Abstract
Measles virus (MV) strain CAM/RB, which was adapted to growth in the brain of newborn rodents, is highly neurovirulent. It has been reported earlier that experimentally selected virus variants escaping from the monoclonal antibodies (MAbs) Nc32 and L77 to hemagglutinin (H) preserved their neurovirulence, whereas mutants escaping MAbs K71 and K29 were found to be strongly attenuated (U. G. Liebert et al., J. Virol. 68:1486–1493, 1994). To investigate the molecular basis of these findings, we have generated a panel of recombinant MVs expressing the H protein from CAM/RB and introduced the amino acid substitutions thought to be responsible for antibody escape and/or neurovirulence. Using these recombinant viruses, we identified the amino acid changes conferring escape from the MAbs L77 (377R→Q and 378M→K), Nc32 (388G→S), K71 (492E→K and 550S→P), and K29 (535E→G). When the corresponding recombinant viruses were tested in brains of newborn rodents, we found that the mutations mediating antibody escape did not confer differential neurovirulence. In contrast, however, replacement of two different amino acids, at positions 195G→R and 200S→N, which had been described for the escape mutant set, caused the change in neurovirulence. Thus, antibody escape and neurovirulence appear not to be associated with the same structural alterations of the MV H protein.
Among the morbilliviruses, measles virus (MV) is associated with an intermediate capacity to cause neurological complications. These include the acute postinfectious measles encephalitis, which develops 2 to 4 weeks after infection, or the late complications, measles inclusion body encephalitis in immunocompromised patients and subacute sclerosing panencephalitis (SSPE), which develops months to years after the initial infection, based on a persistent MV infection (reviewed in reference 3). In late stages of SSPE, massive amounts of MV antigen can be detected in inclusion bodies in various neural cell types (1). SSPE is characterized by a restriction of the viral envelope protein expression as a consequence of mutational, transcriptional, and translational alterations (1, 5). An additional constraint is exerted by the high concentration of antiviral antibodies present in the cerebrospinal fluid of SSPE patients. Tissue culture experiments demonstrated that virus-neutralizing antibodies downregulate not only viral gene expression but also transcription and can completely suppress viral replication (2, 39). Similar results have been obtained in vivo using Lewis rats (22, 38).
Suckling rodents have successfully been used as animal models (predominantly mice and rats) for different forms of MV-induced encephalitis (21, 23, 38). Transgenic mice which express CD46, one of the MV receptors (7, 27), have also been used to induce MV-induced encephalitis (15, 26, 31). However, for development of the acute encephalitis following infection of suckling rats with the rodent-adapted MV strain CAM/RB, or mice with the HNT (hamster neurotropic) strain, the transgenic expression of receptors such as CD46 appears not to be necessary (23, 24, 32, 35). After intracerebral infection with CAM/RB (RB indicates passage in rat brain), 1- to 14-day-old Lewis rats develop a lethal acute measles encephalitis, whereas older animals develop a subacute measles encephalitis (23).
Antiviral antibodies may lead to a restriction of the viral gene expression but also to the selection of escape variants. When monoclonal antibodies (MAbs) are used experimentally to select escape variants, resulting viruses with altered hemagglutinin (H) protein structures might induce differential pathogenicity in animals. This was observed with escape variants selected in the presence of the MAbs L77, Nc32, K71, and K29 recognizing four different epitopes on MV H (20). Variant CAM/RB viruses escaping the MAbs L77 and Nc32 were neurovirulent, whereas viruses escaping the MAbs K29 and K71 appeared to have lost neurovirulence. The H genes of these viruses have been sequenced elsewhere (20). However, because of the number of amino acid changes in this gene and the possibility that changes in other genes also affect the specific phenotype, the molecular basis of the antibody escape and neurovirulence could not be unequivocally determined in earlier experiments. The generation of recombinant MVs has opened the way to make definitive linkages between mutations introduced experimentally into the viral genome and specific phenotypes (30). We therefore assessed, using recombinant MVs, the influence of directed mutations in the H gene on antibody escape and neurovirulence. After intracerebral injection into suckling C57BL/6 mice, a recombinant virus, expressing the H gene of CAM/RB (EdtagCAMH), induced neurological disease, and MV antigen was found in neurons and neuronal processes of the hippocampus, frontal and olfactory cortices, and neostriatum (9). However, the neurovirulence of EdtagCAMH was partially reduced compared to that of the natural strain CAM/RB. Thus, the results indicated that the H protein, albeit an important determinant of neurovirulence, is not the sole determinant and that other viral genes contribute to the observed virus-induced central nervous system disease.
In this study, we generated and tested recombinant viruses expressing single and combined mutations putatively mediating escape from the anti-H MAbs, which were suggested to be associated with neurovirulence (20). We successfully proved the role of mutations in mediating escape from four anti-H MAbs but found, surprisingly, that antibody escape and neurovirulence in suckling Lewis rats are associated with different alterations in the H protein and that phenotypes are not linked.
MATERIALS AND METHODS
Antibodies, cells, and propagation of viruses.
The MAbs L77, Nc32, K71, and K29 (anti-MV H [20]); anti-MV fusion (F) protein (A504); and anti-MV nucleocapsid protein (F227) were produced from hybridomas using RPMI medium containing 10% FCS and purified over protein G-Sepharose in our laboratory. The fluorescein isothiocyanate (FITC)-conjugated and phycoerythrin (PE)-conjugated rabbit anti-mouse immunoglobulin antibodies and streptavidin-FITC were purchased from DAKO GmbH, Hamburg, Germany. Vero cells were cultured in minimum essential medium containing 5% FCS, and BJAB cells were cultured in RPMI medium containing 10% FCS. CAM/RB, the rodent-adapted CAM strain of MV, was passaged by intracerebral infection of brains of 1-day-old rats and reisolation of virus after 4 days postinfection (p.i.). MV strain Edmonston; Edtag-based recombinants; CAM/Vero (Vero cell-passaged CAM strain); and tissue culture-selected escape mutants CAM/L77, CAM/Nc32, CAM/K71, and CAM/K29 escaping the MAbs L77, Nc32, K71, and K29, respectively, were propagated on Vero cells. For virus production, cells were infected at a multiplicity of infection (MOI) of 0.01, and virus was harvested when maximum giant cell formation was observed by one cycle of freezing-thawing and two centrifugations to pellet cell debris. Supernatants were stored at −80°C.
