Abstract
Replication-competent adenoviruses are being investigated as potential anticancer agents. Exclusive virus replication in cancer cells has been proposed as a safety trait to be considered in the design of oncolytic adenoviruses. From this perspective, we have investigated several adenovirus mutants for their potential to conditionally replicate and promote the killing of cells expressing human papillomavirus (HPV) E6 and E7 oncoproteins, which are present in a high percentage of anogenital cancers. For this purpose, we have employed an organotypic model of human stratified squamous epithelium derived from primary keratinocytes that have been engineered to express HPV-18 oncoproteins stably. We show that, whereas wild-type adenovirus promotes a widespread cytopathic effect in all infected cells, E1A- and E1A/E1B-deleted adenoviruses cause no deleterious effect regardless of the coexpression of HPV18 E6E7. An adenovirus deleted in the CR2 domain of E1A, necessary for binding to the pRB family of pocket proteins, shows no selectivity of replication as it efficiently kills all normal and E6E7-expressing keratinocytes. Finally, an adenovirus mutant deleted in the CR1 and CR2 domains of E1A exhibits preferential replication and cell killing in HPV E6E7-expressing cultures. We conclude that the organotypic keratinocyte culture represents a distinct model to evaluate adenovirus selectivity and that, based on this model, further modifications of the adenovirus genome are required to restrict adenovirus replication to tumor cells.
Among human cancers, one of the most distinctive features of anogenital carcinomas is its virological nature. Numerous reports have established the causal association of human papillomavirus (HPV) infections with squamous carcinomas and adenocarcinomas of the anogenital tract (9, 49). Over 95% of cervical tumors contain integrated HPV sequences that belong to the oncogenic HPV group (HPV16, -18, and related types) (49) and consistently express papillomavirus early 6 and 7 oncogenes (38, 42). E6 and E7 oncoproteins interact with the master cell cycle regulatory proteins, p53 and pRB, respectively (17, 36), and are able to immortalize primary keratinocytes in vitro and cause cell transformation in cooperation with other oncoproteins, such as activated Ras (34, 37, 44). Furthermore, continuous expression of E6 and E7 oncoproteins is necessary for the maintenance of the transformed phenotype. These properties have made E6 and E7 targets of various experimental therapeutics, including vaccination-, antisense RNA-, and ribozyme-based approaches (4, 11, 41).
Although a virus-based therapeutic approach for the treatment of cancer is not a novel concept, recent years have witnessed with increasing attention the design of new oncolytic adenoviruses (for a review, see reference 1). Since the discovery of adenovirus in 1953, the knowledge about adenovirus biology and the interaction with its hosts has helped identify the need to introduce safety traits, such as the restriction of adenovirus replication to tumor cells. In this regard, two major approaches have been developed to achieve tumor-specific replication, namely, controlling adenovirus early gene expression by a tumor-specific antigen promoter (2, 35, 53) and introducing viral genomic deletions that affect viral protein functions dispensable in cancer cells (8, 18). Despite the conceptual validity of these approaches, demonstrating replication selectivity has proven to be difficult due, in part, to the incomplete knowledge of all viral protein functions and to the lack of appropriate models to study adenovirus replication in a physiological setting.
Based on functional similarities described for certain HPV and adenovirus proteins, we believe that a window of opportunity exists in the design of replication-selective adenovirus for HPV-associated neoplasias. In particular, both oncogenic papillomaviruses and adenoviruses have evolved similar mechanisms to usurp the cell cycle regulation in order to facilitate viral DNA replication (15, 39). They do so by targeting the same cell cycle regulators, pRB and related pocket proteins, p107 and p130, through the adenovirus E1A and HPV E7 proteins (12). E1A is the first transcription unit to be expressed upon adenovirus infection of cells in culture and is required for cell cycle mobilization and transactivation of other virus early promoters. The E1A polypeptides encoded by the 12S and 13S mRNAs share sequence homology with the HPV E7 protein in the conserved regions (CR) 1 and 2, the same domains that bind and inactivate pRB and related pocket proteins (16). Most importantly, E1A 12S and HPV E7 can transactivate the adenovirus E2 promoter in reporter assays in vitro (7, 32, 33), as well as cellular genes involved in S-phase entry (48), whose promoters contain E2F binding sites. This is mediated by the release of the pRB-histone deacetylase complexes from transcription factor E2F recruited to the promoters (10). However, neither HPV E7 nor E1A 12S can transactivate adenovirus E3 and E4 promoters (32), for which the CR3 domain present only in the E1A 13S product is required. Efficient promoter activation by the 13S product requires cooperation between the CR1 and CR3 domains, since E1A mutants containing large CR1 deletions, affecting binding to both pRB and p300, activate viral early promoters poorly (50). In this context, the HPV16 E7 protein has been shown to restore the transcriptional activity of CR1 deletion adenovirus mutants (51). Therefore, it is conceivable that certain replication-defective adenovirus mutants may be complemented by HPV oncogenes, thus restricting their replication to HPV-positive cancer cells.
In the present report, we have studied the replication of several E1 mutants in organotypic cultures of primary human keratinocytes (hereafter referred to as raft cultures). In the absence of a relevant animal model to study human adenovirus replication, we chose to perform this study in the raft system rather than in submerged cultures, because (i) commonly employed submerged cultures may have different properties from epithelial tissues in vivo (30), (ii) the ability of adenovirus to replicate varies upon the epithelial tissue origin and differentiation stage (5), (iii) the raft culture is a close representation of the tissue architecture found in the cervix (14), and (iv) we can take advantage of the squamous differentiation-dependent HPV biology to investigate adenovirus replication in cells that do or do not express E6E7 genes simultaneously in one tissue.
To recapitulate the differentiation-dependent expression of HPV18 E6E7 oncogenes in this model, we have employed a recombinant retrovirus carrying an HPV18 E6E7 expression cassette under the control of the native HPV18 enhancer and E6 promoter located in the upstream regulatory region (URR) (13). Under physiological conditions, this promoter is repressed in the basal and parabasal strata of a fully stratified epithelium but is upregulated upon squamous differentiation into spinous and granular cells (30). Thus, E6E7 expression occurs in differentiated cells as opposed to basal cells such that E7 reestablishes S phase in a subset of postmitotic keratinocytes (13). By using this model, replication of various adenoviruses can be examined in normal proliferating basal or parabasal cells, in postmitotic, differentiated cells, and in differentiated cells expressing HPV18 E6 and E7. Here we describe the pattern of replication of several adenovirus E1 mutants in the raft culture system.
MATERIALS AND METHODS
Adenoviruses.
