Abstract
Cytomegalovirus gene UL114, a homolog of mammalian uracil-DNA glycosylase (UNG), is required for efficient viral DNA replication. In quiescent fibroblasts, UNG mutant virus replication is delayed for 48 h and follows the virus-induced expression of cellular UNG. In contrast, mutant virus replication proceeds without delay in actively growing fibroblasts that express host cell UNG. In the absence of viral or host cell UNG expression, mutant virus fails to proceed to late-phase DNA replication, characterized by rapid DNA amplification. The data suggest that uracil incorporated early during wild-type viral DNA replication must be removed by virus or host UNG prior to late-phase amplification and encapsidation into progeny virions. The process of uracil incorporation and excision may introduce strand breaks to facilitate the transition from early-phase replication to late-phase amplification.
Uracil incorporation into DNA arises through misincorporation of dUTP by DNA polymerase (2, 51, 54) or from spontaneous deamination of cytosine creating U:G base pair mismatches that resolve into A:T transition mutations upon further rounds of replication (23, 44). To avoid the potential mutagenenic impact of uracil, free-living organisms such as Escherichia coli, yeast, and human beings encode a uracil-DNA glycosylase (UNG) that excises this base from DNA (20, 33, 37, 52). A homolog of the mammalian enzyme is encoded by all poxviruses and herpesviruses, including cytomegaloviruses (CMV), and is a highly conserved in evolution (8, 25, 38, 39, 50). CMV UL114 is the most highly conserved open reading frame in mammalian herpesviruses and retains approximately 40% identity with the major UNG expressed in human cells. Interestingly, although the major form of UNG seems to be dispensable in free-living organisms because of backup uracil-excising activities (3, 6, 35), viral UNG mutants are impaired in their ability to replicate efficiently under certain conditions (8, 25, 38, 39, 50). CMV UNG substitution mutant RC2620 was previously shown to replicate poorly in permissive human fibroblasts (HF cells) due to a delay in viral DNA accumulation (38). This phenotype suggested a specialized role for uracil excision during viral DNA replication.
DNA replication in CMV and other herpesviruses is thought to proceed as a biphasic process (22). Origin-specific initiation on a circularized input genome leads to an early, theta mechanism that later undergoes a switch to a rolling-circle form of replication (1, 16, 17, 22, 40, 48, 49). Rolling-circle replication is responsible for the bulk of viral DNA produced during infection. This switch is believed to be a requisite step in replication, but little is known about the process or how it is regulated (22). Viral DNA replication is a highly recombinagenic process (7, 41, 55), with late replication, in particular, accompanied by the accumulation of complex branched DNA structures (43, 45). Any role for viral UNG in these processes remains poorly understood and is complicated by the fact that the viral enzyme is dispensable for replication of herpes simplex virus type 1 (HSV-1) in cultured cell lines but essential for viral pathogenesis in mice (32). With the exception of hyperthermophilic archaea (13), uracil is itself efficiently incorporated and recognized as a template for the incorporation of adenine by all DNA polymerases. Furthermore, the observations that DNA replication proceeds normally in E. coli, yeast, and mammals even in the absence of UNG (3, 6, 35) demonstrates that excess uracil is not detrimental to either DNA replication or cell viability.
Several models have sought to explain the unique requirement of UNG in viral DNA replication. In poxviruses, where the host UNG cannot substitute for the viral protein due to the cytoplasmic site of viral replication, viral UNG plays an essential role in replication, possibly in association with other proteins of the replication complex (8). In the herpesviruses HSV-1 and CMV, viral UNG is not required for replication. Because of the nuclear site of herpesvirus replication, cellular UNG may substitute for viral UNG (32, 38). Based on the altered in vitro binding characteristics of the HSV-1 origin-binding protein to a uracil-substituted origin of replication, Focher and colleagues proposed that UNG may facilitate removal of uracil before initiation factors bind to this region (11). This work suggested that UNG may play a role early in DNA replication. Alternatively, UNG could function later, during the switch from the theta to the rolling-circle form of replication that amplifies the viral genome. Base excision may contribute to high levels of recombination (7, 41, 55) that generates complex branched DNA structures (43, 45) in infected cells. Uracil excision or some more direct role carried out by UNG might make free ends available for recombination and might facilitate the early-to-late switch in replication. Mammalian UNG is known to interact directly with the replication fork proteins, DNA polymerase α, replication protein A, and proliferating nuclear antigen (36, 42) and to localize with replication complexes in cells (19, 21, 36). Similarly, CMV UNG associates with the viral DNA polymerase accessory protein ppUL44 (M. N. Prichard, unpublished results), raising the possibility that viral UNG may as well be intimately associated with the viral replication machinery during productive infection.
We set out to distinguish between different models and to better understand the functional role of UNG in CMV DNA replication. We identified growth conditions that affect UNG mutant virus replication and have characterized both the uracil content and genomic integrity of the parental laboratory strain and mutant viruses during the infection cycle.
MATERIALS AND METHODS
Cells and virus.
Primary human HF cells and human embryonic lung fibroblasts were maintained in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% NuSerum I (Collaborative Research Inc.), penicillin G (100 U/ml) streptomycin sulfate (100 μg/ml), l-arginine, (0.58 mg/ml), l-glutamine (1.08 mg/ml), and l-asparagine (180 μg/ml). PA317 cells (14) were maintained in medium supplemented with 10% fetal calf serum. Human CMV strain AD169varATCC, here referred to as AD169, was obtained from the American Type Culture Collection and cultured as previously described (26, 47). The recombinant human CMV UNG mutant RC2620, derived from strain AD169varATCC, was described previously (38).
Plasmids.
Plasmid pGEM-3Zf/UNG1A was described previously (32). Plasmid pON2260 contains a 7.36-kbp EcoRI-XhoI fragment from cosmid pCM1007 (10), representing nucleotides 119499 to 126856 of the published AD169 strain sequence (4), inserted between EcoRI and SalI sites of pGEM−3Zf+ (Promega). Plasmid pON2159 contains a 1.78-kbp EcoRI fragment (nucleotides 163071 to 164853 of the AD169 genome) encoding the viral UNG ORF in the MfeI site of pWZLNeo (30).
Infection under serum starvation conditions.
