Abstract
Chromosomal heteromorphisms (CHs) are morphological variations predominantly found in constitutive heterochromatic regions of the genome, primarily composed of tandemly repetitive sequences of satellite DNA. Although not completely devoid of genes, these regions are typically not transcribed into proteins and lack obvious phenotypic impact. Nonetheless, their clinical importance is increasingly under scrutiny, with several studies aiming to assess their influence on human diseases and susceptibilities, especially as they are seemingly part of the long noncoding RNAs in certain tissues. This article summarizes the classification methods of human heterochromatic CHs documented in the literature over the last two decades. Multiple scoring systems have been identified, and previous approaches for CH assessment and reporting in genetic diagnosis have shown inconsistencies. Owing to the current heterogeneity in the classification of CHs, data analysis may be biased, impacting the quality of clinical reports and human genetic research. This review highlights the need for a universal scoring system, which is essential for scientific reproducibility and the accurate identification and clinical evaluation of human CHs.
Subject terms: Cytogenetics, Structural variation, Genomic analysis
Standardizing chromosomal heteromorphisms for better genetic insights
In 2004, the Human Genome Project revealed the complete human DNA sequence, showing that most of our genome consists of noncoding and repetitive DNA sequences. Researchers reviewed various classification systems for chromosomal heteromorphisms to address this knowledge gap. They conducted a literature review using PubMed/MEDLINE, focusing on studies from 2000 to 2024 that described or used classification systems for CHs. This review included 18 studies and identified three main classification methods: Linear Measurement, Twice the Size of the Homologous, and 16p Size Comparison. The results showed inconsistencies in evaluating CHs due to different methodologies and lack of standard sizes for heterochromatic regions. Researchers concluded that standardizing CH classification is crucial for accurate genetic diagnosis and scientific reproducibility. Future research should focus on developing clear guidelines and advanced detection technologies to better understand CHs and their clinical implications. "This summary was initially drafted using artificial intelligence, then revised and fact-checked by the author."
Introduction
In 2004, the Human Genome Project provided comprehensive information on the human DNA sequence and redefined paradigms1. The majority of our genome consists of noncoding and/or repetitive DNA sequences, which underlie the heteromorphisms and polymorphisms identified at the chromosomal and molecular levels2,3. These sequences can be organized tandemly or as single repeat units spread throughout the genome and can be categorized according to the size and motif of the repeats, such as microsatellite, minisatellite and satellite DNAs3,4. Satellite DNA families (I, II, III, α, ß and γ) cover approximately 10% of the human genome and are often organized into higher-order repeat units (HORs), particularly in the centromeric regions of eukaryotic chromosomes5. HOR structures influence regulatory, replication, and gene expression processes, and the specific composition of their monomers plays a crucial role in defining a unique centromere identity, which also contributes to population diversity6.
In human karyotypes, variant heterochromatic portions of repetitive DNA have been recognized and documented for decades7. These regions consist of highly repetitive sequences, composed of micro- and minisatellites; α-, β- and other satellite DNAs; and transposable elements, which vary in extent from five to a few hundred base pairs8. Microscopically visible variations in these sections are commonly observed during standard cytogenetic examinations. Defined as chromosomal heteromorphisms (CHs), these alterations include differences in size, morphology and/or banding patterns of the constitutive heterochromatin segments. CHs are usually inherited and are typically located in 1) the centromeric regions of any of the human chromosomes; 2) the short arms, satellites, or stalks of the acrocentric chromosomes; or 3) the pericentromeric region of chromosomes 1, 9, and 16 and the long arm of Y chromosome9. Although not devoid of genes, these segments are generally not actively transcribed or expressed and are regarded as genetically inert10,11. CHs present in 2–5% of the general population are considered mitotically stable normal variants, and clinical practice cytogenetic guidelines recommend not reporting them12–14.
Despite having been widely accepted as benign, several studies concerning CHs have yielded conflicting results regarding their impact on human health15. Numerous authors have proposed an association between the presence of heteromorphic variants and clinical phenotypes such as infertility, neurodegenerative disorders or cancer predisposition4,7,16–18. The etiological mechanisms behind these correlations remain elusive; however, scientific studies suggest a potential impact of CHs on genome stability and modulation. The prevailing hypotheses point toward a likely effect of CHs on meiotic chromosome pairing and segregation and genome regulation, by either modulating or inhibiting the expression of specific genes through position-effect variegation8,11,19,20. Previous studies have revealed that satellite DNAs are transcribed as long noncoding RNAs (lncRNAs) in advanced cancer, under cellular stress and during embryogenesis (contributing to cellular specification and differentiation), highlighting the influence of repetitive DNA in biological processes21–23.
