Abstract
Chronic demyelination and oligodendrocyte loss deprive neurons of crucial support. It is the degeneration of neurons and their connections that drives progressive disability in demyelinating disease. However, whether chronic demyelination triggers neurodegeneration and how it may do so remain unclear. We characterize two genetic mouse models of inducible demyelination, one distinguished by effective remyelination and the other by remyelination failure and chronic demyelination. While both demyelinating lines feature axonal damage, mice with blocked remyelination have elevated neuronal apoptosis and altered microglial inflammation, whereas mice with efficient remyelination do not feature neuronal apoptosis and have improved functional recovery. Remyelination incapable mice show increased activation of kinases downstream of dual leucine zipper kinase (DLK) and phosphorylation of c-Jun in neuronal nuclei. Pharmacological inhibition or genetic disruption of DLK block c-Jun phosphorylation and the apoptosis of demyelinated neurons. Together, we demonstrate that remyelination is associated with neuroprotection and identify DLK inhibition as protective strategy for chronically demyelinated neurons.
Subject terms: Myelin biology and repair, Diseases of the nervous system, Oligodendrocyte
The mechanisms that trigger neurodegeneration in demyelinating disease are unclear. Here, the authors find that impaired remyelination induces a DLK-mediated loss of retinal ganglion cells (RGCs), and that efficient remyelination or DLK inhibition block RGC death.
Introduction
Remyelination is the regenerative process by which new myelin sheaths are produced in the CNS, typically via the differentiation of oligodendrocytes (OLs) from oligodendrocyte precursor cells (OPCs)1,2, or to a limited extent by OLs that survive the demyelinating insult3–5. In the inflammatory demyelinating disease multiple sclerosis (MS), remyelination is often incomplete6,7, resulting in chronic demyelination of axons. Chronic demyelination and the loss of OLs deprive neurons of crucial metabolic and trophic support and is hypothesized to leave them vulnerable to subsequent degeneration8–10. With disease chronicity, a progressive loss of neurons and their axons in MS drives permanent disability11,12. While several studies have found an association between poor remyelination efficiency and neurodegeneration in MS13,14, there is a paucity of experimental evidence demonstrating that impaired oligodendrogenesis and remyelination failure lead to neurodegeneration. To date, no study has selectively targeted the OL lineage to induce demyelination and determined the extent of neurodegeneration in the context of subsequent successful or failed remyelination. Additionally, the molecular mechanisms critical for neurodegeneration following demyelination remain unclear
Here, we compare two rodent models of genetic demyelination, which feature either successful or impaired remyelination. By contrasting these two rodent models, we find that mice with successful remyelination do not feature axonal degeneration and neuronal apoptosis in the visual system, whereas mice unable to remyelinate have fewer axons and increased retinal ganglion cell (RGC) apoptosis. Mice with failed remyelination have elevated phosphorylation of kinases downstream of DLK in the optic nerve and phosphorylation of the transcription factor c-Jun in RGC nuclei. Pharmacological inhibition or genetic disruption of DLK block both the phosphorylation of c-Jun and RGC apoptosis. We propose that effective remyelination of the axon improves neuronal survival by suppressing DLK-mediated signaling.
Results
Myelin regulatory factor (Myrf) knockout from both OPCs and mature OLs results in genetic demyelination and impaired remyelination
A rodent model that inhibits remyelination in a cell-selective manner would permit an understanding of the extent and mechanisms by which prolonged demyelination damages neurons. Myrf is an essential transcription factor for myelin gene transcription15,16, and its deletion from mature OLs using Plp1-CreERT mice (Myrffl/fl Plp1 CreERT; hereto referred to as MyrfΔiPlp1) results in CNS-wide demyelination17. Reasoning that extending the knockout of Myrf to OPCs would prevent remyelination18, we crossed the Myrffl/fl line to the pan-OL lineage Sox10-CreERT19 line (Myrffl/fl Sox10 CreERT; MyrfiSox10) (Fig. 1a). Dosing of both MyrfΔiPlp1 and MyrfΔiSox10 mice with tamoxifen at eight weeks of age (Fig. 1b) results in progressive ataxia, hindlimb tremor, and paresis (Supplementary Fig. 1a, b) coincident with CNS-wide demyelination (Supplementary Fig. 1c, d). In MyrfΔiPlp1 mice these symptoms peaked by 10–12 weeks post-treatment and gradually improved, as previously published17,20. In contrast, MyrfΔiSox10 mice had reduced mobility, weight loss and after 12 weeks post tamoxifen developed seizures, necessitating their euthanasia (Supplementary Fig. 1a, b).
