Abstract
Hereditary spinocerebellar degenerations (SCDs) is an umbrella term that covers a group of monogenic conditions that share common pathogenic mechanisms and include hereditary spastic paraplegia (HSP), cerebellar ataxia, and spinocerebellar ataxia. They are often complicated with axonal neuropathy and/or intellectual impairment and overlap with many neurological conditions, including neurodevelopmental disorders. More than 200 genes and loci inherited through all modes of Mendelian inheritance are known. Autosomal recessive inheritance predominates in consanguineous communities; however, autosomal dominant and X-linked inheritance can also occur. Sudan is inhabited by genetically diverse populations, yet it has high consanguinity rates. We used next-generation sequencing, genotyping, bioinformatics analysis, and candidate gene approaches to study 90 affected patients from 38 unrelated Sudanese families segregating multiple forms of SCDs. The age-at-onset in our cohort ranged from birth to 35 years; however, most patients manifested childhood-onset diseases (the mean and median ages at onset were 7.5 and 3 years, respectively). We reached the genetic diagnosis in 63% and possibly up to 73% of the studied families when considering variants of unknown significance. Combining the present data with our previous analysis of 25 Sudanese HSP families, the success rate reached 52–59% (31–35/59 families). In this article we report candidate variants in genes previously known to be associated with SCDs or other phenotypically related monogenic disorders. We also highlight the genetic and clinical heterogeneity of SCDs in Sudan, as we did not identify a major causative gene in our cohort, and the potential for discovering novel SCD genes in this population.
Subject terms: Genetics research, Disease genetics
Introduction
Hereditary forms of spastic paraplegia (HSP), cerebellar ataxia, and spinocerebellar ataxia are distinct but overlapping clinical entities caused by related mechanisms that encompass a continuum of phenotypes [1–3]. We refer hereafter to these disorders as hereditary spinocerebellar degenerations (SCDs).
SCDs are characterized clinically by ataxia and/or spasticity complicated, in some cases, by other neurological or extra-neurological manifestations [1, 2]. They have more than 220 subtypes that afflict ~1:10,000 individuals worldwide with evident phenotypic and genetic heterogeneity and clinical overlap with other neurogenetic conditions such as intellectual disabilities, motor neuron diseases, encephalopathies, or neurodevelopmental disorders [4, 5]. The advent of next-generation sequencing (NGS) has markedly boosted SCDs diagnosis in recent years by the identification of a multitude of causative mutations in a large variety of disease-causing genes [4]. Dysfunction of mitochondria, ion channels, and cellular metabolism are the main altered functions by the pathogenic variants in these genes in addition to the abnormal expansions of nucleotide repeats [4].
Sudan is an East-African country with complex genetic and population structures [6]. This complexity stemmed from the linguistic and cultural differences between its ethnic groups acting in parallel with other, sometimes opposing, population genetic forces, e.g., consanguinity, admixture, and migration [6–9]. For instance, 67% of marriages in some parts of the country are consanguineous (42% first-degree cousins and 25% fifth-degree consanguinity and above) [10].
In a previous study, we screened 25 Sudanese families with HSP for mutations in 68 HSP genes using NGS targeted gene panel and reached a genetic diagnosis in 28% of these families [11]. In the current study, we investigated 38 novel Sudanese families with SCDs using a combination of candidate gene approaches, NGS targeted gene panel screening, and whole-exome sequencing (WES). We documented the studied patients’ clinical presentations and compared the diagnostic utility of the approaches in the two studies.
Subjects and Methods
Patients recruitment and interviews
We included a total of 90 patients from 38 Sudanese families in this study with the following inclusion criteria:
Patients presenting with symptoms, signs, and/or history suggestive of SCD.
Non-genetic causes that can mimic neurological illnesses that resemble SCD due to pregnancy- or birth-related insults, as well as toxic exposures, have been excluded through interview of the family members. MRI, when available, excluded tumor or compressions of CNS structures.
Participants from the family (patients and at least two healthy subjects) or their guardian (in the case of participants below 18 years old or patients with intellectual disabilities), agreed to participate in the study. We also examined and samples healthy subjects such as the parents, siblings, second-degree relatives, and/or third-degree relatives, in priority order depending on their availability, in order to help in the variant filtering. They had to be older than patients’ age at disease onset.
Sudanese by descent.
Presence of multiple cases affected in the same family, or sporadic case from a consanguineous marriage, to increase the probability to identify genetic causes.
Four out of the 38 families were screened in our previous study without reaching a genetic diagnosis [11]. Index cases were recruited from multiple neurology and pediatrics neurology clinics in Khartoum, the capital of Sudan that gather most tertiary hospitals and specialized clinics in the country. Most families originated from outside the city. Patients and families were interviewed and examined at the Department of Biochemistry, Faculty of Medicine, University of Khartoum, Sudan; the Pediatric Neurology Clinics, Soba University Hospital, Sudan; or the families’ residences in the capital of Sudan, Khartoum, or other Sudanese cities. The diagnosis protocol followed the EUROSPA/SPATAX clinical criteria (https://spatax.wordpress.com/downloads/). We collected 2 ml of saliva from the patients and healthy-related subjects using Oragene®•DNA (OG-500 and OG-575) kits (DNA Genotek Inc., Ottawa, ON, Canada).
The strategy of genetic studies
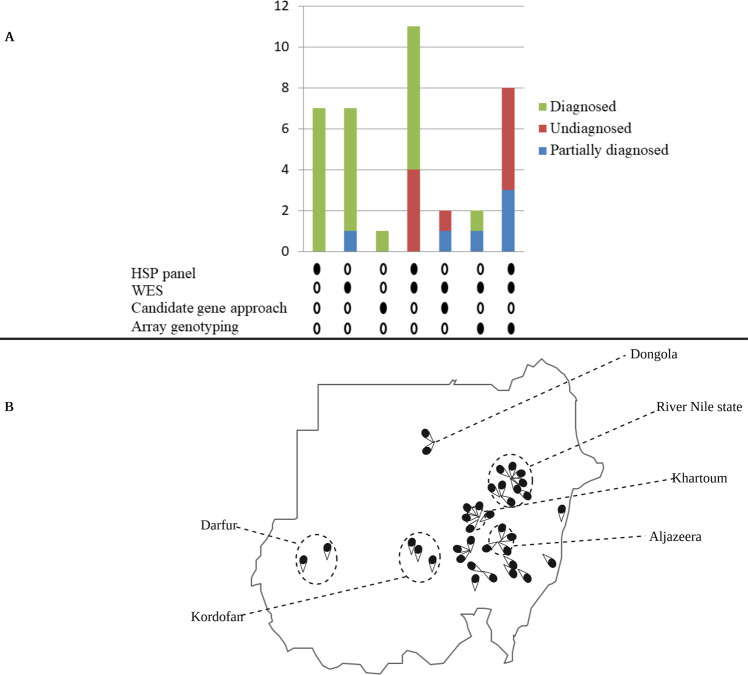
In this study, most families were studied using more than one diagnostic modality. We used various combinations of different genetic approaches, including NGS targeted gene panel screening, WES, candidate gene approach, and array genotyping (Fig. 1A). Initially, our experimental design was to screen the patients for mutations in selected genes depending on their phenotypes and to subsequently perform WES in negative cases. Later, with the drop in the costs of WES, we skipped the screening steps, except in patients with suspected repeat expansions based on clinical and inheritance data. Array genotyping was mainly used for homozygosity mapping and detection of copy number variations (CNVs). The presence of CNVs was also tested through coverage analysis in NGS data (gene panel and WES). Twenty-six families were investigated initially using HSP-targeted NGS gene panel (HSP panel) screening. Of these, eleven families were further investigated using WES and eight using WES and array genotyping. Eleven other families were directly investigated using WES, without HSP panel screening. Candidate gene approach was used in three families, two that were screened for repeat expansion-associated autosomal dominant spinocerebellar ataxias, and one for Friedreich’s ataxia repeat expansion.
