treatment at least five years before the study, with nearly half having completed it over a decade prior.
Among the analytical assessment, significant differences were noted in levels of sFLC, in both κ and λ, as well as in sum κ + λ, with all values significantly lower in the “Suspected-PID” group than in the “SID” group. This finding aligns with prior research identifying sum κ + λ as a critical biomarker for CVID diagnosis [17, 18]. The disparity was particularly pronounced for λ light chains (median 17.00 vs. 9.10, p < 0.001) and sum κ + λ (median 36.00 vs. 19.10, p < 0.001) (Fig. 1), more than for κ light chains (median 16.90 vs. 10.40, p = 0.020).
Fig. 1.
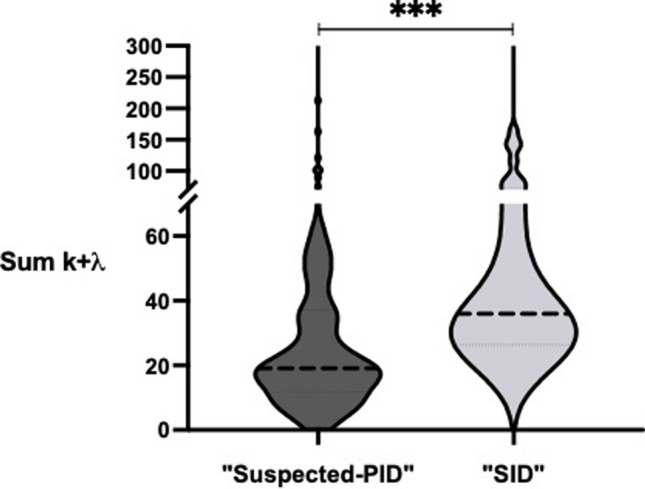
Comparison of serum Free Light Chain sum k + λ distribution and median levels between “Suspected-PID” group and “SID” group. Statistically significant difference with p < 0.001 is represented with ***. Figure made with GraphPad Prism 9
Additionally, similar to patterns observed in some CVID cases, differences in the percentages of switch-memory B cells (smB) were significantly lower in the “Suspected-PID Group” compared to the “SID Group” (median 0.00% vs. 7.80%, p < 0.0103). However, total B-cell counts did not show significant differences. Leukocyte counts were also significantly higher in the “Suspected-PID Group” compared to the “SID Group” (median 8,000 vs. 6,000, p = 0.0275), though neither group exhibited leukocytosis.
Although not statistically significant, certain trends observed in our data merit further exploration in larger cohorts. The prevalence of autoimmune disorders and malabsorption were markedly higher in the “Suspected-PID Group” compared to the “SID Group” (30.84% vs. 17.96%, p = 0.1816 for autoimmune disorders; 29.63% vs. 15.38%, p = 0.1264 for malabsorption). These conditions are typically linked to an increased risk of malignancy in PID patients, suggesting a potential underlying immunological mechanism that warrants further investigation. Additionally, B-CLPD complete remission rates were higher in the “Suspected-PID Group” (61.54% vs. 44.12%, p = 0.12), though this difference did not achieve statistical significance. These findings suggest a possibly distinct clinical course in patients classified as having a suspected PID. Conversely, rates of relapses and secondary neoplasms showed no differences between the groups, nor did the incidence of recurrent or severe respiratory tract infections post-cancer diagnosis.
Interestingly, the rate of recurrent respiratory infections increased slightly in the “Suspected-PID Group” from 51.46% prior to diagnosis to 61.54% afterwards. In the “SID Group,” rates of recurrent infections after the onset of hematological malignancy surged to match those of the “Suspected-PID Group” (from 2.56% prior to diagnosis to 69.23% post-diagnosis) (Table 1.).
Machine Learning Model Accurately Predict Underlying Primary Immunodeficiency
Considering only the significant variables obtained, we then used machine learning for the development of a tree decision model. A recursive partitioning (rpart) algorithm was applied, obtaining two main variables that could aid in the task of discriminating between “Suspected-PID Group” and “SID group” (Fig. 2). The variables, in order or relevance were: sum κ + λ as the first node of the decision tree; and history of severe or recurrent infection during childhood as the second node.
Fig. 2.