Fig. 1. MyrfΔiSox10 mice undergo CNS demyelination with limited remyelination.
a Transgenic strategy to induce demyelination with the aim of permitting (MyrfΔiPlp1 mice) or inhibiting (MyrfΔiSox10 mice) remyelination. b Timeline of tamoxifen administration, demyelination and remyelination in MyrfΔiPlp1 and MyrfΔiSox10 mice. c Electron micrographs of the optic nerve of Myrffl/fl, MyrfΔiPlp1 and MyrfΔiSox10 mice. d Quantification of the percentage of myelinated axons in the optic nerve. ****p < 0.0001 and **p = 0.0017. Week 10 Myrffl/fl n = 7, Week 10/20 MyrfΔiPlp1 n = 3, MyrfΔiSox10 n = 5 mice. e Optic nerve cross sections stained for MBP (myelin) and BCAS1 (new OLs/myelin). f Percentage of optic nerve that is BCAS1+. ***p = 0.0003 and *p = 0.0104. Week 10 Myrffl/fl n = 14, MyrfΔiPlp1 n = 7, MyrfΔiSox10 n = 8, Week 20 Myrffl/fl n = 7, MyrfΔiPlp1 n = 6 mice. g Representative CAPs from the optic nerve. h CAP latency during de- and remyelination. Week 10 MyrfΔiPlp1 relative to Myrffl/fl **p = 0.0069 and Week 20 MyrfΔiPlp1 relative to Myrffl/fl **p = 0.0012, *p = 0.0342, ****p < 0.0001. Week 10 Myrffl/fl n = 12, MyrfΔiPlp1 n = 5, MyrfΔiSox10 n = 5, and week 20 Myrffl/fl n = 4, MyrfΔiPlp1 n = 4 mice. i CAP area during de- and remyelination. MyrfΔiPlp1 relative to Myrffl/fl at 10 weeks *p = 0.0287 and 20 weeks *p = 0.0205, ***p = 0.0010. Week 10 Myrffl/fl n = 12, MyrfΔiPlp1 n = 5, MyrfΔiSox10 n = 5, week 20 Myrffl/fl n = 4, MyrfΔiPlp1 n = 5 mice. j VEPs measured over time. **p = 0.0025, *p = 0.0372. Myrffl/fl n = 13, MyrfΔiPlp1 n = 8, MyrfΔiSox10 n = 7 mice. One-way ANOVA with Tukey’s post hoc for (d), and used for week 10 in (h–j). Kruskal–Wallis with Dunn’s test for (f). Week 20 comparisons used Student’s t test for (h) and with Welch correction for (i). Mann–Whitney U test was used for week 20 comparison in (f). All statistical tests are two-sided. Scale bar is 1 µm in (c), 50 µm in (e). NA not applicable. Error bars are SEM. Source data for this Figure are provided as a Source Data file. a created in BioRender. Duncan (2024) BioRender.com/a35m876. b created in BioRender. Duncan (2023) BioRender.com/g44t478. Schematic in g created in BioRender. Duncan (2023) BioRender.com/h38j796. Schematic in j created in BioRender. Duncan (2023) BioRender.com/v92t428.
We took advantage of the optic nerve, a structure comprised of RGC axons that are nearly all myelinated to compare the remyelination potential of each mouse line (Fig. 1c). By 10 weeks post tamoxifen, only 44.7 ± 9.5% of axons in the optic nerves of MyrfΔiPlp1 mice were myelinated, with the majority of the myelin sheaths present being thin (g-ratio >0.80), characteristic of remyelination (Supplementary Fig. 1e–h). By 20 weeks post tamoxifen, the degree of myelination increased to 75% of the axons in the optic nerve of MyrfΔiPlp1 mice (Fig. 1d). In contrast, only 1.8 ± 0.6% of axons MyrfΔiSox10 mice were myelinated at 10 weeks post tamoxifen with almost no signs of thinly myelinated axons (Fig. 1c, d). Consistent with a failure of remyelination in the MyrfΔiSox10 line, MyrfΔiPlp1 but not MyrfΔiSox10 mice showed elevated expression of BCAS1, a marker of newly-formed OLs and their myelin21, at 10 weeks post tamoxifen (Fig. 1e, f).
To assess the functional consequences of remyelination failure, we performed compound action potential (CAP) recordings of the optic nerve (Fig. 1g). There is an increased CAP onset latency and reduced CAP area in both demyelinated mouse lines (Fig. 1h–i). However, MyrfΔiPlp1 mice show improved conduction speeds relative to MyrfΔiSox10 mice at 10 weeks post tamoxifen (Fig. 1g, h). Similarly, visual evoked potentials, sensitive to remyelination efficacy22, are slower in MyrfΔiSox10 mice relative to MyrfΔiPlp1 mice, and MyrfΔiPlp1 mice return to non-demyelinated control levels by 20 weeks post tamoxifen (Fig. 1j). In summary, MyrfΔiPlp1 mice feature rapid and effective remyelination, which is associated with greater functional and electrophysiological recovery, whereas MyrfΔiSox10 mice have little remyelination and worsened functional outcomes.
New oligodendrocytes are unable to fully mature in MyrfΔiSox10 mice
To better understand the cellular changes seen in each model of demyelination, we next performed single-nuclei RNA sequencing (snRNA-seq) of the optic nerves of Myrffl/fl, MyrfΔiPlp1, and MyrfΔiSox10 mice (Fig. 2a). A total of 49,806 nuclei passed quality control and were sorted into 14 distinct clusters based on well-characterized markers (Fig. 2b–d). Both MyrfΔiPlp1 and MyrfΔiSox10 mice show a near complete loss of the mature OL (MOL) cluster (Fig. 2e, f), consistent with efficient recombination and near absence of the developmentally-generated OLs in both lines. MyrfΔiPlp1 mice had increased proportions of both the newly-formed OL (NFOL, characterized by high Enpp6, Bcas1 and Tcf7l2) and myelin-forming OL (MFOL, characterized by high expression of Mobp, Mbp and other myelin protein transcripts) populations. Together, this supports active oligodendrogenesis and remyelination in MyrfΔiPlp1 mice (Fig. 2f). In contrast, MyrfΔiSox10 mice formed virtually no NFOLs or MFOLs and lacked expression of myelin protein transcripts (Mobp, Mog, Mag) and critical genes for OL function like Anln and Trf (Fig. 2g). The MyrfΔiSox10 mice form committed oligodendrocyte precursors (COPs), which are an intermediate differentiation state characterized by the downregulation of OPC markers Pdgfrα and Cspg4 and the expression of Nkx2.2 and Fyn (Supplementary Fig. 2c). However, these cells in the MyrfΔiSox10 mice lack the characteristic expression of Gpr17 and cluster separately (COP2) from the COPs seen in MyrfΔiPlp1 mice (COP1) (Supplementary Fig. 2a–c). COP1 cells are transcriptionally similar to NFOLs whereas COP2 share many transcripts with the knockout OL (KOOL) cluster (Supplementary Fig. 2c).