In addition to the relevance of clinical and genetic assessments of heteromorphic variants, CHs offer a valuable resource for evolutionary and population research because they are inherited in a Mendelian pattern and have low mutation rates. Therefore, CHs can serve as markers and can be used to identify and determine genetic linkages4.
Fully understanding CHs is crucial for both basic and clinical research, yet heterochromatic regions cannot be easily sequenced and aligned. As a result, CHs remain poorly documented in human genome browsers and are still exclusively identified using (molecular) cytogenetic methods16,24. Although these variations have been identified for many years, there is no consensus within the cytogenetic scientific community regarding the “normal” sizes of CHs or the adoption of a unique classification system for their evaluation9,15. Since the Paris Cytogenetic Conference in 1971, in which the first CH assessment scale was established, several classification methods have been proposed25–28. However, cytogenetic guidelines do not establish a standard scoring system, and approaches for evaluating CHs and their clinical and functional roles are inconsistent9,15.
This article provides a short review of the various classification systems for human heterochromatic CHs reported in the literature over the last two decades. Although “euchromatic variants” are also the result of large euchromatic copy number variations (CNVs), these variants are not considered in this review28. The aim is to identify, describe and critically evaluate different methodological approaches to improve and standardize the classification of human CHs in the future.
Materials and Methods
Searching strategy
The literature review was performed using the PubMed/MEDLINE electronic database. The search was restricted to articles written in the English language and published between 1 January 2000 and 30 April 2024. The key concepts used for the search in the title and/or abstract of all papers were “chromosome/chromosomal heteromorphisms”, “chromosome/chromosomal polymorphisms”, “heterochromatin variants/polymorphisms”, “size polymorphisms” and “heteromorphisms classification”. An additional “human species” filter was applied. The Boolean operators used were “AND”, “OR” and “NOT”.
Study selection: inclusion and exclusion criteria
With respect to the inclusion criteria, we selected only studies that described or used, as a methodological tool, classification systems for human heterochromatic CHs. Additionally, data suggesting a scoring system from a book by Thomas Liehr in 2014 were included28. All the articles were screened on the basis of their titles and abstracts to identify those meeting the inclusion criteria. The full text of each selected paper was subsequently assessed. Papers that evaluated human CHs but omitted the classification system were excluded.
Data extraction
The following information was extracted from the selected articles: (1) study identification (author and year); (2) article type; (3) scoring system; (4) banding technique; and (5) chromosomes assessed.
Results
Review of the literature
A total of 31 articles and their own source28 were identified and screened for inclusion. Fourteen papers were excluded because of the omission of a classification system. Among the 18 selected publications, several scoring systems with numerous variations each were identified. To clarify and simplify the scientific discussion, we grouped scoring systems into three main types on the basis of the use of different methodologies, rating levels and specific chromosome/chromosomal sections to which each system could be applied (1 - CHs at the centromeres of all chromosomes; 2 - CHs at the short arms of the acrocentric chromosomes; 3 - CH susceptible regions of chromosomes 1, 9, 16 and Y; and 4 - specific pericentric inversions). Twelve papers described/used the “twice the size of the homologous” method (TSH), three described/used the “16p size comparison” model (16p SC) and two described/used the “linear measurement” method (LM). One of the articles does not fit into any of these three main groups and therefore will not be categorized29. Information on the articles used in this review is provided in Table 1.
Table 1.
List of reviewed articles that met the stipulated inclusion criteria, ordered by year of publication.