Fig. 1. Genetic tools and geographical origins.
Genetic tools used for investigating our families and their utility (A) and the geographical origin of the families (B). A More than one genetic diagnostic approach was used in 23 out of 38 families, including all the undiagnosed families. Twenty-six families were investigated initially using HSP-targeted NGS gene panel (HSP panel) screening. Of these, eleven families were further investigated using WES and eight using WES and array genotyping. Eleven other families were directly investigated using WES, without HSP panel screening. Candidate gene approach was used in three families, two that were screened for repeat expansion-associated autosomal dominant spinocerebellar ataxias, and one for Friedreich’s ataxia repeat expansion. Sanger sequencing (not shown) was used for testing the segregation of all the identified candidate variants, except repeats expansion variants. HSP panel, hereditary spastic paraplegia next-generation sequencing targeted gene panel; WES, whole-exome sequencing. The families partially diagnosed relate to families where only a fraction of the patients was diagnosed. The numbers on the y-axis indicate the number of families; the filled circles indicate the used genetic tool. B The figure shows regions, states, or cities in Sudan from which the studied families originated (each pin-drop represents a single family).
DNA extraction and quality check
We extracted DNA from saliva following the prepIT®.L2P manual protocol provided by the manufacturer (DNA Genotek). DNA quantity (Absorbance at 260 nm) and quality (check of the high molecular weight DNA, absorbance ratio 260/280 and 260/320) were checked using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA), a Qubit® fluorometer (Promega, Madison, WI, USA), and standard agarose gel electrophoresis.
Next-generation panel screening of HSP genes
Fifty µl of patients’ DNA solution at a concentration of 50 ng per dl were sent for NGS panel screening at the genotyping and sequencing core facility of the Paris Brain Institute - ICM, Paris, France. A double capture enrichment strategy was used (Roche NimbleGen® SeqCap® Ez, USA). Sequencing was done on the MiSeq® platform (Illumina, CA, USA). Detailed methodology and bioinformatics analysis are available in previous reports [11, 12]. We systematically searched for point variations and genomic rearrangements. We targeted a median depth coverage > 100, and variants affecting positions with coverage < 30 were reanalyzed by Sanger sequencing if located in convincing causative genes based on the clinical presentation and inheritance mode.
Whole-exome sequencing
Twenty µl of DNA solution at a concentration of 20 ng per dl were sent for WES at the genotyping and sequencing core facility of the Paris Brain Institute - ICM, Paris, France. Exons were captured on the genomic DNA using the SeqCap® EZ MedExome Kit (Roche, IN, USA), followed by massively parallel sequencing on a Novaseq® 6000 sequencer (Illumina, CA, USA). Except for aligning reads to the hg37 version of the human genome (NCBI) using Burrows-Wheeler Aligner software, we processed exome data up to the calling of variants using the Genome AnalysisToolkit software (GATK) following the GATK4 best-practice pipeline.
We annotated and prioritized variants using software included in VarAFT annotation and filter tool [13]. Data analysis and variants filtration were carried out based on the minor allele frequency, the variant’s effect, and in silico prediction. We filtered all variants with allele frequencies < 0.0001 in the GnomAD genome database. First, we examined variants with predicted major structural effects; nonsense, stop loss, frameshift, and canonical splice site variants. After checking for loss of function variants, we examined missense variants annotated as pathogenic by Sift and Polyphen software [14, 15] and non-frame-shift variants. To verify that we had not missed strong candidate variants due to our conservative frequency filter, we repeated the analysis using a frequency cut-off of 0.001 in the GnomAD genome database. In this study, we focused the analysis to Online Inheritance in Man (OMIM) disease-related genes (https://www.omim.org/) and recently published ataxia or HSP-causative genes with strong evidence from the literature. When multiple affected relatives were processed from the same family, they were analyzed together according to the suspected inheritance mode and then individually to take into account possible phenocopies. Genomic rearrangements were tested using PennCNV-1.0.5 [16].
Sanger sequencing
Primers were designed using Primer3 Plus software [17]. DNA was amplified on a GeneAmp® PCR System 9700 (Thermo Fisher, MA, USA). We checked the quantity and quality of PCR products, including product size and off-target amplification, using the Caliper®LabChip GX System and its related software (PerkinElmer, MA, USA) according to the manufacturer’s protocol. Sanger sequencing was then done at the labs of Eurofins Genomics (Germany) using the Big Dye Chemistry in an ABI3730 automated sequencer (Applied Biosystems, Thermo Fisher Scientific, USA) using the procedures recommended by the manufacturer on the PCR product. Sequencing files (ABI format) were then visualized and analyzed using Sequence Scanner Software® v2.0 (Thermo Fisher Scientific, USA).
Array genotyping
Two hundred nanograms of genomic DNA from participating members of the families F5, F41, F54, F65, F70, F73, F74, F75, F80, F81, and F85 were sent for genotyping at the Pitié-Salpêtrière Post-Genomic Platform (P3S), Paris, France. Genotyping was performed on Illumina Infinium OmniExpress-24vl-3-A1 array, which contained ~ 710,000 SNP markers. Raw data were analyzed at the P3S platform using GenomeStudio™ Software. Runs of homozygosity were performed using version 1.07 of Plink software [18] to prioritize the variants in WES analysis. Candidate pathogenic copy number variants (CNV) were searched using PennCNV-1.0.5 software [16].
Repeats expansion detection
Genomic DNA from patients with clinical presentations and pedigree structures suggestive of dominant spinocerebellar ataxias (F49 and F65) or Friedreich’s ataxia (F38) were screened for repeats expansion using specific PCR-based approaches at the genetics departments of the Pitié-Salpêtrière Hospital and University Hospital of Montpellier, France, respectively. From the dominant spinocerebellar ataxias, we screened for pathogenic DNA repeat expansions in the SCA genes ATXN1 (SCA1), ATXN2 (SCA2), ATXN3 (SCA3), CACNA1A (SCA6), ATXN7 (SCA7), TBP (SCA17), and ATN1 (DRPLA) using a multiplex PCR amplification followed by capillary electrophoresis in a 3730 ABI sequencer (Applied Biosystems). The FRDA gene-associated repeat was amplified by a repeat-primed PCR approach.
Results
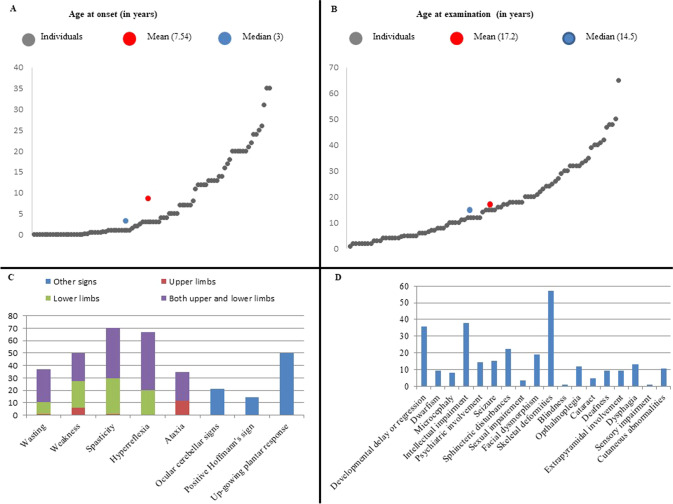
We studied 38 families (90 sampled affected patients), each including at least one patient manifesting features of SCDs. The studied families originated from multiple regions in Sudan, though the distribution is markedly skewed towards the central parts of the country. More than one-fifth of the families (23.6%) originated from a single state in central Sudan, the River Nile state (Fig. 1B). The number of affected males and females in our cohort was approximately equal (53% males vs. 47% females). However, the patients’ age at examination distribution was less homogenous; most patients were less than 18 years old. The mean and median patients’ ages-at-examination were 17.24 (SD = 13.97) and 14.5 years, respectively (Fig. 2B).