Fig. 2. Differentiation of OPCs is blocked at the COP stage following demyelination in MyrfΔiSox10 mice.
a Schematic of approach used to isolate and sequence nuclei from the optic nerve. b Uniform manifold approximation and projection (UMAP) of 49,806 nuclei. COP committed OL precursor cells, NFOL newly-formed OL, MFOL myelin-forming OLs, MOL mature OLs, KOOL knockout OLs, VLMC vascular and leptomeningeal cells, ABC arachnoid barrier cells. c Dot plot showing cluster-specific markers. d UMAP displaying expression of key markers of the OL lineage (Sox10), OPCs (Pdgfra), COP1s (Gpr17), NFOLs (Tcf7l2), MFOLs (Mobp) and MOLs (Anln, Mobp). e UMAP of OL lineage cell nuclei broken down by genotype. f Stacked bar graph showing the proportions of oligodendroglial cell clusters across genotypes. g Violin plots for selected transcripts expressed in OL lineage cells across genotypes. h Optic nerve cross sections stained with the OL lineage marker OLIG2, along with PDGFRα (expressed in OPCs) and CC1 (in OLs). i Quantification of the density of OLIG2+ oligodendrocyte lineage cells. **p = 0.0013 and ****p < 0.0001. j Graph of the density of OLIG2+PDGFRα+ OPCs. **p = 0.0016, ****p < 0.0001 and nsp = 0.2529. k Quantification of OLIG2+CC1+ OLs. *p = 0.0397 and ****p < 0.0001. Week 10 Myrffl/fl n = 14, MyrfΔiPlp1 n = 5, MyrfΔiSox10 n = 6, and Week 20 Myrffl/fl n = 5, MyrfΔiPlp1 n = 4 mice for (i–k). One-way ANOVA with Tukey’s post hoc for week 10 pairwise comparisons in (i, j) with Kruskal–Wallis with Dunn’s test in (k). Week 20 comparisons used Student’s t test in (i, j) and Mann–Whitney U in (k). All statistical tests are two-sided. Error bars are SEM. Scale bar is 50 µm in (h). NA not applicable. ns not statistically significant. Source data for this Figure are provided as a Source Data file. a created in BioRender. Duncan (2024) BioRender.com/t20c114.
One unexpected observation is the formation of a distinct cluster of cells in the two Myrf conditional knockout lines, referred to as KOOLs. KOOLs express OL-enriched transcription factors like Zfp536 and St18 but also OPC/COP transcription factors like Sox6, Zeb1, and Klf6 (Supplementary Fig. 3a). KOOLs also fail to robustly express key myelin genes or OPCs markers like Cspg4 and Pdgfra (Supplementary Fig. 3a–d). KOOLs are produced in similar proportions in both MyrfΔiPlp1 and MyrfΔiSox10 when examined as the percentage of nuclei by snRNAseq (Supplementary Fig. 3f) or using SYT4 immunohistochemistry (Supplementary Fig. 3e, g, h). The KOOL population shares some markers with demyelination/disease-associated OLs present in cuprizone and Alzheimer’s disease, including Col5a3, Serpina3n, Klk8 and Cdkn1a23–25 transcripts indicative of damage (Supplementary Fig. 3a–d). The presence of KOOLs at 10 weeks post tamoxifen suggests that OL lineage cells may persist for some time following Myrf ablation before undergoing apoptosis.
To validate the snRNAseq results, we performed immunohistochemistry on optic nerves (Fig. 2h). Total OL lineage cells (OLIG2+) declined selectively in MyrfΔiSox10 mice (Fig. 2i) in large part due to the reduction of mature OLs (OLIG2+CC1+, Fig. 2k). In MyrfΔiPlp1 mice, OL density initially declined at 10 weeks post tamoxifen but recovers by 20 weeks back to control levels, suggesting new OL genesis. To confirm this, we administered 5’-ethynyl-2’-deoxyuridine (EdU) in the drinking water to label dividing OPCs and their differentiated progeny (Supplementary Fig. 4a). We found few new (EdU+) OLs in MyrfΔiSox10 mice, whereas there was elevated OL genesis in MyrfΔiPlp1 mice that is sufficient to restore the total density of OLs to control levels by 20 weeks post tamoxifen (Fig. 2k and Supplementary Fig. 4c). The density of OLIG2+PDGFRα+ OPCs that incorporated EdU, as well as the total density of OLIG2+PDGFRα+ cells, increased in both MyrfΔiPlp1 and MyrfΔiSox10 relative to Myrffl/fl controls (Fig. 2h, j and Supplementary Fig. 4b), but did not differ between MyrfΔiPlp1 and MyrfΔiSox10 mice, nor did expression of transcripts indicative of proliferation (Supplementary Fig. 4e). These data indicate OPC proliferation is not altered in the absence of Myrf. Collectively, immunohistochemical and sequencing data demonstrate Myrf knockout from OL lineage cells in MyrfΔiSox10 mice results in loss of OLs, with OPCs unable to differentiate beyond the COP level and generate new OLs.