Construction and rescue of recombinant viruses.
All full-length, mutagenized constructs were assembled in an H gene insertion vector (pMVins-H2), the construction of which has been previously described (8a, 9). Briefly, this vector contains two unique restriction sites (PacI and AatII) which permit the directional cloning of complete H genes obtained by PCR amplification using the H-specific, PacI- or Aat II-containing oligonucleotides uniH+ and uniH2−. We have used this vector to generate plasmid p(+)MVCAMH, which contains the H gene of CAM/RB in the Edmonston background. The p(+)MVCAMH plasmid was used as the template to construct a set of full-length MV plasmids containing the nucleotide changes specific to the MAb escape mutations previously identified (20). Overlapping oligonucleotides (24-mers) were synthesized with the appropriate point mutations. Two separate PCRs were performed using the proofreading DNA polymerase Pwo (Boehringer). The 5′ portion of the H gene was amplified using uniH+ and the mutagenic (antigenome-sense) oligonucleotide. The 3′ portion of the gene was amplified using the mutagenic (genome-sense) oligonucleotide and uniH2−. PCR products were gel purified from 1% agarose blocks using a dialysis membrane, and the DNA was phenol extracted and ethanol precipitated. The DNA fragments were mixed in equimolar amounts, and 100 ng of the mixture was added to a standard PCR in the absence of primers. The thermocycler was paused at the beginning of cycle 11 (94°C), during which time primers uniH+ and uniH2− were added. Full-length mutagenized H genes were amplified during the remaining 25 cycles. Following restriction with PacI and AatII, the H genes were inserted into similarly cleaved pMVins-H2 to generate the full-length MV constructs. Alternatively, full-length H genes were amplified by reverse transcription-PCR (RT-PCR) from total RNA isolated from infected Vero cells as described previously (9). Plasmids were sequenced by dideoxynucleotide chain termination (ABI Prism) using MV H-specific primers to verify that the mutagenesis was successful. This also confirmed that the PCR had not introduced nonspecific mutations into the H genes. Recombinant viruses were rescued from these constructs following Lipofectin-mediated transfection of MVA-T7-infected HeLa cells as previously described (9).
Sequencing of the H genes from recombinant viruses.
To determine whether the nucleotide exchanges which were introduced in the recombinants are correct and whether there are additional exchanges in the H genes of the escape mutants, the H genes were sequenced. Total RNA from infected BJAB cells (48 h) was prepared with Qiagen Spin columns as recommended by the manufacturer. The RNA was reverse transcribed with SuperScript II (GIBCO/BRL). The H gene of the virus was amplified by PCR using the primers H62 (upper) (5′-ATCCACAATGTCACCACAACG-3′) and H26 (lower) (5′-TATGCCTGATGTCTGGGTGA-3′). The amplified cDNAs of the H genes were used as templates in the sequencing reaction (ABI-PRISM Bigdye Terminator Cycle Sequencing Ready Reaction kit; Applied Biosystems) using further H-specific primers (H30, 5′-GATAGGGAGTACGACTTCAG-3′; H31, 5′-TTGAAGTAGGTGTTATCAGAA-3′; H32, 5′-CACCATTGAAGGATAACAGGA-3′; H33, 5′-AGGTGGATGGTGATGTCAAA-3′; H25, 5′-GAAGTATCGTAGGTTGCCA-3′; H24K, 5′-CCACTCGGGATTCTCGCAGAG-3′; H23K, 5′-GGATTTCTGATAACACCTA-3′; H20, 5′-CCTTGACCTGATGCTCGATTG-3′; and H27, 5′-TGACATCATGTGATTGGTTC-3′). The analysis was performed using an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer).
Antibody escape assays.
To quantify the antibody escape by flow cytometry, BJAB cells were infected with the natural variants or recombinant viruses and analyzed by double staining with the various MAbs to H and a MAb to the fusion protein (F) as an internal control of expression of the envelope proteins. BJAB cells were infected at an MOI of 1 and incubated for 2 to 3 days at 37°C. The cells were washed with fluorescence-activated cell scanner (FACS) buffer (calcium- and magnesium-free phosphate-buffered saline containing 0.4% bovine serum albumin and 0.01 M NaN3) and incubated for 1 h with the primary antibodies recognizing H (K29, K71, Nc32, and L77), washed twice and treated for 1 h with secondary antibody (PE-conjugated anti-mouse antibody), washed, blocked for 10 min with mouse serum and then treated for 1 h with the biotinylated anti-F MAb A504, washed, and incubated for 1 h with streptavidin-FITC. The fluorescence signals were determined by flow cytometry using a FACScan (Becton Dickinson) cell sorter. The F-specific signals of the cells infected with various virus variants were used as standards for the expression of the viral envelope proteins and equalized for comparison of the H-specific signals obtained with different MAbs. The H-specific signals of EdtagCAMH-infected cells for each H antibody were set to 100%, and the signals of cells infected with virus variants were then expressed in relation to this.
Vero cells were infected with the recombinant viruses in order to observe the antibody escape microscopically. Cells were grown on glass coverslips to 80% confluency and infected at an MOI of 0.01. Infected cells were incubated for 48 h at 37°C. After this time, the coverslips were rinsed in phosphate-buffered saline, and the cells were fixed for 5 min in ice-cold acetone. Expression of the H protein was examined using the selecting MAb. An additional H-specific MAb or rabbit polyclonal antiserum was used as a positive control for H expression. An indirect immunofluorescence assay was performed as previously described, and samples were examined by confocal laser scanning microscopy (10).