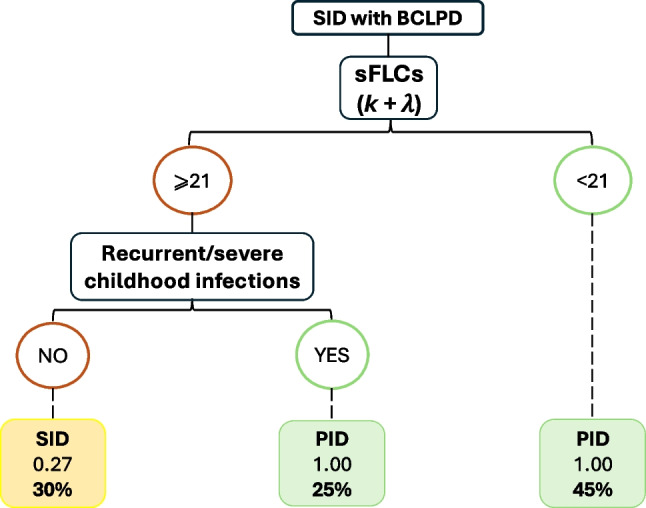
Tree decision model for early detection of “Suspected PID” patients with diagnosis of SID. Model created through Rpart algorithm
The predictive performance of the tree decision model, which was the best predictive model to discriminate between the two groups, achieved the highest accuracy of 91.8%, sensitivity of 89.5%, and specificity of 100%, with positive predictive value of 100%. Due to the limited sample size, we performed bootstrap method, which is the most efficient method for estimation of internal validity of a predictive logistic regression model in small sample cohorts, resulting in stable and nearly unbiased estimates of performance [19]. Consequently, the model demonstrated a robust accuracy within a 95% confidence interval of 87–97%, sensitivity ranged from 87 to 100%, and specificity between 53% and 100%. This high degree of accuracy underscores the model’s utility in identifying PID at the onset of B-CLPD.
Features of Non-Hodgkin Lymphomas within “PID-suspected” Group and “SID” Group
The proportion of NHL cases was not different between both groups, with 47.2% in the “PID-suspected Group” compared to 31.7% in the “SID Group” (p = 0.3). Given that NHL patients underwent similar treatment protocols, they constituted a more homogeneous subgroup for comparative analysis. Table 2 outlines the clinical and immunological differences observed between the two groups in the 37 studied variables.
Table 2.
Comparative analysis of key clinical and immunological variables in suspected PID versus SID within NHL patients
| Variable | “PID-suspected NHL group” No. = 52 |
“SID NHL Group” No. = 13 |
P-value |
|---|---|---|---|
| Childhood recurrent&severe infections | 21 (40.4%) | 0 (0%) | 0.005 |
| Infections prior to NHL diagnosis | 23 (44.2%) | 1 (7.7%) | 0.01 |
| Infections after NHL diagnosis | 30 (57.7%) | 4 (30.8%) | 0.08 |
| Malabsorptive syndrome | 17 (32.7%) | 2 (15.4%) | 0.22 |
| Second primary neoplasia | 15 (28.8%) | 2 (15.4%) | 0.32 |
| Family history of B cell neoplasms | 8 (15.4%) | 0 (0%) | 0.13 |
| Serum IgM at NHL diagnosis (mg/dL) |
56.12 ± 44.54 40 |
225.23 ± 19.79 56 |
< 0.0001 |
| Sum kappa + lambda (mg/dL) |
23.48 ± 15.06 15 |
33.00 ± 12.00 23 |
0.03 |
| Class-switched memory B cells (%) |
2.06 ± 5.29 0 |
6.33 ± 2.12 9 |
0.006 |
The data are presented as the number of patients (percentage). For laboratory values, the data are expressed as mean ± standard deviation (SD) and median
Follow-up after last cycle of rituximab was 10.05 ± 7.77 years (median, 8 years) for the “PID-suspected Group” compared to 10.45 ± 3.53 years (median, 12 years) in the “SID Group” (p = 0.3). There were no statistically significant differences in time interval since Rituximab treatment between the groups, with 37.27% of “Suspected-PID” and 24.39% of “SID” having received it more than 5 years prior, and 21.82% of “Suspected-PID” and 12.19% of “SID” having received it at least 10 years prior. Again, the most significant clinical variables were the occurrence of severe or repeated infections early in life or years before the BCLPD (Table 2). Interestingly, there was a trend of more frequent infections in the suspected-PID NHL than in the SID group (57.7% versus 30.8%, p = 0.08), more common malabsorptive syndrome and second primary neoplasia, and family history of BCLPD, although not significant probably due to the small sample size. Serum IgM at BCLPD diagnosis, sum κ + λ and switched memory B cells were all significantly lower in “Suspected-PID NHL” compared to “SID NHL”. (Table 2).