HF cells (3 × 106) were seeded into 90-mm-diameter tissue culture plates. Once monolayers were confluent, culture medium was replaced with medium supplemented with 0.2% NuSerum and maintained under these conditions for 72 h. Following this treatment, HF cells were infected with parental AD169 or mutant RC2620 at a multiplicity of infection (MOI) of 5, using 0.2% NuSerum-supplemented medium. At the indicated times postinfection, infected monolayers were rinsed twice with phosphate-buffered saline, collected by trypsinization, and counted. The cell suspension was pelleted at 1,000 × g in a tabletop centrifuge for 5 min and stored at −20°C until the time course was completed.
PFA inhibition and release.
Following serum starvation as described above, monolayers were infected with AD169 or RC2620 at an MOI of 5 in medium supplemented with 0.2% NuSerum and phosphonoformate (PFA) at 300 μg/ml. Following adsorption for 1 h, the inoculum was replaced and infected cell monolayers were maintained in PFA-containing medium until 72 h postinfection (hpi). At 72 hpi, PFA inhibition was reversed by extensive rinsing followed by addition of PFA-free medium supplemented with 0.2% NuSerum. At various times after reversal, infected cell monolayers were rinsed with phosphate-buffered saline, trypsinized, counted, sedimented by centrifugation, and stored at −20°C until the time course was completed.
Isolation of viral DNA and Southern analysis.
Frozen infected cell pellets were suspended in Tris-EDTA containing 0.5% sodium dodecyl sulfate and 0.5 mg of proteinase K per ml and incubated at 55°C overnight. Viral nucleic acids were subjected to phenol-chloroform extraction with the addition of phase-lock gel (5 Prime→3 Prime) and then ethanol precipitated. Viral DNA from 105 infected cell equivalents was digested with BamHI, separated on a 0.7% agarose gel, transferred to nitrocellulose membranes, and hybridized with the indicated 32P-radiolabeled probe. The results were quantitated by exposure to a PhosphorImager and analyzed using ImageQuant software (Molecular Dynamics). The data were expressed as a ratio of signal intensity at each time point compared to intensity at 24 hpi.
Transcript analysis.
Total cellular RNA was purified using Trizol reagent as recommended by the manufacturer (Gibco BRL). For RNA blot analysis, 10-μg RNA samples were separated by electrophoresis on denaturing formaldehyde–1% agarose gels, transferred to BrightStar-Plus nylon membranes (Ambion), and hybridized with the indicated biotinylated riboprobes as specified by manufacturer (Ambion).
To prepare a specific probe, cellular UNG was PCR amplified from pGEM-3Zf/UNG1A using the primer set UNG F1 (5′ATGATCGGCCAGAAGACG3′) plus T7 UNG R1 (5′TAATACGACTCACTATAGGGATGATATGGATCCTGTCC3′) or UNG F2 (5′CATGGACCTAATCAAGC3′) plus T7 UNG R2 (5′TAATACGACTCACTATAGGGGCTCCTTCCAGTCAATGGG3′). Psoralen-biotin-labeled UNG antisense riboprobes were then generated from the PCR products by in vitro transcription using T7 RNA polymerase as suggested by the manufacturer (Ambion). Bound biotin-labeled probes were detected with streptavidin conjugated with alkaline phosphatase and developed with a chemiluminescent substrate for the enzyme (Ambion).
HL114 cell construction.
PA317 cells were transfected with pON2159 by the calcium phosphate method (5). The transiently produced retrovirus was used to transduce UL114 expression in low-passage primary human embryonic lung fibroblasts (28). Infected cell cultures were selected with 400 μg of Geneticin (G418; Gibco BRL) per ml commencing at 24 hpi and continuing for 10 days.
CMV virion isolation.
HF or HL114 cells were infected with AD169 or RC2620 at an MOI of 0.01. Four days after the cells exhibited 100% cytopathic effect, infected cell supernatants were harvested and cleared of cell debris by centrifugation at 3,300 rpm in a Beckman J2-21C centrifuge for 30 min at 4°C. Virion particles were sedimented by centrifugation at 28,000 rpm using a SW28 rotor in a Beckman ultracentrifuge for 1 h at 4°C, suspended in medium without serum, and stored at −80°C.
Uracil content assessment on alkaline denaturing gels.
Viral DNA was isolated from AD169- or RC2620-infected cells under serum starvation conditions at the indicated times postinfection. For each time point, 2-μg samples of viral DNA in 25 μl of Tris-EDTA were either treated with E. coli 4U UNG (New England Biolabs) or mock treated for 2 h at 37°C prior to the addition of 0.1 M NaOH to cleave alkali-labile, apyrimidinic sites created by uracil excision. The samples were then separated on a 0.5% alkaline denaturing agarose gel and hybridized overnight with nick-translated (Gibco BRL), 32P-radiolabeled CMV AD169 virion DNA. Results were quantitated by exposure to a PhosphorImager and analyzed using ImageQuant software. The average lengths of UNG-treated and mock-treated viral DNA fragments were then compared. Assay sensitivity was determined with UV-irradiated viral DNA as described in the text. The irradiated DNA was then processed as described above except that where indicated, T4 endonuclease V (TEV; New England Biolabs) was used to specifically cleave pyrimidine dimer sites of DNA.
RESULTS
Viral UNG is required for efficient CMV replication in confluent cells.
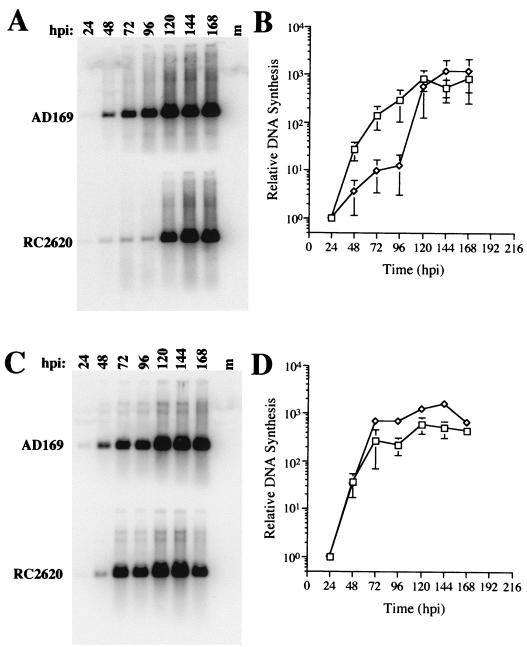
CMV UNG mutant RC2620, carrying a substitution mutation in UL114, was previously shown to lack UNG function and exhibit delayed viral DNA synthesis compared to control virus (38). During the preparation of virus stocks, we observed that the mutant grew slower in confluent HF cell cultures than in subconfluent cultures, suggesting that the requirement for viral UNG was dependent on the growth status of the infected cell. First, we compared the rates of DNA accumulation in control AD169 and mutant RC2620 virus-infected contact-inhibited, serum-starved cells. DNA blot analysis (Fig. 1A and B) revealed that mutant viral DNA levels in confluent, resting cells were lower than control virus levels at 24, 48, 72, and 96 h hpi. Mutant virus DNA levels became equivalent to control virus levels only at 120 hpi and later over the time course of infection. Although mutant viral DNA levels were low over the first 96 hpi, some DNA synthesis was clearly detectable during this period. This observation suggested that the early phase of viral DNA replication had occurred, but transition to late-phase replication had been delayed in the absence of viral UNG.