With the exception of AdΔ24 (18), which was grown in A549 cells, Adwt300 (25), Addl312 (40), and Adβgal (3) were grown in 293 cells and purified by CsCl gradient centrifugation as previously described (19). To minimize the expansion of E1A-positive recombinants that could generate from growth in 293 cells, Addl312, Adβgal, and CB016 viruses were never propagated over four passages. CB016 virus was generated as follows. A derivative of plasmid pXC1 (containing the left 5,766 bp of adenovirus 5 [Ad 5]) (Microbix, Hamilton, Ontario, Canada) was made by religation after digestion with XbaI and NdeI. Then, a deletion spanning amino acids 27 to 80 of E1A was introduced by using an oligonucleotide (5′-CAG CTG ATC GA GAG CTC ACT TTT CCG CCG-3′) flanking the deleted sequence as instructed in the Transformer site-directed mutagenesis kit (Clontech, Palo Alto, Calif.). An EcoRI-BspEI fragment was recovered from the deleted plasmid and cloned in the same sites in pXC1-Δ24 shuttle vector containing a deletion from amino acids 122 to 129 of E1A (18). The resulting plasmid (pCB016) was cotransfected with pBHG10 (Microbix, Hamilton, Ontario, Canada) in 293 cells. Plaques were expanded and viral DNA was extracted by a spermine-based method and sequenced. A plaque showing the correct E1A sequence was further expanded for CsCl banding. Virus stocks were titered by plaque assay in 293 cells.
Recombinant retroviruses and raft cultures.
Organotypic raft cultures of neonatal foreskin keratinocytes were essentially prepared as described elsewhere (30). Recombinant Moloney murine leukemia retroviruses were used to transduce HPV18 E7 or E6E7 into the raft cultures and have been described elsewhere (13). These vectors expressed the neomycin resistance gene from the simian virus 40 promoter, and the HPV genes from the native HPV URR. Retroviruses were generated in the Am12 amphotrophic packaging cell line, and culture supernatant was used to infect keratinocytes for 4 h. The next day, selection of transduced cells with 250 μg of G418 (Life Technologies, Inc., Rockville, Md.) per ml was conducted for 2 days. The antibiotic was removed, and the cells were allowed to recover for another 2 to 4 days. Prior to being transferred onto dermal equivalents, keratinocytes were infected with adenovirus as follows. To calculate the adenovirus dose accurately, the keratinocytes transduced by HPV-containing retroviruses or normal keratinocytes were plated at the same density in multiple wells. On the day of infection, one well was trypsinized and the cell number was determined. The other wells were each exposed for 4 h to adenoviruses at a multiplicity of infection (MOI) of 1 PFU/cell in serum-free keratinocyte medium. Upon transfer onto dermal equivalents, a small cell suspension aliquot was placed in an eight-well chamber slide for adenovirus E1A immunostaining (Adwt300, AdΔ24, and CB016) or X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining (Adβgal) 24 h later. These assays revealed that about 90% of keratinocytes had been infected. Thus, there appeared to be a higher percentage of E1A-positive cells than expected from MOI of 1 based on virus titers determined in 293 cells. Raft cultures were allowed to differentiate for the time indicated in the text, and bromodeoxyuridine (BrdU) was added to a final concentration of 50 μg/ml for the last 12 h of culture. The cultures were then harvested by a 3-h formalin fixation and paraffin embedding. Sections (4 μm) were stained with hematoxylin and eosin (H-E) according to standard procedures. To detect β-galactosidase activity, cultures were fixed in 2% formaldehyde–0.2% glutaraldehyde for 1 h and then incubated with X-Gal (1 mg/ml) in phosphate-buffered saline (PBS) containing 5 mM potassium ferricyanide–5 mM potassium ferrocyanide–2 mM MgCl2 overnight at 37°C before being paraffin-embedded.
Immunoprecipitation and Western blot.
SiHa cells grown in 10-cm culture dishes were infected with Adwt300 or CB016 at an MOI of 20. At 15 h after infection, cells were scraped off the plates, and cell pellets were lysed in 50 mM Tris (pH 8.0), 5 mM EDTA, 0.1% Triton X-100, and 250 mM NaCl for 30 min on ice. Lysates were cleared by centrifugation and then immunoprecipitated with 0.5 μg of anti-E1A mouse monoclonal antibody M73 (Oncogene Research, Boston, Mass.), plus 20 μl of protein A/G-agarose (Santa Cruz Biotech, Santa Cruz, Calif.) for 3 h at 4°C. Immunoprecipitates were washed three times in cold lysis buffer and resuspended in electrophoresis sample buffer. Sample aliquots were electrophoresed in 7.5 or 12% acrylamide gels, transferred to polyvinylidene difluoride membranes, and immunoblotted with either anti-E1A M73 monoclonal antibody (Oncogene Research), mouse anti-Rb monoclonal antibody, rabbit anti-p107 polyclonal antibody, or rabbit anti-p300 polyclonal antibody (Santa Cruz Biotech) in Tris-buffered saline (TBS; 20 mM Tris [pH 7.6], 137 mM NaCl) containing 0.2% Tween 20 at a concentration of 1 μg/ml for 1 h at room temperature (RT). Membranes were washed three times in TBS–0.1% Tween 20 and incubated with the appropriate peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin antibodies (Amersham Pharmacia, Piscataway, N.J.) at a 1:200 dilution in TBS–0.2% Tween 20 for 1 h at RT. Membranes were washed and developed by enhanced chemiluminescence (Amersham).
Immunofluorescence.
For antigen retrieval, sections were deparaffinized, rehydrated, and treated with 10 mM citrate buffer (pH 6.0) at 95°C for 10 min. For double detection of BrdU and E1A, antibody reactivity to E1A was first revealed with the anti-E1A mouse monoclonal antibody M73 (Oncogene) at a final concentration of 2 μg/ml, followed by Alexa 594-coupled goat anti-mouse secondary antibody (Molecular Probes, Eugene, Oreg.) at a 1:200 dilution. A final incubation was performed with fluorescein isothiocyanate (FITC)-labeled anti-BrdU monoclonal antibody (1 μg/ml) (Boehringer Mannheim, Indianapolis, Ind.). For hexon staining, sections were incubated with goat polyclonal anti-Ad2 hexon antibody (Chemicon, Temecula, Calif.) at a 1:300 dilution, followed by incubation with Alexa 588-conjugated donkey anti-goat secondary antibody (Molecular Probes) at a 1:200 dilution. All sections were mounted with Gel/Mount (Biomeda). The photomicrographs in Fig. 3 and 6 were captured with either a Texas red or FITC filter in an Olympus IX70 inverted fluorescence microscope. Individual images were merged by means of the Adobe Photoshop 5.5 application program.
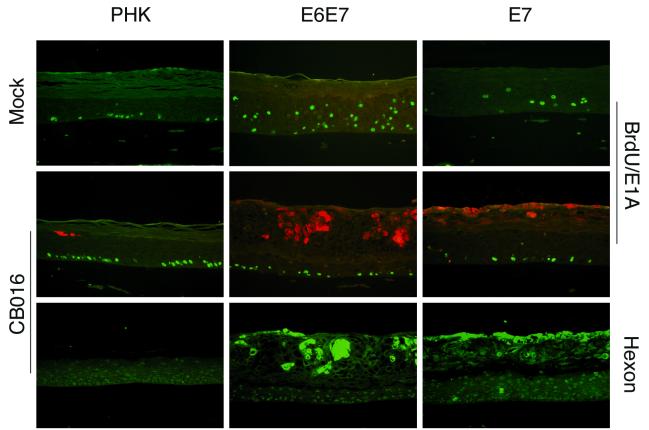
FIG. 3.
