Abstract
Multiomic profiling of single cells by sequencing is a powerful technique for investigating cellular diversity. Existing droplet-based microfluidic methods produce many cell-free droplets, underutilizing bead barcodes and reagents. Combinatorial indexing on microplates is more efficient for barcoding but labor-intensive. Here we present Overloading And unpacKing (OAK), which uses a droplet-based barcoding system for initial compartmentalization followed by a second aliquoting round to achieve combinatorial indexing. We demonstrate OAK’s versatility with single-cell RNA sequencing as well as paired single-nucleus RNA sequencing and accessible chromatin profiling. We further showcase OAK’s performance on complex samples, including differentiated bronchial epithelial cells and primary retinal tissue. Finally, we examine transcriptomic responses of over 400,000 melanoma cells to a RAF inhibitor, belvarafenib, discovering a rare resistant cell population (0.12%). OAK’s ultra-high throughput, broad compatibility, high sensitivity, and simplified procedures make it a powerful tool for large-scale molecular analysis, even for rare cells.
Subject terms: Transcriptomics, RNA sequencing, Microfluidics, Chromatin
Single-cell sequencing approaches need to balance sensitivity, throughput and experimental complexity. Here the authors combine droplet-based microfluidics and combinatorial indexing to develop OAK, a versatile method for ultra-high throughput single-cell multiomic profiling.
Introduction
The technological landscape of single-cell sequencing is rapidly evolving, encompassing newly developed methods1–4 that offer an unprecedented view of cellular heterogeneity. This technical evolution is fueled by the need to achieve more precise cell type or state identification, capture rare cell states or cellular lineages, and conduct comprehensive perturbation screens for new drug target discovery, all of which have steered technological development toward analyzing a greater number of cells at a reduced cost.
Droplet-based microfluidic approaches co-encapsulate a barcoded bead and a cell within an emulsion to enable parallel analysis of thousands of individual cells5–7. These methods constitute an important advancement in streamlining high-throughput single-cell sequencing. However, the low cell concentration required to minimize the number of multi-cell droplets leads to a large number of cell-free droplets and a largely underutilized barcoding capacity. Alternatively, combinatorial indexing on microwell plates8–13 provides a strategy for barcoding over 100,000 cells. However, this ultra-high throughput approach comes with long and laborious protocols, involving multiple rounds of splitting and pooling cells for indexing.
Inspired by the strengths and limitations of these two families of single-cell sequencing methods, we develop OAK which utilizes the Chromium microfluidic system to replace micro-well plates in the first step of split-and-pool for combinatorial indexing, to achieve both elevated throughput and experimental simplicity. With OAK we measure gene expression, accessible chromatin, and antibody conjugated oligonucleotides, either separately or jointly. Using OAK, we perform paired single-nucleus RNA sequencing (snRNA-Seq) and single-nucleus Assay for Transposase Accessible Chromatin with sequencing (snATAC-Seq) on complex retinal tissue. Furthermore, we conduct a lineage tracing experiment capturing RNA and lineage barcodes for over 400,000 cells, revealing the longitudinal response of melanoma cells to a RAF inhibitor, belvarafenib.
Results
Principles and performance of OAK
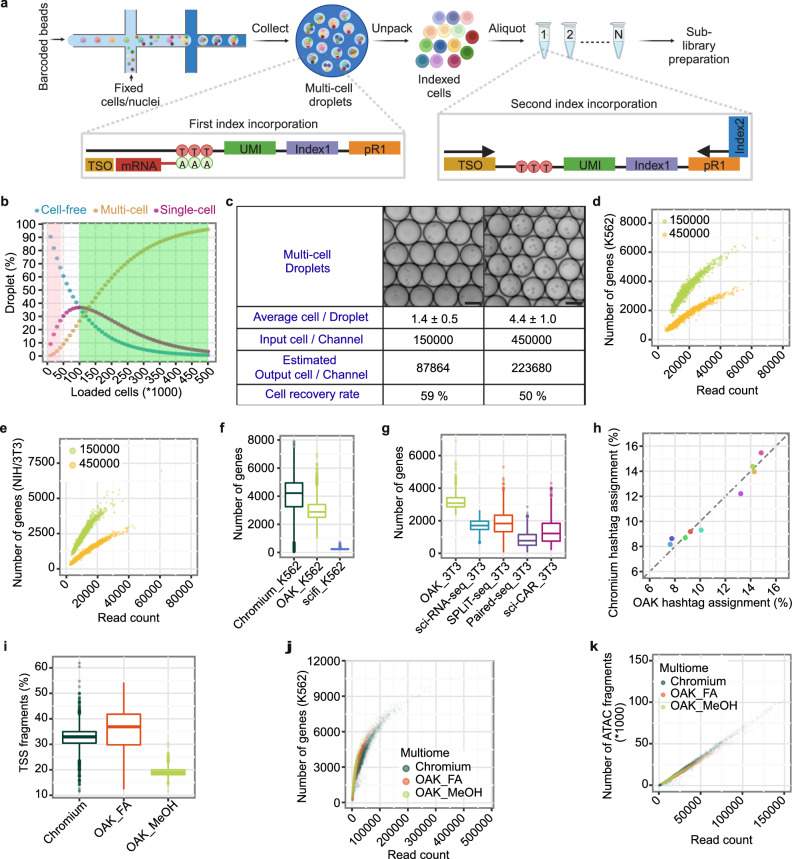
OAK relies on fixed cells or nuclei which serve as individualized reaction chambers for two rounds of indexing (Fig. 1a). The first round is performed in droplets, for which we utilized a commercially available system, the Chromium system by 10x Genomics. In this and other droplet-based single-cell sequencing systems5–7,14, conventionally cells are loaded at a low concentration to minimize the possibility of encapsulating multiple cells within a single droplet. Based on the Poisson distribution, this is estimated to result in over 80% of droplets devoid of a cell (Fig. 1b), leaving their barcoding potential untapped and the majority of reagents unused. To efficiently utilize the droplets, we overloaded the microfluidic chip in the Chromium system, resulting in reduced percentage of cell-free droplets, concomitant with the increase of single- and multi-cell droplets (Fig. 1b). To resolve single cells in multi-cell droplets, after the first round of indexing mediated by in-situ reverse transcription of mRNA, we unpacked droplets by breaking emulsions and retrieving the fixed cells (Supplementary Fig. 1a). As a result, encapsulated cells are recovered, mixed, and then randomly distributed into multiple aliquots. The number of aliquots can be tuned based on the scale of cell loading and the number of droplets generated by the microfluidic system, in order to achieve a desirable theoretical multiplet rate (Supplementary Fig. 1b). Each aliquot receives a unique secondary index integrated to each cDNA molecule that already carries primary indexes coming from droplets (Fig. 1a). A desired number of aliquots are then converted into sub-libraries for sequencing (see “Methods”). In the sequencing data, the combination of the initial droplet index and the secondary index is used to identify single cells.
Fig. 1. Principle and performance of OAK in single cell profiling of multiple molecular modalities.
a Schematic of OAK’s scRNA-Seq workflow. mRNA hybridizes with poly-dT oligos within droplets in fixed cells or nuclei. Following reverse transcription and emulsion break, cells/nuclei are pooled and re-distributed into aliquots for secondary indexing via PCR. TSO: template switch oligo. pR1: primer binding sequence for TrueSeq Read 1. Schematic created in BioRender. Darmanis, S. (2023) BioRender.com/w63n572b Simulation of droplets containing zero (blue), one (magenta), and more than one (yellow) cell, at various cell numbers loaded per channel. Pink and green indicate cell number ranges in regular Chromium and OAK, respectively. c OAK results at different cells per channel. Image scale bars: 75 µm. Mean cells per droplet with 95% confidence margins of error are presented. d-e, Number of genes in K562 (d) and NIH/3T3 cells (e) vs. reads per cell. Green: 150,000 cells loaded; yellow: 450,000 cells loaded, same as in c. f-g, Number of genes in K562 (f) and NIH/3T3 cells (g). ~15000 reads per cell. Boxplots: center lines are medians; limits denote Q1 (lower) and Q3 (higher) quartiles; whiskers extend to 1.5 times the interquartile range (IQR) or last data points if within limits. K562: regular Chromium NextGEM 3’ RNA-Seq, n = 6022; OAK scRNA-Seq with 150,000 cells loaded, n = 3647; scifi-RNA-seq15, n = 1617. NIH/3T3: OAK scRNA-Seq with 150,000 cells loaded, n = 691; sci-RNA-seq9, SPLiT-seq11, sci-CAR10, and Paired-seq13, n = 868 each. h Percentage of human bronchial epithelial cells assigned to each sample hashtag (n = 9) by standard Chromium and OAK. Each dot is a sample hashtag. i Percentage of fragments overlapping TSS. Chromium: Chromium’s standard Multiome ATAC + Gene Expression, n = 4484; OAK_FA: OAK’s multiome with formaldehyde (FA), n = 1835; OAK_MeOH: OAK’smultiome with methanol (MeOH), n = 2903. Boxplots: center lines are medians; limits denote Q1 and Q3; whiskers extend to 1.5 times IQR or last data points if within limits. j–k, Number of genes (j) and ATAC fragments (k) using OAK with FA or MeOH, or Chromium’s standard multiome. Source data are provided as a Source Data file.