| Data Source | Article Type | Classification System | Banding Technique | Assessed Chromosomes |
|---|---|---|---|---|
| Yasseen et al.30 | Original Research | LM | C | 1 9 16 |
| Yasseen et al.31 | Original Research | LM | C | Y |
| Madon et al.34 | Original Research | TSH | G, Q | 9 13 14 15 21 22 Y |
| Sahin et al.32 | Original Research | TSH + G group SC* | G, C, NOR | 1 9 13 14 15 16 21 22 Y |
| Minocherhomji et al.38 | Original Research | TSH | G, C, DAPI | 1 9 13 14 15 16 21 22 Y |
| Martínez et al. 2012 | Original Research | F group SC* | G | Y |
| Akbas et al. 2012 | Original Research | TSH + G group SC* | G, C, NOR | 1 9 13 14 15 16 21 22 Y |
| Guo et al.35 | Original Research | TSH | G | 1 9 13 14 15 16 21 22 Y |
| Mierla et al.39 | Original Research | TSH | G | 1 9 13 14 15 16 21 22 Y |
| Liehr 2014 | Book | 16p SC + 18p SC** | G | 1 9 13 14 15 16 21 22 Y |
| Liehr 2016 | Review | 16p SC + 18p SC** | G | 1 9 13 14 15 16 21 22 Y |
| Zhu et al.37 | Original Research | TSH + G group SC* | G, C, NOR | 1 9 13 14 15 16 21 22 Y |
| Rawal et al.33 | Original Research | TSH | G | 1 9 13 14 15 21 22 Y |
| Karaca et al.15 | Original Research | 16p SC + TSH*** | G | 1 9 13 14 15 16 21 22 Y |
| Li et al.19 | Original Research | TSH | G, C, NOR | 1 9 13 14 15 16 21 22 |
| Chakraborty et al.40 | Original Research | TSH | G, NOR | 1 9 13 14 15 16 21 22 Y |
| Rodriguez et al.41 | Original Research | TSH | G | 1 9 13 14 15 16 21 22 Y |
| Mottola et al.18 | Original Research | TSH | G | 9 |
LM linear measurement method, TSH twice the size of the homologous method, 16p SC 16p size comparison model.
*F or G group size comparison models are exclusively used for the Y chromosome.
**The 18p size comparison model is exclusively used for the acrocentric chromosomes.
***TSH method is exclusively used for the acrocentric chromosomes.
Linear measurement method
Introduced in 1978 by Balicek et al., the LM technique determines the precise length of the constitutive heterochromatin regions on chromosomes 1, 9, 16, and Y27. On the basis of a five-step assessment scale of C-band lengths proposed at the Paris Conference, the authors ascribed exact numerical limits to each category. The determination of the C-band length is conducted directly from an image using linear measurements, with the absolute dimensions expressed in micrometers. Table 2 presents the proposed limits for each chromosome. Originally used for paternity testing, the LM methodology was documented in two of the selected articles, where it was applied to evaluate the influence of CHs on male infertility27,30,31.
Table 2.
Five-step evaluation scale of C-band length limits (in units of 10−7 m) for chromosomes 1, 9, 16 and Y as proposed by Balicek et al.
| Chromosome | Very small | Small | Intermediate | Large | Very Large |
|---|---|---|---|---|---|
| 1 | −7.2 | 7.3–10.0 | 10.1–15.4 | 15.5–18.4 | 18.5- |
| 9 | −6.2 | 6.3–8.3 | 8.4–13.6 | 13.7–15.5 | 15.6- |
| 16 | −3.8 | 3.9–5.3 | 5.4–8.9 | 9.0–10.7 | 10.8- |
| Y | −8.0 | 8.1–8.6 | 8.7–12.2 | 12.3–12.9 | 13.0- |
Twice the size of the homologous method
This methodology is based on direct comparison of the size of the heterochromatic regions between homologous chromosomes of the same metaphase spread. On autosomes, variants must be at least twice or half the size of the corresponding region on its homolog and are reported as “h + ” or “h-”, respectively. The prominent satellites and stalks of the acrocentric chromosomes are referred to as “ps + ” and “pstk + ”, respectively. The length of the Y chromosome is compared with that of G-group chromosomes and reported as “Yqh + /-”, if larger or smaller, respectively. Pericentric inversions are described as “inv”. This evaluation system can be used directly in metaphases with G-banding or after selective banding, such as the C and NOR (nucleolar organizing region using silver stain) techniques. In 12 of the analyzed papers, this methodology was generally used as a tool to assess the clinical impact of CHs on human fertility18,19,32–41.