Immunofluorescence analysis of adenovirus early and late proteins in day 10 CB016-infected normal (PHK) and HPV18 E6E7- and HPV18 E7-transduced raft cultures (same experiment as in Fig. 2). BrdU and hexon are revealed by green fluorescence, while E1A is revealed by red fluorescence. BrdU and E1A were detected by double immunofluorescence on the same section. Hexon immunofluorescence corresponds to the same field of a serial section. Magnification, ×200.
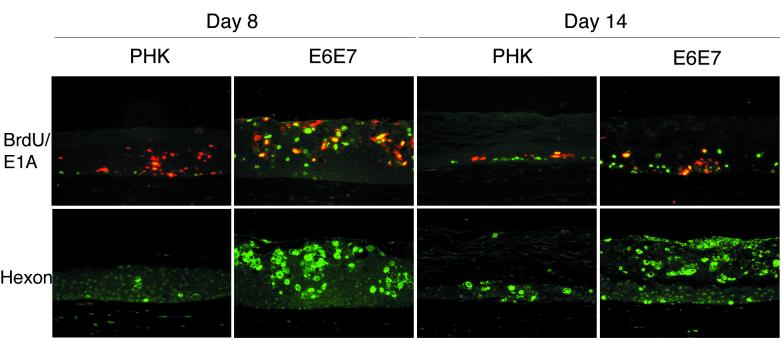
FIG. 6.
Time course of adenovirus infection in CB016-infected normal (PHK) and HPV18 E6E7-transduced raft cultures (same experiment as in Fig. 5). Immunofluorescence staining for BrdU, E1A, and hexon was performed as for Fig. 3. Colocalization of BrdU (green) and E1A (red) in the same cell yields a yellow fluorescence signal. Magnification, ×200.
Quantification of adenovirus genome copies.
Raft cultures were infected with adenovirus at an MOI of 0.1 and processed on day 8 of culture. Raft cultures were washed once with PBS and separated from the dermal equivalent by gently peeling off the epithelium. Total genomic DNA was purified from these cultures by using the DNeasy Qiagen kit, and recovered in a volume of 200 μl. Real-time PCR (LightCycler; Roche) was performed with oligonucleotides to the E4 region. The sequences of the primers used were as follows: forward E4 primer, 5′-TGACACGCATACTCGGAGCTA-3′; reverse E4 primer, 5′-TTTGAGCAGCACCTTGCATT-3′; and probe, 5′-AAGCTTGTTGCATGGGCGGCG-3′. For quantification, a standard curve with a known number of viral DNA copies from 0 to 108 spiked into genomic DNA from a mock-infected raft culture was used. In parallel, the amount of genomic DNA per raft was quantified based on the determination of actin gene copies. The primers sequences were as follows: forward actin primer, 5′-CAGCAGATGTGGATCAGCAAG-3′; reverse actin primer, 5′-CTAGAAGCATTTGCGGTGGAC-3′; and probe, 5′-AGGAGTATGACGAGTCCGGCCCCTC-3′. A standard curve was performed with known amounts of human genomic DNA (Clontech) from 200 to 0 ng.
RESULTS
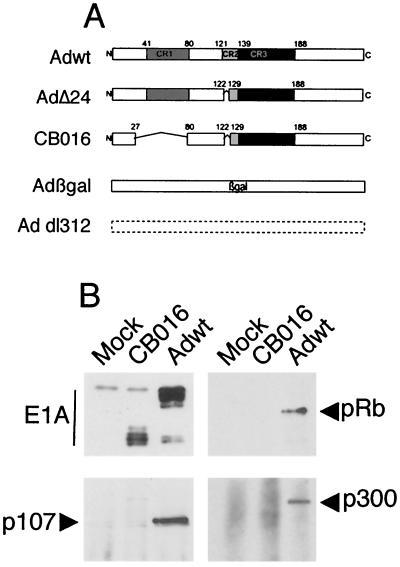
Figure 1A depicts the E1A region of the adenoviruses used in this study. With the exception of Adβgal, which carries a deletion of the entire E1 region, the remaining adenoviruses contain total or partial deletions of E1A, while retaining E1B. Adβgal harbors a substitution of E1A and E1B with a β-galactosidase expression cassette under the control of the cytomegalovirus promoter (3). Addl312 has a complete deletion of E1A from nucleotides 448 through 1349 of Ad5 (40). AdΔ24 is deleted of E1A amino acids 122 to 129 within the CR2 domain (18). CB016 harbors two deletions of amino acids 27 to 80 and 122 to 129, affecting the CR1 and CR2 domains, respectively. As demonstrated by immunoprecipitation of E1A and E1A-bound proteins from infected SiHa cells, CB016 expresses a truncated set of E1A products that are unable to bind p300, pRB, and p107, in contrast to wild-type E1A (Fig. 1B).
FIG. 1.
(A) Structure of the E1A regions of the adenovirus used in this study (for clarity, the E1A 13S-derived polypeptide is shown). (B) Immunoprecipitation of E1A and E1A-bound cellular proteins. SiHa cells were mock infected or infected with CB016 or Adwt (20 PFU/cell), and E1A was immunoprecipitated in nondenaturing conditions. E1A immunoblotting reveals a truncated set of E1A protein species for CB016 as opposed to Adwt; coprecipitation of pRB, p300, and p107 by CB016 E1A is abrogated.
To investigate the outcome of raft culture infection with the adenovirus mutants described above, primary human foreskin keratinocytes were isolated and transduced with a retroviral vector expressing HPV18 oncoproteins, as described in Materials and Methods. Normal (untransduced) primary human keratinocytes and E6E7-transduced keratinocytes were infected with the various adenoviruses at an MOI of 1 PFU/cell. The cells were then transferred onto a dermal equivalent and allowed to differentiate at the air-medium interface for 10 days. To monitor E7-mediated reactivation of S phase, cultures were labeled with BrdU 12 h prior to harvesting by fixation in formalin. Cultures were paraffin embedded and sectioned for histological and biochemical analyses. Figure 2 shows the histology of control raft cultures and cultures infected with retroviruses, adenoviruses, or both.