First, to assess the impact of cell overloading, we performed parallel experiments where channels on the microfluidic chip were loaded with 150,000 and 450,000 methanol-fixed cells respectively (Fig. 1c). After sequencing a subset of cells from each experiment, we projected that by sequencing all aliquots we could potentially recover 87,864 cells from the 150,000-cell loading (59% recovery), and 223,680 cells from the 450,000-cell loading (50% recovery) (Fig. 1c, Supplementary Table 1). This represents a high efficiency of cell recovery compared to previously published ultra-high throughput methods11,13,15 (Supplementary Fig. 1c). At the same sequencing depth per cell, more genes per cell were detected when 150,000 cells were loaded compared to 450,000 cells (Figs. 1d, e). The input cells consisted of a 1:1 mixture of a mouse (NIH/3T3) and a human (K562) cell line, enabling us to identify collision events when a mouse and a human cell share the same combinatorial indices. When loading 150,000 cells, we detected 3.3% cells to be mix-species multiplets, indicating an overall multiplet rate of 6.6% to include the unobservable multiplets within the same species (Supplementary Fig. 1d). This overall multiplet rate closely aligns with the theoretical expected collision rate for the number of secondary indexing aliquots (n = 12) generated (Supplementary Fig. 1b, Supplementary Table1). At the higher loading of 450,000 cells, while we recovered a higher number of cells (Fig. 1c), the overall multiplet rate was 10.6% (Supplementary Fig. 1d). In summary, OAK is flexible to operate with a broad spectrum of loaded cell quantities. The choice on the number of cells to load should be guided by research objectives, balancing between detection sensitivity and cell yield.
Next, we benchmarked OAK to the widely used 10x Genomics’ Chromium NextGEM 3’ v3.1 scRNA-Seq procedure (standard Chromium)7. From the 150,000 cells loaded, OAK demonstrated an over eightfold increase in throughput (Fig.1c) compared to the standard Chromium procedure, which recovers up to 10,000 cells per channel. With a matched sequencing depth of ~15,000 reads per cell, OAK detected a mean of 3014 genes per cell for the K562 cell line, while the standard Chromium detected 3,905 genes indicating a mild reduction in sensitivity by OAK (Fig. 1f). Further investigation into the gene detection difference revealed that reduced detection primarily occurred for the lowly expressed genes (Supplementary Fig. 1e). In addition, OAK exhibited a lower percentage of reads that map to mitochondrial DNA (Supplementary Fig. 1f), which is likely attributed to the methanol-fixation and its membrane permeabilization effect that led to partial loss of mitochondria as well as cytoplasmic RNA. This was supported by the higher percentage of reads mapping to intronic regions (Supplementary Fig. 1g), which is a feature shared by snRNA-Seq and combinatorial indexing methods12,16. Such intronic molecules are also indicative of transcript abundance, since their counts, quantified using unique molecular identifiers (UMIs), are highly correlated with the counts of exonic molecules from the same gene (spearman correlation = 0.65, Supplementary Fig. 1h). Overall, a strong correlation between OAK and the standard Chromium method was observed in terms of mean number of UMIs across cells for each gene (Spearman correlation coefficient = 0.92, Supplementary Fig. 1i). We also compared OAK with previously published ultra-high throughput single-cell methods, including sci-RNA-seq9, SPLiT-seq11, sci-CAR10, Paired-seq13, and scifi-RNA-seq15. OAK outperformed these methods by providing higher sensitivity as measured by number of genes (Figs. 1f, g) and transcripts (UMIs) detected per cell (Supplementary Figs. 1j, k).
Leveraging ultra-high throughput for sample multiplexing
Since OAK enables profiling of hundreds of thousands of cells, it is suitable for experiments that aim to process many different samples, donors and conditions. In such experiments, cell hashing with barcoded antibodies is frequently used for sample multiplexing as it enables pooling of cells from different sources for single-cell profiling, streamlining workflows and reducing experimental cost17. Cell hashing by itself enables moderate overloading, as multiplets can be identified by the mix of hashing antibodies present; however, unlike in OAK, data from these multiplet droplets is not usable. To take advantage of OAK’s ultra-high throughput combined with sample multiplexing, we evaluated antibody hashing using human bronchial epithelial cells differentiated in transwell plates. We used the same sample of antibody stained cells and processed it with OAK and standard Chromium, and asked whether cell assignment was comparable. In the OAK workflow, cells were fixed in methanol after staining, whereas in the standard Chromium workflow the cells were not fixed. We sequenced 4 out of 22 aliquots obtaining 8096 cells, and projected that the total recovery from OAK would be 44,528 cells if all aliquots were sequenced (Supplementary Table1). We found that 80% of cells were assigned a hashtag identity in OAK, compared to 81% in the standard Chromium. Furthermore, we found a strong correlation (Pearson correlation coefficient = 0.98, Fig. 1h) in the abundance of each hashtag between OAK and standard Chromium. We then clustered cells based on gene expression (Supplementary Fig. 1l). After cell annotation, all expected cell types were present in both data sets at very similar proportions (Supplementary Fig. 1m). Therefore, OAK was compatible with the cell hashing approach for sample multiplexing, and furthermore did not introduce any biases in cellular composition.
Flexibility in multimodal single cell profiling
We next investigated whether OAK can perform paired profiling of transcriptome and chromatin accessibility. Since the beads from the Chromium Next GEM Single Cell Multiome kit readily provide barcoding capacity for both mRNA and ATAC fragments, only adjustments in secondary indexing primers were necessary to make OAK compatible with the Chromium multiome workflow (Supplementary Fig. 1n). In order to identify a suitable fixative for paired snRNA-Seq and snATAC-Seq, we evaluated methanol and formaldehyde fixation with K562 cells. Compared to formaldehyde fixation, methanol fixation led to a lower transcription start site (TSS) fragment percentage in the sequencing data (Fig. 1i), likely due to methanol’s chromatin denaturing effect. Formaldehyde fixation generated high quality gene expression data (Fig. 1j) and chromatin accessibility data for K562 cells (Figs. 1i, k). These results underscore OAK’s adaptability in supporting multiple molecular modalities.
Paired snRNA-Seq and snATAC-Seq for human retinal cells
A common scenario in collecting single-cell data from tissue, is that the most abundant cell types are orders of magnitude higher than the rarest cell types. A couple of examples include recovering neurons from the enteric nervous system, where they represent less than 1% of colon cells18, or tuft cells which represent 0.2% of dissociated lung cells19. The human retina is another example of a primary tissue with high cellular heterogeneity and disproportionate representation of various cell types. Effectively capturing all cellular subtypes is key to understanding the health and disease of the eye; however, the overwhelming presence of rod photoreceptors (around 60% of cells) often impedes the efficient recovery of other cells, such as retinal ganglion cells (less than 1%). Some techniques can be employed for depleting rod cells, however this adds experimental complexity and may unintentionally affect representation of other cell types, for example depletion of bipolar cells20. Utilizing a method such as OAK to perform paired snRNA-Seq and snATAC-Seq on retinal samples enables generation of large-scale high-resolution data from these precious samples, which are obtained only from careful dissection of post-mortem donations.