Genetic Screening for Inborn Errors of Immunity in the “Suspected-PID” Group Compared to the PID Group
Genetic screening of 59 patients of the “Suspected-PID Group” revealed the presence of, at least, one genetic variant in 69.49% (41 patients in total); and, at least, one genetic variant related to IEI in 66.10% (39/59). Ten (23.73%) of them carried more than one genetic variant.
Following the American College of Medical Genetics and Genomics (ACMG) guidelines, variants were classified as benign, likely benign, variants of uncertain significance (VUS), likely pathogenic, and pathogenic. A total of 60 genetic variants were discovered. We found 36 VUS (60%) and 24 likely-pathogenic/pathogenic variants (40%). All of them were heterozygous. According to the latest classification of IEI [1], 85% (51/60) of the variants could be classified into one of the ten categories of IEI (Table 3). The most represented groups were ‘Combined Immunodeficiencies’, with or without syndromic features (18/51, 35.29%), and ‘Diseases of immune dysregulation’ (11/51, 21.57%), followed by ‘Predominantly antibody deficiencies’ (8/51, 15.69%) and ‘Congenital defects of phagocyte number or function’ (8/51, 13.73%). There were also 2 variants of ‘Autoinflammatory disorders’, 3 related to ‘Bone Marrow failure’ and 1 Phenocopy of IEI (Table 3). Interestingly, 11 of the 60 variants (18.33%) were related to DNA repair processes defects.
Table 3.
Genetic variants related to IEIs in SID patients with “Suspected PID”. Variants that can be autosomic dominant (AD) are represented with *
| Classification of IEI (Tangyee et al.) | Genetic variants found in SID patients with Suspected-PID (N = 59) | |
|---|---|---|
| LIKELY/PATHOGENIC | VUS | |
|
1. Immunodeficiencies affecting cellular and humoral immunity 2. Combined immunodeficiencies with associated or syndromic features |
PMS2 (2), SKIV2L (1), PRKDC (1), DNMT3B (1), ATM (1) | ERCC6L2 (3), STAT3* (2), POLE (2), FOXN1*(1), PRKDC (1), JAK3 (1), DOCK8 (1), IL7R (1) |
| 3. Predominantly antibody deficiencies | TNFRSF13B*(2), TRNT1 (1), | TNFRSF13B*(2), MSH6(1), CD19 (1), PIK3CD (1) |
| 4. Diseases of immune dysregulation | PRF1 (2), STXBP2* (2), TET2 (1) | LYST (2), PDCD1 (1), LRBA (1), BACH2* (1) |
| 5. Congenital defects of phagocyte number or function | CLPB (1), NCF1 (1), SBDS (1) | SBDS (2), GATA2* (1), NCF1 (1) |
| 6. Defects in intrinsic and innate immunity | - | MYD88 (1) |
| 7. Autoinflammatory disorders | - | PLCG2*(1), MEFV* (1) |
| 8. Complement deficiencies | - | - |
| 9. Bone Marrow failure | CTC1 (2) | CTC1 (1) |
| 10. Phenocopies of IEI | - | NRAS (1) |
*Figures could be in color, especially, Fig. 1, but it is not necessary
To increase the reliability of these results, we compared them with the genetic results of 17 adult definite PID patients, in whom exome was performed and where malignancy had been previously excluded. A total of 23 variants were found in 18 genes. 78.26% were VUS and 21.74% likely-pathogenic/pathogenic variants. All of them were heterozygous. Also alike the “Suspected-PID”, a high percentage (94.44% (17/18)) of the gene variants could be classified into one of the categories of IEI, with the most represented being ‘Predominantly antibody deficiencies’ (5/17, 29.41%), ‘Combined Immunodeficiencies’, with or without syndromic features (4/17, 23.53%) and ‘Autoinflammatory disorders’ (4/17, 23.53%). There was 1 variant related to ‘Immune dysregulation’ (5.88%), and 3/17 (17.65%) in ‘Congenital defects of phagocyte number or function’ (Supplementary Table 2).