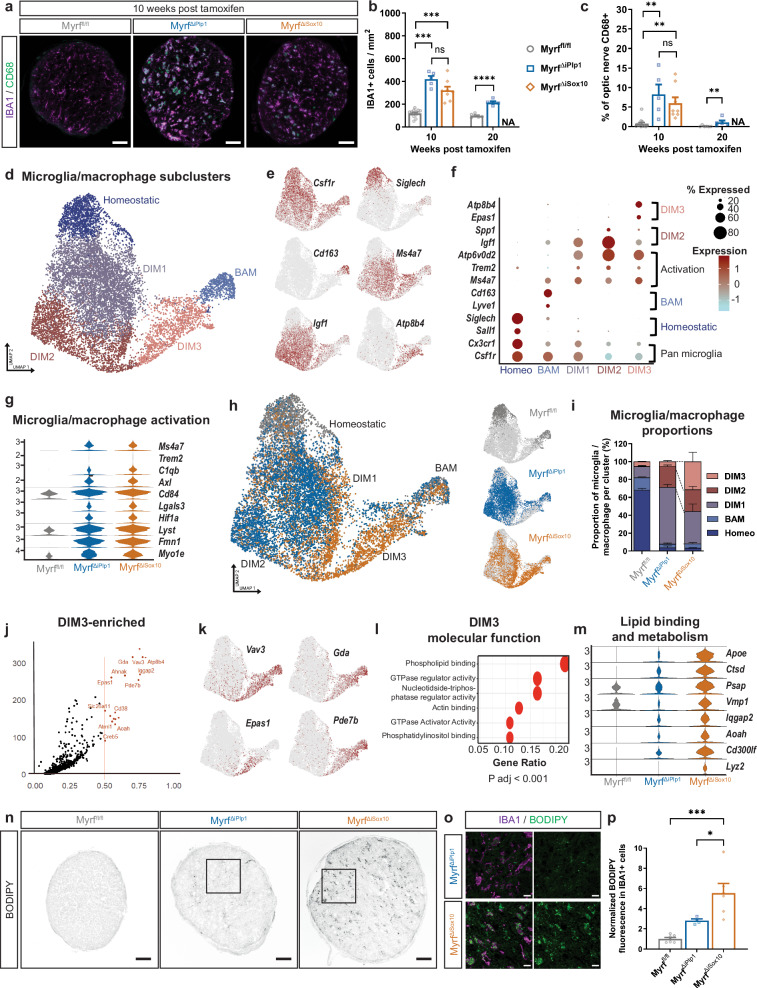
Remyelination failure is correlated with expansion of a microglia/macrophage population characterized by increased expression of lipid binding and metabolism transcripts
We next determined the influence of remyelination failure on the neuroinflammatory response. Demyelination in both MyrfΔiPlp1 and MyrfΔiSox10 mice is associated with an increase in microglia/macrophage density (Fig. 3a, b), astrogliosis (Supplementary Fig. 5a–e) and sparse T cell infiltration (Supplementary Fig. 6a). However, the densities of IBA1+ microglia/macrophages, and CD68 expression did not differ between MyrfΔiPlp1 and MyrfΔiSox10 mice (Fig. 3a–c). Densities of CD3+, CD3+CD4+, and CD3+CD8+ T cells also do not differ between MyrfΔiPlp1 and MyrfΔiSox10 mice (Supplementary Fig. 6b–e). Astrocytes in MyrfΔiSox10 mice did show a marked upregulation of Lcn2, Slc39a14, and Cp transcripts, encoding proteins critical for the uptake and detoxification of iron (Supplementary Fig. 5e). We examined oxidative damage using an antibody for oxidized phosphatidylcholines but did not see an increase in MyrfΔiPlp1 and MyrfΔiSox10 mice (Supplementary Fig. 5f, g). Upregulation of these transcripts in astrocytes may, therefore, be an adaptive change to reduce oxidative stress in response to the loss of OLs, the major iron-storing cells of the CNS26.