We transposed 100,000 formaldehyde-fixed peripheral retinal nuclei for overloading (see “Methods”). We recovered snATAC-Seq data from 42,632 nuclei, and snRNA-Seq data from 46,487 nuclei, with an overlap for 40,691 nuclei (Supplementary Table2). In parallel we ran a standard Chromium multiome workflow on unfixed nuclei from the same sample and retrieved snATAC-Seq data from 5,655 nuclei, snRNA-Seq from 6510 nuclei, with an overlap of 5551 nuclei. In OAK snRNA-Seq data we observed a mean of 1,666 genes per cell, compared to 2029 genes in the standard Chromium data (Supplementary Fig. 2a). In OAK snATAC-Seq data we observed a mean of 12,539 fragments per cell compared to 14,217 in standard Chromium data (Supplementary Fig. 2b). In the snATAC-Seq data we observed a mean transcription start site (TSS) enrichment of 14.71 (Supplementary Fig. 2c) and an expected fragment distribution pattern (Supplementary Fig. 2d).
Using the snRNA-Seq data we clustered and annotated the main cell types of the retina based on known marker genes (Supplementary Fig. 2e, Fig. 2a). With a single donor sample, we obtained thousands of rod, cone, Müller glia, amacrine and bipolar cells, as well as hundreds of horizontal cells, astrocytes and retinal ganglion cells, representing the major cell types of the retina20–22. We used the snRNA-Seq annotations with the OAK snATAC-Seq data (Supplementary Fig. 2f) to call open chromatin regions (OCRs) in each cell subtype (Fig. 2b, Supplementary Fig. 2g). We found unique OCR signatures even for the least abundant cell types, including retinal ganglion cells and astrocytes23. Peaks called were primarily in intronic and promoter regions as expected (Supplementary Fig. 2h). Looking in more detail at the chromatin peaks in specific cell types we observed differential chromatin accessibility in ARR3 in cone cells (Fig. 2c), and DOK5 in DB5 bipolar cells (Fig. 2d), consistent with previous findings21.
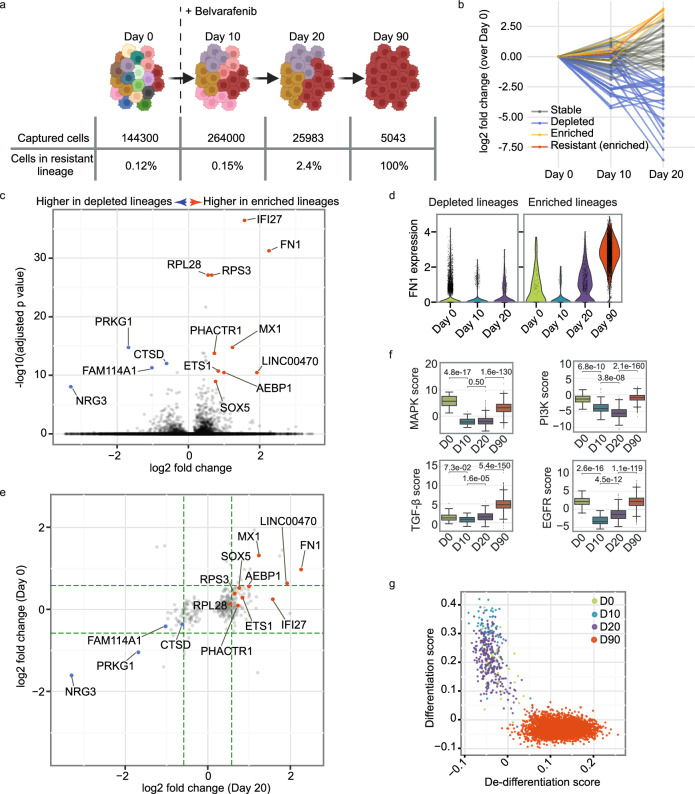
Fig. 2. OAK paired snRNA-Seq and snATAC-Seq on the human peripheral retina.
a Uniform Manifold Approximation and Projection (UMAP) of snRNA-Seq data with table of number and percentage of each cell type. The color of the dot in the table indicates the position in the UMAP. b Heatmap displaying OCRs in each cell type. c Chromatin tracks in major cell types for the genomic region spanning the ARR3 gene with a Ridge plot (expression values are normalized and log transformed) indicating gene expression of ARR3 from snRNA-Seq data. d Chromatin tracks in bipolar cell types for the genomic region including the TSS of DOK5, with a Ridge plot indicating DOK5 expression level. e Significant transcription factors by weighted gene activity for each major cell type. Differential activity was determined using a one-sided t-test adjusted for multiple comparisons. Source data are provided as a Source Data file.
Utilizing paired snRNA-Seq and snATAC-Seq data, we identified putative candidates for master regulators in the different cell types using Epiregulon24 (Fig. 2e). Epiregulon infers regulatory elements to target genes based on correlated gene expression and chromatin accessibility in clustered cells, matching these elements to known transcription factor binding sites from repositories of public CHIP-Seq data. As a proxy for the strength of the interaction, Epiregulon uses the correlation between transcription factor expression and target gene expression. We plotted the activity for each transcription factor based on the expression of target genes combined with the strength of regulation. We identified elevated BLIMP1/PRDM1 regulation activity in cone cells (Fig. 2e), previously found to be transiently expressed in developing photoreceptors, likely preventing bipolar cell fate25. Another example of expected transcription factor activity is of the ONECUT1 and ONECUT2 paralogs activated downstream of PAX6 (Fig. 2e), previously found to be important in the differentiation and maintenance of horizontal cells26. Many functional roles of transcription factors in the human retina have been identified by studying early development in analogous animal models or in organoids27. Multiomic data generated from post-mitotic cells, as obtained from this retinal sample, offers an intriguing window into ongoing regulation of gene activity decades after initial differentiation events. Obtaining this type of data is especially valuable when considering potential treatments for age-related eye diseases.
Melanoma resistance to RAF inhibitor belvarafenib
Understanding therapy response and resistance in cancer is crucial for improving treatment outcomes. Belvarafenib is a pan-RAF inhibitor with clinical activity in melanoma28. Resistance to belvarafenib arises spontaneously in IPC-298 cells at low frequency28. To track emergence of these rare events that could be as infrequent as 0.1%, processing a substantial cell population is necessary to ensure sufficient representation of the resistant lineages at baseline. By leveraging the high-throughput capabilities of OAK and a lineage tracing technique29, we examined the transcriptomic response of IPC-298 melanoma cells to a 90-day treatment course with vemurafenib at multiple time points including Day 0, Day 10, Day 20, and Day 90.
We transduced IPC-298 cells with a lentivirus-based library containing 100,000 unique barcode sequences for lineage tracing29. A subsample of 1000 transduced cells, each expected to carry a unique lineage barcode, was expanded. Prior to belvarafenib treatment (Day 0), we collected transcriptomic profiles and lineage barcodes from 144,300 methanol-fixed cells (Fig. 3a). The representation of each lineage within the single-cell data displayed a strong correlation with the quantity of reads in bulk sequencing data (Spearman correlation coefficient = 0.93, Supplementary Fig. 3a), confirming accurate lineage recovery with OAK. Furthermore, as the sequenced population of cells increased, the extent of correlation between the single cell data and bulk data also increased (Supplementary Fig. 3b), emphasizing the benefit of sampling a high number of cells in systems with such a high lineage diversity.
Fig. 3. OAK single-cell lineage tracing and transcriptome profiling for melanoma cells during belvarafenib treatment.
a Diagram of lineage tracing experiment. IPC-298 cells labeled with lineage barcodes were sampled for scRNA-Seq on Days 0, 10, 20, and 90. Belvarafenib treatment commenced following Day 0 subculture collection. Schematic created in BioRender. Darmanis, S. (2023) BioRender.com/l09z998b Fold change in cell count for each lineage at each time point. Cell counts from Day 0 served as the baseline. Enriched (yellow) includes lineages with over tenfold increase from Day 0 to Day 20. Resistant (enriched) refers to the lineage enriched on Day 20 and resistant on Day 90. Stable refers to lineages neither depleted nor enriched. c Volcano plot depicting differentially expressed genes on Day 20 between depleted and enriched lineages. P values are calculated by the Wilcoxon rank-sum method (two-sided) with the benjamini-hochberg correction method. Genes with adjusted p values lower than 1e-8 and log2 fold changes beyond ±0.5 are labeled. d Violin plots for FN1 expression level (normalized and log-transformed) in cells within depleted and enriched lineages. e Fold changes between the depleted and the enriched lineages, with specific genes labeled the same as in (d). Green dashed lines denote ±1.5-fold changes. f PROGENy pathway scores for each cell at Day0 (n = 42), Day 10 (n = 59), Day 20 (n = 275), and Day 90 (n = 4827) within the resistant lineage. P values are calculated using the Mann-Whitney-Wilcoxon test (two-sided) with Bonferroni adjustment. Boxplots’ center lines represent medians. Boxplots: center lines are medians; limits denote Q1 and Q3; whiskers extend to 1.5 times IQR or last data points if within limits. g De-differentiation and differentiation scores for cells within the resistant lineage. Source data are provided as a Source Data file.