Discussion
The idea that patients with immunodeficiency to B-CLPD may suffer from a late-onset PID is an increasingly on-date debate. Due to clinical similarities of both entities, a mild-symptomatology spectrum and the absence of a thorough clinical immunodeficiency-focused examination, many PID could be hidden until debuting with a malignant lymphoproliferative disorder. Indeed, PID are associated with higher risk of developing hematological malignant processes than general population [2, 20]. Therefore, identifying late-onset PID among patients with B-CLPD and immunodeficiency could have major importance for early infection prevention, and ultimately, to aim for a better prognosis with more targeted or adjusted therapies, beyond current protocols, when available, to potentially improve prognosis and reduce iatrogenic effects. For example, certain mutations in the context of APDS or Familial Hemophagocytic Lymphohistiocytosis (FHL) could be treated with specific targeted therapies or protocols, but most genetic variants do not currently have targeted therapies. In this work, we found 10 clinical and immunological variables that discriminated between “Suspected-PID” and SID patients. Furthermore, through a robust tree decision model, two of those variables: “sum κ + λ” and “childhood infections”, identified PID with high accuracy.
In our cohort, according to the model, 70% of the patients that were thought to be SID could be reclassified as “Suspected-PID”. We acknowledge that the initial 70% prevalence of ‘Suspected PID’ might appear disproportionate given the known prevalence rates of PID versus SID. However, the initial classification was based on rigorous clinical and immunological criteria of CVID diagnosis as outlined by the ESID Registry [13]. This classification may overlap SID patients with infectious manifestations years prior to the overt cancer. The relevance of our AI regression algorithm is that it did not rely solely on these criteria. Instead, it selected a combination of vertical criteria (childhood recurrent or severe infections) and horizontal criteria (undetectable sFLC) to give a more consistent approach that may indicate a cohort skewed towards hidden PID, underscoring the complexity of differentiating between PID and SID and stressing the critical need for a comprehensive clinical and immunological evaluation at the time of B-CLPD diagnosis. The observed high prevalence of suspected PID might be influenced by a referral bias, as hematologists often refer patients who present with significant infectious complications.
Even though the number of genetic variants identified to be related to PID pathogenesis is increasing exponentially, to date, PID diagnosis still relies heavily on clinical criteria. Likewise, in our study, the weight of recurrent or severe infections early in life was the second best classifier. We also acknowledge the potential importance of certain clinical variables, like the increased frequency of autoimmune disorders and malabsorption [20, 21] in the “Suspected PID Group”. While these findings did not reach statistical significance, this may be attributed to our limited sample size. These conditions are traditionally linked to PID, which can lead to poorer outcomes and an increased risk of neoplasia [5]. Nonetheless, they should be further tested in larger cohorts to verify their potential diagnostic role.
Our study revealed that patients in the “Suspected-PID Group” not only exhibited higher rates of complete remission but also demonstrated greater requirements of Rituximab for cancer control, and of IgRT for infection prevention. Altogether, this suggests that these patients may exhibit a different clinical behavior due to higher immunogenicity while also being more vulnerable to immunodepletion and infection-related complications. In response, their treatment protocols may necessitate adjustments or the incorporation of targeted therapies. Conventional aggressive anti-tumor therapies could exacerbate the immune state of “Suspected-PID” patients, heightening their susceptibility to infections and subsequent organ damage, aligning with the increased demand for IgRT observed in our research. Historically, cancer treatments primarily aimed at reducing malignant cell populations. Recently, attention has shifted towards reverting the immunosuppressed tumor microenvironment with novel immunotherapies. Nonetheless, the potential systemic immunodeficiency in patients has not been sufficiently considered. Understanding the variables that distinguish these groups can help tailor diagnostic and treatment strategies to the specific needs of patients, potentially leading to better outcomes.
An intriguing and original finding was the presence of a family history of lymphoid malignancies, which is challenging to interpret, and raises the possibility of an enrichment of other genetic defects associated with cancer, rather than directly supporting a PID diagnosis. However, this observation could also point to the presence of driver germline variants within the family, particularly at the intersection between PID and cancer/lymphoproliferation. A significant proportion of genetic variants related to IEI, including those in our cohort, are linked to a higher susceptibility to lymphoproliferation. Based on this, we hypothesize that a higher frequency of familial lymphoid malignancies may indicate an underlying genetic defect contributing to PID and cancer, though this remains speculative and requires further investigation in larger cohorts and additional studies.