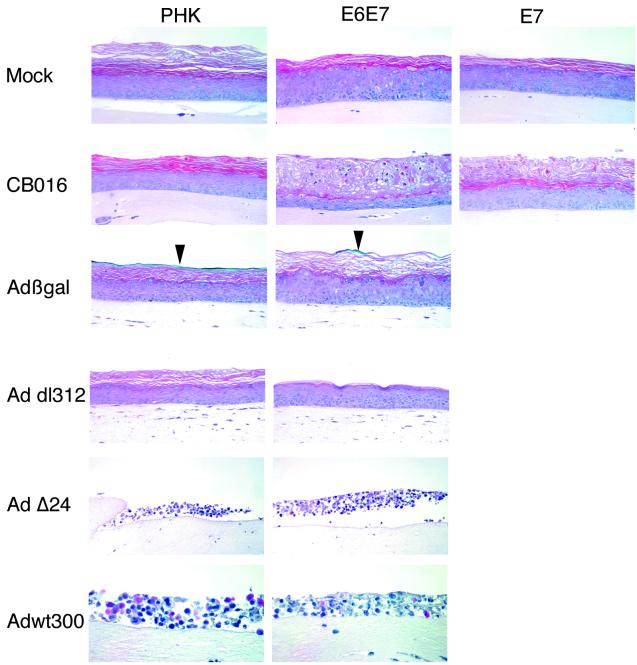
FIG. 2.
Effects of adenovirus infection in raft cultures of normal PHK and HPV18 E6E7- and E7-expressing keratinocytes. H-E staining of day 10 raft culture sections is shown. Untransduced keratinocytes and keratinocytes acutely transduced with retrovirus expressing HPV18 E6E7 or E7 from the native HPV18 URR were infected with the indicated adenoviruses at an MOI of 1 prior to being transferred to dermal equivalents (see Materials and Methods) and allowed to stratify for 10 days. Adβgal-infected cultures were stained with X-Gal prior to H-E staining. Blue X-Gal staining is indicated by arrowheads. Magnification, ×200 (except Adwt [×400]).
Wild-type Ad5 induces a widespread cytopathic effect in all infected cultures.
Adwt300 infection caused a widespread cytopathic effect in all the cultures with or without HPV18 E6E7 (Fig. 2). The epithelium appeared disorganized, and condensed cell nuclei were abundant. The mere existence of a dead epithelium indicated that replication of Adwt300 did not completely prevent epithelium stratification in the early phases of raft culture development or, alternatively, that Adwt requires epithelium stratification and differentiation to give rise to a productive infection.
Adenovirus mutants harboring entire deletions of E1A or E1A-E1B have no effect on raft culture development, regardless of HPV E6E7 expression.
Albeit generally considered replication deficient, E1A and E1A-E1B-deleted adenoviruses have been shown to replicate in certain conditions, such as in cells with E1A-like functions (26, 40, 43). To test the hypothesis that such functions may be provided by the HPV18 oncoproteins, we infected normal and E6E7-transduced raft cultures with these adenovirus mutants (Fig. 2). Neither Addl312 nor Adβgal infection induced any cytopathic effect in the infected raft cultures, regardless of the expression of E6E7. To substantiate Adβgal infection, we monitored the expression of the β-galactosidase transgene by X-Gal staining. In uninfected raft cultures, no β-galactosidase activity was detected (not shown), as reported previously (30). In the Adβgal-infected culture, it was detected only in the enucleated outer squames (Fig. 2). We believe that these residual signals correspond to the expression of β-galactosidase in cells that did not divide extensively before transiting to the surface and undergoing terminal differentiation. In contrast, cells that divided many times before differentiation would have diluted adenovirus genomes and diluted β-galactosidase activity that would be too low to detect. Indeed, we did not detect Adβgal genomes by in situ hybridization with hexon-directed DNA probes in the normal or E6E7 raft cultures (data not shown). Taken together, these results indicate that at the MOI of 1 PFU/cell used in these experiments, adenoviruses deleted of the entire E1A region cannot be complemented for replication in E6E7-expressing raft cultures.
An adenovirus mutant harboring a deletion of the pRB-binding domain of E1A exhibits no selectivity of replication.
Since the presence of the entire E1A provides no selectivity of replication and a complete deletion of E1A cannot be complemented by HPV18 E6E7 in the raft cultures, we hypothesized that smaller deletions affecting distinct functional domains may confer a restricted replication which can be complemented by E6E7. AdΔ24, which has a deletion of amino acids 122 to 129 in E1A CR2, has been proposed as a selective oncolytic agent in the treatment of gliomas and other cancers (18, 20). We therefore studied whether this small deletion could confer a more HPV-specific phenotype in the raft cultures. Upon infection at an MOI of 1 PFU/cell, H-E staining of the resulting raft culture revealed a widespread cytopathic effect similar to the one induced by Adwt300 in normal and in HPV E6E7-expressing cultures (Fig. 2), indicating lack of replication selectivity.
CB016 preferentially replicates in HPV18 E6E7 expressing cultures.
The fact that the CR2 deletion in AdΔ24 was not sufficient to confer selective replication prompted us to delete additional E1A functions that could be complemented by HPV proteins. Transcomplementation of E1A amino terminal deletions by HPV16 E7 has been described (51). This deletion affects the CR1 domain, and therefore the binding to pRB and p300 but not to p107 and p130, a function that resides in the CR2 domain. Because E7 can also bind to these pocket proteins, we hypothesized that CR1 and CR2 are both dispensable for adenovirus replication in the presence of HPV E7. Consequently, we generated adenovirus mutant CB016 (Fig. 1) and investigated its behavior in the keratinocyte raft culture.
In contrast to Adwt300, AdΔ24, Addl312, and Adβgal, CB016 showed distinct patterns of replication in the normal versus the E6E7-transduced raft cultures. In normal cultures, CB016 produced virtually no cytopathic effect in the main central portion of the cultures (Fig. 2) except some localized effect in the edge of the cultures (not shown). However, in E6E7-transduced cultures, cytotoxicity was widespread and confined to the differentiated strata normally comprising spinous and granular cells. These areas appeared disorganized and contained many enucleated cells. Some remaining nuclei appeared condensed and pycnotic. The basal and parabasal strata were spared from this cytopathic effect, in agreement with the spatial distribution of E6E7 expression (24).
To ascertain that the histopathology observed in CB016-infected raft cultures was attributed to adenovirus replication and progeny production, we performed immunofluorescence staining to detect BrdU, adenovirus E1A protein, and adenovirus late protein hexon (Fig. 3). In mock-infected normal cultures, incorporation of BrdU was confined exclusively to the basal or parabasal proliferating cells. In normal CB016-infected cultures, E1A and hexon expression was scarce, with the exception of the margin of the cultures in which it was more pronounced (not shown). In uninfected but E6E7-transduced cultures, incorporation of BrdU was detected in a subset of both basal and suprabasal cells, as described previously (24). In CB016-infected, E6E7-transduced raft cultures, adenovirus E1A and hexon proteins were localized within the suprabasal lesion previously identified by hematoxylin-eosin staining. Furthermore, DNA in situ hybridization of the adenovirus genome confirmed the presence of adenovirus DNA in the remaining nuclei of the cells within that area, which also stained positive for hexon (not shown). However, only cells in the basal or parabasal strata incorporated BrdU. We conclude that the adenovirus had specifically replicated in and killed the E6E7-expressing differentiated suprabasal cells prior to exposure to BrdU. Taken together, these results indicate that CB016 replication is complemented in HPV18 E6E7-expressing keratinocytes.