16p size comparison model
Originally developed in 1978 by Verma et al. for C banding, size heteromorphisms were classified into levels using 16p as a reference standard26. This methodology evaluates the “qh” regions on chromosomes 1, 9, 16 and Y by comparing their size with the short arm of chromosome 16 at the same G- or C-banded metaphase spread. Subsequently, heterochromatic regions can be scored as levels 1 to 5 on the basis of their length relative to the length of 16p (Table 3) or can be classified as “qh + ” variants if they are greater than 16p and “qh-” variants if they are less than half of 16p. TSH methodology is often used to assess the sizes of the short arms, stalks and satellites of acrocentric chromosomes. In 1977, Verma et al. suggested 18p as a reference standard for evaluating the sizes of the short arms of acrocentric chromosomes (“p + /−” variants) (Fig. 1)42. In 2016, Liehr suggested the diameter of a chromatid from the same metaphase as a size reference to evaluate the centromeric regions9. The 16p SC model was documented in three of the selected sources, where it was applied as a research and diagnostic tool9,15,28.
Table 3.
Heterochromatin region size levels, adapted from Verma et al. and detailed by Karaca et al.15.
| Level | 16p size ratio | Size classification |
|---|---|---|
| 1 | 0.5 < x 16p | Very small |
| 2 | ≥ 0.5–1 x 16p | Small |
| 3 | > 1–1.5 x 16p | Intermediate |
| 4 | > 1.5–2 x 16p | Large |
| 5 | > 2 x 16p | Very large |
Fig. 1. The 16p size comparison model adapted by Liehr (2016).
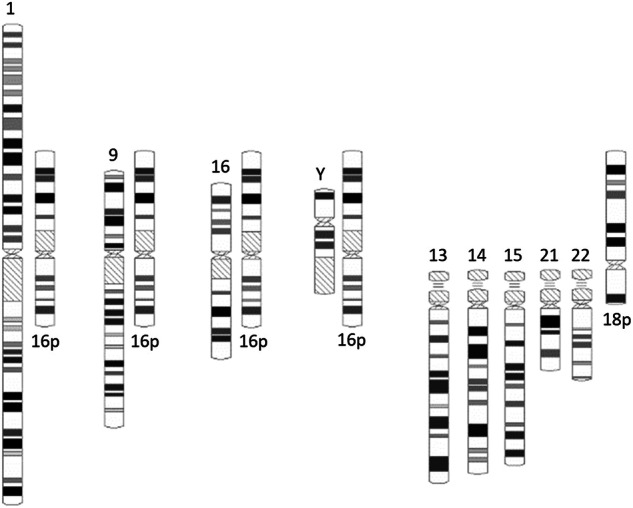
In this CH classification, the heteromorphic regions of chromosomes 1, 9, 16 and Y are compared to the short arm of chromosome 16, and the short arms of all the acrocentric chromosomes are compared to the short arm of chromosome 18. For the purpose of demonstration, the ideograms of chromosomes 16 and 18 are shown upside down next to each heteromorphic region as references.
Discussion
Constitutive heterochromatin comprises approximately 45% of the human genome, with chromosomal heteromorphic regions constituting at least 10% of the latter10,17. Each individual not only possesses a distinctive DNA primary sequence but also carries a unique combination of these heterochromatin variants. In the human karyotype, the major CHs are located at 1q12, 9q12, 13pter-q11, 14pter-q11.1, 15pter-q11.1, 16q11.2, 19p12-q12, 21pter-q11.1, 22pter-q11.1 and Yq1243. The reported incidence of each subgroup of CHs may be subject to variations on the basis of the ethnic origin of the population and the cytogenetic methodologies and observation criteria applied28. At the Paris Cytogenetic Conference in 1971, a system for evaluating human CHs was proposed for the first time. The lengths of the heterochromatin regions were visually categorized into five groups on the basis of their size in C-banding and intensity levels in Q-banding: very small, small, intermediate, large, and very large44. Several CH classification methods have been presented, and cytogenetic variants have been classified by position, size and staining intensity, using diverse selective banding techniques, such as CBG (C-bands by barium hydroxide using Giemsa), QFQ (Q-bands by fluorescence using quinacrine), NOR and DAPI (distamycin A/DAPI banding)45. CHs described in the early stages have been summarized in previous Human Cytogenetic/Genomic Nomenclature Conferences. In recent decades, CHs have also started to be classified using standard G-banding and are documented in the actual version of the International System of Cytogenomic Nomenclature (ISCN)46.