When considering analytical biomarkers, very few of them have been proved to discriminate between PID diagnoses yet. sFLC, and specifically, sum κ + λ, however, may serve as a valuable biomarker in distinguishing between primary and secondary hypogammaglobulinemia, particularly in the context of CVID versus SID [17, 22, 23]. In our study, we found statistically significant differences in sFLC κ and λ concentrations, and in sum κ + λ between “Group Suspected-PID” and SID patients, and in NHL subgroup analysis, which could validate the use of sum κ + λ in SID populations. Serum concentration of λ was more significant than κ concentration, probably due to clonality tends to be more frequent in κ chain. During normal B-cell development, κ is produced in higher quantities than λ. Although not yet demonstrated, it could also be possible that, in impaired B-cell development, λ production is affected earlier, making it a more sensitive biomarker than κ. Interestingly, the sum κ + λ measurement was taken after patients had undergone anti-tumor therapies, including anti-CD20 therapies, with extended follow-up, particularly in the subanalysis of NHL. Despite this, the “Suspected PID NHL” maintained very low concentrations.
One potential limitation that reflects the real-world experience is that 61.76% of “Suspected-PID Group” vs. 38.24% of SID had been on an anti-CD20 therapy, making it difficult to discriminate the cause of the underlying B-cell defect. Normal B-cell reconstitution is described to occur from 6 to 9 months to, approximately, 2 years after therapy [24]. Most suspected PID patients failed to achieve B-cell reconstitution even after a decade, suggesting a potential intrinsic B-cell defect potentially triggered by prior therapies, as hypothesized for other conditions [25–27]. This persistent deficit may have disrupted the normal B-cell reconstitution process, resulting in sustained small B-cell depletion and below range serum κ + λ levels. However, even though these results should be verified in further studies, we believe that testing B-cell parameters in these patients before undergoing therapy could be essential for their identification.
Despite these patients share some similarities, just as it is known about PIDs, the great variability of their clinical and immunological presentations can make it a very challenging task to reach a suited diagnosis, and even more, when they present with the diagnosis of a malignant disorder. In that sense, AI tools could aid clinicians reduce the “noise” of the vast clinical spectrum to reach an early diagnosis. We developed an AI model of diagnosis which is based on the most significant variables creating a regression tree decision model that seems to accurately target patients with a high suspicion of late-onset PID at time of malignancy diagnosis. Three quarters of our cohort could be identified as possibly having a PID with this model, highlighting the importance of identifying late-onset PID in the setting of B-CLPD with the help of AI. Unlike conventional statistical methods, the AI approach does not rely on assumptions about data distribution, allowing for a more robust analysis. AI went deeper into variable interactions, handled unobserved constructs by selecting a regression tree model out of different models. It also addressed measurement errors and conducted confirmatory factor analysis using bootstrapping for internal validation.
Accordingly, AI could help address genetic studies in those PID-suspected patients, allowing the study of genetic variants susceptible of more targeted therapies. In this study, we found that a high proportion up to 66.10% of 59 “Suspected-PID Group” patients carried, at least, one genetic variant related to IEI, according to Tangye SG et al. [1], supporting the hypothesis that PID and SID may fall below the same genetic and clinical spectrum. More than a third of them were related to combined immunodeficiencies, followed by immune dysregulation, and antibody deficiencies. Moreover, almost 20% of the variants related to combined immunodeficiencies and predominant antibody deficiencies, were also implicated in defects of DNA repair mechanisms. In line with that described for lymphomagenesis mechanisms in PID [4, 28], our data point to shared genetic defects related predominantly to immunosurveillance, intrinsic DNA repair and to B-cell biology intersecting PID and B-cell lymphoproliferative disorders. A broad spectrum of variants was observed, involving major signaling pathways that can impede the normal clearance of microorganisms and transformed cells, disrupt antibody production, and contribute to increased genetic instability and mutational load, as well as to chronic inflammation and lymphoproliferation. This underscores the intertwined nature of immunodeficiency and immune dysregulation and B-cell cancer. However, according to current literature, the heterozygous nature of the variants alone might not fully explain the clinical outcomes we see in our patients. Nonetheless, it is plausible that the delayed clinical presentation of the malignancy may be attributable to the heterozygosity of the genetic variants, possibly suggesting that these germinal genetic predispositions may require the cumulative effects by added somatic mutations and environmental triggers in a multi-hit process to manifest clinically [29]. It is likely, that if these