FIG. 1.
Viral DNA accumulation in parental AD169- and mutant RC2620-infected cells. Shown are DNA blot hybridizations and corresponding PhosphorImager analyses of AD169 (□) and mutant RC2620 (◊) viral DNA accumulating in confluent, serum-starved HF cells (A and B) or subconfluent monolayers (C and D) of HF cells. Blots were probed with 32P-radiolabeled pON2260, specific for CMV nucleotides 122699 to 124902, to assess the level of viral DNA. Error bars (B and D) represent standard deviation of the geometric mean of three replicate samples. m, mock-infected DNA.
When this analysis was undertaken in subconfluent, actively dividing cells, mutant RC2620 viral DNA synthesis proceeded with similar kinetics as the control virus and even reached slightly higher levels (Fig. 1C and D). Thus, the UNG mutant phenotype that was observed so clearly in quiescent cells was not observed in proliferating cells, suggesting that a cellular factor(s) present in these cells complemented viral UNG activity. Previous work has shown that human UNG is activated during the cell cycle (15, 34, 46) and is highly expressed in proliferating cells (15). Based on these observations, we speculated that the human UNG present in actively replicating cells may be responsible for the efficient mutant virus replication.
Complementation of CMV UNG mutant follows induction of human UNG transcript expression.
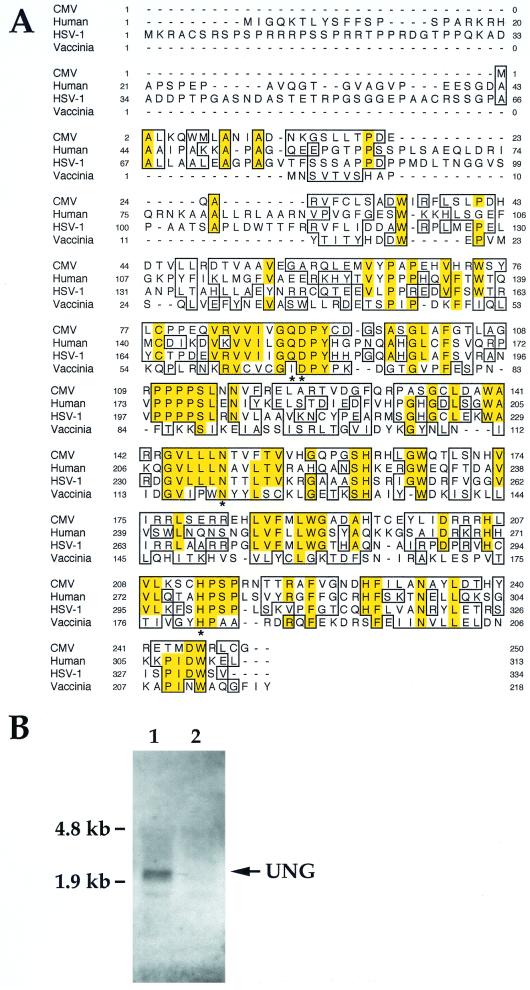
The sequence similarity between human UNG and CMV UL114 extends across all known essential regions (Fig. 2A), consistent with a role for the cellular enzyme in complementing mutant virus growth. To confirm that cellular UNG was expressed differently in proliferating cells compared to confluent cells, we subjected total RNA isolated from cells maintained under both conditions to blot hybridization analysis using a human UNG probe (Fig. 2B). As expected (15), UNG transcript levels were higher in subconfluent, dividing cells than in confluent, serum-starved cells, suggesting a correlation with their ability to support UNG mutant virus replication.
FIG. 2.
(A) Sequence alignment for UNG proteins encoded by viruses (CMV, HSV-1, and vaccinia virus) and humans. Regions of identity are shaded; regions of similarity are shown in blocks. Asterisks denote the residues which are known to abrogate human UNG activity (29). (B) RNA blot of total cellular RNA (10 μg/lane) isolated from uninfected, actively dividing cells (lane 1) and confluent, serum-starved cells (lane 2), hybridized with a biotinylated antisense riboprobe to human UNG. The arrow indicates the position of the UNG transcript; positions of the rRNA subunits are indicated on the left. rRNA was equivalent in all lanes (data not shown).