Animal infections and histology.
Timed pregnant Lewis rats were purchased from Harlan-Winkelmann (Borchem, Germany). One- to two-day-old pups were infected intracerebrally in the left hemisphere with 20 μl of virus suspension (1 × 104 to 5 × 104 PFU). Body weights of animals were measured at days 0, 3, 5, 7, and 10 p.i. Animals were infected for no longer than 10 days, anesthetized, and sacrificed by decapitation. The brains were removed and fixed in 4% paraformaldehyde in 0.2 M phosphate buffer (pH 7.4) for at least 2 days before frontal brain sections were embedded in paraffin. Tissue sections were routinely stained for hematoxylin-eosin and Luxol-fast blue. In brains infected with a neurovirulent recombinant, cortical and infracortical necrosis occurred in the left hemisphere and frequently also in the right hemisphere, revealing numerous pycnotic nuclei and a perifocal edema.
An immunohistology assay for detection of MV nucleocapsids was performed using a MAb (F227; 2 μg/ml). In brief, slides were rehydrated and pretreated for 10 min in a microwave oven (600 W) in citrate buffer (pH 6.0). Endogenous peroxidase was blocked by incubation with 1% hydrogen peroxide. Nonspecific binding sites were blocked by normal swine serum. F227 was then applied overnight at 4°C. Antibody binding was visualized with biotinylated secondary antibody (rabbit anti-mouse), streptavidin and biotinylated horseradish peroxidase complex (StreptABComplex/HRP; DAKO GmbH), and diaminobenzidine (Fluka, Neu-Ulm, Germany) as chromogen. Sections were counterstained with Mayer's hemalaun.
RESULTS
Cloning and rescue of recombinant MVs expressing H antibody escape mutations.
In order to study the potential linkage between antibody escape and neurovirulence, a set of recombinant viruses (summarized in Table 1) was generated based on EdtagCAMH (9) and the sequences of MAb escape mutants selected from CAM/RB (20). Mutations were introduced into the CAM/RB H gene by overlap PCR using mutagenic primers. All mutagenized H genes were inserted into the plasmid vector (pMVins-H2) to generate full-length recombinant constructs (Fig. 1). These were transfected into HeLa cells using the host-range-adapted helper virus MVA-T7 system (9). Recombinant viruses were rescued 6 to 10 days posttransfection. Total RNA was isolated from virus-infected cells, and H genes were amplified by RT-PCR. The mutagenized portions were sequenced to ensure that the mutations were stably retained in the recombinant viruses, and no spurious mutations were identified. Recovery of the recombinant viruses was reproducible in three independent transfections, they all grew to equivalent titers (1 × 105 to 5 × 105 PFU/ml/106 cells) at similar rates, and no differences in plaque size were observed for Vero cells.
TABLE 1.
Antibody escape and neurovirulence of MV recombinants expressing the putative escape mutations in their H proteins
| Recombinant virus | Amino acid at position:
|
Additional change(s) at amino acid position | Escape from anti-H MAb
|
Neurovirulence | |||
|---|---|---|---|---|---|---|---|
| Putative | Detecteda
|
||||||
| 195 | 200 | IF | FACS (%) | ||||
| Edtag (Edmonston H) | R | N | None | − | − | No | |
| EdtagCAMH | G | S | None | − | − | Yes | |
| EdtagCAMH-1 | G | S | 377P→Q | L77 | − | 59 | Yes |
| EdtagCAMH-2 | G | S | 378M→K | L77 | − | 82 | Yes |
| EdtagCAMH-3 | G | S | 377P→Q | L77 | + | 98 | Yes |
| 378M→K | |||||||
| EdtagCAMH-4 | G | S | 388G→S | Nc32 | + | 98 | Yes |
| EdtagCAMH-5 | G | S | 395E→K | K71 | − | 27 | Yes |
| EdtagCAMH-6 (=EdtagCAMH/Vero) | R | N | None | − | − | No | |
| EdtagCAMH-7 | R | N | 377P→Q | L77 | − | 84 | No |
| EdtagCAMH-8 | R | N | 378M→K | L77 | − | 90 | No |
| EdtagCAMH-9 | R | N | 377P→Q | L77 | + | 97 | No |
| 378M→K | |||||||
| EdtagCAMH-10 | R | N | 388G→S | Nc32 | + | 93 | No |
| EdtagCAMH-11 | R | N | 395E→K | K71 | − | − | No |
| EdtagCAMH-12 | R | N | 395E→K | K71 | + | 89 | No |
| 492E→K | |||||||
| 550S→P | |||||||
| EdtagCAMH-13 | R | N | 535E→G | K29 | + | 88 | No |
Escape from the corresponding MAb was detected by microscopy. +, escape; −, no escape detected; IF, immunofluorescence by laser scanning microscopy; FACS; flow cytometry. Given percentages of escape were determined by flow cytometry in comparison to the interaction of a certain MAb with CAMH (Fig. 4).
FIG. 1.
Location of antibody-selected escape mutations in the H protein of CAM/RB and construction of recombinant viruses. The H genes of the rodent-adapted MC CAM/RB (a) escape mutants were sequenced, and the amino acid alterations were identified (b). Plasmid p(+)MVCAMH, which contains the wild-type CAM/RB H gene (c), was used as the template to introduce back mutations into the gene by overlap PCR. The 3′ fragment of the gene was amplified using uniH2− and the required mutagenic primer (d). The 5′ fragment was amplified using uniH+ and the complementary mutagenic primer (e). The PCR products were joined by overlap PCR (f) to generate a full-length H gene which was restricted with PacI and AatII and inserted into the insertion vector p(+)MVins-H2 (g). Steps d and e were omitted when full-length H genes were amplified directly from total RNA by RT-PCR.