In this retrospective analysis spanning the past 20 years, we identified several reported systems for identifying and classifying human CHs with numerous variations each. We attribute these discrepancies to the emergence of new techniques, the lack of evaluation standards, and the limited information provided in the scientific literature and databases. We should also add that the size of “normal” heterochromatin blocks on each chromosome has not been established, which also makes its correct assessment difficult. The ideograms displayed in different versions of the ISCN and genome browsers fail to elucidate this issue28. Importantly, visualizing CHs properly is difficult, as it is a nonautomated task that depends on experienced cytogeneticists.
To streamline and simplify the scientific discourse, we categorized scoring systems into three primary types distinguished by their methodological approaches, rating levels and assessed chromosome/chromosomal sections (linear measurement, twice the size of the homologous and 16p size comparison model) (Table 4). The Linear Measurement method is the least reported but likely the most accurate, as it assigns exact numerical measurement values. However, extra C-banding is always required and only evaluates the heteromorphisms of 4 chromosomes (1, 9, 16 and Y). The occurrence of different degrees of chromosomal resolution is also a critical point of this methodology. The Twice the Size of the Homologous method is by far the most extensively used and reported system for evaluating CHs. This straightforward methodology can be readily applied in routine G-banding procedures. This technique is effective for all chromosomal resolutions, as comparisons between homologs are conducted within the same metaphase spread. This system assesses all heteromorphic regions in all the chromosomes; however, as heteromorphisms can be present simultaneously in both homologs, this size comparison methodology does not identify and report multiple variants. The 16p Size Comparison model is a simple methodology that can be effectively performed in C-banding or in routine G-banding procedures. This method is efficient for all chromosomal resolutions, as it compares the CHs with the 16p of the same metaphase. However, this comparison model only establishes the upper limit for the size of heterochromatic regions and assesses the heteromorphisms of only 4 chromosomes (1, 9, 16 and Y). To evaluate the hetermorphisms of acrocentric chromosomes, it is necessary to use an additional model (18p SC or TSH for short arms, stalks and satellites). For the centromeric regions, the diameter of a chromatid from the same metaphase is used as a size reference. The combination of the 16p, 18p and chromatid SC models likely represents the most efficient approach to identify and evaluate all the main variable chromosomal sections, as suggested by Liehr in 20169. This publication also suggests minimum size limits to establish the “p-” and “qh-” variants.
Table 4.
Categorization of scoring systems, applications and limitations.
| Scoring System Models | Banding and Methodology | Assessed Regions | Advantages | Limitations |
|---|---|---|---|---|
| Linear Measurement | CBG image measurement | Pericentromeric 1, 9, 16 and distal Yq | Scoring scale with exact numerical limits | Extra C-banding; not effective for all chromosomal resolutions; only evaluates 4 chromosomes |
| Twice the Size of the Homologous | GTG/CBG/NOR observation and comparison | All observable heteromorphic regions | GTG evaluation of all chromosomes; effective for all chromosomal resolutions | Failure to identify variants when present in both homologous |
| 16p Size Comparison | GTG/CBG observation and comparison | Pericentromeric 1, 9, 16 and distal Yq | GTG evaluation; effective for all chromosomal resolutions | Only evaluates 4 chromosomes |
| Trio (16p + 18p + chromatide) Size Comparison | GTG observation and comparison | All observable heteromorphic regions | GTG evaluation of all chromosomes; effective for all chromosomal resolutions |
Currently, there is no chromosome banding assessment system that is effective and comprehensive for evaluating all cytogenetically visible heteromorphic regions. In addition to the well-known heterochromatic blocks present on chromosomes 1, 9, 16, and Y and the acrocentric regions, the centromeric and pericentromeric regions of all chromosomes display variability that must be accurately evaluated. Without an exact description and molecular characterization of all repetitive regions susceptible to heteromorphic changes, such variants can easily be confused with clinically significant gains or losses of chromosomal material43. For example, variations in the short arms of acrocentric chromosomes can be due to translocation with Yq12 or with other acrocentric p-arms, amplification or loss of satellite DNA and/or NOR28,47. Therefore, only by establishing clear protocols to characterize putative CHs will it be possible to determine which CHs may hint at possible cryptic (unbalanced) translocations, insertions or euchromatic variants, such as chromosome 9 as 9 ph+ and amplification at 9q1348,49.