Next, we collected samples on Day 10 and Day 20 of belvarafenib treatment (Fig. 3a). Five lineages demonstrated over tenfold increase in their relative abundance from Day 0 to Day 20, and therefore were categorized as enriched lineages that are drug tolerant (Fig. 3b). Conversely, 61 lineages, each representing less than 1% of Day 20’s total cells, were defined as depleted lineages (Fig. 3b). After Day 20, as the number of cells continued to decrease, we observed the emergence of a belvarafenib-resistant clone among the five enriched lineages (Supplementary Fig. 3c). This clone underwent expansion from a single colony on the plate, and accounted for all of the captured cells on Day 90 (Fig. 3a). Consistent with previous characterization of belvarafenib resistance28, this resistant clone only accounted for 0.12% of the cells on Day 0 (Fig. 3a). Thus, cells of that lineage could only be captured with sufficient representation through massive-scale sampling techniques such as OAK. Specifically, for Day 0, we performed stepwise sequencing of sub-libraries, until enough cells from this lineage were recovered for downstream analysis (Supplementary Fig. 3d), and sufficient lineage diversity was achieved (Supplementary Fig. 3e).
To interrogate the transcriptional features associated with drug-tolerance, we looked for marker genes that distinguish the enriched and the depleted lineages on Day 20. We found Fibronectin 1 (FN1) among the overexpressed genes in the enriched lineages (Fig. 3c). Fibronectin-rich extracellular matrix has been shown to provide tolerance for melanoma cells in BRAF inhibition30. Moreover, FN1 has been shown to be associated with a mesenchymal phenotype31 in melanoma cells. Interestingly, the epithelial mesenchymal transition (EMT) hallmark gene set32 emerged as one of the features for Day 90 within the resistant lineage (Supplementary Fig. 3f). In addition, in the depleted lineages FN1 levels remained stable along the course of belvarafenib treatment, while in the enriched lineages the gain of this mesenchymal marker was already observed on Day 20 (Fig. 3d). Moreover, the longitudinal nature of our experiment enabled us to probe for potential pre-existing transcriptional differences between the enriched and depleted lineages. We observed that many of the differentially expressed genes on Day 20 showed differences in expression levels as early as Day 0 (Fig. 3e). This indicates that distinct lineages may possess inherent transcriptional programs for responding to belvarafenib treatment. Furthermore, sustained exposure to belvarafenib led to amplification of selective pre-existing differences, as exemplified by increased fold changes on Day 20 in some of the most differentially expressed genes, such as FN1 and NRG3 (Fig. 3e).
Belvarafenib directly inhibits kinase activity of the RAF kinases, which are responsible for MAPK pathway activation downstream of an oncogenic NRAS mutation in the IPC-298 cells28. To assess how the resistant clone adapted to belvarafenib treatment, we specifically compared the activity of the MAPK pathway and other related pathways within the resistant lineage across different time points. We observed an initial downregulation of MAPK, PI3K, and EGFR pathway signatures at early time points, which suggested an initial response to belvarafenib. However, on Day 20 we noticed a rebound of EGFR pathway activity (Fig. 3f). During the same time frame, we observed activation of the transforming growth factor-β (TGF-β) pathway (Fig. 3f), which is a known driver of resistance against MAPK pathway inhibitors in melanoma cells33. Furthermore, from Day 20 to Day 90 we observed significant rebound of MAPK and PI3K pathway activities (Fig. 3f), suggesting that reactivation of these pathways may be essential for the establishment of the resistant phenotype.
TGF-β is known to induce EMT34 and de-differentiation in melanoma33,35. Given a mesenchymal-like state suggested by FN1 upregulation (Fig. 3c), increased TGF-β signaling (Fig. 3f) as early as Day 20, and the enrichment of EMT hallmark genes on Day 90 (Supplementary Fig. 3f), we examined whether the resistant cells switched to a less differentiated state in response to belvarafenib. Despite the initial shift towards a more differentiated melanocyte-like state on Day 10 (Fig. 3g), the resistant cells ultimately reverted to an undifferentiated state (Fig. 3g), resembling the state transitions seen in patient-derived BRAF mutant melanoma cell lines that accompany RAF inhibitor resistance36,37.
In summary, our data suggest a progression of transcriptomic alterations along the development of belvarafenib resistance. Initial tolerance is associated with activation of EGFR and TGF-β signaling as well as FN1 upregulation. This is followed by MAPK and PI3K pathway reactivation and a shift towards an undifferentiated state, thereby promoting the expansion of the resistant cells. OAK’s ultra-high throughput and stepwise sequencing capability render it an exceptionally suitable tool for investigating transcriptomic signatures within rare cell populations that lead to drug resistance in cancer.
Discussion
OAK combines droplet microfluidics with combinatorial indexing, enabling ultra-high throughput and multimodal single-cell profiling. Our study underscores OAK’s versatility across diverse experimental designs and modalities, including scRNA-Seq, sample multiplexing, and paired profiling of snRNA-Seq and snATAC-Seq. Moreover, with minor adjustments in the secondary indexing primers and library preparation, broad compatibility can be expected within the full spectrum of applications offered by the Chromium platform, encompassing immune profiling, cell surface protein detection and CRISPR perturbations. In addition, the experimental feature of distributing a large number of cells into multiple aliquots enables sequencing of each sub-library separately. Such stepwise sequencing allows the sequencing of a smaller number of cells for quality assessment prior to embarking on large-scale sequencing. Furthermore, sub-libraries provide the opportunity to sequence the number of cells desired for analysis, while preserving unprocessed ones for future data acquisition. Finally, OAK data processing is compatible with analysis pipelines that have been developed for the prevailing commercial Chromium platform. This aspect facilitates a seamless integration of OAK into researchers’ existing data processing workflows.
OAK presents multiple opportunities for reducing cost. First, overloading a single microfluidic channel enables more efficient utilization of costly reagents, including the barcoding beads. Secondly, in contrast to other combinatorial indexing methods8–12,15,38, OAK avoids a substantial upfront investment in synthesizing plates of indexing oligos or assembling pre-indexed transposome for the ATAC modality - thereby also streamlining benchwork. Thirdly, unlike some overloading methods that identify and discard multi-cell droplets without being able to recover single cells encapsulated within17,39–41, OAK is able to resolve single cells in multi-cell droplets, maximizing the usage of sequencing data. In summary ultra-high throughput, extensive versatility across different molecular modalities, experimental convenience, and cost efficiency distinguish OAK from alternative technologies in the field.
The single-cell sequencing field is undergoing rapid transformation and growth. Recent examples include innovations like 10x Genomics’ GEM-X and Flex products, both exhibiting superior throughput compared to the regular Chromium products. However, these products are currently unable to perform paired snRNA-Seq and snATAC-Seq. Nevertheless, it is expected that OAK will be adaptable to these evolving platforms, thereby leveraging improvements in droplet generation technologies to deliver even higher throughput. Although in this study, our focus rests primarily on validating OAK on the Chromium platform, we expect OAK to be also compatible with other droplet systems that make use of releasable barcoding primers from microspheres, such as the inDrops system6 and Hydrop system14.
In addition to various cell lines and primary tissues analyzed in this study, we have also tested OAK with human brain samples and human peripheral blood mononuclear cells (PBMCs). While OAK successfully generates high-quality data for joint scRNA-Seq and scATAC-Seq from brain samples, the current protocol faces limitations in generating high-complexity libraries for PBMCs. This may arise from the fact that more fragile cells are more sensitive to the strong detergents present in droplets. As such, optimizing the fixation and detergent usage presents a future direction for technological development, in order to broaden OAK’s applicability to a wider array of sample types.