We also analyzed the effect of HPV18 E7 expression alone (13) on the replication of CB016. Primary human keratinocytes transduced with a retrovirus expressing HPV18 E7 from the HPV18 URR and then infected with CB016 also exhibited signs of adenovirus-induced cytopathic effect in the upper strata (Fig. 2 and 3). However, this effect was less pronounced than in the E6E7-transduced cultures. Not only the basal or parabasal but also the majority of the spinous cells were spared. Whether this more selective cytopathic effect is due to a lower level of E7 expression remains to be determined. However, it has been a consistent observation that a higher percentage of the differentiated cells reenter S phase in the E6E7-transduced cultures than those transduced by E7 alone (28) (also see Fig. 3). Alternatively, a possible role of E6 in complementation of CB016 replication cannot be ruled out since E6 has been recently shown to bind the transcriptional coactivator p300 (31, 54), which is also a target of adenovirus E1A.
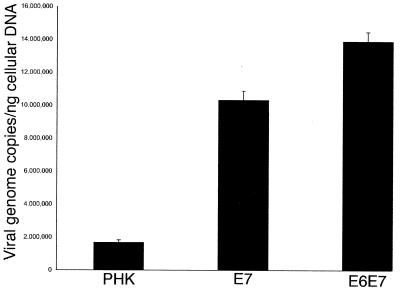
To quantify the E6E7-dependent differences in adenovirus replication, we performed real-time PCR of the adenovirus genome from total genomic (viral plus cellular) DNA isolated from whole infected rafts. For this particular experiment, and in order to avoid massive virus-induced cytotoxicity which could mask the differences (see Fig. 5), raft cultures were infected with adenovirus at an MOI of 0.1 and processed on day 8 of culture. Figure 4 represents the number of normalized copies of adenovirus genomes per nanogram of cellular genomic DNA. E6E7- and E7-expressing cultures contained approximately eight and six times more adenovirus genomes, respectively, than normal primary human keratinocytes (PHKs). Thus, the calculated viral genome copy numbers correlated well with the observed cytopathic effect assessed by histology.
FIG. 5.
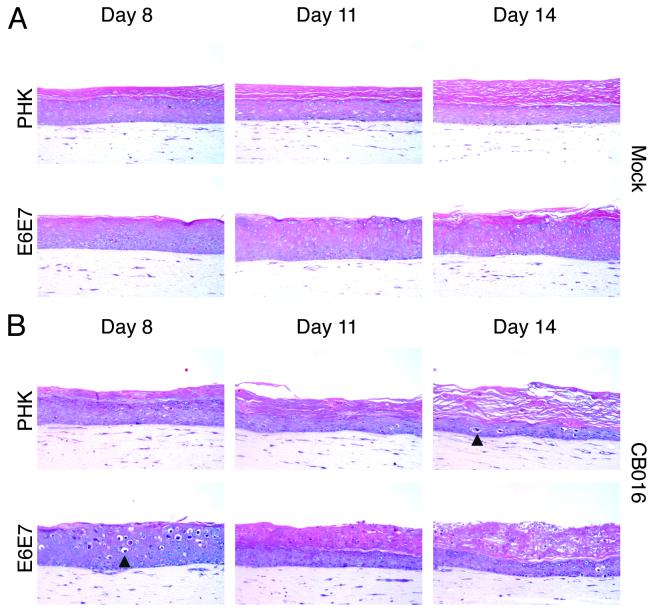
Time course of adenovirus infection in CB016-infected normal (PHK) and HPV18 E6E7-transduced raft cultures. (A) H-E staining of day 8, day 11, and day 14 uninfected control culture sections. (B) H-E staining of day 8, day 11, and day 14 CB016-infected culture sections. Arrowheads point to representative cells showing ballooning degeneration secondary to adenovirus replication. Magnification, ×200.
FIG. 4.
Quantification of adenoviral genomes in CB016-infected raft cultures. The numbers of adenovirus genome copies are expressed per nanogram of cellular genomic DNA. The error bars represent the standard deviation of the assay.
CB016 replicates in normal raft cultures with a delayed kinetics relative to HPV18 E6E7-expressing cultures.
Finally, we examined the temporal replication of CB016 in the raft cultures. For this purpose, three CB016-infected normal or E6E7-transduced culture replicas were harvested at days 8, 11, and 14 postinfection. Uninfected raft cultures were harvested and morphologically evaluated on the same time points. As shown in Fig. 5A, the uninfected normal cultures revealed a time-dependent thinning of the epithelium, along with increased orthokeratosis. We believe this is a consequence of the gradual decrease of the keratinocyte proliferating capacity. In contrast, uninfected E6E7-transduced cultures maintained a constant epithelium thickness, with only slight orthokeratosis. We suggest that this is due to a delay in terminal differentiation or programmed cell death brought about by the expression of E6E7 oncoproteins.
When assessing CB016-infected normal cultures (Fig. 5B), we found that day 8 cultures displayed a properly stratified and differentiated epithelium with no apparent virus-induced toxicity. By days 11 and 14, the infected epithelia became increasingly thinner as in the uninfected controls. By day 14, however, some spinous cells showed an evident cytopathic effect characterized by ballooning degeneration (condensed cell nuclei and vacuolated cytoplasm). In contrast to normal cultures, E6E7-transduced, CB016-infected cultures (Fig. 5B) showed cytopathic effects as early as day 8. At this time point, a fully differentiated epithelium was observed and revealed scattered suprabasal cells with morphological evidence of ballooning. Strikingly, by day 11, the epithelium suffered a dramatic morphological change. The lower half of the epithelium remained well stratified and differentiated, while the upper half had apparently become heavily keratinized yet still retaining nuclear structures. This histopathology remained almost unchanged through day 14, although an increase in ballooned cells was observed in the lower strata. We conclude that this histology is the result of adenovirus-induced cell death.
To monitor the expression of early and late adenovirus proteins over time, an immunofluorescence analysis was performed (Fig. 6). This analysis revealed the presence of scattered E1A-positive cells across day 8 normal raft cultures, with little concomitant hexon expression. In contrast, day 8 E6E7-transduced raft cultures showed large areas of E1A expression in the upper strata. Colocalization of BrdU and E1A appeared in the majority of E1A-positive cells, indicating that the viral and/or cellular DNA was being actively replicated at the time of BrdU labeling. Hexon-positive cells were concomitantly detected in the same area. On day 14, normal raft cultures showed the presence of basal proliferating BrdU-positive cells along with some E1A-hexon-positive basal and suprabasal cells. In E6E7-transduced raft cultures, E1A and BrdU-positive cells were confined to the lower strata. Hexon signal, however, was mostly concentrated in the lesions developed in the upper layers. Taken together, these results indicate that productive infection of CB016 in the E6E7-transduced cultures initially occured in the differentiated strata where HPV oncogenes are expressed. Adenovirus-induced cell death appears to have precluded terminal differentiation. With time, infection by local high concentration of newly released virus may have spread to the lower strata. Our results also suggest that CB016 replication is not completely abrogated in normal keratinocytes, resulting in a delayed and reduced cytopathic effect.