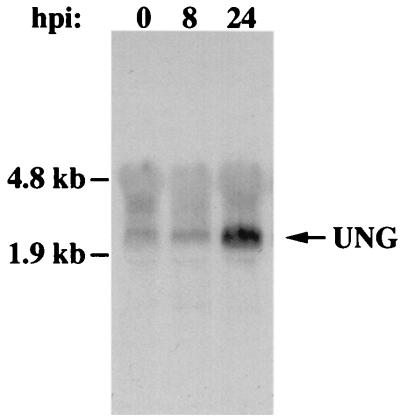
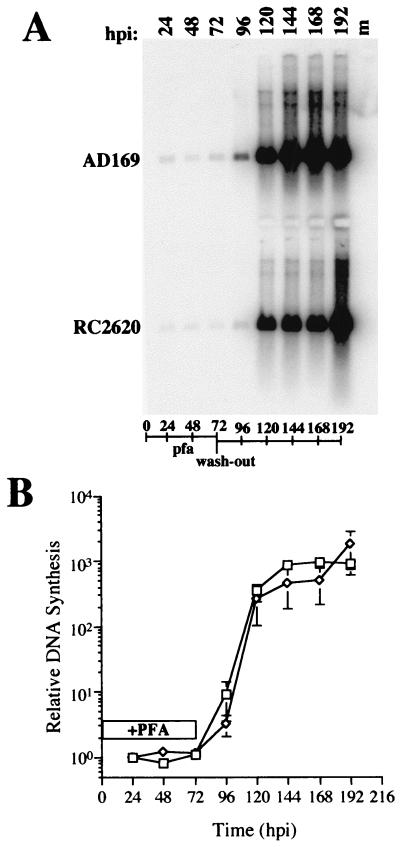
The impact of CMV infection on cellular UNG transcript levels was also investigated in confluent HF cells collected at 0, 8, and 24 hpi. Parental virus was found to exhibit a marked induction of host cell UNG transcript by 24 hpi (Fig. 3) that continued through later times of infection (data not shown). A similar pattern was observed in UNG mutant virus-infected cells (data not shown), indicating that cellular UNG expression was induced at later times following viral infection, independent of viral UNG expression. These results were consistent with the idea that cellular UNG may complement mutant virus and promote the abrupt increase in mutant virus DNA replication between 96 and 120 hpi in confluent cells (Fig. 1A and B). To address this possibility further, we investigated mutant and parental viral DNA accumulation under conditions where viral DNA replication was reversibly inhibited for the initial 72 h of infection. This block would allow expression of CMV-induced host genes, including UNG. If cellular UNG was able to complement mutant virus replication, DNA replication of mutant and parental virus would be expected to rise in parallel when the block was released. Serum-starved HF cells were infected with mutant RC2620 or control AD169 virus and maintained in the presence of the viral DNA polymerase inhibitor PFA for 72 h. At this time, the inhibitor was washed out and infection was allowed to proceed in the absence of drug for an additional 120 h until 192 hpi, when cells were collected for DNA blot analysis with a CMV DNA-specific probe. Autoradiographic and PhosphorImager analyses of this blot are shown in Fig. 4. As expected, neither mutant nor control viral DNA levels changed during the PFA block; however, UNG mutant virus DNA accumulated as rapidly as AD169 following release. Mutant viral DNA accumulated to maximum levels within a 2-day period similar to that observed with parental virus in the absence of any drug block (Fig. 1). The UNG mutant defect was completely reversed under these conditions, consistent with a role for host cell UNG in place of the viral enzyme. Given that proliferating cells support unhindered replication of the UNG mutant and that confluent cells support replication of this mutant only after the UNG transcript was induced, UNG appears to be very important for late phase viral DNA replication.
FIG. 3.
RNA blot analysis of human UNG transcript during CMV infection. Total cellular RNA was isolated from uninfected (0 hpi) and AD169 virus-infected HF cells at 8 and 24 hpi, separated by gel electrophoresis (10 μg/lane), and transferred to a nylon membrane. The blot was hybridized with a biotinylated antisense riboprobe to human UNG; the arrow indicates the position of the cellular UNG transcript. Positions of the rRNA subunits are indicated on the left. rRNA was equivalent in all lanes (data not shown).
FIG. 4.
Viral DNA accumulation in parental AD169 and mutant RC2620-infected cells in the presence of the DNA replication inhibitor PFA and following the reversal of the PFA block at 72 hpi (wash-out). (A) DNA blot of AD169 and mutant RC2620 viral DNA probed with 32P-radiolabeled pON2260. (B) PhosphorImager analysis of AD169 (□) and RC2620 (◊). Duration of PFA treatment is indicated by an open box (+PFA). Error bars represent standard deviation of the geometric mean of three replicate samples. m, mock-infected DNA.
Uracil content in virion DNA.
Our observations suggested that cellular or viral UNG suffices to provide a function that allows viral DNA amplification to proceed at late times in infection. One possible explanation for the prolonged growth cycle of mutant RC2620 was that uracil present in input viral DNA inhibited binding of replication initiation factors to the viral origin of replication, as suggested in another herpesvirus replication system (11). We evaluated the level of uracil incorporation into CMV DNA isolated from parental and mutant progeny virus particles collected from either normal HF or HL114 cell culture supernatants. HL114 cells expressed constitutive levels of viral UNG and were capable of complementing the growth defect of mutant virus (Prichard, unpublished).
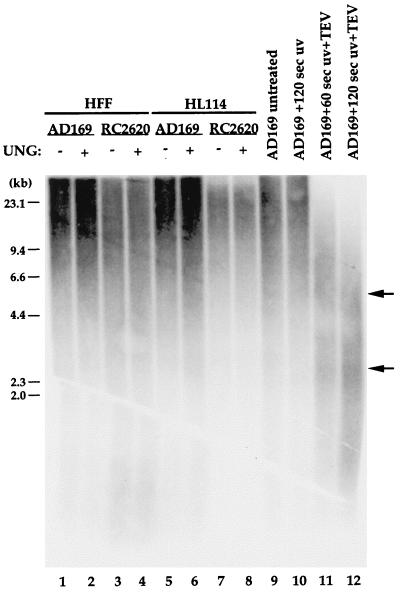
To assess the uracil load in CMV DNA, we collected virions from cells infected at low MOI (0.01), isolated virion DNA, and treated isolated DNA with purified E. coli UNG followed by alkaline hydrolysis to introduce nicks at sites of uracil incorporatation into DNA. The length distribution of DNA fragments arising from UNG treatment was compared to that in untreated DNA samples by alkaline denaturing agarose gel electrophoresis. The results of this assay are shown in Fig. 5 (lanes 1 to 8). We did not observe any detectable differences in the size distribution of parental or mutant viral DNA regardless of whether viruses were propagated on HF or HL114 cells. The majority of UNG-treated viral DNA migrated close to the wells and exhibited a size range of greater than 10 kbp. Virion DNA isolated from mutant RC2620 passaged on HF cells was observed to contain small DNA species (approximately 0.5 kb) in both mock-treated and UNG-treated samples. It is unclear whether these fragments arise as a result of site-specific cleavage; however, we have also observed a similar species in control virion DNA on occasion. As a control, viral DNA samples were UV irradiated at 40 or 80 J/m2, doses that have previously been shown to induce cyclobutane pyrimidine dimers at frequencies of one in 3 kb or one in 6 kb, respectively (18, 24). Subsequent (TEV) treatment of these samples revealed DNA fragments migrating at the expected sizes, which validated the assay sensitivity (Fig. 5, lanes 9 to 12). Comparison of the UNG-treated lanes with the UV-irradiated lanes indicated that uracil is rare in virion DNA, with less than one uracil residue incorporated per 10 kb of DNA (the limit of detection by this assay). The low level of uracil present in mutant DNA argues against the idea that UNG is strictly required to repair misincorporated uracil. This result contrasts somewhat with our previous determination that approximately 0.1% of thymidines in parental virion DNA were substituted with uracil and that there were about threefold-greater levels of uracil in UNG mutant virions (38). Measured either way, levels of uracil in virions were low, and may have been further influenced by the use of a different MOIs or cells for infection. The low amounts of uracil incorporation in infecting virus particles suggests that UNG is not expressed exclusively to repair damaged viral DNA prior to the initiation of early rounds of DNA replication.