When the H gene sequences of the natural escape variants were determined, they all contained at least two nucleotide changes in comparison to the H gene of CAM/RB at positions 603 and 619, corresponding to two amino acid substitutions at positions 195 (G→R) and 200 (S→N), respectively. Since these exchanges were present in all variants, they could not be responsible for the antibody-specific escape of the variants. These two shared mutations were not present in a subset of recombinant EdtagCAMH mutants, recombinants 1 to 5 (G and S at positions 195 and 200, respectively), and were introduced in recombinants 6 to 13 (R and N at positions 195 and 200, respectively) as a basis for the additional mutations putatively mediating antibody escape (Table 1).
Several amino acid alterations in the MAb escape variants were detected which were assumed, but not formally proven, to be associated with the antibody escape. The panel of recombinant viruses was designed to reflect these observations. The first four recombinant viruses (recombinants 1 to 4) were generated based on escape mutants isolated using MAbs L77 and Nc32. Two mutations (377P→Q and 378M→K) were selected using L77, and these were introduced both individually and together into the H gene of CAM/RB to assess the contribution of each mutation to the L77 antibody escape. A single mutation (388G→S) was selected using Nc32. Four further recombinant viruses (recombinants 7 to 10) were generated to include the same mutations putatively involved in antibody escape as in recombinants 1 to 4 in addition to the shared mutations 195G→R and 200S→N. This subset of viruses permitted us to assess the effects of the shared mutations on neurovirulence and antibody escape.
The published sequence of the H gene of the escape variant CAM/K29 did not reveal a specific difference in comparison to the CAM H gene. At the time, this necessitated explanations based on a potential role for the 195G→R and/or 200S→N mutation or the involvement of interacting mutations in the F or M protein which were not sequenced in the study (20). In the present investigation, we sequenced this gene again and found a previously unrecognized nucleotide exchange at position 1624 (A→G) corresponding to an amino acid change at position 535 (E→G). In addition, resequencing of the H gene of variant CAM/K71 revealed previously unrecognized mutations at the positions 1494 (G→A) and 1668 (T→C) corresponding to amino acid substitutions at positions 492 (E→K) and 550 (S→P), respectively.
Two recombinant viruses were generated by overlap PCR mutagenesis based on the initial sequence of the K71 escape mutant. Both included the single mutation 395E→K. The first, recombinant 5, did not contain the two shared mutations, 195G→R and 200S→N, whereas recombinant 11 contained these alterations. The additional mutations, which were identified in this study in the MAb escape mutants selected by K71 and K29, were widely separated in the H genes. To facilitate the generation of the full-length MV cDNA, complete H genes were amplified by RT-PCR from total RNA using uniH+ and uniH2− (Fig. 1). Three mutations (395E→K, 492E→K, and 550S→P) are present in the escape mutant selected using K71, and these were introduced into the Edmonston background by amplification of the H gene from the escape mutant to generate the recombinant virus 12. The same approach was used to generate recombinant 13, which contains the mutation present in the escape mutant selected using K29 (535E→G). These two viruses, therefore, also contain the shared mutations 195G→R and 200S→N. All of these viruses and their respective mutations are described in Table 1, and the general cloning strategy is outlined in Fig. 1.
Determination of antibody escape.
Immunofluorescence was used to examine H protein expression in Vero cells infected with the recombinant viruses using the four anti-H MAbs L77, Nc32, K71, and K29. Representative micrographs are shown in Fig. 2. Recombinant viruses 1 and 2 were still recognized by L77 (Fig. 2B and D); however, introduction of the double mutation in recombinant 3 ablated antibody binding (Fig. 2F). These observations were identical when EdtagCAMH mutants 7 to 9 were examined (data not shown). Recombinants 4 and 10 no longer bound MAb Nc32 (Fig. 2H and data not shown), and recombinants 12 and 13 no longer bound MAbs K71 and K29, respectively (Fig. 2J and L).
FIG. 2.
Analysis of recombinant escape mutants by confocal microscopy. Vero cells were infected with the parental and recombinant viruses at an MOI of 0.01. Cells were fixed and examined by confocal laser scanning microscopy for immunoreactivity, and micrographs represent 8- to 10-μm-deep composite optical sections. The viral H proteins were detected with different MAbs and secondary Cy3-conjugated antibodies (green), and nuclei were counterstained with propidium iodide (red); all images were obtained in double excitation mode. EdtagCAMH mutants were mutant 1 (K71 [A] and L77 [B]), recombinant 2 (K71 [C] and L77 [D]), recombinant 3 (K71 [E] and L77 [F]), recombinant 4 (L77 [G] and Nc32 [H], recombinant 12 (Nc32 [I] and K71 [J]), and recombinant 13 (K71 [K] and K29 [L]). Nonselecting control antibodies were used to demonstrate H protein expression in the recombinant viruses (A, C, E, G, I, and K), and the selecting antibodies were used to determine if the mutated H proteins were no longer bound (B, D, F, H, J, and L). Expression of the H gene in the parental viruses was examined as controls: Edtag (Nc32 [M]), CAM/RB (Nc32 [N]), EdtagCAMH (Nc32 [O]), and EdtagCAMH/Vero (L77 [P]). Magnification, ×160.
The influence of single amino acid changes on the interaction with MAbs was examined quantitatively by flow cytometry. As a preliminary experiment, we established the method using natural escape variants. To achieve comparable signals, cells infected with the different viruses were all double stained with anti-F and anti-H antibodies. The signals detected with anti-F MAb were used as standards for normalization, and the signals obtained with anti-H MAbs with CAM-infected cells were set to 100%. The signals of the variant viruses were then expressed as percentages of the CAM H signal. The antibodies, from which certain variants were supposed to escape, did not at all interact with the corresponding infected cells (Fig. 3). Thus, this method can be used for the quantification of antibody interaction with the mutant H proteins.
FIG. 3.