Fig. 3. Remyelination failure in MyrfΔiSox10 mice is associated with microglia/macrophages with elevated transcription of lipid metabolism genes and accumulated neutral lipids.
a Optic nerve cross-sections stained with IBA1 for microglia/macrophages and the lysosomal marker CD68. b IBA1+ cell quantification in the optic nerve. MyrfΔiPlp1 relative to Myrffl/fl at week 10 ***p = 0.0005 and MyrfΔiSox10 relative to Myrffl/fl at week 10 ***p = 0.0010, nsp = 0.1173 ****p < 0.0001. c Quantification of the percent of the optic nerve occupied by CD68 staining. MyrfΔiPlp1 relative to Myrffl/fl at week 10 post tamoxifen (**p = 0.0030), MyrfΔiPlp1 relative to Myrffl/fl at week 20 post tamoxifen (**p = 0.0087) and MyrfΔiSox10 relative to Myrffl/fl at 10 weeks post tamoxifen **p = (0.0038), nsp > 0.9999. Week 10 Myrffl/fl n = 13, MyrfΔiPlp1 n = 5, MyrfΔiSox10 n = 8 and week 20 Myrffl/fl n = 6, MyrfΔiPlp1 n = 5 mice for (b, c). d UMAP plot of reclustered microglia/macrophage nuclei identifying five annotated subclusters (homeostatic microglia, barrier associated macrophages (BAM), demyelination induced microglia/macrophages (DIM 1–3). N = 11,402 nuclei. e UMAP of key subcluster transcripts enriched within microglial/macrophage population; pan microglial/macrophage marker (Csf1r), homeostatic microglia (Siglech), BAMs (Cd163), DIMs (Ms4a7), DIM2 (Igf1) and DIM3 (Atp8b4). f Dot plot showing expression of sub-cluster specific markers. g Violin plots for selected transcripts associated with activation in microglia/macrophage nuclei. h UMAP of microglia/macrophage lineage cell nuclei broken down by genotype. i Stacked bar graph of microglia/macrophage subcluster composition by genotype. j Volcano plot of key enriched transcripts in DIM3. Log2(fold change) >0.5 adjusted p < 0.05. Wilcoxon rank-sum test. k UMAPs of genes enriched in DIM3 cluster. l DIM3 gene ontology top six terms for molecular function. Two-sided Wilcoxon rank-sum test with padj <0.05. m Violin plots showing expression of select lipid binding and metabolism genes in the microglia/macrophage lineage by genotype. n BODIPY fluorescence in the optic nerve of Myrffl/fl, MyrfΔiPlp1 and MyrfΔiSox10 mice at 10 weeks post tamoxifen. o Magnified region of the optic nerve from MyrfΔiPlp1 and MyrfΔiSox10 demonstrating the majority of BODIPY signal colabels with IBA1. p BODIPY fluorescence in the optic nerve. *p = 0.0258 and ***p = 0.001. Myrffl/fl n = 7, MyrfΔiPlp1 n = 4, MyrfΔiSox10 n = 6 mice. One-way Welch’s ANOVA with Dunnett’s T3 post hoc for week 10 in (b), Kruskal–Wallis with Dunn’s test for 10 week comparison in (c) and one-way ANOVA with Tukey’s post hoc in (p). For week 20 comparisons Student’s t test was used in (b) and Mann–Whitney U in (c). All statistical tests are two-sided. Error bars are SEM. Scale bars 50 µm in (a, n) and 10 µm in (o). NA not applicable, ns not statistically significant. Source data for this Figure are provided as a Source Data file.
To more closely assess the microglial/macrophage response following remyelination failure in MyrfΔiSox10 mice, we subclustered microglia from our snRNAseq dataset (Fig. 3d). We annotated five microglia clusters with three of these clusters (demyelination-induced microglia/macrophage 1-3; DIM1-3) enriched in both MyrfΔiPlp1 and MyrfΔiSox10 mice following demyelination. DIM1-3 are distinguished from homeostatic or barrier-associated macrophages (BAMs) by the presence of elevated activation markers, including Ms4a7, Axl and Atp6v0d2 (Fig. 3e, f). The expression of these transcripts does not differ between MyrfΔiPlp1 and MyrfΔiSox10 mice, suggesting broadly similar activation (Fig. 3g). DIM2 microglia/macrophages are characterized by elevated levels of Igf1 and Spp1, which have previously been found in a subpopulation of microglia along axon tracts27 (Fig. 3f). DIM3 is the only population that is increased in MyrfΔiSox10 mice relative to the MyrfΔiPlp1 line (Fig. 3h, i). This population is characterized by transcripts including Vav3, Gda, Epas1 and Pde7b (Fig. 3j, k). Gene set enrichment analysis on DIM3 enriched transcripts revealed the top term to be “phospholipid binding” (Fig. 3l); accordingly, there is an upregulation of lipid binding and metabolism genes in MyrfΔiSox10 microglia nuclei (Fig. 3m). When myelin is phagocytosed by microglia/macrophages, it undergoes lysosomal processing to produce cholesterol and fatty acids, which can be stored in lipid droplets and act as a reservoir for efflux28. BODIPY, a marker of neutral lipids present in lipid droplets29, accumulates in IBA1+ cells in MyrfΔiSox10 relative to MyrfΔiPlp1 (Fig. 3n–p). In summary, while both MyrfΔiPlp1 and MyrfΔiSox10 mice show similar microglia/macrophage numbers, a subset of microglia/macrophages in MyrfΔiSox10 mice are characterized by upregulated lipid metabolism and binding transcripts and have increased lipid storage in the absence of remyelinating OLs.