Fig. 2. Clinical overview of the cohort.
A Patients’ age-at-examination. The mean age-at-examination was 17.2 years. B Age-at-onset of the SCDs in our patients. The mean age-at-onset was 7.54 years. C Signs detected during patients’ examination. The percentages of patients with pyramidal and cerebellar signs are shown. The majority of our patients presented with pyramidal features. D Features complicating the SCDs phenotype in our cohort. Skeletal deformities, intellectual impairment, and developmental delay and/or regression are the most common features complicating the SCDs phenotype in our cohort.
Disease phenotypes
Most patients had an early-onset disease; the mean and median ages-at-onset were 7.44 (SD = 9.11) and three years, respectively (Fig. 2A). Most of the patients in our cohort had spasticity (70%). Limb ataxia was noted only in ~30% of the patients, while ocular cerebellar signs were noted in 20% (Fig. 2C). A pure SCDs phenotype was noted in ~14% of the patients, nine presented with a pure HSP while four presented with pure cerebellar ataxia. The most common features complicating the SCD phenotype in our patients were skeletal deformities, developmental delay or regression, and intellectual impairment (Fig. 2D). Table 1 summarizes the clinical presentation and the genetic diagnosis in each family where appropriate, and detailed phenotype of all the patients in our cohort are available per patient in the Supplementary Table. The diversity of clinical association did not allow us to distinguish a frequent phenotype that could have been analyzed separately as a whole. However, several families presented with similar clinical presentations, such as families F63 and F84, but they finally appeared to segregate mutations in different genes.
Table 1.
Overview of the clinical presentation of the families in our cohort with the OMIM corresponding identity deduced from the genetic results.
| Family code | Clinical features observed in patients | OMIM phenotype (MIM ID) | Gene |
|---|---|---|---|
| F8 | Cerebellar ataxia, cataract, and hypotonia | Marinesco-Sjogren syndrome (# 248800) | SIL1 |
| F27 | Stunted growth, dysmorphic features, microcephaly, cerebellar ataxia, and retinitis pigmentosum | Cockayne syndrome A (# 216400) | ERRC8 |
| F31 | Cerebellar ataxia and developmental delay | Cerebellar ataxia, mental retardation, and disequilibrium syndrome 1 (# 224050) | VLDLR |
| F38 | Cerebellar ataxia | Friedreich ataxia (# 229300) | FXN |
| F41 | Cafe-au-lait spots, cerebellar ataxia, and skeletal deformities | Neurofibromatosis, type 1 (# 162200) | NF1 |
| F49 | Cerebellar ataxia, spasticity, and dysarthria | Machado-Joseph disease (# 109150) | ATXN3 |
| F53 | Spastic limbs, wasting, and low-set ears | Spastic paraplegia 11, autosomal recessive (# 604360) | SPG11 |
| F54 | Developmental delay and spastic limbs | Spastic paraplegia 48, autosomal recessive (# 613647) | AP5Z1 |
| F57 | Spastic limbs, skeletal deformities, cerebellar ataxia, wasting, and weakness | Spastic ataxia, Charlevoix-Saguenay type (# 270550) | SACS |
| F59 | Intellectual disability, spastic limbs, and skeletal deformities | Spastic paraplegia 45, autosomal recessive (# 613162) | NT5C2 |
| F61 | Regressed developmental milestones, spastic limbs, cerebellar ataxia, and ophthalmoplegia | Spastic paraplegia 35, autosomal recessive (# 612319) | FA2H |
| F62 | Stunted growth, intellectual disability, spastic limbs, dysarthria, wasting, and cerebellar ataxia | Cockayne syndrome B (# 133540) | ERCC6 |
| F63 | Intellectual disability, aggression, dysmorphic features, squint, skeletal deformities, and spastic limbs (ref. 19) | Mental retardation, autosomal recessive 36 (# 615286) | ADAT3 |
| F66 | Developmental delay, spastic limbs, and weakness | Novel phenotype in a known gene not associated previously with SCD | DMXL2 (VUS) |
| F67 | Cerebellar ataxia, epilepsy, and learning disability | Ceroid lipofuscinosis, neuronal, 7 (# 610951) | MFSD8 (VUS) |
| F68 | Abnormal gait, spasticity, and cerebellar ataxia | Spastic paraplegia 35, autosomal recessive (# 612319) | FA2H |
| F70 | Weakness, wasting, dysphagia, developmental delay/regression, and unsteady gait | Leukodystrophy, hypomyelinating, 7, with or without oligodontia and/or hypogonadotropic hypogonadism (# 607694) | POLR3A |
| F76 | Developmental delay, spasticity, wasting, and skeletal deformities | Spastic paraplegia 54, autosomal recessive (# 615033) | DDHD2 |
| F78 | Spasticity, cerebellar ataxia, weakness, intellectual disability, and ears of the lynx sign on MRI | Spastic paraplegia 15, autosomal recessive (# 270700) | ZFYVE26 |
| F79 | Developmental delay, dysmorphic features, abnormal gait, autistic features, and behavioral disturbances (ref. 19) | Mental retardation, autosomal recessive 38 (# 615516); spinocerebellar ataxia, X-linked 1 (# 302500) | HERC2/ ATP2B3 (VUS) |
| F80 | Developmental delay, spastic limbs, and skeletal deformities | Spastic paraplegia 45, autosomal recessive (# 613162) | NT5C2 |
| F81 | Developmental delay, spastic limbs, and epilepsy (ref. 19) | Novel phenotype in a known gene not associated previously with SCD. | CCDC82 |
| F82 | Global developmental delay, microcephaly, dysmorphic features, epilepsy, and spastic lower limbs | Mental retardation, X-linked, syndromic, turner type (# 309590) | HUWE1 |
| F83 | Early-onset pure hereditary spastic paraplegia (ref. 19) | Spinocerebellar ataxia 40 (# 616053) | CCDC88C |
| F84 | Developmental delay/regression, microcephaly, squint, and generalized spasticity | Mucolipidosis IV (# 252650) | MCOLN1 |
| F85 | Deafness and mutism, mild cerebellar ataxia, spasticity | Deafness, autosomal recessive 84a (# 613391) | PTPRQ (VUS) |
| FM3 | Global developmental delay, spastic limbs, and skeletal deformities (ref. 27) | Neurodevelopmental disorder with microcephaly, hypotonia, and variable brain anomalies (# 617481) | PRUNE1 |
| FM6 | Spasticity and ocular cerebellar signs (ref. 39) | Leukoencephalopathy with brainstem and spinal cord involvement and lactate elevation (# 611105) | DARS2 |
| The undiagnosed families | |||
| F5 | Spasticity and cerebellar ataxia | Not applicable | |
| F46 | Skeletal deformities and cerebellar ataxia | Not applicable | |
| F74 | Weakness, myopathic faces, contractures, and hypophonia | Not applicable | |
| Families with mutations in candidate novel genes | |||
| F7 | Pure hereditary spastic paraplegia | Unpublished | |
| FM2 | Cerebellar ataxia, spasticity, epilepsy, dysphagia, extrapyramidal features, and brain MRI white matter abnormalities. | Unpublished | |
| F65 | Late-onset ataxia and spasticity | Unpublished | |
| F69 | Febrile convulsions, global developmental delay, spastic limbs, and skeletal deformities (ref. 30) | Spastic Paraplegia 86, autosomal recessive (# 619735) | ABHD16A |
| F73 | Slurred speech, gait abnormalities, and epilepsy | Unpublished | |
| F75 | Weakness and spasticity | Unpublished | |
| F77 | Global developmental delay, spasticity, and convulsions | Unpublished | |
OMIM Online inheritance in man. More detailed clinical information is provided in the supplementary table.