In summary, we developed a single-cell multiomic profiling method, OAK, which empowers extensive characterization of complex tissues and cellular systems, while maintaining a streamlined and cost-efficient experimental approach. We anticipate that OAK will readily scale with ongoing advances in droplet generation platforms, and will be flexible to accommodate measurement of additional molecular modalities.
Methods
Ethical statement
The described studies comply with ethical regulations of Genentech. Institutional approval and the written, informed consent for the collection of donor eyes to be used for research purposes was obtained from the University of Utah, and conformed to the tenets of the Declaration of Helsinki. All tissue was de-identified in accordance with HIPPA privacy rules.
Cell culture and single-cell suspension preparation
K562 cells (ATCC number CCL-243) were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) with 10% fetal bovine serum (FBS). NIH/3T3 cells (ATCC number CRL-1658) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% FBS. IPC-298 cells (DSMZ number ACC 251) were cultured in RPMI medium with 10% FBS, 2 mM L-glutamine, and 1% penicillin/streptomycin. Cells were incubated at 37 °C with 95% Air and 5% CO2. TrypLE™ Express (Thermo Fisher Scientific 12604013) was used to detach adherent cells from culture flasks. Harvested cells were washed twice with phosphate-buffered saline (PBS) with 0.04% Bovine albumin Fraction V (Thermo Fisher Scientific 15260037), and resuspended with PBS to achieve single-cell suspensions.
Culture and staining of normal human bronchial epithelial cells
Normal human bronchial epithelial cells (Lonza CC-2540, Epithelix) were differentiated in transwell plates at an air-liquid interface. Cells were dissociated with accutase and then washed twice with PBS/1% Bovine Serum Albumin (BSA). Nine sample wells of cells were resuspended in 50 µL PBS/1% BSA and each stained with 1 µL of TotalSeq-A antibody (Biolegend, Clone LNH-94; 2M2. TotalSeq™-A0251 anti-human Hashtag 1 Antibody, Catalog number: 394601; TotalSeq™-A0252 anti-human Hashtag 2 Antibody, Catalog number: 394603; TotalSeq™-A0253 anti-human Hashtag 3 Antibody, Catalog number: 394605; TotalSeq™-A0254 anti-human Hashtag 4 Antibody, Catalog number: 394607; TotalSeq™-A0256 anti-human Hashtag 6 Antibody, Catalog number: 394611; TotalSeq™-A0257 anti-human Hashtag 7 Antibody, Catalog number: 394613; TotalSeq™-A0259 anti-human Hashtag 9 Antibody, Catalog number: 394617; TotalSeq™-A0262 anti-human Hashtag 12 Antibody, Catalog number: 394623; TotalSeq™-A0263 anti-human Hashtag 13 Antibody, Catalog number: 394625). Incubation was at 4 °C for 20 minutes. Cells were washed 3x in PBS/1% BSA and then all wells were pooled together. Pooled cells were stained with Sytox Green for 5 minutes at room temperature before sorting for live cells on a Sony SH800S into PBS.
Retinal tissue nuclei preparation
Human donor eye collection was followed according to a standardized protocol42. In brief, the isolated peripheral retinal sample from an 80-year old male was obtained within a 6-hour post-mortem interval, defined as death-to-preservation time, in collaboration with the Utah Lions Eye Bank. The sample was placed in a cryotube and flash frozen in liquid nitrogen prior to storage at −80 °C. The sample was dissociated by douncing ten times in a glass homogenizer in 1 mL ice cold NIM4 buffer (9.9 mL NIM1 [250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris Buffer in nuclease-free water], 10 µL 100 mM DTT, 1 cOmplete Mini protease inhibitor cocktail tablet (11836153001, Roche), 100 µL 10% Triton X-100, 100 µL RNAseIN, 100 µL SUPERasin) and incubated on ice for 10 min. Tissue homogenate was centrifuged at 450 G for 5 min 4 °C. Supernatant was removed and 500 µL ice-cold wash buffer (400 µL salt buffer [200 µL 1 M Tris pH 7.4, 40 µL 5 M NaCl, 40 µL 5 M NaCl, 60 µL 1 M MgCl2, 1.7 mL nuclease-free water], 4 µL 100 mM DTT, 40 µL 10% Tween 20, 800 µL 5% RNAse-free BSA, 100 µL RNAse inhibitor, 2.66 mL nuclease-free water) was added to the nuclei and the sample pipet mixed five times. The nuclei were passed through a 40 μm filter then counted.
OAK scRNA-Seq
Methanol-fixed cells were used to generate data in results sections: Principles and performance of OAK, Leveraging ultra-high throughput for sample multiplexing, and Melanoma resistance to RAF inhibitor belvarafenib. Single-cell suspension in 400 µl PBS was transferred to a 2 mL round-bottom tube and fixed by adding 1600 µL chilled methanol drop by drop with gentle stirring. Cells were then incubated at −20 °C for 30 min. After fixing, cells were placed on ice for 5 min and then pelleted at 1000 G for 5 min at 4 °C in a pre-cooled swinging bucket centrifuge. Supernatant was removed and the pellet was resuspended with appropriate volume of resuspension buffer to target 30,000 cells/µl or higher. Resuspension buffer is composed of 3X saline sodium citrate (SSC) (Invitrogen, 115557044), 1-2% BSA, 0.2 U/µL Protector RNAse inhibitor (Roche, 03335402001), and 1 mM DTT in nuclease-free water. Cells were counted and a desired number of cells (typically 150,000) were loaded per channel. Other reagents were used for loading according to standard 10x Genomics’ Chromium 3’ RNA-Seq protocol. After droplet generation, reactions were transferred to microfuge tubes for reverse transcription at 53 °C for 45 min. Immediately after reverse transcription, the droplets were unpacked by adding 125 µL recovery agent to break the emulsion. After phase separation, the aqueous phase, containing recovered fixed cells, was transferred to a 2 mL microfuge tube. 800 µL 3X SSC was added to the cell suspension. The cells were spun at 650 G at 4 °C for 5 min. Supernatant was carefully removed. 1 mL 3X SSC was added to the cell pellet with gentle tapping on the tube to dislodge the pellet. Cells were spun again at 650 G at 4 °C for 5 min. The pellet was resuspended in 215 µL 3X SSC with gentle pipette mixing. A 10 µl solution was used for cell counting to estimate the number of cells per aliquot. The remaining solution was evenly distributed into multiple aliquots by PCR Strip Tubes (typically 20 aliquots per 150,000 cells loaded to aim for 4000 cells per aliquot). The aliquots were immediately stored at −80 °C until ready for the next step.
From the frozen aliquots, researchers can select how many of them to process into sub-libraries for sequencing. This allows for an increasing number of cells to be sequenced in a stepwise manner. To prepare sequencing libraries, a desired number of aliquots were heated to 80 °C for 5 min to aid release of 1st strand cDNA. DynabeadsTM Silane Viral NA kit (ThermoFisher, 37011D) was used to purify 1st strand cDNA according to manufacturer’s instructions. The cDNA was eluted in 35 µL of the elution buffer. For cDNA amplification PCR, a TSO recognition primer (AAGCAGTGGTATCAACGCAGAGT) and a primer that adds a secondary index (e.g., with barcode underlined: AATGATACGGCGACCACCGAGATCTACACAACGTGATACACTCTTTCCCTACACGACGCTCTTCCGATCT) were used. For capturing antibody-derived fragments in cell hashing experiments, a single relevant primer can be added, for example, the HTO primer in the case of TotalSeqA hashing. cDNA from multiple aliquots (typically 2-4) can be pooled for sub-library construction by following the standard Chromium protocol, except in the library PCR where a partial P5 primer (AATGATACGGCGACCACCGAGA) is used alongside an i7 index primer (e.g., with barcode underlined: CAAGCAGAAGACGGCATACGAGATCGCATGTTACGTGACTGGAGTTCAGACGTGT. 10x Genomics library kits (Chromium Single Cell 3ʹ Library Kit v3, PN-1000095 and Chromium Single Cell 3ʹ Feature Barcode Library Kit, PN-1000079) can be purchased independently to provide enough reagents for construction of the OAK sub-libraries. For a cost-effective alternative for generating libraries from the antibody-derived fragment component, cDNA and indexing primers can be ordered and used with KAPA HiFi HotStart ReadyMix (Roche Diagnostics, KK2601).