variants were homozygous, patients might have been diagnosed earlier in life with classical pediatric IEI. CVID, the most common adult PID, can manifest from adolescence to late adulthood. While the prevalence of monogenic causes in CVID patients is estimated to be 25–30% [30], most cases could involve digenic or oligogenic variants, genetic modifiers, epigenetic factors, or somatic mutations acquired over time. Interestingly, the array of monogenic variants linked to CVID-like disorders is broad, encompassing over fifty variants that produce a similar clinical and immunological phenotype [31]. These variants also demonstrate significant overlaps with the somatic variants of NHL [32]. We argue that similar mechanisms might apply to our “Suspected-PID” patients, which could account for the presence of only heterozygous variants. Indeed, we could reinforce this idea by comparing those findings to the genetic results of our definite PID patients. Just as the “Suspected-PID” group, we found that a high percentage of the PID patients (64.71%) carried, at least, one genetic variant, being 94.44% of them present in genes related to IEIs. All of them were heterozygous as well, and also, similarly, they mostly belonged to the categories of ‘Predominantly antibody deficiencies’ and ‘Combined Immunodeficiencies’, with or without syndromic features. On the contrary, ‘Autoinflammatory disorders’ were more represented, and variants related to ‘Immune dysregulation’ were less observed than in the “Suspected-PID”. Interestingly, variants of ‘Combined Immunodeficiencies’ were more frequent in the “Suspected-PID” than in definite PID, possibly suggesting a higher risk of malignancy development due to defects in cellular immunosurveillance, further suggesting our hypothesis regarding family history of cancer malignancy.
It should be stated that a significant limitation of our study is the availability of genetic data exclusively from the “Suspected-PID” group with none from the “SID” group as comparison. However, these results stress that the identification of pathogenic variants confirming PID in previously SID patients could be of paramount importance in applying more personalized management [6]. It is imperative to accurately identify these patients at diagnosis to tailor personalized therapeutic strategies and follow-up plans, ensuring a holistic approach that addresses both the oncological and immunological challenges. Some examples of a more personalized management could be: the inclusion of periodic viral load tracing, as viral infections may worsen their prognosis and increase the risk of more malignant transformations in certain PID patients; specific therapy protocols, as for FHL or even specific targeted therapy in cases of APDS. Also, familial counseling should be included in those cases where genetic variants are identified. An intriguing observation from our study is the heightened familial incidence of hematological malignancies, particularly in the NHL subgroup among suspected-PID patients. This suggests that earlier detection of genetic variants could expedite the diagnosis of patients and potentially reduce the incidence of B-CLPD in cases involving IEIs and late-onset PID.
Beyond the immediate clinical scope, the distinctions among the groups based on specific variables point to opportunities for further research into the mechanisms driving these differences, as well as the development of new diagnostic tools or therapeutic approaches.
Overall, the decision tree model’s insights into the variables that discriminate among “Group PID”, and “Group SID” highlight the complexity of the cohort’s health profiles and the need for detailed analysis at time of lymphoproliferative diagnosis to support effective patient diagnosis, treatment and, hopefully, aid in their prognosis.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Supplementary Table 1. Variables studied in the B-CLPD patients’ cohort. (JPG 151 KB)
Supplementary file2 Supplementary Table 2. Genetic variants related to IEIs in PID patients. Variants that can be Autosomic dominant (AD) are represented with *. (JPG 160 KB)
Acknowledgements
We are immensely grateful to the patients and controls that made this study possible. The authors would like to acknowledge the nursing staff and in particular to Marta Ortíz Pica of the Hospital Day Care of the Hospital Clínico San Carlos, for their excellent care of our patients.
Author Contributions
MPO: performed the conceptualization, methodology, formal analysis, investigation, writing of the original draft, review and editing. TGG: participated in the conceptualization, validation, visualization of the study. AJH: did part of the methodology, software analysis, and performed the formal analysis. JMG: validated the study and helped providing resources. MPG, MDM, ÁV, EMH, APR, EFM: performed investigation, helped with resources. CPL, APC, MP, MMM, EAM, EB, BI, FM, MCC: provided resources, and validated the study. AC, CJG: collaborated in the formal investigation. JOG, BGS, YGC, MFA, CBC, RPD: did conceptualization and supervision. NR: performed validation, visualization and supervision. SSR: performed the conceptualization, methodology, investigation, writing of the original draft and the review and editing of the final manuscript, as well as, the supervision and project administration. All authors reviewed the manuscript.