Human heteromorphisms are clearly a controversial topic in genetic diagnosis, and current cytogenetic guidelines do not recommend reporting these variants. The latest version of the ISCN (2020) clarified and reinforced that heteromorphic variants should not be included in the final clinical report. Instead, they should be reserved for the description text that supports the report, where variants may be useful for distinguishing between distinct cell lines or clones14. However, this practice is not consensual in clinical cytogenetics or in the scientific community43. In a survey conducted by Brothman et al. among cytogeneticists regarding reporting practices for CHs, the responses were not unanimous. Nevertheless, there is a clear consensus on which regions are recognized as heteromorphic and which should be included in reports. The majority of cytogeneticists advocated mentioning pericentric inversions and rare CHs in the final report46. In fact, the importance of systematic evaluation and characterization of these variants has been reinforced by several authors, both in the clinical and evolutionary context2,4,8,12,28,50. However, the inconsistency in the methodologies and classification systems used to evaluate CHs does not allow accurate reports15. For example, the ISCN (2020) suggests a nomenclature for heterochromatic variants of acrocentric short arms that is practically never used in the literature, potentially indicating that this nomenclature is not well clarified or established.
In daily practice, cytogeneticists often rely on parental studies, additional laboratory analyses, and thorough literature reviews to determine whether heteromorphic findings are benign or pathological46. This task proves to be problematic, especially in prenatal diagnosis, as there are no defined workflows, and supporting publications are scarce, leading to low-quality reports. Moreover, it is crucial to highlight that relying exclusively on one methodological approach might not offer comprehensive solutions to a particular heteromorphism28. Specifically, it is obvious that cytogenetic banding alone is not able to resolve the nature of all CHs; in many cases, fluorescence in situ hybridization is needed to clarify which CH is present in a specific case (see, e.g., CHs of chromosome 9)16. The ongoing advancements in computational techniques and new algorithms for identifying repetitive DNA sequences are promising but remain ineffective. The limited length of next-generation sequencing (NGS) reads and significant sequencing error rates in third-generation sequencing (TGS) make it difficult to develop efficient algorithms, imply high detection costs, and provide unsuccessful results8. Considering the highly repetitive nature of heterochromatic regions and the challenges associated with sequencing them, molecular methodologies are unlikely to readily replace cytogenetic analyses of CHs51. Therefore, reevaluating the importance of establishing clear cytogenetic guidelines and a universal scoring system is imperative.
CHs constitute a natural part of genetic variation; however, they can exert significant effects on gene and phenotype expression. Understanding CHs is crucial for gaining insights into genome evolution, gene regulation and disease mechanisms. Nevertheless, CHs remain a challenge in genetic diagnostics and are often not adequately assessed or reported in routine cytogenetic analysis. The present study provides a comprehensive understanding of the diverse methodologies and classification systems for heterochromatic CHs outlined in the literature and exposes the inconsistencies and difficulties in correctly evaluating and reporting these variants. Currently, the literature and databases still lack descriptions of the standard sizes attributed to heterochromatic regions and the exact characterization and documentation of numerous human CHs. The absence of a universal scoring system hinders the accurate identification of CHs, preventing the scientific community from assigning a precise value to their impact on genetic diagnosis. This lack of standardization also compromises scientific reproducibility, potentially leading to erroneous conclusions. Therefore, it is crucial to establish robust standardization in the evaluation of human CHs and carefully consider the advantages and disadvantages of reporting these variants. Such efforts can be achieved only through cooperation and collaboration within the global cytogenetic scientific community. Furthermore, it is crucial to highlight that the use of multiple techniques to address the same issue often uncovers the harmless or pathological essence of the variation. Ultimately, we emphasize the importance of strengthening global scientific research on human heterochromatic regions and their variations, as well as advanced detection technologies, to unravel their intricate structures, functions, interactions and clinical implications.
Acknowledgments
Springer Nature Publisher Group kindly provided Article Processing Fee support for the publication of this article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Miga, K. H. Centromeric satellite DNAs: hidden sequence variation in the human population. Genes10, 352 (2019). [DOI] [PMC free article] [PubMed]
- 2.Gemmell, N. Repetitive DNA: genomic dark matter matters. Nat. Rev. Genet22, 1 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Levy, B. & Warburton, P. E. Molecular dissection of heteromorphic regions. In Atlas of Human Chromosome Heteromorphisms (ed. Wyandt, HET, VS (eds)) (Springer, Dordrecht, 2004).