FACS analysis of natural escape mutants. The relative expression of the H protein on the surface of BJAB cells infected with CAM/RB and the natural escape variants CAM/Nc32, CAM/K71, CAM/L77, and CAM/K29 was determined by double staining and flow cytometric analysis. The relative signals obtained after staining of the cells with MAbs L77, Nc32, K71, and K29 are shown. Signals detected by anti-F antibody A504 were taken as standards for the glycoprotein expression, and signals obtained by the four anti-H antibodies were set in relation to the signals of CAM-infected cells (=100%). The MAbs do not recognize cells infected with their respective escape variants.
The interactions of the four anti-H MAbs L77, Nc32, K71, and K29 with the surface of cells infected with the recombinant viruses expressing the putative escape mutations were quantified by FACS (Fig. 4). The EdtagCAMH mutants 1, 2, 7, and 8 had less interaction, and recombinants 3 and 9 did not interact with MAb L77 (Fig. 4A), recombinants 4 and 10 did not interact with MAb Nc32 (Fig. 4B), recombinant 12 did not interact with MAb K71 (Fig. 4C), and recombinant 13 did not interact with MAb K29 (Fig. 4D). Thus, the mutations leading to the escape from all four MAbs have been unequivocally identified. The results of the analysis of antibody interactions with mutant H proteins are summarized in Table 1.
FIG. 4.
FACS analysis of recombinant escape mutants. The expression of the H and F proteins on the surface of BJAB cells infected with the EdtagCAMH mutants 1 to 13 (bars 1 to 13, respectively) and EdtagCAMH (bar 14) was determined by double staining with anti-H (L77, Nc32, K71, and K29) and anti-F MAbs and flow cytometric analysis (Fig. 2). The signal intensity of EdtagCAMH-infected cells was set to 100% (bar 14), and the intensities of the other H signals are presented in relation to this signal.
Analysis of the neurovirulence of recombinants.
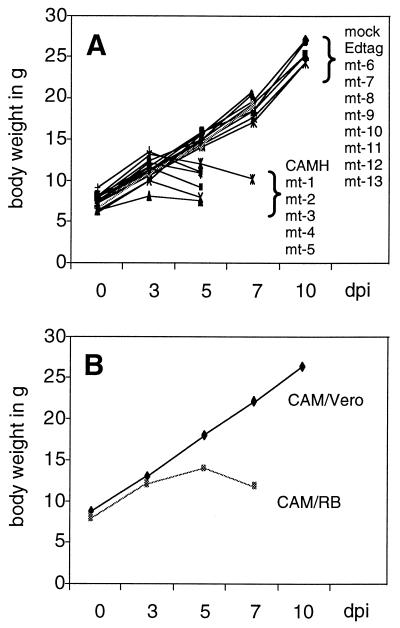
In order to analyze the neurovirulence of the recombinants, we infected 1- to 2-day-old suckling Lewis rats intracerebrally with approximately 2 × 104 PFU of the viruses. As a disease parameter, we measured the body weight of the infected animals, which closely reflects the degree of infection. Infection with Edtag did not induce a detectable disease in the animals, whereas infection with EdtagCAMH caused strong signs of disease leading to death within approximately 5 days (Fig. 5A). Thus, as found with mice (9), replacement of Edmonston H by the CAMH gene alone is sufficient to cause neurovirulence in suckling rats. The EdtagCAMH mutants 1 to 5 caused a disease similarly as strong as that caused by EdtagCAMH, leading to death between days 5 and 7 p.i. In contrast, recombinants 6 to 13 were not neurovirulent, and animals survived and were apparently healthy (Fig. 5A). Thus, unexpectedly, the mutations mediating the antibody escape did not cause differential neurovirulence, whereas the presence of amino acids G and S at positions 195 and 200, respectively, in EdtagCAMH and the recombinants 1 to 5 correlated with the induction of the lethal brain disease.
FIG. 5.
Analysis of the body weight of cerebrally infected rats. One- to two-day-old pups were infected intracerebrally in the left hemisphere with 20 μl of virus suspension. Body weights of animals were measured at days 0, 3, 5, 7, and 10 p.i. Data for mock infection (medium) and infection with the recombinants Edtag, EdtagCAMH, and mutants 1 to 13 (approximately 2 × 104 PFU) are shown in panel A. The body weight development of rodent-adapted CAM/RB (2 × 103 PFU)- and cell culture-adapted CAM/Vero (2 × 104 PFU)-infected suckling rats is shown in panel B. The animals infected with the neurovirulent virus CAM/RB and the recombinants EdtagCAMH and mutants 1 to 5 died between days 5 and 7. dpi, day postinfection.
Immunohistological analysis of the viral spread in infected rat brains revealed corresponding results. In the brains of all animals infected with neurovirulent recombinants (EdtagCAMH and recombinants 1 to 5) at days 3 and 5 p.i., numerous nucleocapsid-positive cells were detected, predominantly in the left hemisphere, where virus had been injected. In contrast, after infection of rat brains with Edtag or recombinants 6 to 13 (R and N at positions 195 and 200, respectively) no or only a few MV nucleocapsid-positive cells were detectable up to day 10 p.i. As examples for the differential neurovirulence of the recombinant viruses, immunohistological staining patterns in brains infected with EdtagCAMH (Fig. 6A and C) and recombinant 6 (Fig. 6B and D) are shown. Results are summarized in Table 1.
FIG. 6.
Immunohistological analysis of brain infections with a neurovirulent and a nonneurovirulent MV recombinant. Staining patterns of MV nucleocapsids in frontal brain sections of rats 5 days p.i. with EdtagCAMH (A and C) and at day 10 p.i. with recombinant 6 (B and D) are shown. Schematic drawings of immunolabeled brain sections (A and B) demonstrate immunoreactivity with MAb F227 against MV N for EdtagCAMH (C) and recombinant 6 (D) as seen with lower magnification. Higher magnifications prove that the majority of neurons contain MV nucleocapsids in a brain infected with EdtagCAMH (C), whereas only a few scattered neurons are immunolabeled in a brain infected with recombinant 6 (D) (bar = 50 μm).