Remyelination is associated with protection from axonal degeneration and neuronal apoptosis
To determine how neuronal integrity is impacted during remyelination failure in MyrfΔiSox10 mice, we assessed the health and integrity of retinal ganglion cells (RGCs) and their axons in the optic nerve. There is an increase in the percentage of axons with accumulated organelles following demyelination in both MyrfΔiPlp1 and MyrfΔiSox10 mice at 10 weeks post tamoxifen, indicative of axonal transport deficits and injury30,31 (Fig. 4a, b). By 20 weeks post tamoxifen in Myrf∆iPlp1 mice, when remyelination is more complete, organelle accumulations are decreased relative to 10 weeks (Fig. 4b). Serum neurofilament light chain (NfL), a biomarker of axonal injury in a variety of diseases including MS32, is increased at 10 weeks post tamoxifen in both MyrfΔiPlp1 and MyrfΔiSox10 mice (Fig. 4c). Consistent with axonal accumulations, NfL levels fall in MyrfΔiPlp1 mice by 20 weeks post tamoxifen and are indistinguishable from controls (Fig. 4c). Despite the presence of accumulations in both lines, quantification of total axonal numbers in the optic nerves reveals that detectable loss of axons relative to myelinated controls is only present in the MyrfΔiSox10 line (Fig. 4d) and MyrfΔiPlp1 mice do not feature axonal loss relative to controls at 10 or 20 weeks post tamoxifen.
Fig. 4. MyrfΔiSox10 mice incur axon loss and have increased neuronal apoptosis relative to remyelinating MyrfΔiPlp1 mice.
a Electron micrographs of the optic nerve demonstrating axons with organelle accumulations in MyrfΔiPlp1 and MyrfΔiSox10 mice. Boxed areas show individual axons with organelle accumulation enlarged to the right. b Quantification of the number of axons with organelle accumulation. ****p < 0.0001, ***p = 0.0004 and *p = 0.0437. n = 5 mice per group. c Serum neurofilament levels measured at 10 and 20 weeks post tamoxifen. ****p < 0.0001, ***p = 0.0004 and nsp = 0.1111. Week 10 Myrffl/fl n = 22, MyrfΔiPlp1 n = 7, MyrfΔiSox10 n = 19, and week 20 n = 4, MyrfΔiPlp1 n = 5 mice. d The total number of axons within the optic nerve. *p = 0.0202. n = 5 mice per group. e Overview of retina with locations of cleaved caspase-3+ cells in the GCL indicated by black X. f RBPMS in the retina across genotypes. g Density of cleaved caspase-3+ cells in retinal flatmounts. MyrfΔiSox10 ****p < 0.0001. h Density of cleaved caspase-3 and RBPMS double-labeled cells. ****p < 0.0001 and **p = 0.0018. Week 10 Myrffl/fl n = 11, MyrfΔiPlp1 n = 7, MyrfΔiSox10 n = 5, Week 12 Myrffl/fl n = 8, MyrfΔiPlp1 n = 4, MyrfΔiSox10 n = 5, and Week 20 Myrffl/fl n = 10, MyrfΔiPlp1 n = 8 mice for (g, h). i Co-labeling between cleaved caspase-3 and RBPMS or AP-2 α/β in the retina. Arrowhead indicates cleaved-caspase-3+RBPMS+ cell. Arrow indicates cleaved caspase-3+AP-2α/β-negative cell. j The majority of cleaved-caspase-3+ in MyrfΔiSox10 mice are RBPMS+ and AP-2α/β-negative. k Total density of RBPMS+ RGCs. Week 10 Myrffl/fl n = 18, MyrfΔiPlp1 n = 7, MyrfΔiSox10 n = 13, Week 12 Myrffl/fl n = 13, MyrfΔiPlp1 n = 8, MyrfΔiSox10 n = 9, and Week 20 Myrffl/fl n = 10, MyrfΔiPlp1 n = 10 mice. One-way ANOVA with Tukey’s post hoc for pairwise comparisons in (b, d). Kruskal–Wallis with Dunn’s test for comparisons at week 10 for (c). For week 20, Student’s t test used in (g, h, k) and Mann–Whitney U for (c). Two-ANOVA with Tukey’s post hoc for comparisons at week 10 and 12 in (g, h, k). All statistical tests are two-sided. Error bars are SEM. Scale bars are 1 µm in (a), 50 µm in (f) and overview images in (i) and insets are 5 µm. Scale bars are 500 µm (e). NA not applicable, ns not statistically significant. Source data for this Figure are provided as a Source Data file.
We next asked whether the cell bodies of RGCs, which project their axons through the optic nerve, undergo apoptosis and loss in MyrfΔiSox10 mice. Retinal flatmounts were stained with cleaved-caspase-3 to detect apoptotic cells (Fig. 4e) and with RBPMS (Fig. 4f), a pan RGC marker33. MyrfΔiSox10 mice have increased apoptosis in the ganglion cell layer (GCL; Fig. 4g) and increased numbers of cleaved caspase-3+ RBPMS+ RGCs (Fig. 4h) relative to both the remyelinating Myrf∆iPlp1 mice and non-demyelinated controls. We found that a portion of cleaved caspase-3 cells do not express RBPMS34, and we examined whether there is apoptosis in other cell types. Apoptotic cells are largely confined to the GCL, which consists of displaced amacrine cells and RGCs. Labeling amacrine cells with AP-2α and AP-2β, we found that only 2.7% ± 1.7% of the cleaved caspase-3+ cells are AP-2α+ or AP-2β+ in Myrf∆iSox10 mice, whereas 55.2 ± 4.8% are RBPMS+ (Fig. 4j). RBPMS is downregulated during injury34, so it seems plausible that the remaining cleaved caspase-3+ cells represent late stage apoptotic RGCs in which the RBPMS protein has been lost (Fig. 4i). The apoptosis of RGCs cannot be attributed to Myrf knockout from neurons, as Sox10-CreERT does not recombine in RGCs and Myrf knockout within RGCs does not directly drive their apoptosis (Supplementary Fig. 7a–g). Neither MyrfΔiPlp1 nor MyrfΔiSox10 mice have a significant reduction of RGC density, likely owing to the rate of apoptosis in MyrfΔiSox10 mice being insufficient to accrue a detectable loss over the time frame of these experiments (Fig. 4k). Taken together, both MyrfΔiSox10 and MyrfΔiPlp1 mice feature axonal swellings consistent with damage following demyelination. However, only MyrfΔiSox10 mice, characterized by remyelination failure, culminate in axon loss relative to Myrffl/fl controls. Remyelination is associated with protection of RGCs from apoptotic cell death in MyrfΔiPlp1 mice.