After QC, sub-libraries with unique secondary index (i5) and i7 index were pooled for sequencing on Illumina platforms, with 28 cycles for Read 1, 10 cycles for i7 index, 8 cycles for i5 index, and 90 cycles for Read 2. Targeted sequencing depth was 20,000 read pairs per cell. Cell counting at the aliquoting step was used to estimate the number of cells expected to recover.
OAK paired snRNA-Seq and snATAC-Seq
Nuclei were centrifuged at 500 G in a 2 mL round-bottom microfuge tube. After removing the supernatant, the pellet was resuspended in a fixative solution of 1 mL calcium-free PBS with 0.3% formaldehyde. During addition of fixative solution, it is important to pipette up and down gently to disrupt the pellet and prevent clumping. Nuclei in fixation solution were placed on ice for 10 min and then centrifuged for 5 min at 500 G at 4 °C. After supernatant was removed, 1.5 mL wash buffer was added. The wash buffer was 10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 1% BSA, 0.1% Tween-20, 1 mM DTT, 1 U/μL RNase inhibitor (Roche, 03335402001) in nuclease-free water. After 5 min 500 G at 4 °C, the supernatant was removed and the nuclei were resuspended in the appropriate volume of nuclei resuspension buffer to target 2,400 nuclei/μL or more. The nuclei resuspension buffer was 1X Nuclei Buffer (from a 20X stock, 10x Genomics, PN2000207), 1 mM DTT, and 1 U/μL RNase inhibitor in nuclease-free water.
After fixation, we typically transpose 75,000-200,000 nuclei, with an expected cell recovery rate of 40%. This recovery rate may be dependent on sample type and quality. We found that TDE1 enzyme (Illumina Tagment DNA Enzyme and Buffer Small Kit, 20034197) could be used for the additional tagmentation reactions required to support processing large numbers of nuclei. Including this additional reagent and the subsequent library preparation reagents, a cost analysis of the OAK multiome protocol ran on the retinal sample came to $ 0.09 per nuclei, versus $ 0.39 per nuclei for the regular Chromium multiome protocol. Each transposition reaction was 15 μL, which is composed of 12,000 nuclei in 5 μL 1X nuclei buffer, 3 μL TDE1 enzyme, and 7 μL ATAC Buffer B (10x Genomics, PN 2000193). To transpose 100,000 nuclei in the retinal profiling experiment, 8 such reactions were prepared. Reactions were incubated at 37 °C for 1 hr. All transposition reactions were combined to a single 2 mL round-bottom microfuge tube and spun at 500 G in a pre-cooled centrifuge at 4 °C for 5 min. Most supernatant was removed, leaving the last 15 μL to resuspend the combined nuclei. All of the transposed nuclei in this 15 μL were used for loading 1 channel. Other reagents were used for loading according to regular 10x Genomics’ Chromium Next GEM Single Cell Multiome protocol. After GEM generation, barcoding, and quenching according to the standard Chromium multiome protocol, 125 μL recovery agent (10x Genomics, PN 220016) was added to break the emulsion. The aqueous layer containing fixed nuclei was carefully transferred to a 2 mL round-bottom microfuge tube. 800 µL 3X SSC was added to the nuclei suspension. The nuclei were spun at 650 G at 4 °C for 5 min. The supernatant was carefully removed. 1 mL 3X SSC was added to the nuclei pellet with gentle tapping on the tube to dislodge the pellet. Nuclei were spun again at 650 G at 4 °C for 5 min. The pellet was resuspended in 215 µL 3X SSC with gentle pipette mixing. A 10 µl solution was used for counting to estimate the number of nuclei per aliquot. The remaining solution was evenly distributed into multiple aliquots by PCR Strip Tubes to aim for 4,000 nuclei per aliquot. The aliquots were immediately stored at −80 °C until ready for sequencing library preparation.
To prepare sequencing libraries, a desired number of aliquots were heated to 80 °C for 5 min to aid the release of 1st strand cDNA and ATAC fragments. DynabeadsTM Silane Viral NA kit (ThermoFisher, 37011D) was used to purify 1st strand cDNA and ATAC fragments according to manufacturer’s instructions. The resulting products were pre-amplified in a 100 μL reaction using 10 cycles with 4 μL pre-amp primers (10x Genomics PN 20002714) and 50 μL of NEBNext High-Fidelity 2X PCR Master Mix (NEB, M0541S). Reactions were cleaned with 1.6X SPRI and eluted in 40 μL EB. 10 μL of the product was used for constructing snATAC-Seq libraries. One PCR reaction was set up for each aliquot, with 0.6 μL of 100 μM partial P5 primer, 50 μL NEBNext High-Fidelity 2X PCR Master Mix (NEB, M0541S), 36.9 μL nuclease-free water and 2.5 μL of 10 μM sample index N (10x Genomics, PN 1000212). The PCR program was 98 °C 30 s, n cycles [98 °C 10 s, 67 °C 30 s, 72 °C 20 s], 72 °C 2 min, held at 4 °C. N is typically recommended cycles for standard Chromium protocol for the number of cells. Extra cycles can be added if ATAC library yield is low. A double-sided size selection was performed as instructed in the standard Chromium protocol. cDNA library amplification, sequencing library construction and sequencer operation were conducted in the same way as described in OAK scRNA-Seq.
For snATAC-Seq libraries, sub-libraries with unique i7 index were pooled for sequencing on Illumina platforms. Targeted sequencing depth was 25,000 read pairs per cell, with 50 cycles for Read 1, 8 cycles for i7 index, 24 cycles for i5 index, and 49 cycles for Read 2. Cell counting at the aliquoting step was used to estimate the number of cells expected to recover.
Sequencing read processing
Illumina Miseq, Nextseq 2000, and NovaSeq 6000 were used for sequencing. Raw sequencing data was demultiplexed by Illumina’s Bcl2Fastq software (v2.20) to resolve reads per OAK sub-libraries. Fastq files for each sub-library were processed with Cell Ranger software v6 (single-cell RNAseq) or Cell Ranger ARC software v2 (paired snRNA-Seq and snATAC-Seq) to generate gene and chromatin fragment counts.
Simulation for cell distribution in droplets
The percentage of having k cells in a droplet is approximated by p(k, λ) = e(-λ) × λk/(k)! based on Poisson distribution, where λ is the loading rate approximated as the number of loaded cells divided by the number of generated droplets.
Multiplet rate theoretical estimation
The expected number of events when more than one cell share the same combinatorial barcodes is N-D + D × [(D − 1)/D]N based on the closed form solution for expected number of collisions in the birthday paradox12, where N is the number of cells loaded and D is the total number of barcode combinations. We used 100,000 as the number of droplets generated per channel on the Chromium microfluidic chip. Hence D was calculated as 100000 × n_aliquot, where n_aliquot is the number of aliquots generated.
Cell recovery calculation
Cell recovery rate for each method is calculated by dividing the number of cells obtained after all rounds of barcoding by the total number of cells used for the first round of barcoding. All experiments presented in this study are used to generate the range of recovery rates for OAK. The rate range for scifi-RNA-seq is based on the multiplication of the recovery rate from the first round of barcoding, as reported in the publication15, with an assumed perfect (65%) cell recovery in the second round by the droplet system. This could result in a slight overestimation of its rates. The rate range for SPLiT-seq is derived from the information regarding Parse Biosciences’ Evercode WT products, which represent the commercial implementation of SPLiT-seq. The rate range for Paired-seq is based on the data reported in the corresponding publication13.
Species-mixing experiment and multiplet rate estimate
In the species-mixing experiment when 150,000 cells (Fig. 1c) were loaded in one channel of the Chromium chip, all cells were unpacked into 12 aliquots. In the experiment when 450,000 cells (Fig. 1c) were loaded in one channel, all cells were unpacked into 40 aliquots. In each experiment, one aliquot was processed to generate a sequencing library. Reads were mapped to a hybrid Human-Mouse reference genome that consists of GRCh38 and mm10. Cells were classified into observed multiplets (human+mouse), mouse cells, and human cells by Cell Ranger software v6. Since the input cells consisted of a 1:1 mixture of a mouse and a human cell line, true multiplet rate was estimated as (observed multiplet rate) × 2 to include those inferred human+human and mouse+mouse multiplets.