FIG. 5.
Uracil load in parental AD169 and mutant RC2620 virus particles. DNA was isolated from AD169 (lanes 1, 2, 5, and 6) and RC2620 (lanes 3, 4, 7, and 8) virions following passage on HF cells (lanes 1 to 4) or CMV UNG-expressing HL114 cells (lanes 5 to 8). Half of the viral DNA from each sample was treated with E. coli UNG to determine uracil levels (lanes 2, 4, 6, and 8). AD169 virion DNA was also treated with UV and TEV as described in Materials and Methods. Reaction products were subjected to alkaline denaturing agarose gel electrophoresis, transferred to a nylon membrane, and probed with 32P-radiolabeled total CMV DNA. The blot is shown overexposed to reveal minor bands. Positions of the molecular weight markers are indicated on the left.
Uracil incorporation and excision during CMV infection.
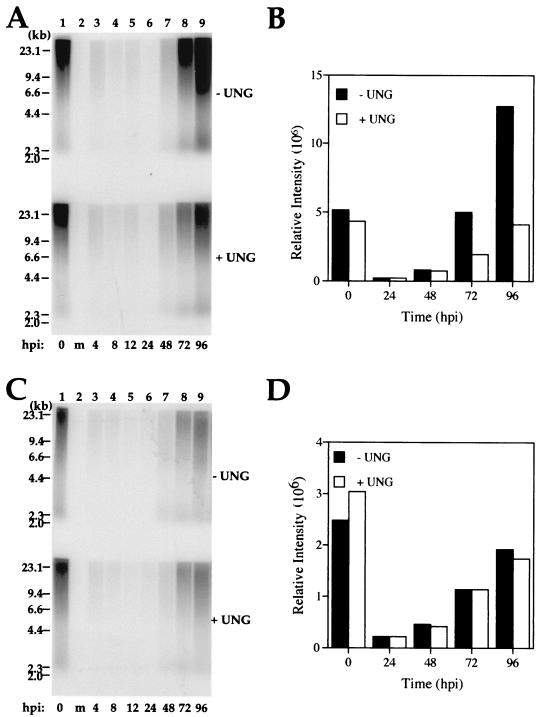
Delayed initiation of UNG mutant viral DNA synthesis at early times of infection was not apparently due to increased uracil content of viral DNA. The behavior of the mutant virus was consistent with a role for UNG-mediated uracil excision in the transition to late-phase viral DNA replication. To determine when UNG exerts its impact on CMV DNA replication, we compared the patterns of uracil incorporation and excision during a time course of mutant RC2620 and control AD169 virus replication in serum-starved cells. Equal amounts of isolated total cellular DNA were denatured and separated on an alkaline denaturing agarose gels and then probed with CMV DNA to determine the size and quantity of viral DNA over the course of the viral replication cycle (Fig. 6). The analysis was performed on DNA either treated with E. coli UNG or left untreated to reveal levels of uracil incorporation. Consistent with the experiments described above, input AD169 DNA exhibited similar patterns of digestion before and after E. coli UNG treatment, confirming that the uracil content of input virus DNA was low (Fig. 6A, lane 1, top and bottom). In the early phase of infection, up to 24 hpi, we observed a gradual loss of viral DNA (Fig. 6A, lanes 3 to 6). Although the total amount of viral DNA present was at the limit of detection at these times, uracil incorporation was not detected. By 72 hpi, when robust DNA amplification had begun in AD169 virus-infected cells, high levels of uracil incorporation were detected and remained detectable through later times (Fig. 6A, lanes 8 and 9; Fig. 6B). Thus, these results demonstrate that although uracil is not abundant in the infecting genome, CMV incorporates significant amounts of uracil beginning at the time of rapid DNA amplification late in infection.
FIG. 6.
Uracil incorporation during parental AD169 and mutant RC2620 virus infection. (A and C) DNA blot analysis of viral DNA isolated from AD169 (A)- and RC2620 (C)-infected HF cells probed with 32P-radiolabeled total viral DNA. Parallel DNA samples from each time point were treated with E. coli UNG (bottom) or left untreated (top). Mock-infected cellular DNA (m) is shown in lane 2 of each blot. Positions of the molecular weight markers are indicated on the left. (B and D) PhosphorImager analyses for selected samples of AD169 (B) and RC2620 (D) viral DNA either treated with E. coli UNG (+ UNG) or left untreated (− UNG) plotted as a function of time.
We next determined whether mutant virus DNA incorporated uracil in a similar manner. We found that input mutant DNA behaved similarly to control virus with or without E. coli UNG treatment (Fig. 6C, lane 1), and we detected a similar gradual loss in the amount of infecting viral DNA up to 24 hpi (Fig. 6C, lanes 3 to 6). During the time that corresponded to the period of rapid viral DNA amplification (48 hpi and later) in the control virus, we observed a much more modest increase of signal with mutant virus (Fig. 6D). The DNA replication in UNG mutant virus-infected cells was not as robust as that in AD169-infected cells at this time, with mutant virus DNA accumulating to approximately 10-fold-lower levels between 24 and 96 hpi (Fig. 1). In contrast to parental virus, we did not detect uracil incorporation in the smaller amounts of mutant viral DNA that was replicated during these times (Fig. 6C, lanes 8 and 9). These results were consistent with an association between rapid DNA amplification and incorporation of uracil that was dependent on UNG expression. Taken together, these results show that the incorporation of uracil into viral DNA late in infection is reduced in the UNG mutant at times when replication fails to keep up with parental virus. This phenotype correlates with a role for UNG in the removal of uracil to promote the transition to efficient late phase genome amplification.