- 4.Liehr, T. Repetitive elements in humans. Int J. Mol. Sci.22, 2072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartley, G. & O’Neill, R. J. Centromere repeats: hidden gems of the genome. Genes10, 223 (2019). [DOI] [PMC free article] [PubMed]
- 6.Lopes, M., Louzada, S., Gama-Carvalho, M. & Chaves, R. Genomic tackling of human satellite DNA: Breaking Barriers through Time. Int. J. Mol. Sci.22, 4707 (2021). [DOI] [PMC free article] [PubMed]
- 7.Sipek, A. Jr et al. Heterochromatin variants in human karyotypes: a possible association with reproductive failure. Reprod. Biomed. Online29, 245–50 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Liao, X. et al. Repetitive DNA sequence detection and its role in the human genome. Commun. Biol.6, 954 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liehr, T. Cytogenetically visible copy number variations (CG-CNVs) in banding and molecular cytogenetics of human; about heteromorphisms and euchromatic variants. Mol. Cytogenet.9, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsano, R. M. & Dimitri, P. Constitutive heterochromatin in eukaryotic genomes: A Mine of Transposable Elements. Cells11, 761 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allshire, R. C. & Madhani, H. D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol.19, 229–44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tempest, H. G. & Simpson, J. L. Why are we still talking about chromosomal heteromorphisms? Reprod. Biomed. Online35, 1–2 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Ralapanawe, M. S. B. et al. A comprehensive analysis of chromosomal polymorphic variants on reproductive outcomes after intracytoplasmic sperm injection treatment. Sci. Rep.13, 1319 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nomenclature, ISCOHC, McGowan-Jordan, J, Hastings, R.J. & Moore, S. ISCN 2020: An International System for Human Cytogenomic Nomenclature (2020), (Karger, 2020).
- 15.Karaca, Y. et al. Co-occurrences of polymorphic heterochromatin regions of chromosomes and effect on reproductive failure. Reprod. Biol.20, 42–47 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Kosyakova, N. et al. Heteromorphic variants of chromosome 9. Mol. Cytogenet.6, 14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liehr, T. Chromosomal heteromorphisms and cancer susceptibility Revisited. Cells11, 3239 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mottola, F., Santonastaso, M., Ronga, V., Finelli, R. & Rocco, L. Polymorphic rearrangements of human chromosome 9 and male infertility: New evidence and impact on spermatogenesis. Biomolecules13, 729 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, S.-J. et al. Chromosomal polymorphisms associated with reproductive outcomes after IVF-ET. J. Assist. Reprod. Genet.37, 1703–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rang, F. J., Kind, J. & Guerreiro, I. The role of heterochromatin in 3D genome organization during preimplantation development. Cell Rep.42, 112248 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Pajares, V. Long non-coding RNA regulation of gene expression during differentiation. Pflug. Arch.468, 971–81 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Ariffen, N. et al. Amplification of different satellite-DNAs in prostate cancer. Pathol. Res Pr.256, 155269 (2024). [DOI] [PubMed] [Google Scholar]
- 23.Hall, L. E., Mitchell, S. E. & O’Neill, R. J. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res.20, 535–46 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Liehr, T. Molecular cytogenetics in the era of chromosomics and cytogenomic approaches. Front. Genet.12, 720507 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamerton, J. L., Klinger, H. P. & Foundation, N. Paris Conference (1971), Supplement (1975), Standardization in Human Cytogenetics, (National Foundation, 1975).
- 26.Verma, R. S., Dosik, H. & Lubs, H. A. Size and pericentric inversion heteromorphisms of secondary constriction regions (h) of chromosomes 1, 9, and 16 as detected by CBG technique in Caucasians: classification, frequencies, and incidence. Am. J. Med. Genet.2, 331–9 (1978). [DOI] [PubMed] [Google Scholar]
- 27.Balíček, P., Žižka, J. & Skalská, H. Variability and familial transmission of constitutive heterochromatin of human chromosomes evaluated by the method of linear measurement. Hum. Genet42, 257–265 (1978). [DOI] [PubMed] [Google Scholar]
- 28.Liehr, T. Benign & pathological chromosomal imbalances, microscopic and submicroscopic copy number variations (CNVs) in genetics and counseling, (Academic Press, 2014).