Hashtag assignment and cell annotation in human bronchial epithelial cells
The Cell Ranger package v6 was used to determine hashtag assignment rate for the human bronchial epithelial cell experiment using a matching antibody-derived tag and gene expression library for each of four (out of 22) OAK aliquots, and for standard comparison data generated in parallel. We imported and merged the data from the multiple OAK aliquots in Seurat, then integrated the OAK and standard scRNA-Seq data with Harmony43 prior to clustering using the Seurat FindClusters function (resolution = 0.6) and assigning cluster identity (Club/Goblet, Basal, Ciliated, Basal cycling, Neuroendocrine or Unknown) based on gene scores for known markers.
Retinal paired snRNA-Seq and snATAC-Seq data analysis
snATAC-Seq and gene expression data from each sub-library were combined using cell ranger-arc aggr with -normalize=none. Gene expression data was first imported into a Seurat (Version 4.3.0.1) object for assessment of snRNA-Seq quality and comprehensive annotation. Cells with > 200 genes and < 10000 genes were retained. Cells were clustered, then marker gene scores were used to validate assignment of clusters to the major known cell types. For cones, horizontal, amacrine and bipolar cells, further sub-clustering was performed prior to annotation and propagation into the master Seurat object. The snRNA-Seq annotations were added as metadata into an ArchR project containing both snATAC-Seq and snRNA-Seq data, based on cell barcodes. For further analysis of snATAC-Seq data, only cells with data passing filters from both modalities were kept.
In ArchR (Version 1.0.2), hg38 was used as the reference genome and barcodes were filtered for TSS enrichment > 4 and nFrags > 1000. Peaks of open chromatin were identified by using ArchR tools. First addReproduciblePeakSet was utilized with MACSr, which uses the MACS3 algorithm for peak-calling44, on the snRNA-Seq annotation, excluding chrMT and ‘chrY, followed by addPeakMatrix. Marker peaks were called using this matrix for each cell type with getMarkerFeatures with options bias = c(“TSSEnrichment”, “log10(nFrags)”) and testMethod = “wilcoxon”. Peaks called were filtered for FDR cutOff of 0.01 and Log2FC ≥ 1. The plotMarkerHeatmap function was used to plot a heatmap of the markers with FDR ≤ 0.001 and Log2FC ≥ 1. The plotBrowserTrack function in ArchR was used to plot example chromatin tracks and peaks for each annotated cell group.
Epiregulon infers regulatory elements to target genes based on correlated gene expression and chromatin accessibility in clustered cells, matching these elements to known transcription factor binding sites from repositories of public ChIP-Seq data. In Epiregulon (Version 1.0.34) we extracted the normalized gene expression counts and peak matrices from the ArchR project, removing unannotated cells, and calculated the peak to gene expression linkages using the LSI snRNA-Seq and snATAC-Seq combined dimensions from ArchR. We annotated the linkages as regulons with known motifs from the human ChIP-Atlas and Encode databases. We further pruned the regulons by setting a correlation test cutoff for for all components (peaks, gene expression and TFs in the same cells) using the following parameters to the pruneRegulon function (test = “chi.sq”, prune_value = “pval”, regulon_cutoff = 0.05 and defined clusters by the major cell types). We used the addWeights function to add an estimate and multiplier for the strength of regulation, using the parameters tf_re.merge = FALSE, method = “corr”. We calculated a score for each regulon using calculateActivity to combine weights with activity of linked genes (mode = “weight”, method = “weightedMean”, exp_assay = “normalizedCounts”, normalize = FALSE). To find the transcription factors with differential regulation activity associated with each major cell type we used the findDifferentialActivity function with parameters, pval.type = “some”, direction = “up” and test.type = “t”). We filtered these by significant transcription factors with an FDR cutoff of 0.05 and a logFC cutoff of 0.1. With this list in hand, we added information on the proportion of cells that have expression of the identified transcription factor in the significant group. We filtered the transcription factors to those that have > 30% expression in the associated cell type. We took the top ten transcription factors with the highest calculated activity in each cell type and ordered them by the proportion of cells expressing within the cell type. We then used the plotBubble function to plot the top seven transcription factors with the highest calculated activity for each cell type.
TraCe-seq cell preparation and scRNA-Seq
TraCe-seq barcode lentivirus was produced in HEK293T cells, and cells were infected at 0.05–0.1 of multiplicity of infection (MOI), and sorted for eGFP expression on a BD Aria Fusion cell sorter to enrich for barcoded cells29. After sorting, 1000 IPC-298 cells were used to form the starting population. This population was expanded for 17 doublings. A subculture was used for the Day 0 experiment, while the rest of cells were treated with 10 µM belvarafenib. Medium containing belvarafenib was replenished twice a week. Subcultures of cells were taken for OAK on Day 0 and Day 10 as lineage diversity was highest before and early in treatment. For Day 0, 2 channels on the Chromium chip were loaded, each with 138,000 cells. 39 aliquots were generated, each contained 3700 cells. 20 aliquots were processed into sub-libraries and sequenced. For Day 10, 3 channels were loaded, each with 180,000 cells. 44 aliquots were generated, each contained 6000 cells. 12 aliquots were processed into sub-libraries and sequenced. The remaining aliquots were stocked in −80 °C for potential future data acquisition. Standard Chromium scRNAseq was performed according to manufacturer’s instructions for cells collected on Day 20 and Day 90 of treatment as lineage diversity dropped.
TraCe-seq single-cell lineage barcode library generation
OAK indexed cDNA libraries from multiple aliquots can be pooled for the generation of lineage barcode libraries. Typically, 7.5 µl cDNA from each aliquot is used and 2 aliquots were pooled for 1 reaction. A semi-nested PCR strategy was used to ensure the specificity of the resulting lineage barcode library. In the first round of PCR, the partial P5 primer and GPF_F1_outer primer (GTGCACTTAGTAAGGACCCAAACG) were used. In the second round of PCR, the partial P5 primer and an i7 indexed GFP_F2_inner primer (e.g., with index underlined:CAAGCAGAAGACGGCATACGAGATCCGCGGTTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGATAACCCTCGGGATGGATGAACTG) were used.
TraCe-seq bulk lineage library generation
Cells from Day 0 were used to amplify the lineage transcripts. The reverse transcription mix was composed of 5 µL Maxima H minus Reverse Transcriptase (Thermo Fisher Scientific EP0753), 20 µL 5X RT buffer, 5 µL dNTP (10 mM each), 1.5 µL TraCe_libABC_end_RT primer (GTGGATCCACCGAACGCAACGCAC, 100 µM), 1.5 µL Protector RNase Inhibitor (Sigma PN 3335399001), 5 µl methanol fixed cells, and 62 µl water. The reaction was incubated at 50 °C for 30 min, followed by 85 °C for 5 min, and held at 4 °C briefly. The product was subsequently amplified by PCR with P5 indexed primer (e.g., with index underlined: AATGATACGGCGACCACCGAGATCTACACGATATCGACGAACGCAACGCACGCACACT) and i7 indexed GFP_F2_inner primer. The SPRISelect beads were used to perform a 0.6X–1.6X double sided size selection for the PCR product.
Drug response curve generation
Cells were seeded at 2000 cells per well in 96-well plate, and were treated with belvarafenib 24 h after seeding. Cells were treated with a 9-point titration (1:3) and DMSO control using the HP D300 drug dispenser. Cell growth was assessed using CellTiter-Glo Luminescent Cell Viability Assays (Promega G7570), and luminescence was read by a 2104 EnVision Multilabel Plate Reader (PerkinElmer) five days after treatment. All cell viability data was collected and calculated for 4 replicates per condition. Data from the DMSO control was set to 100%. Nonlinear regression curves were generated by GraphPad Prism to fit the viability data.