DISCUSSION
Previously, UNG mutant CMV was found to exhibit a prolonged replication cycle that corresponded to a delay in DNA replication (38). Here we show that mutant virus replication is slowed significantly because of a need for UNG to remove uracils from viral DNA prior to or during robust viral DNA amplification that takes place late in replication. UNG mutant and control virus DNA carried similar low levels of uracils at the beginning of infection. Uracil incorporation increased during the transition to the late phase of infection in control virus. High levels of uracil incorporation appear to be part of the rapid DNA amplification process during infection, raising the intriguing possibility that uracil turnover controlled the transition from early- to late-phase CMV DNA replication. The UNG mutant virus appeared to begin DNA replication at the same time as parental virus but failed to make the transition to late phase until levels of induced cellular UNG complemented the defect. The fact that the initial stages of mutant virus replication proceeded normally, but the transition to high-level viral DNA amplification was compromised until host cell UNG was induced, implicates host UNG function in this process. Thus, our studies suggest that virus-induced host UNG may replace viral UNG and provide a key viral replication function in particular cell types during natural infection.
It is thought that herpesvirus DNA replication occurs as a biphasic process involving a theta mechanism early in replication and proceeding to rolling-circle form late in replication during which the bulk of viral DNA is synthesized (1, 16, 17, 22, 40, 48, 49). Much of the information on replication forms during herpesvirus infection has been derived from studies of HSV-1. CMV shares a common set of core replication fork proteins and a similar replication mechanism (22, 27). Although the most highly conserved of the herpesvirus-common functions, UNG has not been ascribed a key role in the herpesvirus replication process. This situation may reflect the fact that studies on most herpesviruses are carried out with malignant cell lines where cellular UNG levels are constitutively high, and thus replication does not rely on the virus-encoded enzyme. An important role for HSV-1 UNG has been suggested by animal studies where mutant virus exhibits an attenuated phenotype for both replication and reactivation from latency in the mouse host (39). These studies are consistent with a role for this enzyme in certain cell types that may be reflected in cultured cells such as HF cells, although this remains to be studied.
Based on in vitro binding studies, Focher and colleagues (11) proposed that HSV-1-UNG-removes uracils from the viral replication origin, allowing better recognition of origin sequences by the initiator protein. In contrast to the predictions of this hypothesis and to earlier studies carried out with UNG mutant RC2620 using high-MOI infection (38), we found that neither parental nor mutant CMV contained significant levels of uracil in virion DNA or in the input DNA that can be detected early during infection. Indeed, the uracil content of either control or UNG mutant virion DNA was very low, representing less than one uracil residue per 10 kb. UNG activity did not appear to be needed for events that lead up to the initiation of replication. The low level of uracil carried into cells with the input viral genome seems to preclude a role for uracils in the early phase of viral DNA synthesis. Uracils incorporated later during replication must be removed prior to packaging viral DNA, and this may be carried out by either viral or induced cellular UNG.
During infection, the uracil content of control and mutant viral DNA remained at similar low levels through 48 hpi. This surprising lack of uracil incorporation in the presence or absence of UNG expression suggests that only the late-phase replication machinery may be affected by this enzyme. We observed a transient increase in uracil incorporation into CMV DNA corresponding to the start of rapid viral DNA amplification that normally occurs at this time. The failure of the viral UNG mutant to enter rapid DNA amplification was associated with a low uracil content. Rapid DNA amplification occurred at a time when significant increases in uracil incorporation can be observed. The fact that the UNG mutant is restricted in its replication at a time when uracil begins to be incorporated into control virus DNA is most consistent with a model where CMV UNG promotes some aspect of the amplification.
The mechanism involved in the transition from early- to late-phase viral DNA replication in any herpesvirus is not known. In the bacteriophage T4 and lambda systems, efficient late-phase DNA amplification occurs following a switch from origin-dependent to recombination-dependent replication (9, 31). Recombination and DNA replication occur at similar times during the herpesvirus life cycle (7, 55), and frequent recombination is thought to produce the complex branched DNA structures that have been observed in studies aimed at resolving HSV-1 replication intermediates (43, 45). These characteristics suggest that the mode of replication seen in the large bacteriophages may occur in herpesviruses as well.
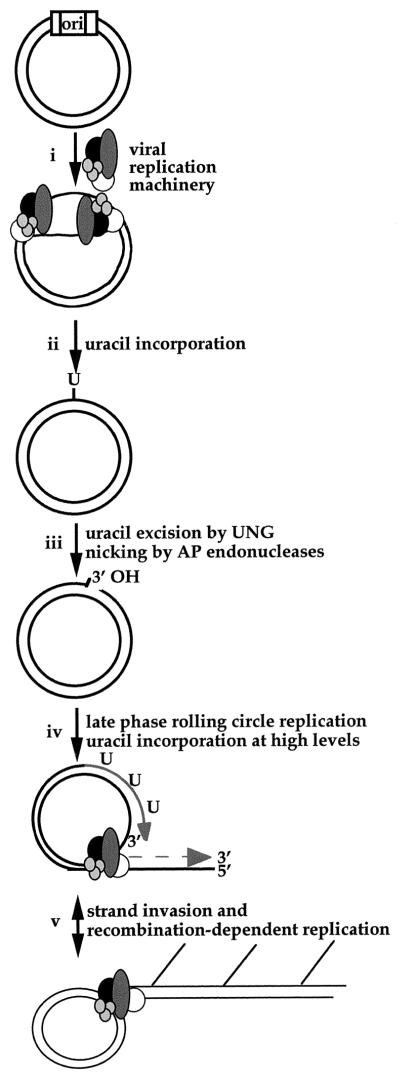
An intriguing possibility suggested by the point of constriction in the CMV UNG mutant replication cycle is that UNG excises uracils from replicating DNA to create sites that serve as substrates for initiation of recombination-dependent replication late in infection (Fig. 7). During the early phase of replication, DNA synthesis has been proposed (22, 27) to initiate in a bidirectional, theta mechanism from an origin of replication (Fig. 7i). During or after the switch to late-phase, rolling-circle DNA replication, incorporated uracils would become substrates for the uracil excision by UNG (Fig. 7ii and iii). This process would lead to free 3′ hydroxyl groups in viral DNA due to cleavage at abasic sites by apurinic/apyrimidinic endonucleases (Fig. 7iii). The induced nicks would occur throughout the genome and would be available to facilitate the transition to a rolling-circle form of DNA replication (Fig. 7iv) in a manner similar to that proposed for bacteriophage lambda (9). UNG-initiated nicks may also serve as substrates for recombination-mediated strand exchange. Strand invasion by single-stranded DNA tails generated by the activities of nucleases or helicases in bacteriophage T4 are thought to promote pathways of recombination-dependent replication during the late-phase DNA amplification in this system (31). By analogy, nicks induced by herpesvirus or mammalian UNG may promote strand invasion and recombination-mediated replication (Fig. 7v), contributing to levels of late-phase amplification. In support of a nicking model of initiation, newly synthesized HSV-1 DNA is known to contain a greater number of fragments than mature virion DNA arising from single-stranded DNA breaks and multiple initiation sites (12, 53). Taken together, these results suggest that nicks generated by UNG may serve a functional role in herpesvirus DNA replication.