Genetic tests results
When focusing the analysis on known genes involved in neurogenetic conditions, we reached a genetic diagnosis in 63% (24/38) of the studied families, possibly 73% (28/38) if including families with variants of uncertain significance (VUS). In most of these families (23/28, 82%), diagnosis concerned all patients of the family that could be tested. The candidate variants in the families F41, F85, F54, F70, and F80 were not identified in all the patients within the family (Table 2 and the Supplementary material). This partial segregation was probably due to the high consanguinity rate that concentrated several disease-causing mutations or non-genetic phenocopies in the same family.
Table 2.
Overview of the genetic data in our patients (full cohort).
| Family code | Patient ID | Consanguinity | Gender | Genetic tests performed | Gene (causative variant in known SCD genes) | Gene (VUS or CN) |
|---|---|---|---|---|---|---|
| F5 | 53 | yes | M | WES, array genotyping | none | none |
| F5 | 56 | yes | M | Array genotyping | none | none |
| F5 | 61 | yes | M | WES, array genotyping | none | none |
| F7 | 85 | yes | M | WES, Sanger seq. validation | none | CN gene |
| F7 | 86 | yes | F | WES, Sanger seq. Validation | none | CN gene |
| F8 | 98 | yes | F | WES, Sanger seq. validation | SIL1 | none |
| F8 | 99 | yes | F | WES, Sanger seq. validation | SIL1 | none |
| F27 | 227 | no | M | WES, Sanger seq. validation | ERCC8 | none |
| F27 | 228 | no | F | WES, Sanger seq. validation | ERCC8 | none |
| F27 | 229 | no | F | WES, Sanger seq. validation | ERCC8 | none |
| F31 | 267 | yes | F | WES, Sanger seq. validation | VLDLR | none |
| F31 | 268 | yes | F | WES, Sanger seq. validation | VLDLR | none |
| F38 | 318 | yes | M | WES; FRDA screening | FRDA expansion | none |
| F38 | 317 | yes | M | Sanger seq. validation | FRDA not tested | none |
| F38 | 319 | yes | F | Sanger seq. validation | FRDA not tested | none |
| F38 | 328 | yes | M | not sampled | not sampled | not sampled |
| F38 | 329 | yes | F | WES | FRDA not tested | none |
| F41 | 351 | yes | F | WES, array genotyping, Sanger seq. validation | NF1 | none |
| F41 | 349 | yes | M | WES, array genotyping, Sanger seq. validation | no segreg of NF1 | none |
| F46 | 380 | yes | M | WES | none | none |
| F46 | 383 | yes | F | Not sampled | Not sampled | Not sampled |
| F49 | 396 | no | M | SCA expansion screening | SCA3 | none |
| F49 | 398 | no | M | SCA expansion screening | SCA3 | none |
| FM2 | 2016 | yes | F | WES, Sanger seq. validation | none | CN gene |
| FM2 | 2008 | yes | M | Sanger seq. validation | none | no segreg of CN gene |
| FM2 | 2013 | yes | F | WES, Sanger seq. validation | none | CN gene |
| FM3 | 2020 | yes | M | WES, microsatelittes genotyping, Sanger seq. validation | PRUNE1 | none |
| FM3 | 2021 | yes | F | Microsatelittes genotyping, Sanger seq. validation | PRUNE1 | none |
| FM3 | 2022 | yes | F | WES, microsatelittes genotyping, Sanger seq. validation | PRUNE1 | none |
| FM6 | 2042 | yes | F | Sanger seq. validation | DARS2 | none |
| FM6 | 2043 | yes | F | WES, Sanger seq. validation | DARS2 | none |
| FM6 | 2044 | yes | F | Sanger seq. validation | DARS2 | none |
| F53 | 417 | no | M | Sanger seq. validation | SPG11 | none |
| F53 | 418 | no | M | Sanger seq. validation | SPG11 | none |
| F53 | 419 | no | M | HSP panel screening, Sanger seq. validation | SPG11 | none |
| F54 | 427 | yes | M | HSP panel, WES, array genotyping, Sanger seq. validation | AP5Z1 | none |
| F54 | 426 | yes | F | WES, array genotyping, Sanger seq. validation | no segreg of AP5Z1 | none |
| F57 | 439 | yes | F | Sanger seq. validation | SACS | none |
| F57 | 440 | yes | M | Sanger seq. validation | SACS | none |
| F57 | 441 | yes | F | HSP panel, Sanger seq. validation | SACS | none |
| F59 | 451 | yes | F | Sanger seq. validation | NT5C2 | none |
| F59 | 452 | yes | M | HSP panel, Sanger seq. validation | NT5C2 | none |
| F59 | 453 | yes | M | Sanger seq. validation | NT5C2 | none |
| F61 | 465 | yes | M | HSP panel, Sanger seq. validation | FA2H | none |
| F61 | 467 | yes | F | Sanger seq. validation | FA2H | none |
| F62 | 470 | yes | M | WES, Sanger seq. validation | ERCC6 | none |
| F62 | 471 | yes | M | HSP panel, WES, Sanger seq. validation | ERCC6 | none |
| F63 | 476 | yes | F | HSP panel, WES, Sanger seq. validation | ADAT3A | none |
| F63 | 477 | yes | F | WES, Sanger seq. validation | ADAT3A | none |
| F65 | 484 | yes | M | Array genotyping, Sanger seq. validation | none | CN gene |
| F65 | 485 | yes | M | SCA expansion screening, WES, array genotyping, Sanger seq. validation | none | CN gene |
| F65 | 486 | yes | M | HSP panel, SCA expansion screening, WES, array genotyping, Sanger seq. validation | none | CN gene |
| F65 | 487 | yes | F | Array genotyping, Sanger seq. validation | none | CN gene |
| F66 | 490 | yes | M | HSP panel, WES, Sanger seq. validation | none | DMXL2 |
| F66 | 493 | yes | M | HSP panel, WES, Sanger seq. validation | none | DMXL2 |
| F67 | 496 | yes | F | WES, Sanger seq. validation | none | MFSD8 |
| F68 | 503 | yes | F | HSP panel, WES, Sanger seq. validation | FA2H | none |
| F68 | 504 | yes | F | HSP panel, WES, Sanger seq. validation | FA2H | none |