Funding
The work is supported by a grant from Pharming.
Data Availability
No datasets were generated or analysed during the current study.
Declarations
Competing Interests
JMG-A is an employee of Health in Code S.L.. The rest of authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Institutional Review Board Statement
Approval for the study was obtained from the hospital institutional research Ethics Committee (19/219-E).
Declaration of Generative AI and AI-assisted Technologies in the Writing Process
Authors disclose that they did not make use of any generative AI and AI-assisted technologies in the writing process.
Footnotes
The original online version of this article was revised: Table 1 carried an erratum. sFLC kappa and lambda values were in switched positions by mistake. The values of PID-suspected group were placed by error in the column of the SID group and vice versa.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/8/2025
The original online version of this article was revised: Table 1 carried an erratum. sFLC kappa and lambda values were in switched positions by mistake. The values of PID-suspected group were placed by error in the column of the SID group and vice versa.
Change history
2/7/2025
A Correction to this paper has been published: 10.1007/s10875-025-01865-3
References
- 1.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022;42(7):1473–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiri A, et al. Inborn errors of immunity and cancer. Biology (Basel). 2021;10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otani IM, et al. Practical guidance for the diagnosis and management of secondary hypogammaglobulinemia: a Work Group Report of the AAAAI primary immunodeficiency and altered Immune Response committees. J Allergy Clin Immunol. 2022;149:1525–60. [DOI] [PubMed] [Google Scholar]
- 4.Riaz IB, Faridi W, Patnaik MM, Abraham RS. A Systematic Review on Predisposition to Lymphoid (B and T cell) Neoplasias in Patients With Primary Immunodeficiencies and Immune Dysregulatory Disorders (Inborn Errors of Immunity). Front Immunol. 2019;16(10):777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood. 2012;119:1650–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballow M, Sánchez-Ramón S, Walter JE. Secondary Immune Deficiency and primary Immune Deficiency crossovers: hematological malignancies and Autoimmune diseases. Front Immunol. 2022;13:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolijn PM, Langerak AW. Immune dysregulation as a leading principle for lymphoma development in diverse immunological backgrounds. Immunol Lett. 2023;263:46–59. [DOI] [PubMed] [Google Scholar]
- 8.Ye X, et al. Genomic characterization of lymphomas in patients with inborn errors of immunity. Blood Adv. 2022;6:5403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voskoboinik I, Trapani JA. Perforinopathy: a spectrum of human immune disease caused by defective perforin delivery or function. Front Immunol. 2013;4:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raphael BJ, Dobson JR, Oesper L, Vandin F. Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine. Genome Med. 2014;6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alaggio R, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: lymphoid neoplasms. Leukemia. 2022;36:1720–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jolles S, Chapel H, Litzman J. When to initiate immunoglobulin replacement therapy (IGRT) in antibody deficiency: a practical approach. Clin Exp Immunol. 2017;188:333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seidel MG, Kindle G, Gathmann B, Quinti I, Buckland M, van Montfrans J, et al., ESID Registry Working Party and collaborators. The European Society for Immunodeficiencies (ESID) Registry Working Definitions for the Clinical Diagnosis of Inborn Errors of Immunity. J Allergy Clin Immunol Pract. 2019;7(6):1763–70. [DOI] [PubMed]
- 14.Jolles S, et al. Treating secondary antibody deficiency in patients with haematological malignancy: European expert consensus. Eur J Haematol. 2021;106:439–49. [DOI] [PubMed] [Google Scholar]