Increased activation of the DLK-mediated MAPKs and c-Jun in RGCs in MyrfΔiSox10 mice with failed remyelination
To understand how chronically demyelinated neurons are damaged in MyrfΔiSox10 mice, we performed bulk RNA-seq on the GCL (Fig. 5a). Laser dissecting the GCL greatly enriched for RGC-specific transcripts relative to the whole retina (Fig. 5b). The GCL of MyrfΔiSox10 mice showed an upregulation of Ecel1, Hrk, and Atf3 relative to Myrffl/fl (Fig. 5c). In situ hybridization of Ecel1 and Hrk confirmed these transcripts are expressed in Rbpms+ RGCs in MyrfΔiSox10 mice (Fig. 5d, e). Notably, these transcripts are known to be upregulated following DLK activation35,36. DLK can mediate a retrograde signal that couples axonal injury to transcriptional changes in the nucleus, and often triggers subsequent apoptosis in the central nervous system35–37. DLK and the related leucine zipper-bearing kinase (LZK)38,39 mediate this retrograde signal by activating downstream mitogen-activated protein kinases (MAPKs) including MAPK kinase 4/7 (MKK4/7) and c-Jun N-Terminal Kinase 2/3 (JNK2/3), resulting in the phosphorylation and activation of the transcription factor c-Jun40–43 (Fig. 5l). Activation of this pathway has not previously been identified after demyelination.
Fig. 5. Activation of the DLK/JNK/c-Jun pathway in chronically demyelinated MyrfΔiSox10 mice.
a Experimental schematic and images of laser microdissection of the GCL of the retina. b Heatmap of expression of RGC-specific transcripts in the micro-dissected GCL samples relative to whole retina. c Heatmap of select transcripts from the GCL known to be activated by c-Jun/DLK signaling compared between genotypes. In situ hybridization in the retina at 10 weeks post tamoxifen with probes against Rbpms and Hrk (d) or Ecel1 (e). Arrowheads indicate double-positive cells. f Retina at 10 weeks post tamoxifen stained with phosphorylated (Ser63) c-Jun. g Quantification of phosphorylated c-Jun cells in the retinae of each genotype. ****p < 0.0001, and MyrfΔiPlp1 relative to Myrffl/fl at 10 (**p = 0.0088) and 20 weeks post tamoxifen (**p = 0.0092). Week 10 Myrffl/fl n = 16, MyrfΔiPlp1 n = 4, MyrfΔiSox10 n = 8, week 12 Myrffl/fl n = 7, MyrfΔiPlp1 n = 3, MyrfΔiSox10 n = 3, and week 20 Myrffl/fl n = 5, MyrfΔiPlp1 n = 7 mice. h Phosphorylated c-Jun costained with RBPMS cells in MyrfΔiSox10 mice. Inlays are of boxed area. Arrowheads indicate colabeled cells. i Representative images of phosphorylated c-Jun stained with AP-2α/β in MyrfΔiSox10 retinae. Inlays are of boxed area. Arrows indicate phosphorylated c-Jun positive cells negative for AP-2α/β. j Quantification of RBPMS and AP-2α/β expression in phosphorylated c-Jun+ cells. k Image of retinal layers stained with phosphorylated c-Jun, RBPMS and DAPI. Inlays are of boxed area. l Schematic of the DLK-mediated MAPK cascade and c-Jun. m Western blot of optic nerves for DLK, pMKK4, MKK4, pJNK, JNK, MOG and β-actin loading control from optic nerves of Myrffl/fl, MyrfΔiPlp1 and MyrfΔiSox10 mice. n Quantification of western blots in MyrfΔiPlp1 mice relative to Myrffl/fl. ***p = 0.0007 and *p = 0.0309. n = 4 per group. o Quantification of western blots in MyrfΔiSox10 mice relative to Myrffl/fl. pMKK4 (*p = 0.0434), pJNK (**p = 0.0072), total JNK (**p = 0.0034) and MOG **p = 0.0038). Myrffl/fl n = 4 and MyrfΔiSox10 n = 3. Scale bars are 50 µm in (a, f, h, i, k), and 5 µm in (d, e), insets in (h, i, k). Two-way ANOVA with Tukey’s post hoc at 10 and 12 weeks post tamoxifen, and Student’s t test to compare groups at 20 weeks post tamoxifen in (g). Student’s t test in (n, o). All statistical tests are two-sided. Error bars are SEM. NA not applicable. Source data for this Figure are provided as a Source Data file. Schematic in a created in BioRender. Duncan (2023). BioRender.com/n39a548. l created in BioRender. Duncan (2023) BioRender.com/g94u157.