| F69 | 508 | yes | F | HSP panel, WES, Sanger seq. validation | none | CN gene (ABHD16A) |
| F69 | 509 | yes | M | HSP panel, WES, Sanger seq. validation | none | CN gene (ABHD16A) |
| F70 | 513 | yes | F | WES, array genotyping, Sanger seq. validation | POLR3A | none |
| F70 | 514 | yes | M | HSP panel, WES, Sanger seq. Validation, array genotyping | no segreg of POLR3A | none |
| F73 | 527 | yes | M | HSP panel, WES, Sanger seq. Validation, array genotyping | none | CN gene |
| F73 | 525 | yes | F | HSP panel, WES, Sanger seq. Validation, array genotyping | none | no segreg of CN gene |
| F74 | 529 | yes | M | Array genotyping, Sanger seq. validation | none | none |
| F74 | 530 | yes | M | HSP panel, WES, Sanger seq. Validation, array genotyping | none | none |
| F74 | 531 | yes | M | HSP panel, WES, Sanger seq. Validation, array genotyping | none | none |
| F74 | 532 | yes | M | Array genotyping, Sanger seq. validation | none | none |
| F74 | 533 | yes | M | Array genotyping, Sanger seq. validation | none | none |
| F74 | 535 | yes | M | Array genotyping, Sanger seq. validation | none | none |
| F74 | 536 | yes | M | Array genotyping, Sanger seq. validation | none | none |
| F75 | 542 | yes | F | HSP panel, WES, Sanger seq. Validation, array genotyping | none | CN gene |
| F75 | 543 | yes | M | HSP panel, WES, Sanger seq. Validation, array genotyping | none | CN gene |
| F76 | 547 | yes | F | HSP panel, Sanger seq. validation | DDHD2 | none |
| F76 | 548 | yes | M | HSP panel, Sanger seq. validation | DDHD2 | none |
| F77 | 550 | yes | F | HSP panel, WES, Sanger seq. validation | none | CN gene |
| F77 | 551 | yes | M | HSP panel, WES, Sanger seq. validation | none | CN gene |
| F78 | 557 | yes | M | HSP panel, Sanger seq. validation | ZFYVE26 | none |
| F78 | AA | yes | F | Sanger seq. validation | ZFYVE26 | none |
| F79 | 568 | yes | M | WES, Sanger seq. validation | none | HERC2/ATP2B3 |
| F80 | 573 | yes | F | WES, array genotyping, Sanger seq. validation | NT5C2 | none |
| F80 | 572 | yes | F | WES, array genotyping, Sanger seq. validation | no segreg of NT5C2 | none |
| F81 | 576 | yes | M | WES, array genotyping, Sanger seq. validation | CCDC82 | none |
| F81 | 577 | yes | F | WES, array genotyping, Sanger seq. validation | CCDC82 | none |
| F82 | 580 | yes | M | WES, Sanger seq. validation | HUWE1 | none |
| F83 | 581 | distant | F | WES, Sanger seq. validation | CCDC88C | none |
| F84 | 588 | yes | M | WES, Sanger seq. validation | MCOLN1 | none |
| F84 | 589 | yes | F | WES, Sanger seq. validation | MCOLN1 | none |
| F85 | 2056 | yes | M | WES, array genotyping, Sanger seq. validation | none | PTPRQ homozygous |
| F85 | 2059 | yes | F | Array genotyping, Sanger seq. validation | none | PTPRQ heterozygous |
M Males, F Females, CN gene Candidate novel gene, WES Whole exome sequencing, FRDA Friedreich ataxia gene (GAA expansion), SCA3 Repeat expansion in the ATXN3 gene, HSP panel Hereditary spastic paraplegia gene panel, no cosegreg no cosegregation of the variant (absent in the patient)
Inheritance patterns
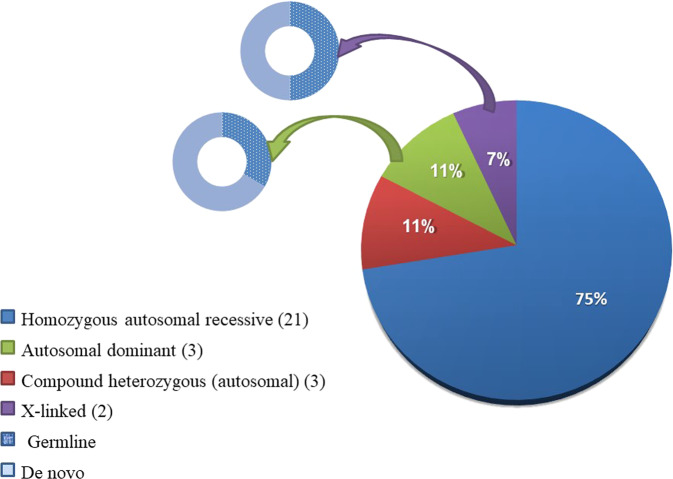
The pattern of inheritance in most of the possibly diagnosed families was an autosomal recessive pattern in 24 of them (Fig. 3); of note, F79 was counted in two inheritance modes as it segregated two likely causative variants with different patterns of inheritance but both possibly contributing to the phenotype as we reported previously [19]). Most autosomal recessive families were segregating homozygous variants (75%), while compound heterozygous variants were observed in 11%. Autosomal dominant inheritance was identified in 11% of the families, and two families showed X-linked inheritance (Fig. 3).
Fig. 3. The pattern of inheritance in the families with mutations in known disease genes.
Compound heterozygous inheritance is separated from homozygous autosomal recessive inheritance to highlight the effect of consanguinity. One family, F79, was counted in two inheritance modes as it segregated two likely causative variants with different patterns of inheritance but both possibly contributing to the phenotype as we reported previously (Ref. 19).
Genetic variants
We identified 31 different variants in known disease genes in this study (Table 3): 26 causative or likely causative variants in 24 families and five variants of unknown significance (VUS) in four families. One variant in FA2H, NM_024306.5:c.674 T > C (p.Leu225Pro), was identified twice in families F61 and F68 who shared a related phenotype. Most of the variants we identified were missense variants (12/31, 39%), followed in frequency by splice-site (23%) and frameshift (19%) variants. We identified pathogenic repeat expansions in two families and nonsense variants in three families. Array genotyping didn’t detect candidate CNV or chromosomal rearrangements. Each variant was validated by VariantValidator version 2.1.1 and classified by the authors after evaluating the entire clinical situation (Supplementary table), segregation analysis (Supplementary material) and the ACMG 2015 classification (Table 3).
Table 3.
Causative variants and variants of uncertain significance (VUS) identified in genes previously known to be associated with neurological phenotypes.