FIG. 7.
Model for late-phase CMV DNA replication using uracil excision activity (explained in the text). The viral lytic origin of replication (ori) and sites of single uracil residues (U) are indicated. The viral replication machinery includes DNA polymerase (ppUL54), polymerase accessory protein (ppUL44), helicase-primase complex (ppUL70-ppUL102-ppUL105), and single-stranded DNA binding protein (ppUL57). AP, apurinic/apyrimidic.
Consistent with the possibility that viral UNG is involved in the switch to late-phase DNA amplification, we found that UNG mutant virus was restricted to low levels of DNA synthesis (approximately 10% of the parental level) under suboptimal culture conditions. Robust levels of viral DNA replication were seen only following induction of cellular UNG. Interestingly, bacteriophage λ red mutants replicate DNA at an abnormally low rate and exhibit a decrease in the total amount of phage DNA synthesized (9). The similarity in phenotypes between λ recombination mutants and RC2620 further suggests a link between recombination and late-phase DNA replication in CMV.
Similar mechanisms of late-phase DNA amplification may also be utilized by other DNA viruses. A vaccinia virus UNG mutant is able to replicate DNA at approximately 2% of the levels of control virus and does not transition to high-level, late-phase DNA amplification (25). The cytoplasmic location of poxvirus DNA replication apparently prevents the host UNG from complementing the defect in the way we have observed with CMV. Such studies reinforce a role for UNG in the early to late-phase DNA replication switch during infection by poxviruses as well as herpesviruses. Viral UNG may be particularly important in quiescent or terminally differentiated, nondividing cells encountered by these viruses in the host.
ACKNOWLEDGMENTS
We thank Shinya Watanabe for helpful discussions and A. Louise McCormick and Philip Hanawalt for critical reading of the manuscript. We also thank Ann Ganesan for kindly providing TEV, Sal Caradonna for plasmid pGEM-3Zf/UNG1A, Denise Galloway for the PA317 packaging cell line, and Garry Nolan for the retroviral vector pWZLNeo.
This work was supported by PHS grant AI20211.
REFERENCES
- 1.Ben-Porat T, Tokazewski S A. Replication of herpesvirus DNA. II. Sedimentation characteristics of newly synthesized DNA. Virology. 1977;79:292–301. doi: 10.1016/0042-6822(77)90356-7. [DOI] [PubMed] [Google Scholar]
- 2.Brynolf K, Eliasson R, Reichard P. Formation of Okazaki fragments in polyoma DNA synthesis caused by misincorporation of uracil. Cell. 1978;13:573–580. doi: 10.1016/0092-8674(78)90330-6. [DOI] [PubMed] [Google Scholar]
- 3.Burgers P M, Klein M B. Selection by genetic transformation of a Saccharomyces cerevisiae mutant defective for the nuclear uracil-DNA-glycosylase. J Bacteriol. 1986;166:905–913. doi: 10.1128/jb.166.3.905-913.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A I, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan B K, Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982;151:750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutch R E, Bruckner R C, Mocarski E S, Lehman I R. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J Virol. 1992;66:277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison K S, Peng W, McFadden G. Mutations in active-site residues of the uracil-DNA glycosylase encoded by vaccinia virus are incompatible with virus viability. J Virol. 1996;70:7965–7973. doi: 10.1128/jvi.70.11.7965-7973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enquist L W, Skalka A. Replication of bacteriophage lambda DNA dependent on the function of host and viral genes. I. Interaction of red, gam and rec. J Mol Biol. 1973;75:185–212. doi: 10.1016/0022-2836(73)90016-8. [DOI] [PubMed] [Google Scholar]
- 10.Fleckenstein B, Muller I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 11.Focher F, Verri A, Verzeletti S, Mazzarello P, Spadari S. Uracil in OriS of herpes simplex 1 alters its specific recognition by origin binding protein (OBP): does virus induced uracil-DNA glycosylase play a key role in viral reactivation and replication? Chromosoma. 1992;102:S67–S71. doi: 10.1007/BF02451788. [DOI] [PubMed] [Google Scholar]
- 12.Frenkel N, Roizman B. Separation of the herpesvirus deoxyribonucleic acid duplex into unique fragments and intact strand on sedimentation in alkaline gradients. J Virol. 1972;10:565–572. doi: 10.1128/jvi.10.4.565-572.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greagg M A, Fogg M J, Panayotou G, Evans S J, Connolly B A, Pearl L H. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc Natl Acad Sci USA. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbert C L, Demers G W, Galloway D A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haug T, Skorpen F, Aas P A, Malm V, Skjelbred C, Krokan H E. Regulation of expression of nuclear and mitochondrial forms of human uracil-DNA glycosylase. Nucleic Acids Res. 1998;26:1449–1457. doi: 10.1093/nar/26.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igarashi K, Fawl R, Roller R J, Roizman B. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J Virol. 1993;67:2123–2132. doi: 10.1128/jvi.67.4.2123-2132.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koehler D R, Courcelle J C, Hanawalt P C. Kinetics of pyrimidine (6–4)pyrimidone photoproduct repair in Escherichia coli. J Bacteriol. 1996;178:1347–1350. doi: 10.1128/jb.178.5.1347-1350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krokan H. Preferential association of uracil-DNA glycosylase activity with replicating SV40 minichromosomes. FEBS Lett. 1981;133:89–91. doi: 10.1016/0014-5793(81)80477-2. [DOI] [PubMed] [Google Scholar]
- 20.Krokan H, Haugen A, Myrnes B, Guddal P H. Repair of premutagenic DNA lesions in human fetal tissues: evidence for low levels of O6-methylguanine-DNA methyltransferase and uracil-DNA glycosylase activity in some tissues. Carcinogenesis. 1983;4:1559–1564. doi: 10.1093/carcin/4.12.1559. [DOI] [PubMed] [Google Scholar]
- 21.Lee K A, Sirover M A. Physical association of base excision repair enzymes with parental and replicating DNA in BHK-21 cells. Cancer Res. 1989;49:3037–3044. [PubMed] [Google Scholar]