Analysis of neurovirulence and H gene sequence of Vero cell-adapted CAM.
The sequence analysis of the H genes of the natural escape variants detected changes at amino acid positions 195 and 200 in all variants, although they varied in their neurovirulent capacity and therefore this mutation was not suggested to be responsible for the neurovirulent capacity of the variants (20). This is in contrast to our findings with the MV recombinants, where mutations at these positions determined neurovirulence. We therefore compared the neurovirulence and the H gene sequences of CAM/Vero, a CAM strain adapted by at least 30 passages on Vero cells, with CAM/RB propagated in rat brain. CAM/Vero was not neurovirulent for suckling rats (Fig. 5B), and its H protein is recognized by all four anti-H MAbs, L77, Nc32, K71, and K29 (data not shown). In CAM/Vero, we found only two nucleotide changes in the H gene, at positions 603 (G→A) and 619 (G→A), resulting in amino acid exchanges at positions 195 (G→R) and 200 (S→N), as found in the natural escape variants and which were introduced in the recombinants 6 to 13 (Table 1). The CAM/Vero H gene sequence is thus identical to that of recombinant 6. This confirmed the results using the recombinant viruses 6 to 13 and demonstrated that amino acids glycine (G) at position 195 and serine (S) at position 200 are decisive for the neurovirulence of the CAM/RB and the recombinant MVs. When we compared the sequences of all known MV H proteins at positions 195 and 200, we found that the neurovirulent rodent-adapted strain CAM/RB is unique in these positions, expressing G and S, respectively.
DISCUSSION
Interaction of the viral envelope proteins with cellular receptors is not only a prerequisite for viral entry but also has great impact on the pathogenesis of virus-induced diseases (37). Single amino acid changes in the envelope proteins of several viruses, including mumps virus (18) and MV (20), were found to have a profound and differential influence on pathogenesis. Alteration of viral envelope proteins may have a variety of pathogenic consequences: single mutations affect neuroinvasiveness of tick-borne encephalitis virus (14, 33, 41), neurovirulence of dengue virus (13) and Japanese encephalitis virus (28), viral persistence in the case of lymphocytic choriomeningitis virus (25), cell-to-cell spread of rabies virus (6, 40), and virus replication efficiency of Sindbis virus (8). Using recombinant MV with a different H protein, we found earlier that replacement of the Edmonston H by CAM/RB H is sufficient to induce neurovirulence in mice (9). As described here, the same holds true for intracerebral infection of suckling Lewis rats. The H protein of CAM/RB is sufficient to confer neurovirulence on recombinant MV based on the viral backbone of the nonneurovirulent strain Edmonston. The exact role of single mutations in antibody escape or neurovirulence could not be determined earlier, because there were several mutations in the H proteins and mutations in other viral genes could not be ruled out as having modulating effects (20). Here, we succeeded in identifying single amino acid mutations which mediate the antibody escape and we could separate them from those mediating neurovirulence. Mutations mediating antibody escape are situated on the outer propellers of the MV H protein according to the Langedijk model of paramyxovirus attachment proteins (19). This part of the molecule is supposed to interact not only with neutralizing antibodies (12, 17, 29) but also with the cellular receptors for MV, CD46 (7, 27) and CD150 (11, 16, 42). Since the escape mutations did not affect the replication of the viruses in Vero cells, the interaction with the receptor CD46 appears not to be altered.
In contrast to the mutations mediating antibody escape, the mutations associated with neurovirulence are situated in the stem 2 region of the H protein. The surface of this area is proposed to be parallel to the vertical axis of the H molecule and could affect interactions with molecules in the same membrane as H (e.g., F). It is not clear what structural consequences the mutation at position 195 (G→R) may have, except that it potentially introduces a new positive charge on the surface of the molecule. The change at position 200 (S→N) generates an additional potential glycosylation site in the CAM/RB H molecule. This sixth potential glycosylation site in the H protein may well cause a structural alteration or mask binding sites, leading to the loss of neurovirulence. The structural alteration might affect the interaction with the unknown cellular attachment receptor in the rat or the fusion helper function of H and, thus, virus entry or cell-to-cell spread. The H protein interaction with further viral proteins such as the matrix-fusion protein complex (M-F) might also influence the viral spread (3, 4). An infectious matrix protein deletion mutant MV exhibited a higher fusogenic capacity than did standard virus and penetrated more deeply into the brain parenchyma in mouse brains (3). Similar results concerning the spread of virus were found with recombinant viruses lacking the cytoplasmic tail of the fusion (F) or hemagglutinin (H) protein, suggesting that the interaction of the M protein with the cytoplasmic parts of these proteins is involved in the regulation of virus-induced cell fusion (4). Interestingly, mutations in the matrix protein and the cytoplasmic domain of the fusion protein have been found in several SSPE brains (1, 5, 36). To prove whether virus entry or cell-to-cell spread is indeed affected by mutation of the glycosylation site at position 200 requires further studies using single mutations at positions 195, 200, and 202 as well as positions 157 and 175 in the CAM/RB background. The latter two must be considered because the mutations in CAM/RB that confer neurovirulence are unique and not present in any other MV lineage or in the other rodent-neurovirulent strain HNT (34).
The sequence analysis of the H genes of the natural escape variants detected changes at amino acid positions 195 and 200 in all variants, in neurovirulent as well as nonneurovirulent viruses (20). Therefore, this mutation initially was not suggested to be responsible for the differential neurovirulent capacity of the variants. Since the variants described above were amplified using Vero cells before RNA was isolated for sequencing, this mutation could have been introduced or selected during the growth in tissue culture as an adaptation to Vero cells. It is known that the CAM/RB strain loses its neurovirulence when propagated several times in tissue culture. The finding that the nonneurovirulent virus CAM/Vero bears only these two mutations at positions 195 and 200 in its H gene in comparison to CAM/RB strongly supports the suggestion that these mutations are selected by Vero cell passage and not for neurovirulence. Our findings using recombinant viruses suggest that the differences in the neurovirulence of the natural escape variants have apparently not been induced by the antibodies used for selecting the escape variants but rather are a consequence of the necessary propagation of the viruses in tissue culture.