| Family code | Inheritance pattern | Gene | Variant | ACMG class for point mutations / Conclusion from other criteria | ACMG criteria |
|---|---|---|---|---|---|
| Families in which the candidate variant explains the disease in all the patients | |||||
| F8 | AR homozygous | SIL1 | NM_001037633.2:c.767 + 1 G > A (splice-site) | Pathogenic | PVS1, PM2, PP1, PP4 |
| F27 | AR compound heterozygous | ERCC8 | NM_000082.4:c.523 T > C (p.Ser175Pro) / NM_000082.4:c.572_574del (p.Ala191del) | Likely pathogenic/likely pathogenic | PM2, PP1, PP3, PP4 / PM2, PP1, PP3, PP4, BP3 |
| F31 | AR homozygous | VLDLR | NM_003383.5:c.2144 G > A (p.Cys715Tyr) | Likely pathogenic | PP1 (moderate), PM2, PP3, PP4 |
| F38 | AR homozygous | FXN | GAA repeat expansion (a single patient tested but phenotype compatible in other patients) | Not applicable/Toxic expansion | |
| F49 | AD | ATXN3 | CAG repeat expansion (73 + /−3 CAG) | Not applicable/Toxic expansion | _ |
| F53 | AR compound heterozygous | SPG11 | NM_025137.4:c.2399del (p.Tyr800Phefs*19) / NM_025137.4:c.6739_6742del (p.Glu2247Leufs*14) | Pathogenic/pathogenic | PVS1, PM2, PM3, PP1/ PVS1, PM2, PP1, PP5 |
| F57 | AR homozygous | SACS | NM_014363.6:c.10444_10447del (p.Leu3482Glnfs*12) | Pathogenic | PVS1, PM2, PP1 |
| F59 | AR homozygous | NT5C2 | NM_001351171.2:c.175 + 1 G > A (splice-site) | Pathogenic | PVS1, PM2, PP1, PP5 |
| F61 | AR homozygous | FA2H | NM_024306.5:c.674 T > C (p.Leu225Pro) | Likely pathogenic | PP1 (strong), PM2, PP3, PP4 |
| F62 | AR homozygous | ERCC6 | NM_000124.4:c.4063–1 G > C (splice-site) | Pathogenic | PVS1, PM2, PP1, PP4, PP5 |
| F63 | AR homozygous | ADAT3 | NM_138422.4:c.430 G > A (p.Val144Met) | Pathogenic | PM2, PP1 (strong), PS3, PP3, PP4, PP5 |
| F66 | AR homozygous | DMXL2 | NM_001174116.3:c.5020 A > C (p.Lys1674Gln) | VUS | PM2, PP1, PP3, BP1 |
| F67 | AR homozygous | MFSD8 | NM_152778.3:c.753 A > G (p.Glu251Glu) (splice site) | VUS | PM2 |
| F68 | AR homozygous | FA2H | NM_024306.5:c.674 T > C (p.Leu225Pro) | Likely pathogenic | PP1 (strong), PM2, PP3, PP4 |
| F76 | AR homozygous | DDHD2 | NM_015214.3:c.985 C > T (p.Arg329*) | Pathogenic | PVS1, PM2, PP5, PP1, PP3 |
| F78 | AR homozygous | ZFYVE26 | NM_015346.4:c.1254dup (p.Cys419Valfs*61) | Pathogenic | PVS1, PM2, PP1, PP3 |
| F79 | AR homozygous / X-linked | HERC2/ATP2B3 | NM_004667.6:c.10855 C > T (p.Pro3619Ser)/ NM_021949.3:c.2086 C > T (p.Arg696Cys) | VUS/VUS | PM2, PP3 / PM2, PP3 |
| F81 | AR homozygous | CCDC82 | NM_001318736.2:c.535 C > T (p.Arg179*) | Pathogenic | PVS1, PM2, PP1 |
| F82 | X-linked (likely de novo) | HUWE1 | NM_031407.7:c.12639 G > A (p.Met4213Ile) | Likely pathogenic | PM1, PM2 |
| F83 | AD (likely de novo) | CCDC88C | NM_001080414.4:c.1993G > A (p.Glu665Lys) | Likely pathogenic | PS3, PM2, PP3 |
| F84 | AR homozygous | MCOLN1 | NM_020533.3:c.514 C > T (p.Arg172*) | Pathogenic | PVS1, PP5, PM2, PP1 |
| FM3 | AR homozygous | PRUNE1 | NM_021222.3:c.132 + 2 T > C (splice-site) | Pathogenic | PVS1, PM2, PP1 |
| FM6 | AR compound heterozygous | DARS2 | NM_018122.5:c.1762C > G (p.Leu588Val) / NM_018122.5:c.563 G > A (p.Arg188Gln) | Likely pathogenic/likely pathogenic | PM2, PP1 (moderate), PP3, PP4, PP5 / PM2, PP1 (moderate), PP3, PP4 |
| Families in which the candidate variant explains the disease only in a proportion of the patients | |||||
| F41 | AD (likely de novo) | NF1 | NM_001042492.3:c.187_188del (p.Lys63Glufs*3) | Pathogenic | PVS1, PM2, PM6 |
| F54 | AR homozygous | AP5Z1 | NM_014855.3:c.1132 G > A (p.Gly378Arg) (splice-site) | Likely pathogenic | PVS1, PM2 |
| F70 | AR homozygous | POLR3A | NM_007055.4:c.1771–7 C > G (splice-site) | Likely pathogenic | PP1 (strong), PS3, PM2, PP5 |
| F80 | AR homozygous | NT5C2 | NM_001134373.3:c.629del (p.Tyr210Serfs*17) | Pathogenic | PVS1, PM2, PP3 |
| F85 | AR homozygous | PTPRQ | NM_001145026.2:c.5893 C > A (p.Pro1965Thr) | VUS | PM2, PP3 |
AR Autosomal recessive, AD Autosomal dominant, PVS1 null variant in a gene where loss of function is a known disease mechanism, PS3 well-established in vitro or in vivo functional studies supportive of a damaging effect on the gene or gene product, PM1 located in a mutational hot spot or well-established critical protein domain without benign variation, PM2 absent or have low frequency in gnomAD public database, PM3 detected in trans with a pathogenic variant, PM6 assumed de novo, PP1 cosegregate with disease in multiple affected family members, PP3 multiple lines of computational evidence support a deleterious effect, PP4 Patients’ phenotype or family history is highly specific for a disease with a single genetic etiology, PP5 reported as deleterious by a reputable source.
Approximately eighty percent of the candidate single nucleotide and insertion/deletion variants located in known disease genes (23/29, excluding the two nucleotide expansions) were either pathogenic or likely pathogenic, according to the ACMG 2015 guidelines for interpreting sequence variations [20].
Additionally, five likely causative variants fitted to the category of VUS but some with convincing evidence of pathogenicity, however. All the candidate deleterious VUS identified in this cohort segregated with the disease and could fit the categories of pathogenic or likely pathogenic variants if additional evidence is identified in the future. The VUS NM_152778.3:c.753 A > G (p.Glu251Glu), identified in family F67, is a synonymous variant but predicted to alter the splicing of MFSD8 (TraP score 0.96; SpliceAI score 0.7) and cause skipping of exon 8. It was absent from the gnomAD v2.1.1 database and from 120 index cases of Sudanese origin with various neurological conditions. The patient presented with intellectual disability, cerebellar ataxia, and epilepsy (Supplementary Table), a phenotype suggestive of, but not exclusive to, neuronal ceroid lipofuscinosis. Furthermore, the patient had cousins who passed away in their early childhood after a similar illness. We considered it as a plausible candidate based on in-silico prediction tools and suggestive phenotype and family history.
The second VUS was in DMXL2, NM_001174116.3:c.5020 A > C (p.Lys1674Gln), and was identified in the two probands from family F66. It is predicted as pathogenic by Sift, Polyphen 2, MutationTaster [21], LRT [22], and Provean [23] and had a CADD score of 28. The variant was not predicted by the Missense3D tool to alter the protein structure, however. On the other hand, the variant was predicted to unmask a splice site inside exon 21 which may affect the mRNA stability and must then be explored in patient’s cells if expressed in leukocytes or fibroblasts. This was not possible, however. Pathogenic mutations in the DMXL2 gene cause the autosomal dominant deafness type 71 (OMIM # 617605), and the autosomal recessive developmental and epileptic encephalopathy type 81 (OMIM # 618663) and polyendocrine-polyneuropathy syndrome (OMIM # 616113) [24–26]. We herein, potentially extended the phenotype of DMXL2 mutations to include complex HSP. Details about the clinical presentations of the previous families with DMXL2 variants are provided in the Supplementary material.
The variant NM_001145026.2:c.5893 C > A (p.Pro1965Thr) in PTPRQ was detected in two adult patients from family F85 who presented with congenital deafness and mutism but at different zygosity state. PTPRQ variants have been reports in AR and AD hearing loss with mildly delayed development (MIM # 617663 & 613391). This variant was also predicted as deleterious by Sift, Polyphen 2, MutationTaster, and Provean. It was absent in the gnomAD v2.1.1 database. We considered it as a VUS.
Details about the VUS identified in family F79 in HERC2 and ATP2B3 were provided in a previous report [19].
Discussion
Diagnosis yield
The Sudanese population is paradoxically characterized by a complex genetic structure and high consanguinity rates [6, 10]. The high level of homozygosity in our cohort was reflected by the predominance of homozygous recessive diseases (75%) and the detection of three established/possible founder variants. Two of these founder variants were in ADAT3 and PRUNE1 genes as we reported previously [19, 27]. The third possible founder variant, NM_024306.5:c.674 T > C (p.Leu225Pro), was in FA2H and was detected in two unrelated families, F61 and F68, that descended from different tribes in Kordofan province, western Sudan. Nevertheless, we also identified autosomal dominant and X-linked (hemizygous) conditions in several families.