- 15.Sánchez-Ramón S, de Gracia J, García-Alonso AM, Rodríguez Molina JJ, Melero J, de Andrés A, et al. EMPATHY group. Multicenter study for the evaluation of the antibody response against salmonella typhi Vi vaccination (EMPATHY) for the diagnosis of Anti-polysaccharide antibody production deficiency in patients with primary immunodeficiency. Clin Immunol. 2016;169:80–4. [DOI] [PubMed] [Google Scholar]
- 16.Ochoa-Grullón J, et al. Evaluation of polysaccharide typhim vi antibody response as a predictor of humoral immunodeficiency in haematological malignancies. Clin Immunol. 2020;210:108307. [DOI] [PubMed]
- 17.Guevara-Hoyer K, et al. Serum free immunoglobulins light chains: a common feature of common variable immunodeficiency? Front Immunol. 2020;11:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerra-Galán T, Palacios-Ortega M, Jiménez-Huete A, Guevara-Hoyer K, Cárdenas MC, Villegas-Mendiola A, et al. An exploratory approach of clinically useful biomarkers of CVID by logistic regression. J Clin Immunol. 2024;44(6):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steyerberg EW, et al. Internal validation of predictive models. J Clin Epidemiol. 2001;54:774–81. [DOI] [PubMed] [Google Scholar]
- 20.Knight AK, Cunningham-Rundles C. Inflammatory and autoimmune complications of common variable immune deficiency. Autoimmun Rev. 2006;5:156–9. [DOI] [PubMed] [Google Scholar]
- 21.Uzzan M, Ko HM, Mehandru S, Cunningham-Rundles C. Gastrointestinal Disorders Associated with Common Variable Immune Deficiency (CVID) and Chronic Granulomatous Disease (CGD). Curr Gastroenterol Rep. 2016;18(4):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Compagno N, Cinetto F, Boscaro E, Semenzato G, Agostini C. Serum free light chains in the differential diagnosis and prognosis of primary and secondary hypogammaglobulinemia: to the editor. J Allergy Clin Immunol. 2015;135:1075-e10776. [DOI] [PubMed] [Google Scholar]
- 23.Scarpa R, Pulvirenti F, Pecoraro A, Vultaggio A, Marasco C, Ria R, et al. Serum Free Light Chains in Common Variable Immunodeficiency Disorders: Role in Differential Diagnosis and Association With Clinical Phenotype. Front Immunol. 2020;11:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ottaviano G, Sgrulletti M, Moschese V. Secondary rituximab-associated versus primary immunodeficiencies: the enigmatic border. Eur J Immunol. 2022;52:1572–80. [DOI] [PubMed] [Google Scholar]
- 25.Ottaviano G, et al. Rituximab Unveils Hypogammaglobulinemia and immunodeficiency in children with Autoimmune Cytopenia. J Allergy Clin Immunol Pract. 2020;8:273–82. [DOI] [PubMed] [Google Scholar]
- 26.Labrosse R, et al. Rituximab-induced hypogammaglobulinemia and infection risk in pediatric patients. J Allergy Clin Immunol. 2021;148:523-e5328. [DOI] [PubMed] [Google Scholar]
- 27.Kano G, Nakatani T, Yagi K, Sakamoto I, Imamura T. Complicated pathophysiology behind rituximab-induced persistent hypogammaglobulinemia. Immunol Lett. 2014;159:76–8. [DOI] [PubMed] [Google Scholar]
- 28.Hauck F, Voss R, Urban C, Seidel MG. Intrinsic and extrinsic causes of malignancies in patients with primary immunodeficiency disorders. J Allergy Clin Immunol. 2018;141:59-e684. [DOI] [PubMed] [Google Scholar]
- 29.Caeser R, et al. Genetic modification of primary human B cells to model high-grade lymphoma. Nat Commun. 2019;10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maffucci P, Filion CA, Boisson B, Itan Y, Shang L, Casanova JL, et al. Genetic Diagnosis Using Whole Exome Sequencing in Common Variable Immunodeficiency. Front Immunol. 2016;7:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ameratunga R, Edwards ESJ, Lehnert K, Leung E, Woon ST, Lea E, Allan C, Chan L, Steele R, Longhurst H. The rapidly expanding genetic spectrum of Common Variable Immunodeficiency–Like disorders. J Allergy Clin Immunol Pr. 2023. 10.1016/j.jaip.2023.01.048. [DOI] [PubMed] [Google Scholar]
- 32.Guevara-Hoyer K, et al. Genomic crossroads between non-hodgkin’s lymphoma and common variable immunodeficiency. Front Immunol. 2022;13:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Supplementary Table 1. Variables studied in the B-CLPD patients’ cohort. (JPG 151 KB)
Supplementary file2 Supplementary Table 2. Genetic variants related to IEIs in PID patients. Variants that can be Autosomic dominant (AD) are represented with *. (JPG 160 KB)
Data Availability Statement
No datasets were generated or analysed during the current study.