- 22.Lehman I R, Boehmer P E. Replication of herpes simplex virus DNA. J Biol Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 23.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 24.Mellon I, Hanawalt P C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 25.Millns A K, Carpenter M S, DeLange A M. The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology. 1994;198:504–513. doi: 10.1006/viro.1994.1061. [DOI] [PubMed] [Google Scholar]
- 26.Mocarski E S, Bonyhadi M, Salimi S, McCune J M, Kaneshima H. Human cytomegalovirus in a SCID-hu mouse: thymic epithelial cells are prominent targets of viral replication. Proc Natl Acad Sci USA. 1993;90:104–108. doi: 10.1073/pnas.90.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mocarski E S, Courcelle C T. Cytomegaloviruses and their replication. In: Knipe D M, Howley P M, editors. Fields virology. New York, N.Y: Lippincott-Raven; 2001. pp. 2629–2673. [Google Scholar]
- 28.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mol C D, Arvai A S, Slupphaug G, Kavli B, Alseth I, Krokan H E, Tainer J A. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 30.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 32.Muller-Weeks S, Mastran B, Caradonna S. The nuclear isoform of the highly conserved human uracil-DNA glycosylase is an Mr 36,000 phosphoprotein. J Biol Chem. 1998;273:21909–21917. doi: 10.1074/jbc.273.34.21909. [DOI] [PubMed] [Google Scholar]
- 33.Myrnes B, Giercksky K E, Krokan H. Interindividual variation in the activity of O6-methyl guanine-DNA methyltransferase and uracil-DNA glycosylase in human organs. Carcinogenesis. 1983;4:1565–1568. doi: 10.1093/carcin/4.12.1565. [DOI] [PubMed] [Google Scholar]
- 34.Nagelhus T A, Slupphaug G, Lindmo T, Krokan H E. Cell cycle regulation and subcellular localization of the major human uracil-DNA glycosylase. Exp Cell Res. 1995;220:292–297. doi: 10.1006/excr.1995.1318. [DOI] [PubMed] [Google Scholar]
- 35.Nilsen H, Rosewell I, Robins P, Skjelbred C F, Daly G, Krokan H E, Lindahl T, Barnes D E. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 36.Otterlei M, Warbrick E, Nagelhus T A, Haug T, Slupphaug G, Akbari M, Aas P A, Steinsbekk K, Bakke O, Krokan H E. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Percival K J, Klein M B, Burgers P M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989;264:2593–2598. [PubMed] [Google Scholar]
- 38.Prichard M N, Duke G M, Mocarski E S. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J Virol. 1996;70:3018–3025. doi: 10.1128/jvi.70.5.3018-3025.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pyles R B, Thompson R L. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J Virol. 1994;68:4963–4972. doi: 10.1128/jvi.68.8.4963-4972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizman B, Borman G S, Kamali-Rousta M. Macromolecular synthesis in cells infected with herpes simplex virus. Nature. 1965;206:1374–1375. doi: 10.1038/2061374a0. [DOI] [PubMed] [Google Scholar]
- 41.Sarisky R T, Weber P C. Requirement for double-strand breaks but not for specific DNA sequences in herpes simplex virus type 1 genome isomerization events. J Virol. 1994;68:34–47. doi: 10.1128/jvi.68.1.34-47.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seal G, Sirover M A. Physical association of the human base-excision repair enzyme uracil DNA glycosylase with the 70,000-dalton catalytic subunit of DNA polymerase alpha. Proc Natl Acad Sci USA. 1986;83:7608–7612. doi: 10.1073/pnas.83.20.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Severini A, Scraba D G, Tyrrell D L. Branched structures in the intracellular DNA of herpes simplex virus type 1. J Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapiro R. Damage to DNA caused by hydrolysis. In: Seeburg E, Kleppe K, editors. Chromosome damage and repair. New York, N.Y: Plenum Press; 1981. pp. 3–18. [Google Scholar]
- 45.Shlomai J, Friedmann A, Becker Y. Replication intermediates of herpes simplex virus DNA. Virology. 1976;69:647–659. doi: 10.1016/0042-6822(76)90493-1. [DOI] [PubMed] [Google Scholar]
- 46.Slupphaug G, Olsen L C, Helland D, Aasland R, Krokan H E. Cell cycle regulation and in vitro hybrid arrest analysis of the major human uracil-DNA glycosylase. Nucleic Acids Res. 1991;19:5131–5137. doi: 10.1093/nar/19.19.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spaete R R, Mocarski E S. Regulation of cytomegalovirus gene expression: α and β promoters are trans activated by viral functions in permissive human fibroblasts. J Virol. 1985;56:135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.St Jeor S, Hutt R. Cell DNA replication as a function in the synthesis of human cytomegalovirus. J Gen Virol. 1977;37:65–73. doi: 10.1099/0022-1317-37-1-65. [DOI] [PubMed] [Google Scholar]
- 49.Stinski M F. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J Virol. 1978;26:686–701. doi: 10.1128/jvi.26.3.686-701.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart D T, Upton C, Higman M A, Niles E G, McFadden G. A poxvirus-encoded uracil DNA glycosylase is essential for virus viability. J Virol. 1993;67:2503–2512. doi: 10.1128/jvi.67.5.2503-2512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tye B K, Lehman I R. Excision repair of uracil incorporated in DNA as a result of a defect in dUTPase. J Mol Biol. 1977;117:293–306. doi: 10.1016/0022-2836(77)90128-0. [DOI] [PubMed] [Google Scholar]
- 52.Varshney U, Hutcheon T, van de Sande J H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung) J Biol Chem. 1988;263:7776–7784. [PubMed] [Google Scholar]
- 53.Wilkie N M. The synthesis and substructure of herpesvirus DNA: the distribution of alkali-labile single strand interruptions in HSV-1 DNA. J Gen Virol. 1973;21:453–467. doi: 10.1099/0022-1317-21-3-453. [DOI] [PubMed] [Google Scholar]
- 54.Wist E, Unhjem O, Krokan H. Accumulation of small fragments of DNA in isolated HeLa cell nuclei due to transient incorporation of dUMP. Biochim Biophys Acta. 1978;520:253–270. doi: 10.1016/0005-2787(78)90225-3. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]