Most of our families originated from the central parts of Sudan. This can be attributed either to differences in the accessibility to the health system and our collaborating clinics or genuine differences in the frequency of genetic diseases between central Sudan populations and other Sudanese populations. We favor the first explanation as other consanguinity-linked genetic diseases, such as sickle cell anemia, are common in non-central parts of the country [10].
All age groups were represented in our cohort, particularly those < 18 years, indicating the degree of care provided to this age group by their families. On the other hand, we have patients with childhood-onset diseases who were first examined after their forties (after decades of disease duration, > 40 years in two patients), epitomizing the long-term odysseys of patients with genetic diseases and underlining the importance of genetic diagnosis for patients’ and families’ satisfaction. Also, the percentages of males and females in our cohort were approximately equal, signifying the absence of gender-based inequalities in the accessibility of care and minimizing the contribution of X-linked dominant inheritance to SCDs in our cohort.
Previously, we screened 25 Sudanese families with HSP for mutations in 68 known HSP genes using NGS targeted gene panel [11]. We reached a genetic diagnosis in 28% of these cases [11], a diagnostic rate very similar to Portuguese [28] and European [12] patients. This last study, (ref. 12), showed that combining HSP panel with subsequent WES increased the diagnosis rate up to 50% when focusing on OMIM disease-related genes. WES used to further identify novel genes was shown to give a diagnostic yield of up to 75% [29]. In the current study, by using multiple genetic approaches, we identified disease-causing variants in known SCDs genes in 63–73% of the studied families. The overall diagnostic success rate if we consider our previous cohort (ref 11) is 52–59% (31–35/59 families). Furthermore, extending the analysis to all genes covered by the exome, we identified variants in novel candidate genes in seven out of the ten remaining families (see Tables 1, 2), potentially raising our diagnostic success rate ceiling to 92% instead of 73%. One of those seven novel causative genes has been reported [30] and the others are under validation and will be reported elsewhere (unpublished data). According to the results of our two studies, most of the major autosomal recessive SCDs genes are present in Sudan (SACS, SPG11, FXN) and some of the major dominant ones as well (e.g., SCA3), but there is no single major gene causing SCDs in Sudan. This might result from the position of Sudan in east Africa, at the frontiers between North Africa, the Middle East, and sub-Saharan Africa.
Lessons for genetic diagnosis of SCD in Sudan
In five families, we could establish the diagnosis in only a portion of the patients or branches since the variants were not segregating in all patients, outlining the need to introduce into the analysis pipeline of Sudanese families an additional step that consists of analyzing the patients individually after excluding the variants shared by multiple patients from the same family.
WES outweighs NGS targeted gene panel in discovering new SCDs genes [4]. However, based on our experience with the Sudanese population, and the experience of others, exome sequencing also significantly outweighs NGS-targeted gene panels in diagnosing known SCDs phenotypes, particularly in complex phenotypes [31]. Furthermore, WES enables the extension of phenotypes previously associated with mutations in certain genes in contrast to conservative NGS-targeted gene panels that target only the phenotype of interest. For instance, we extended the phenotypes associated with mutations in CCDC82 and CCDC88C in the current Sudanese cohort by using WES. CCDC82 was reported previously to cause an intellectual disability syndrome [32, 33]. We expanded the CCDC82-linked phenotype to include spastic paraplegia [19]. Later, another report of a patient of Pakistani origin confirmed that spasticity is part of the CCDC82-linked syndrome [34]. Similarly, we expanded the presentation of heterozygous mutations in CCDC88C to include early-onset pure spastic paraplegia [35]. Before, heterozygous gain-of-function CCDC88C mutations were only associated with spinocerebellar ataxia SCA40 [36]. In this report we also potentially extended the phenotype of DMXL2- and PTPRQ-linked disorders to include complex HSP.
In our opinion, the higher diagnostic success rate of WES overrides its technical difficulties when compared to NGS-targeted gene panel upon studying diseases with overlapping phenotypes like SCDs, particularly when considering the increasing technical feasibility of WES [37]. However, WES is less efficient for rearrangement detection than panels of genes, usually optimized for such discovery, as discussed (ref, 12). An issue in SCDs is the detection of nucleotide repeat expansions that require independent specific techniques but there are improvements of some algorithm for such quest in WES data and in genome sequencing [38].
In conclusion, up-to-now, SCDs in Sudan are caused by multiple genes; none of them significantly predominate over the others. The use of multiple genetic approaches that included WES enhanced the diagnosis of known SCDs phenotypes and the potential discovery of new SCDs genes.
Supplementary information
Acknowledgements
We are grateful to the genotyping and sequencing core facility of the Brain Institute (ICM, Paris, France) and the Pitié-Salpêtrière Post-Genomic Platform (P3S, Paris, France) for doing the next-generation sequencing and array genotyping, respectively. We thank Nicolas Auger, Claire-Sophie Davoine, Mélanie Papin and Lena Guillot-Noel for their advice and Prof. Michel Koenig, Dr. Marine Guillaud-Bataille, and Dr. Guillaume Banneau for their help in nucleotide repeat expansion detection.
Author contributions
AY, AEA, LEOE, and GS designed the study. AY, AAAH, INM, MAH, MAS, SME, HES, AEMN, MAA, ME, FYO, AMB, SOMAT, EEB, MK, AEA, and LEOE evaluated the patients. AY, RA1; FA, RA2, SE, MAM, IZME, ZO, HM, MOEM, AAE, EOEM, AKMAA, EAAA, EE, BKH, ASIAA, LS, MN, OMTE, TEAE, AE, ESAA, MF, KFA, MA, and LEOE collected the samples and relevant data and performed the experiments. AY and GS interpreted the results. MEI, AEA, LEOE, and GS supervised the study. AY and GS drafted the manuscript. GS obtained funds for implementing the study procedures. All authors critically revised and approved the final version of the manuscript and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This study was financially supported by the European Union (grant n°779257 - Solve-RD from the Horizon 2020 Research and Innovation Program, to GS), the Association Connaitre les Syndromes Cérébelleux (to GS) and by the ASL-HSP-France patient association (to GS). AY was supported by the Ministry of Higher Education, Sudan and the French Embassy, Sudan. MAS was supported by Researchers Supporting Project, King Saud University, Riyadh, Saudi Arabia (project number RSP-2020/38). Open access funding provided by Karolinska Institute.
Data availability
Some details of the participants, including family pedigrees and natural history, have been removed from this report to ensure anonymity and comply with the journal’s standards. Further information on segregation analysis and data supporting the findings of this study are available from the corresponding authors upon reasonable requests. All novel variants and VUS have been submitted to ClinVar (submission number SUB12076628).
Competing interests
GS received a grant from BIOGEN (Cambridge, USA) unrelated to this work, and the other authors declare that they have no competing interests.
Ethical approval
This study was approved by The Ethical Committee of Medical Campus, University of Khartoum, Sudan, and The Ethical Committee of the National University, Sudan (approval number NU-RECG200). All procedures in this study were performed per the 1975 Helsinki declaration and its later amendments. Informed written consent forms for participation were obtained from all participants or their guardians. DNA was anonymized and this report does not contain pedigree or full genealogical data to preserve this anonymization.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ashraf Yahia, Email: ashraf.yahia@ki.se.
Giovanni Stevanin, Email: giovanni-b.stevanin@inserm.fr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-023-01344-6.