DISCUSSION
Adenovirus-mediated oncolysis is a rapidly growing field with the potential for conceptualization of new agents (1). So far, two distinct strategies have been described to achieve tumor-specific adenovirus replication. The first utilizes transcriptional control of early viral genes with tumor-specific antigen promoters. The second incorporates genomic deletions that affect functions dispensable in cancer cells. We have searched for a new strategy to restrict adenovirus replication to HPV-positive cells, on the basis of the functional complementation of replication-defective Ad mutants by HPV oncoproteins. Based on the cellular proteins that they bind to, as well as the mechanisms involved in mobilizing the cell cycle or preventing cellular apoptosis, functional homologues for E6 and E7 have been identified in the adenovirus as products of the E1B and E1A transcriptional units, respectively. We have taken advantage of these similarities to study the potential of several adenovirus mutants as selective cytolytic agents.
One of the main issues that the field of adenovirus-mediated oncolysis faces is the lack of experimental animal models to study selectivity of replication and toxicity in normal human tissues, especially those surrounding the target tumors. An alternative to this deficiency has been the employment of cultured cell lines and primary cultures. However, these cultures may not display a well-differentiated phenotype and only provide a bidirectional mode of adenovirus spread. For this reason, the use of organotypic cultures of human keratinocytes to investigate the replication selectivity of adenoviruses may provide information on virus-associated toxicity in a three-dimensional setting. This fact, coupled to the multiple differentiated cell phenotypes found in a stratified squamous epithelium, which can closely regulate adenovirus replication, makes the raft cultures system unique in the assessment of adenovirus mode of replication and propagation.
The HPV URR shows a differentiation-dependent pattern of activation (47). Interestingly, the cytolytic pattern generated by Adwt infection, with the appearance of a pseudostratified epithelium, indicates that adenovirus replication also depends on tissue differentiation. Earlier reports have suggested that adenovirus replication depends on keratinocyte differentiation (6, 47). One possible explanation is that the activation of viral early promoters, such as E1, may be dependent on keratinocyte differentiation, as is the case for papillomaviruses. In this regard, the E1 regulatory region has been reported to contain enhancers that are differentiation dependent (21). In addition, preliminary results indeed suggest that the E1A promoter becomes transcriptionally active upon keratinocyte differentiation in upper spinous cells (F. Noya et al., unpublished results).
The deletion of the CR2 domain found in AdΔ24 (or mutant dl922) has been found to confer selectivity on the basis of its reduced replication in growth-arrested versus proliferating primary epithelial cells and cancer cell lines (18, 20). In our model, this mutant exhibits a behavior similar to the Adwt and is cytotoxic to normal raft cultures even at a low MOI. This observation suggests that, in this system, the inability to bind pRB and related pocket proteins can be somehow compensated for by other adenoviral proteins. In this regard, an adenovirus mutant containing a similar CR2 deletion has been shown to initiate cellular DNA synthesis, by mechanisms presumably involving binding to p300, a function that is preserved in AdΔ24 (22). More recently, the E4 open reading frame 6 or 7 protein has been shown to bind pRB and induce virus DNA replication and cell cycling (29). Therefore, adenovirus has alternative pathways to facilitate viral and cellular DNA replication in normal raft cultures. The discrepancy with the previous published reports may be due to the distinct culture systems and certain characteristics that can only be revealed in organotypic cultures.
In contrast, we have not detected replication of adenovirus mutant dl312 or Adβgal in this model in the absence or in the presence of E6E7. Although several studies have shown that E1A deletion mutants can be transcriptionally activated by cellular proteins with E1A-like functions (26, 43) and that HPV E6 and E7 share functional similarities to adenovirus E1B and E1A, our study shows that these adenoviral functions cannot be provided by cellular proteins or by HPV oncoproteins in the raft culture system. Preliminary experiments performed at an MOI of 10 PFU/cell have confirmed this observation (data not shown). However, infection at a 100 MOI caused a deterioration of both normal and HPV E6E7-expressing epithelia. We believe this is due to uncontrolled early adenovirus gene expression and replication, which can occur at high MOI (27, 40, 52).
Therefore, among the E1A mutants studied, only a CR1-CR2 double-deleted E1A adenovirus mutant can preferentially replicate in and kill HPV18 E6E7-expressing keratinocytes. The pattern of viral DNA replication, late protein expression, and virus-associated cell toxicity correlates with HPV E6E7 expression. Of note is the time-dependent onset of cytopathicity associated with CB016 infection. Ballooning degeneration observed in day 8 E6E7-expressing cultures represents an early cytopathic effect that occurs after complete epithelium stratification and differentiation. The absence of this phenotype in the day 8 normal cultures indicates complementation of CB016 by HPV18 E6E7. Interestingly, this kind of virus-induced histopathology has also been reported in keratinocyte raft cultures as a result of herpes virus type 1 infection (46). In contrast, later time points revealed a morphologically abnormal, E6E7-transduced epithelium in which adenovirus hexon protein, and presumably whole adenovirions, are abundant. We believe this may be the result of massive adenovirus replication and lateral spread, which in turn causes cell death along with acute morphological changes.
A recent report has established that the expression of the coxsackie-adenovirus receptor (CAR) correlates with the undifferentiated state of oropharyngeal keratinocytes, being detected in the basal strata but undetectable in the suprabasal layers (23). This suggests that adenovirus would require the infection of basal cells, which are normally unexposed and inaccessible, in order to give rise to a productive infection in a stratified squamous epithelium. In the present study, adenovirus infection was performed in submerged keratinocyte cultures, a mostly undifferentiated cell population, prior to stratification and differentiation. We speculate that after the infection of basal undifferentiated cells, the adenovirus genome is carried over across the culture as cells stratify. Upon cell differentiation, the adenovirus E1A gene is transactivated by specific cellular factors and adenovirus replication takes place. In the case of Adwt, replication is unrestricted in normal cultures, resulting in profound epithelial disorganization and widespread tissue disintegration. As for CB016, despite E1A transactivation in differentiated cells, the particular deletions introduced in this gene significantly reduce the efficiency of replication in normal raft cultures. In contrast, in E6E7-transduced cultures, cell differentiation also results in activation of E6E7 transcription, a condition promoting efficient CB016 replication. Because of the reported absence of CAR in the differentiated strata, we speculate that other receptors may contribute to the lateral spread of the virus in the upper layers. Nevertheless, the distribution of CAR in our model, especially in cells expressing E6E7 oncoproteins, warrants further investigation.