On the basis of recombinant viruses, it is clear that these two mutations are the only difference between the recombinant EdtagCAMH, which is neurovirulent, and mutant 6, which is nonneurovirulent, and that antibody escape and neurovirulence are not linked in this model.
ACKNOWLEDGMENTS
We thank F. Dimpfel, S. Löffler, and P. Haddock for technical support.
This work was supported by the Deutsche Forschungsgemeinschaft, the Wellcome Trust (grant 047245), and the European Social Fund.
REFERENCES
- 1.Baczko K, Lampe J, Liebert U G, Brinckmann U, ter Meulen V, Pardowitz I, Budka H, Cosby S L, Isserte S, Rima B K. Clonal expansion of hypermutated measles virus in a SSPE brain. Virology. 1993;197:188–195. doi: 10.1006/viro.1993.1579. [DOI] [PubMed] [Google Scholar]
- 2.Barrett P N, Koschel K, Carter M, ter Meulen V. Effect of measles virus antibodies on a measles SSPE virus persistently infected C6 rat glioma cell line. J Gen Virol. 1985;66:1411–1421. doi: 10.1099/0022-1317-66-7-1411. [DOI] [PubMed] [Google Scholar]
- 3.Cathomen T, Mrkic B, Spehner D, Drillien R, Naef R, Pavlovic J, Aguzzi A, Billeter M A, Cattaneo R. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 1998;17:3899–3908. doi: 10.1093/emboj/17.14.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cathomen T, Naim H Y, Cattaneo R. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol. 1998;72:1224–1234. doi: 10.1128/jvi.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter M A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietzschold B, Wiktor T J, Trojanowski J Q, Macfarlan R I, Wunner W H, Torres-Anjel M J, Koprowski H. Differences in cell-to-cell spread of pathogenic and apathogenic rabies virus in vivo and in vitro. J Virol. 1985;56:12–18. doi: 10.1128/jvi.56.1.12-18.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dörig R E, Marcil A, Chopra A, Richardson C D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 8.Dropulic L K, Hardwick J M, Griffin D E. A single amino acid change in the E2 glycoprotein of Sindbis virus confers neurovirulence by altering an early step of virus replication. J Virol. 1997;71:6100–6105. doi: 10.1128/jvi.71.8.6100-6105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Duffy I. Analysis of measles virus glycoproteins. Ph.D. thesis. Belfast, Northern Ireland: The Queen's University of Belfast; 2000. [Google Scholar]
- 9.Duprex W P, Duffy I, McQuaid S, Hamill L, Schneider-Schaulies J, Cosby L, Billeter M, ter Meulen V, Rima B. The H gene of rodent brain-adapted measles virus confers neurovirulence to the Edmonston vaccine strain. J Virol. 1999;73:6916–6922. doi: 10.1128/jvi.73.8.6916-6922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duprex W P, McQuaid S, Hangartner L, Billeter M A, Rima B K. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J Virol. 1999;73:9568–9575. doi: 10.1128/jvi.73.11.9568-9575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erlenhoefer C, Wurzer W J, Löffler S, Schneider-Schaulies S, ter Meulen V, Schneider-Schaulies J. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J Virol. 2001;75:4499–4505. doi: 10.1128/JVI.75.10.4499-4505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlier D, Varior-Krishnan G, Devaux P. CD46-mediated measles virus entry: a first key to host-range specificity. Trends Microbiol. 1995;3:338–345. doi: 10.1016/s0966-842x(00)88972-6. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu K, Tadano M, Men R, Lai C-J. Mutational analysis of a neutralisation epitope on the dengue type 2 virus (DEN-2) envelope protein: monoclonal antibody resistant DEN-2/DEN-4 chimeras exhibit reduced mouse neurovirulence. Virology. 1996;224:437–445. doi: 10.1006/viro.1996.0550. [DOI] [PubMed] [Google Scholar]
- 14.Holzmann H, Stiasny K, Ecker K, Kunz C, Heinz F X. Characterization of monoclonal antibody-escape mutants of tick-borne encephalitis virus with reduced neuroinvasiveness in mice. J Gen Virol. 1997;78:31–37. doi: 10.1099/0022-1317-78-1-31. [DOI] [PubMed] [Google Scholar]
- 15.Horvat B, Rivailler P, Varior-Krishnan G, Cardoso A, Gerlier D, Rarourdin-Combe C. Transgenic mice expressing human measles virus (MV) receptor CD46 provide cells exhibiting different permissivities to MV infections. J Virol. 1996;70:6673–6681. doi: 10.1128/jvi.70.10.6673-6681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu E C, Iorio C, Sarangi F, Khine A A, Richardson C D. CDw150 (SLAM) is a receptor for a lymphotropic strain of measles virus and may account for the immunosuppressive properties of this virus. Virology. 2001;279:9–21. doi: 10.1006/viro.2000.0711. [DOI] [PubMed] [Google Scholar]
- 17.Hummel K B, Bellini W J. Localization of monoclonal antibody epitopes and functional domains in the hemagglutinin protein of measles virus. J Virol. 1995;69:1913–1916. doi: 10.1128/jvi.69.3.1913-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kövamees J, Rydbeck R, Örvell C, Norrby E. Haemagglutinin-neuraminidase (HN) amino acid alterations in neutralisation escape mutants of Kilham mumps virus. Virus Res. 1983;17:117–129. doi: 10.1016/0168-1702(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 19.Langedijk J P M, Daus F J, van Oirschot J T. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J Virol. 1997;71:6155–6167. doi: 10.1128/jvi.71.8.6155-6167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebert U G, Flanagan S G, Löffler S, Baczko K, ter Meulen V, Rima B K. Antigenic determinants of measles virus hemagglutinin associated with neurovirulence. J Virol. 