- 29.Romo Martinez, E. et al. Y-SNP haplogroups related to the Yqh+ heteromorphism in the Mexican northwestern population. J. Genet.91, 297–302 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Yasseen, A. A. & Al-Khafaji, S. M. The significance of autosomal C-band size polymorphism in male infertility. Saudi Med J.23, 1473–1477 (2002). [PubMed] [Google Scholar]
- 31.Yasseen, A. & Alkhafaji, S. The role of Y chromosome C-band size polymorphism in male infertility with a reference to their effect on the total length of the chromosome. Saudi Med J.25, 452–455 (2004). [PubMed] [Google Scholar]
- 32.Sahin, F. I. et al. Chromosome heteromorphisms: an impact on infertility. J. Assist Reprod. Genet.25, 191–5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawal, L., Kumar, S., Mishra, S. R., Lal, V. & Bhattacharya, S. K. Clinical manifestations of chromosomal anomalies and polymorphic variations in patients suffering from reproductive failure. J. Hum. Reprod. Sci.13, 209–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madon, P. F., Athalye, A. S. & Parikh, F. R. Polymorphic variants on chromosomes probably play a significant role in infertility. Reprod. Biomed. Online11, 726–32 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Guo, T. et al. The role of male chromosomal polymorphism played in spermatogenesis and the outcome of IVF/ICSI-ET treatment. Int. J. Androl.35, 802–9 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Akbaş, H. et al. Chromosome heteromorphisms are more frequent in couples with recurrent abortions. Genet Mol. Res.11, 3847–51 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Zhu, J. J. et al. C-banding and AgNOR-staining were still effective complementary methods to indentify chromosomal heteromorphisms and some structural abnormalities in prenatal diagnosis. Mol. Cytogenet12, 41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minocherhomji, S. et al. A case-control study identifying chromosomal polymorphic variations as forms of epigenetic alterations associated with the infertility phenotype. Fertil. Steril.92, 88–95 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Mierla, D. & Stoian, V. Chromosomal polymorphisms involved in reproductive failure in the romanian population. Balk. J. Med. Genet.15, 23–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakraborty, A., Kar, S., Mohapatra, P. C. & Banerjee, B. A case-control study identifying the frequency and spectrum of chromosomal anomalies and variants in a cohort of 1000 couples with a known history of recurrent pregnancy loss in the eastern region of India. J. Hum. Reprod. Sci.14, 422–430 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez, F., Cruz, M. & Requena, A. Impact of parental chromosomal polymorphisms on the incidence of congenital anomalies and perinatal complications in a cohort of newborns conceived after ICSI + PGT-A. Reprod. Biol. Endocrinol.20, 145 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verma, R. S., Dosik, H. & Lubs, H. A. Size variation polymorphisms of the short arm of human acrocentric chromosomes determined by R-banding by fluorescence using acridine orange (RFA). Hum. Genet38, 231–234 (1977). [DOI] [PubMed] [Google Scholar]
- 43.Liehr, T. Copy number variations - is there a biological difference between submicroscopic and microscopically visible ones? OA Genet.1, 2 (2013). [Google Scholar]
- 44.Paris Conference (1971), supplement (1975) Standardization in human cytogenetics. Cytogenet. Cell Genet.15, 203–238. [PubMed]
- 45.Verma, R. S. & Dosik, H. Human Chromosomal Heteromorphisms: Nature and Clinical Significance. In International Review of Cytology, 62 (eds. Bourne, GH, Danielli, JF & Jeon, KW) 361–383 (Academic Press, 1980). [DOI] [PubMed]
- 46.Brothman, A. R. et al. Cytogenetic heteromorphisms: survey results and reporting practices of giemsa-band regions that we have pondered for years. Arch. Pathol. Lab Med.130, 947–949 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Fuchs, S., Lisfeld, J., Kankel, S., Person, L. & Liehr, T. The acrocentric part of der(Y)t(Y;acro)(q12;p1?2) contains D15Z1 sequences in the majority of cases. Hum. Genome Var.8, 32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starke, H. et al. Homologous sequences at human chromosome 9 bands p12 and q13-21.1 are involved in different patterns of pericentric rearrangements. Eur. J. Hum. Genet10, 790–800 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Tsan, S. et al. About cryptic acrocentric pericentromeric abnormalities in infertile. OBM Genet.05, 135 (2021). [Google Scholar]
- 50.Anton, E. et al. Chromosome heteromorphisms: do they entail a reproductive risk for male carriers? Asian J. Androl.22, 544–546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heng, E., Thanedar, S. & Heng, H. H. Challenges and opportunities for clinical cytogenetics in the 21st century. Genes14, 493 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]