Our results show that, despite the preferential replication of CB016 in E6E7 expressing keratinocytes, replication is not completely abrogated in normal cultures. Rather, this virus has a slow replication kinetics and low virus yield that culminates in evident cytopathicity on day-14 normal cultures. The finding that the margin of the cultures frequently display virus-induced cytopathicity may reflect that, relative to cells away from the growing margin, these cells have distinct growth properties that affect virus replication.
Collectively, our observations suggest that careful modifications of the adenovirus genome are warranted to further restrict replication to HPV E6E7 cells. In this regard, other genomic deletions affecting E4 ORF6 and ORF7, whose protein has recently been shown to interact with pRB (29), could be combined with the CB016 deletions. Additionally, controlling transcription of viral genes with exogenous promoters may add a different level of selectivity. It is possible, however, that extensive modifications of viral transcription and/or viral protein functions could alter the oncolytic ability or potency of the resulting virus. Therefore, a compromise between safety (selectivity) and efficacy (potency) will be necessary to achieve the highest therapeutic index. Another line of improvement that could be superimposed on CB016 is at the level of receptor binding, either to restrict infection to tumor cells or to broaden its tropism and avoid the resistance of tumors that have lost CAR expression (45).
In summary, complementation of adenovirus replication by viral oncoproteins responsible for HPV-induced carcinogenesis in combination with the study of adenovirus replication in a human organotypic model are novel additions to the field of conditionally replicative adenoviruses with broad therapeutic implications. An imminent therapeutic application for an adenovirus mutant, such as CB016, would be as an oncolytic agent in HPV-associated cancer. Given the role of HPV E6 and E7 proteins in the productive phase of HPV infections, the range of therapeutic applications may be extended to include HPV-associated premalignant lesions (i.e., dysplasias) and benign lesions induced by oncogenic and nononcogenic HPV types. Further studies on the complementation for other adenovirus mutants or by oncoproteins of other HPV types are warranted.
ACKNOWLEDGMENTS
We thank Brenda Gossage for excellent technical assistance in the large-scale preparation of the adenoviruses used in this study and Ge Jin for paraffin-embedding and sectioning of the raft cultures. We are also grateful to Gene Siegal for histological examination of the sections.
This work was supported by the U.S. Department of Defense (PC 970193 and PC 991018), NIH grant CA83821, grant CA86881-01, the CapCure Foundation, NIH grant CA36200, and grant DE/CA11910. We thank Thomas Broker for use of the Digital Imaging Microscopy Facility, which was established with funds provided in large measure by the UAB Health Services Foundation and by grant DE/CA11910.
REFERENCES
- 1.Alemany R, Balagué C, Curiel D T. Replicative adenoviruses for cancer therapy. Nat Biotechnol. 2000;18:723–727. doi: 10.1038/77283. [DOI] [PubMed] [Google Scholar]
- 2.Alemany R, Lai S, Lou Y C, Jan H Y, Fang X, Zhang W W. Complementary adenoviral vectors for oncolysis. Cancer Gene Ther. 1999;6:21–25. doi: 10.1038/sj.cgt.7700001. [DOI] [PubMed] [Google Scholar]
- 3.Alemany R, Ruan S, Kataoka M, Koch P E, Mukhopadhyay T, Cristiano R J, Roth J A, Zhang W W. Growth inhibitory effect of anti-K-ras adenovirus on lung cancer cells. Cancer Gene Ther. 1996;3:296–301. [PubMed] [Google Scholar]
- 4.Alvarez-Salas L M, Cullinan A E, Siwkowski A, Hampel A, DiPaolo J A. Inhibition of HPV-16 E6/E7 immortalization of normal keratinocytes by hairpin ribozymes. Proc Natl Acad Sci USA. 1998;95:1189–1194. doi: 10.1073/pnas.95.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aneskievich B J, Taichman L B. Epithelium-specific response of cultured keratinocytes to infection with adenovirus type 2. J Investig Dermatol. 1988;91:309–314. doi: 10.1111/1523-1747.ep12475641. [DOI] [PubMed] [Google Scholar]
- 6.Aneskievich B J, Taichman L B. Evidence for two points of restriction in the expression of adenovirus type 2 in cultured epidermal keratinocytes. J Virol. 1988;62:4365–4368. doi: 10.1128/jvi.62.11.4365-4368.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bagchi S, Raychaudhuri P, Nevins J R. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A trans-activation. Cell. 1990;62:659–669. doi: 10.1016/0092-8674(90)90112-r. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 9.Bosch F X, Manos M M, Muñoz N, Sherman M, Jansen A M, Peto J, Schiffman M H, Moreno V, Kurman R, Shah K V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 10.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 11.Breitburd F, Coursaget P. Human papillomavirus vaccines. Semin Cancer Biol. 1999;9:431–444. doi: 10.1006/scbi.1999.0147. [DOI] [PubMed] [Google Scholar]
- 12.Chellappan S, Kraus V B, Kroger B, Münger K, Howley P M, Phelps W C, Nevins J R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng S, Schmidt-Grimminger D C, Murant T, Broker T R, Chow L T. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 1995;9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 14.Chow L T, Broker T R. In vitro experimental systems for HPV: epithelial raft cultures for investigations of viral reproduction and pathogenesis and for genetic analyses of viral proteins and regulatory sequences. Clin Dermatol. 1997;15:217–227. doi: 10.1016/s0738-081x(97)00069-2. [DOI] [PubMed] [Google Scholar]
- 15.Chow L T, Broker T R. Small DNA tumor viruses. In: Nathanson N, editor. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 267–301. [Google Scholar]
- 16.Dyson N, Guida P, Münger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyson N, Howley P M, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 18.Fueyo J, Gomez-Manzano C, Alemany R, Lee P S, McDonnell T J, Mitlianga P, Shi Y X, Levin V A, Yung W K, Kyritsis A P. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene. 2000;19:2–12. doi: 10.1038/sj.onc.1203251. [DOI] [PubMed] [Google Scholar]
- 19.Graham F L, Prevec L. Manipulation of adenovirus vectors. In: Murray E J, editor. Methods in molecular biology. Vol. 7. Clifton, N.J: The Humana Press; 1991. pp. 109–128. [DOI] [PubMed] [Google Scholar]
- 20.Heise C, Hermiston T, Johnson L, Brooks G, Sampson-Johannes A, Williams A, Hawkins L, Kirn D. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat Med. 2000;6:1134–1139. doi: 10.1038/80474. [DOI] [PubMed] [Google Scholar]
- 21.Herbst R S, Pelletier M, Boczko E M, Babiss L E. The state of cellular differentiation determines the activity of the adenovirus E1A enhancer element: evidence for negative regulation of enhancer function. J Virol. 1990;64:161–172. doi: 10.1128/jvi.64.1.161-172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howe J A, Bayley S T. Effects of Ad5 E1A mutant viruses on the cell cycle in relation to the binding of cellular proteins including the retinoblastoma protein and cyclin A. Virology. 1992;186:15–24. doi: 10.1016/0042-6822(92)90057-v. [DOI] [PubMed] [Google Scholar]