1994;68:1486–1493. doi: 10.1128/jvi.68.3.1486-1493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liebert U G, Hashim G A, ter Meulen V. Characterization of measles virus-induced cellular autoimmune reactions against myelin basic protein in Lewis rats. J Neuroimmunol. 1990;29:139–147. doi: 10.1016/0165-5728(90)90156-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liebert U G, Schneider-Schaulies S, Baczko K, ter Meulen V. Antibody-induced restriction of viral gene expression in measles encephalitis in rats. J Virol. 1990;64:706–713. doi: 10.1128/jvi.64.2.706-713.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebert U G, ter Meulen V. Virological aspects of measles virus-induced encephalomyelitis in Lewis and BN rats. J Gen Virol. 1987;68:1715–1722. doi: 10.1099/0022-1317-68-6-1715. [DOI] [PubMed] [Google Scholar]
- 24.Love A, Norrby E, Kristensson K. Measles encephalitis in rodents: defective expression of viral proteins. J Neuropathol Exp Neurol. 1986;45:258–267. [PubMed] [Google Scholar]
- 25.Matloubian M, Somasundaram T, Kolhekar R, Selvakumar R, Ahmed R. Genetic basis of viral persistence: single amino acid change in the viral glycoprotein affects ability of lymphocytic choriomeningitis virus to persist in adult mice. J Exp Med. 1990;172:1043–1048. doi: 10.1084/jem.172.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mrkic B, Pavlovic J, Rulicke T, Volpe P, Buchholz C J, Hourcade D, Atkinson J P, Aguzzi A, Cattaneo R. Measles virus spread and pathogenesis in genetically modified mice. J Virol. 1998;72:7420–7427. doi: 10.1128/jvi.72.9.7420-7427.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naniche D, Varior-Krishnan G, Cervoni F, Wild T F, Rossi B, Rabourdin-Combe C, Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni H, Barrett A D. Attenuation of Japanese encephalitis virus by selection of its mouse brain membrane receptor preparation escape variants. Virology. 1998;241:30–36. doi: 10.1006/viro.1997.8956. [DOI] [PubMed] [Google Scholar]
- 29.Patterson J B, Scheiflinger F, Manchester M, Yilma T, Oldstone M B A. Structural and functional studies of the measles virus hemagglutinin: identification of a novel site required for CD46 interaction. Virology. 1999;256:142–151. doi: 10.1006/viro.1999.9644. [DOI] [PubMed] [Google Scholar]
- 30.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter M A. Rescue of measles virus from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rall G F, Manchester M, Daniels L R, Callahan E M, Belman A R, Oldstone M B. A transgenic mouse model for measles virus infection of the brain. Proc Natl Acad Sci USA. 1997;94:4659–4663. doi: 10.1073/pnas.94.9.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rammohan K W, McFarland H F, Bellini W J, Gheuens J, McFarlin D E. Antibody-mediated modification of encephalitis induced by hamster neurotropic measles virus. J Infect Dis. 1983;147:546–550. doi: 10.1093/infdis/147.3.546. [DOI] [PubMed] [Google Scholar]
- 33.Rey F, Heinz F X, Mandl C, Kunz C, Harrison S C. The envelope glycoprotein from tick-borne encephalitis at 2A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 34.Rima B K, Earle J A P, Baczko K, ter Meulen V, Carabana J, Caballero M, Celma M L, Fernandez-Munoz R. Sequence divergence of measles virus haemagglutinin during natural evolution and adaptation to cell culture. J Gen Virol. 1997;78:97–106. doi: 10.1099/0022-1317-78-1-97. [DOI] [PubMed] [Google Scholar]
- 35.Roos R P, Griffin D E, Johnson R T. Determinants of measles virus (hamster neurotropic strain) replication in mouse brain. J Infect Dis. 1978;137:722–727. doi: 10.1093/infdis/137.6.722. [DOI] [PubMed] [Google Scholar]
- 36.Schmid A, Spielhofer P, Cattaneo R, Baczko K, ter Meulen V, Billeter M A. Subacute sclerosing panencephalitis is typically characterized by alterations in the fusion protein cytoplasmic domain of the persisting measles virus. Virology. 1992;188:910–915. doi: 10.1016/0042-6822(92)90552-z. [DOI] [PubMed] [Google Scholar]
- 37.Schneider-Schaulies J. Cellular receptors for viruses: links to tropism and pathogenesis. J Gen Virol. 2000;81:1413–1429. doi: 10.1099/0022-1317-81-6-1413. [DOI] [PubMed] [Google Scholar]
- 38.Schneider-Schaulies S, Liebert U G, Baczko K, Cattaneo R, Billeter M, ter Meulen V. Restriction of measles virus gene expression in acute and subacute encephalitis of Lewis rats. Virology. 1989;171:525–534. doi: 10.1016/0042-6822(89)90622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider-Schaulies S, Liebert U G, Segev Y, Rager-Zisman B, Wolfson M, ter Meulen V. Antibody-dependent transcriptional regulation of measles virus in persistently infected neural cells. J Virol. 1992;66:5534–5541. doi: 10.1128/jvi.66.9.5534-5541.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seif I, Coulon P, Rollin P E, Flamand A. Rabies virulence: effect on pathogenicity and sequence characterization of rabies virus mutations affecting site III of the glycoprotein. J Virol. 1985;53:926–934. doi: 10.1128/jvi.53.3.926-934.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiasny K, Allison S L, Marchler-Bauer A, Kunz C, Heinz F X. Structural requirements for low-pH-induced rearrangements in the envelope protein of tick-borne encephalitis virus. J Virol. 1996;70:207–212. doi: 10.1128/jvi.70.11.8142-8147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatsuo H, Ono N, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]