TraCe-seq data analysis
Cells were assigned to a lineage when the UMI count for one lineage barcode was at least two-fold higher than the other ones detected in the given cell. Single-cell gene expression matrix was analyzed with Scanpy (v1.9.1)45. Gene set enrichment for MSigDB’s hallmark sets32 was performed with decoupleR (v1.8.0)46. MAPK, EGFR, PI3K, and TGF-β pathway scores were generated with PROGENy (v1.18.0)47. P values were calculated using the Mann-Whitney-Wilcoxon test (two-sided) with Bonferroni adjustment. Genes representing differentiation and dedifferentiation states were based on an established melanoma four-stage differentiation model48. The melanocytic, transitory-melanocytic, transitory, and neural crest-like-transitory signatures were grouped as the differentiation signature. The undifferentiated, undifferentiated-neural crest-like, and neural crest-like signatures were grouped as the de-differentiation signature. The signature scores were generated by Scanpy’s tl.score_genes function.
Statistics & Reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank the donor and the donor’s family who contributed the retinal sample for this study. Human donor eye collection was supported by the Macular Degeneration Foundation, Inc. (Henderson, NV, USA); the Carl Marshall Reeves & Mildred Almen Reeves Foundation, Inc. (Fenton, MO, USA), Ira G. Ross and Elizabeth Olmsted Ross Endowed Chair, and by NEIH/NEI: 5R01EY031209-04. The content is solely the responsibility of the authors and does not necessarily represent official views of the U.S. Government. We acknowledge the single-cell sequencing community at Genentech for collaborations, discussions, and sequencing support, including Ahmet Kurdoglu, Qixin Bei, Manching Ku, Jie Liu, Daniel Le, Ashley Byrne, William Stephenson, Vasu Kameswaran, Bence Daniel, Jay Leone, Shiqi Russell Xie, Diana Wu, Katie Geiger-Schuller, Ana Meireles, Kristel Dorighi, Aviv Regev, Xiaosai Yao, David Garfield, Luz Orozco, Natalie Fox, Jack Kamm, Lyndsay Murrow and Jennie Lill. Schematics used in this manuscript were created with BioRender.com.
Author contributions
B.W. conceived and developed OAK. B.W. and H.M.B. optimized the methods. B.W., H.M.B., X.Y., and S.D. designed the study. M.D., H.C., L.A.O., and I.K.K. facilitated and provided the retinal sample. H.M.B. and A.S. conducted the experiments on the retinal sample, and H.M.B. analyzed the retinal data. H.M.B., E.V.N., C.Eidenschenk and C.Everett conducted the experiments on the in vitro differentiation bronchial samples and H.M.B. and S.D. analyzed the data. B.W. and X.Y. performed the melanoma cell lineage tracing experiments, and B.W. analyzed the sequencing data. C.C. and J.L. generated sequencing libraries. M.S., J.M.L., A.X-M., N.P., and Y.L. performed sequencing. B.W., H.M.B., X.Y., S.D., and Z.M. wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Linas Mazutis, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Sequencing data generated in this study have been deposited to NCBI Sequence Read Archive under accession number PRJNA1046517. External datasets are publicly available from GEO: scifi-RNA-seq GSE168620, sci-RNA-seq GSE98561, SPLiT-seq GSE110823, Paired-seq GSE130399, sci-CAR GSE117089. Source data are provided with this paper.
Code availability
The code used to analyze data is available in the GitHub repository: https://github.com/bingwu2017/OAK_manuscript.git49.
Competing interests
B.W., H.M.B., X.Y., A.S., C.Eidenschenk, C.Everett, E.V.N., H.C., C.C., J.L., M.S., J.M.L., A.X.M., N.P., Y.L., Z.M. and S.D. are currently employees of Genentech and shareholders of Roche. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zora Modrusan, Email: modrusan@gene.com.
Spyros Darmanis, Email: darmanis@gene.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53227-z.
References
- 1.Ogbeide, S., Giannese, F., Mincarelli, L. & Macaulay, I. C. Into the multiverse: advances in single-cell multiomic profiling. Trends Genet38, 831–843 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Zhu, C., Preissl, S. & Ren, B. Single-cell multimodal omics: the power of many. Nat. Methods17, 11–14 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Kashima, Y. et al. Single-cell sequencing techniques from individual to multiomics analyses. Exp. Mol. Med.52, 1419–1427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandereyken, K., Sifrim, A., Thienpont, B. & Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet.24, 494–515 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun.8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cusanovich, D. A. et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science361, 1380–1385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma, S. et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell183, 1103–1116.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu, C. et al. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol.26, 1063–1070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rop, F. V. D. et al. Hydrop enables droplet-based single-cell ATAC-seq and single-cell RNA-seq using dissolvable hydrogel beads. eLife11, e73971 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datlinger, P. et al. Ultra-high-throughput single-cell RNA sequencing and perturbation screening with combinatorial fluidic indexing. Nat. Methods18, 635–642 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol.38, 737–746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol.19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drokhlyansky, E. et al. The human and mouse enteric nervous system at single-cell resolution. Cell182, 1606–1622.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barr, J. et al. Injury-induced pulmonary tuft cells are heterogenous, arise independent of key Type 2 cytokines, and are dispensable for dysplastic repair. eLife11, e78074 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, Q. et al. A multi-omics atlas of the human retina at single-cell resolution. Cell Genom.3, 100298 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orozco, L. D. et al. A systems biology approach uncovers novel disease mechanisms in age-related macular degeneration. Cell Genom.3, 100302 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan, W. et al. Cell atlas of the human fovea and peripheral retina. Sci. Rep.10, 9802 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, S. K. et al. Quality assessment and refinement of chromatin accessibility data using a sequence-based predictive model. Proc. Natl Acad. Sci.119, e2212810119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wlodarczyk, T. et al. Epiregulon: Inference of single-cell transcription factor activity to dissect mechanisms of lineage plasticity and drug response. Preprint at bioRxiv 10.1101/2023.11.27.568955 (2023).
- 25.Brzezinski, J. A., Lamba, D. A. & Reh, T. A. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development137, 619–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klimova, L., Antosova, B., Kuzelova, A., Strnad, H. & Kozmik, Z. Onecut1 and Onecut2 transcription factors operate downstream of Pax6 to regulate horizontal cell development. Dev. Biol.402, 48–60 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Wahle, P. et al. Multimodal spatiotemporal phenotyping of human retinal organoid development. Nat. Biotechnol. 1–11 10.1038/s41587-023-01747-2 (2023). [DOI] [PMC free article] [PubMed]
- 28.Yen, I. et al. ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma. Nature594, 418–423 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Chang, M. T. et al. Identifying transcriptional programs underlying cancer drug response with TraCe-seq. Nat. Biotechnol.40, 86–93 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Hirata, E. et al. Intravital imaging reveals how BRAF Inhibition generates drug-tolerant microenvironments with High Integrin β1/FAK Signaling. Cancer Cell27, 574–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, B. et al. Fibronectin 1 promotes melanoma proliferation and metastasis by inhibiting apoptosis and regulating EMT. OncoTargets Ther.12, 3207–3221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon, A. et al. The molecular signatures database hallmark gene set collection. Cell Syst.1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, C. et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature508, 118–122 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Xu, J., Lamouille, S. & Derynck, R. TGF-β-induced epithelial to mesenchymal transition. Cell Res19, 156–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, J. H. et al. Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat. Commun.11, 1897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su, Y. et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl Acad. Sci.114, 13679–13684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fallahi‐Sichani, M. et al. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de‐differentiated state. Mol. Syst. Biol.13, 905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lareau, C. A. et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol.37, 916–924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGinnis, C. S. et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods16, 619–626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol.36, 89–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods17, 615–620 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Owen, L. A. et al. The utah protocol for postmortem eye phenotyping and molecular biochemical analysis. Investig. Ophthalmol. Vis. Sci.60, 1204–1212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y. et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol.9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badia-i-Mompel, P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv.2, vbac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schubert, M. et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun.9, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsoi, J. et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell33, 890–904.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, B. et al. Overloading And unpacKing (OAK) - droplet-based combinatorial indexing for ultra-high throughput single-cell multiomic profiling. bingwu2017/OAK_manuscript: Initial publication. Zenodo 10.5281/zenodo.13844576 (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated in this study have been deposited to NCBI Sequence Read Archive under accession number PRJNA1046517. External datasets are publicly available from GEO: scifi-RNA-seq GSE168620, sci-RNA-seq GSE98561, SPLiT-seq GSE110823, Paired-seq GSE130399, sci-CAR GSE117089. Source data are provided with this paper.
The code used to analyze data is available in the GitHub repository: https://github.com/bingwu2017/OAK_manuscript.git49.