References
- 1.Synofzik M, Schüle R. Overcoming the divide between ataxias and spastic paraplegias: Shared phenotypes, genes, and pathways. Mov Disord. 2017;32:332–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parodi L, Coarelli G, Stevanin G, Brice A, Durr A. Hereditary ataxias and paraparesias: Clinical and genetic update. Curr Opin Neurol. 2018;31:462–71. [DOI] [PubMed] [Google Scholar]
- 3.Manto M, Gandini J, Feil K, Strupp M. Cerebellar ataxias: An update. Curr Opin Neurol. 2020;33:150–60. [DOI] [PubMed] [Google Scholar]
- 4.Yahia A, Stevanin G. The history of gene hunting in hereditary spinocerebellar degeneration: Lessons from the past and future perspectives. Front Genet. 2021;12:638730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruano L, Melo C, Silva MC, Coutinho P. The Global epidemiology of hereditary ataxia and spastic paraplegia: A systematic review of prevalence studies. Neuroepidemiology 2014;42:174–83. [DOI] [PubMed] [Google Scholar]
- 6.Ibrahim ME. Genetic diversity of the Sudanese: insights on origin and implications for health. Hum Mol Genet. 2021;30:R37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollfelder N, Schlebusch CM, Günther T, Babiker H, Hassan HY, Jakobsson M. Northeast African genomic variation shaped by the continuity of indigenous groups and Eurasian migrations. Tishkoff SA, editor. PLoS Genet. 2017;13:e1006976. [DOI] [PMC free article] [PubMed]
- 8.Dobon B, Hassan HY, Laayouni H, Luisi P, Ricaño-Ponce I, Zhernakova A, et al. The genetics of East African populations: A Nilo-Saharan component in the African genetic landscape. Sci Rep. 2015;5:9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babiker HMA, Schlebusch CM, Hassan HY, Jakobsson M. Genetic variation and population structure of Sudanese populations as indicated by 15 Identifiler sequence-tagged repeat (STR) loci. Investig Genet. 2011;2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daak AA, Elsamani E, Ali EH, Mohamed FA, Abdel-Rahman ME, Elderdery AY, et al. Sickle cell disease in western Sudan: Genetic epidemiology and predictors of knowledge attitude and practices. Tropical Med Int Health. 2016;21:642–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsayed LEO, Mohammed IN, Hamed AAA, Elseed MA, Johnson A, Mairey M, et al. Hereditary spastic paraplegias: Identification of a novel SPG57 variant affecting TFG oligomerization and description of HSP subtypes in Sudan. Eur J Hum Genet. 2016;25:100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Méreaux JL, Banneau G, Papin M, Coarelli G, Valter R, Raymond L, et al. Clinical and genetic spectra of 1550 index patients with hereditary spastic paraplegia. Brain 2022;145:1029–37. [DOI] [PubMed] [Google Scholar]
- 13.Desvignes JP, Bartoli M, Delague V, Krahn M, Miltgen M, Béroud C, et al. VarAFT: A variant annotation and filtration system for human next-generation sequencing data. Nucleic Acids Res. 2018;46:W545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, et al. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yahia A, Ayed I, ben, Hamed AA, Mohammed IN, Elseed MA, Bakhiet AM, et al. Genetic diagnosis in Sudanese and Tunisian families with syndromic intellectual disability through exome sequencing. Ann Hum Genet. 2022;86:181–94. [DOI] [PubMed] [Google Scholar]
- 20.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6. [DOI] [PubMed] [Google Scholar]
- 22.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi Y, Chan AP. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015;31:2745–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tata B, Huijbregts L, Jacquier S, Csaba Z, Genin E, Meyer V, et al. Haploinsufficiency of Dmxl2, Encoding a Synaptic Protein, Causes Infertility Associated with a Loss of GnRH Neurons in Mouse. PLoS Biol. 2014;12:e1001952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maddirevula S, Alzahrani F, Al-Owain M, Al Muhaizea MA, Kayyali HR, AlHashem A, et al. Autozygome and high throughput confirmation of disease genes candidacy. Genet Med. 2019;21:736–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen DY, Liu XF, Lin XJ, Zhang D, Chai YC, Yu DH, et al. A dominant variant in DMXL2 is linked to nonsyndromic hearing loss. Genet Med. 2017;19:553–8. [DOI] [PubMed] [Google Scholar]
- 27.Koko M, Yahia A, Elsayed LE, Hamed AA, Mohammed IN, Elseed MA, et al. An identical‐by‐descent novel splice‐donor variant in PRUNE1 causes a neurodevelopmental syndrome with prominent dystonia in two consanguineous Sudanese families. Ann Hum Genet. 2021;85:186–95. https://onlinelibrary.wiley.com/doi/10.1111/ahg.12437. [DOI] [PubMed] [Google Scholar]
- 28.Morais S, Raymond L, Mairey M, Coutinho P, Brandão E, Ribeiro P, et al. Massive sequencing of 70 genes reveals a myriad of missing genes or mechanisms to be uncovered in hereditary spastic paraplegias. Eur J Hum Genet. 2017;25:1217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science (1979). 2014;343:506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yahia A, Elsayed LEO, Valter R, Hamed AAA, Mohammed IN, Elseed MA, et al. Pathogenic variants in ABHD16A cause a novel psychomotor developmental disorder with spastic paraplegia. Front Neurol. 2021;12:720201. https://pubmed.ncbi.nlm.nih.gov/34489854/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coutelier M, Hammer MB, Stevanin G, Monin ML, Davoine CS, Mochel F, et al. Efficacy of exome-targeted capture sequencing to detect mutations in known cerebellar ataxia genes. JAMA Neurol. 2018;75:591–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harripaul R, Vasli N, Mikhailov A, Rafiq MA, Mittal K, Windpassinger C, et al. Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol Psychiatry. 2018;23:973–84. [DOI] [PubMed] [Google Scholar]
- 33.Riazuddin S, Hussain M, Razzaq A, Iqbal Z, Shahzad M, Polla DL, et al. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol Psychiatry. 2017;22:1604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauer G, Buchert R, Haack TB, Harting I, Gutschalk A. CCDC82 frameshift mutation associated with intellectual disability, spastic paraparesis, and dysmorphic features. Clin Genet. 2022;102:80–1. [DOI] [PubMed] [Google Scholar]
- 35.Yahia A, Chen ZS, Ahmed AE, Emad S, Adil R, Abubaker R, et al. A heterozygous mutation in the CCDC88C gene likely causes early-onset pure hereditary spastic paraplegia: a case report. BMC Neurol. 2021;21:78. https://bmcneurol.biomedcentral.com/articles/10.1186/s12883-021-02113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsoi H, Yu ACS, Chen ZS, Ng NKN, Chan AYY, Yuen LYP, et al. A novel missense mutation in CCDC88C activates the JNK pathway and causes a dominant form of spinocerebellar ataxia. J Med Genet. 2014;51:590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwinski P, Ong CK, Ling MHT, Poh YM, Khan AM, Ong HS. Advancing personalized medicine through the application of whole exome sequencing and big data analytics. Front Genet. 2019;10:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Depienne C, Mandel JL. 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? Am J Hum Genet. 2021;108:764–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yahia A, Elsayed L, Babai A, Salih MA, El-Sadig SM, Amin M, et al. Intra-familial phenotypic heterogeneity in a Sudanese family with DARS2-related leukoencephalopathy, brainstem and spinal cord involvement and lactate elevation: A case report. BMC Neurol. 2018;18:175. https://www.ncbi.nlm.nih.gov/pubmed/30352563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Some details of the participants, including family pedigrees and natural history, have been removed from this report to ensure anonymity and comply with the journal’s standards. Further information on segregation analysis and data supporting the findings of this study are available from the corresponding authors upon reasonable requests. All novel variants and VUS have been submitted to ClinVar (submission number SUB12076628).