- 23.Hutchin M E, Pickles R J, Yarbrough W G. Efficiency of adenovirus-mediated gene transfer to oropharyngeal epithelial cells correlates with cellular differentiation and human coxsackie and adenovirus receptor expression. Hum Gene Ther. 2000;11:2365–2375. doi: 10.1089/104303400750038471. [DOI] [PubMed] [Google Scholar]
- 24.Jian Y, Van Tine B A, Chien W M, Shaw G M, Broker T R, Chow L T. Concordant induction of cyclin E and p21cip1 in differentiated keratinocytes by the human papillomavirus E7 protein inhibits cellular and viral DNA synthesis. Cell Growth Differ. 1999;10:101–111. [PubMed] [Google Scholar]
- 25.Jones N, Shenk T. Isolation of deletion and substitution mutants of adenovirus type 5. Cell. 1978;13:181–188. doi: 10.1016/0092-8674(78)90148-4. [DOI] [PubMed] [Google Scholar]
- 26.La Thangue N B, Rigby P W. An adenovirus E1A-like transcription factor is regulated during the differentiation of murine embryonal carcinoma stem cells. Cell. 1987;49:507–513. doi: 10.1016/0092-8674(87)90453-3. [DOI] [PubMed] [Google Scholar]
- 27.Lieber A, He C Y, Kirillova I, Kay M A. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J Virol. 1996;70:8944–8960. doi: 10.1128/jvi.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noya F, Chien W-M, Broker T R, Chow L T. p21cip1 degradation in differentiated keratinocytes is abrogated by costabilization with cyclin E induced by human papillomavirus E7. J Virol. 2001;75:6121–6134. doi: 10.1128/JVI.75.13.6121-6134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connor R J, Hearing P. The E4–6/7 protein functionally compensates for the loss of E1A expression in adenovirus infection. J Virol. 2000;74:5819–5824. doi: 10.1128/jvi.74.13.5819-5824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker J N, Zhao W, Askins K J, Broker T R, Chow L T. Mutational analyses of differentiation-dependent human papillomavirus type 18 enhancer elements in epithelial raft cultures of neonatal foreskin keratinocytes. Cell Growth Differ. 1997;8:751–762. [PubMed] [Google Scholar]
- 31.Patel D, Huang S M, Baglia L A, McCance D J. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phelps W C, Bagchi S, Barnes J A, Raychaudhuri P, Kraus V, Munger K, Howley P M, Nevins J R. Analysis of trans activation by human papillomavirus type 16 E7 and adenovirus 12S E1A suggests a common mechanism. J Virol. 1991;65:6922–6930. doi: 10.1128/jvi.65.12.6922-6930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phelps W C, Yee C L, Münger K, Howley P M. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell. 1988;53:539–547. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 34.Pirisi L, Yasumoto S, Feller M, Doniger J, Dipaolo J A. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. J Virol. 1987;61:1061–1066. doi: 10.1128/jvi.61.4.1061-1066.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez R, Schuur E R, Lim H Y, Henderson G A, Simons J W, Henderson D R. Prostate attenuated replication competent adenovirus (ARCA) CN706: a selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 1997;57:2559–2563. [PubMed] [Google Scholar]
- 36.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 37.Schlegel R, Phelps W C, Zhang Y L, Barbosa M. Quantitative keratinocyte assay detects two biological activities of human papillomavirus DNA and identifies viral types associated with cervical carcinoma. EMBO J. 1988;7:3181–3187. doi: 10.1002/j.1460-2075.1988.tb03185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 39.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 40.Shenk T, Jones N, Colby W, Fowlkes D. Functional analysis of adenovirus-5 host-range deletion mutants defective for transformation of rat embryo cells. Cold Spring Harbor Symp Quant Biol. 1979;44:367–375. doi: 10.1101/sqb.1980.044.01.041. [DOI] [PubMed] [Google Scholar]
- 41.Shillitoe E J, Kamath P, Chen Z. Papillomaviruses as targets for cancer gene therapy. Cancer Gene Ther. 1994;1:193–204. [PubMed] [Google Scholar]
- 42.Smotkin D, Wettstein F O. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc Natl Acad Sci USA. 1986;83:4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spergel J M, Chen-Kiang S. Interleukin 6 enhances a cellular activity that functionally substitutes for E1A protein in transactivation. Proc Natl Acad Sci USA. 1991;88:6472–6476. doi: 10.1073/pnas.88.15.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storey A, Pim D, Murray A, Osborn K, Banks L, Crawford L. Comparison of the in vitro transforming activities of human papillomavirus types. EMBO J. 1988;7:1815–1820. doi: 10.1002/j.1460-2075.1988.tb03013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki K, Fueyo J, Krasnykh V, Reynolds P N, Curiel D T, Alemany R. A conditionally replicative adenovirus with enhances infectivity shows improved oncolytic potency. Clin Cancer Res. 2001;7:120–126. [PubMed] [Google Scholar]
- 46.Syrjanen S, Mikola H, Nykanen M, Hukkanen V. In vitro establishment of lytic and nonproductive infection by herpes simplex virus type 1 in three-dimensional keratinocyte culture. J Virol. 1996;70:6524–6528. doi: 10.1128/jvi.70.9.6524-6528.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taichman L B, Reilly S S, LaPorta R F. The role of keratinocyte differentiation in the expression of epitheliotropic viruses. J Investig Dermatol. 1983;81:137S–140S. doi: 10.1111/1523-1747.ep12540909. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi Y, Rayman J B, Dynlacht B D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 49.Walboomers J M, Jacobs M V, Manos M M, Bosch F X, Kummer J A, Shah K V, Snijders P J, Peto J, Meijer C J, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 50.Wong H K, Ziff E B. Complementary functions of E1A conserved region 1 cooperate with conserved region 3 to activate adenovirus serotype 5 early promoters. J Virol. 1994;68:4910–4920. doi: 10.1128/jvi.68.8.4910-4920.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong H K, Ziff E B. The human papillomavirus type 16 E7 protein complements adenovirus type 5 E1A amino-terminus-dependent transactivation of adenovirus type 5 early genes and increases ATF and Oct-1 DNA binding activity. J Virol. 1996;70:332–340. doi: 10.1128/jvi.70.1.332-340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y, Nunes F A, Berencsi K, Gonczol E, Engelhardt J F, Wilson J M. Inactivation of E2a in recombinant adenoviruses improves the prospect for gene therapy in cystic fibrosis. Nat Genet. 1994;7:362–369. doi: 10.1038/ng0794-362. [DOI] [PubMed] [Google Scholar]
- 53.Yu D C, Sakamoto G T, Henderson D R. Identification of the transcriptional regulatory sequences of human kallikrein 2 and their use in the construction of calydon virus 764, an attenuated replication competent adenovirus for prostate cancer therapy. Cancer Res. 1999;59:1498–1504. [PubMed] [Google Scholar]
- 54.Zimmermann H, Degenkolbe R, Bernard H U, O'Connor M J. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73:6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]