To determine whether this pathway has elevated activation in the chronically demyelinated RGCs, we examined the phosphorylation of c-Jun in retinal flatmounts (Fig. 5f). The retinae of MyrfΔiSox10 mice showed a large increase in the density of phosphorylated c-Jun+ cells relative to both remyelinating MyrfΔiPlp1 and Myrffl/fl control mice (Fig. 5g). Nearly all these phosphorylated c-Jun+ cells co-labeled with RBPMS (Fig. 5h–j), with virtually no colocalization in AP-2α/β+ amacrine cells (Fig. 5i–j) and phosphorylation of c-Jun was largely confined to the GCL (Fig. 5k). In MyrfΔiSox10 mice, the onset of c-Jun phosphorylation is coincident with decreased myelination and OL density at 8 weeks post tamoxifen, indicating a tight temporal relationship between demyelination and c-Jun phosphorylation in RGCs (Supplementary Fig. 8a–g). Inflammatory demyelination in experimental autoimmune encephalomyelitis (EAE) also results in the phosphorylation of c-Jun in RGCs (Supplementary Fig. 9a, c, d). Phosphorylation of c-Jun is not confined to RGCs, as spinal neurons also had elevated expression of phosphorylated c-Jun in MyrfΔiSox10 mice (Supplementary Fig. 10b, d).
We next examined if MAPKs downstream of DLK are induced following remyelination failure in MyrfΔiSox10 mice. Both the phosphorylation of JNK and MKK4 are increased in the optic nerves of MyrfΔiSox10 mice at 10 weeks post tamoxifen (Fig. 5m–o), whereas neither kinase has detectable increases in phosphorylation in MyrfΔiPlp1 mice (Fig. 5n). Although total DLK levels are not altered in MyrfΔiSox10 mice relative to controls, the apparent molecular weight of the protein increases, consistent with post-translational modifications such as palmitoylation previously demonstrated after axonal injury44,45. In summary, MyrfΔiSox10 mice have increased phosphorylation of MAPKs, c-Jun and transcription of genes associated with axonal injury and stress.
Pharmacological inhibition of DLK blocks apoptosis of demyelinated RGCs
To determine if DLK-mediated signaling is necessary to drive the activation of downstream c-Jun signaling and apoptosis in neurons following chronic demyelination, we treated demyelinated MyrfΔiSox10 mice with the DLK inhibitor GNE-351146 for 3 days via oral gavage (Fig. 6a). This regimen has been shown to be effective at blocking c-Jun phosphorylation after optic nerve crush36 and led to average serum levels over 4 µM (Fig. 6b). At ten weeks post tamoxifen, retinal flatmounts were stained with phosphorylated c-Jun, or cleaved caspase-3 (Fig. 6c, d). Treatment of MyrfΔiSox10 mice with GNE-3511 returned the phosphorylation of c-Jun to baseline levels observed in myelinated Myrffl/fl littermates (Fig. 6e). Likewise, cleaved-caspase-3+ cells and cleaved-caspase-3+RBPMS+ levels return to baseline, indicating near-complete protection from apoptosis of RGCs following acute GNE-3511 treatment (Fig. 6f, g). Thus, DLK inhibition completely blocks c-Jun phosphorylation and apoptosis of chronically demyelinated RGCs.
Fig. 6. Pharmacological inhibition of DLK reduces c-Jun phosphorylation and blocks neuronal apoptosis in demyelinated MyrfΔiSox10 mice.

a Schematic of the DLK MAPK cascade and timeline of GNE-3511 administration. b Serum levels of GNE-3511 following 3 days of oral gavage. c Representative retinal flatmount images of vehicle and GNE-3511 treated mice stained with phosphorylated c-Jun and RBPMS at 10 weeks post-tamoxifen. d Overview of retinas with locations of cleaved caspase-3+ cells in the ganglion cell layer (GCL) indicated by black X. e Quantification of phosphorylated c-Jun+ cells in vehicle or GNE-3511-treated retinae. ****p < 0.0001. Vehicle Myrffl/fl n = 8, MyrfΔiSox10 n = 11 and GNE-3511 Myrffl/fl n = 6, MyrfΔiSox10 n = 9. f Cleaved caspase-3 density in vehicle or GNE-3511-treated retinae. ***p = 0.0002 and ****p < 0.0001. g Cleaved caspase-3+RBPMS+ cell density in vehicle or GNE-3511-treated retinae. Vehicle-treated MyrfΔiSox10 relative to GNE-treated MyrfΔiSox1 mice at 10 weeks post-tamoxifen (p = 0.0005) and relative to Myrffl/fl controls (vehicle, p = 0.0008, GNE-3511, p = 0.0001). Vehicle Myrffl/fl n = 7, MyrfΔiSox10 n = 7 and GNE-3511 Myrffl/fl n = 6, MyrfΔiSox10 n = 6 mice in (f, g). Two-way ANOVA with Tukey’s post hoc for individual pairwise comparisons in (e–g). All statistical tests are two-sided. Error bars are SEM. Scale bars are 50 µm in (c) and 500 µm in (d). Source data for this Figure are provided as a Source Data file. a created in BioRender. Duncan (2023) BioRender.com/h30g439.
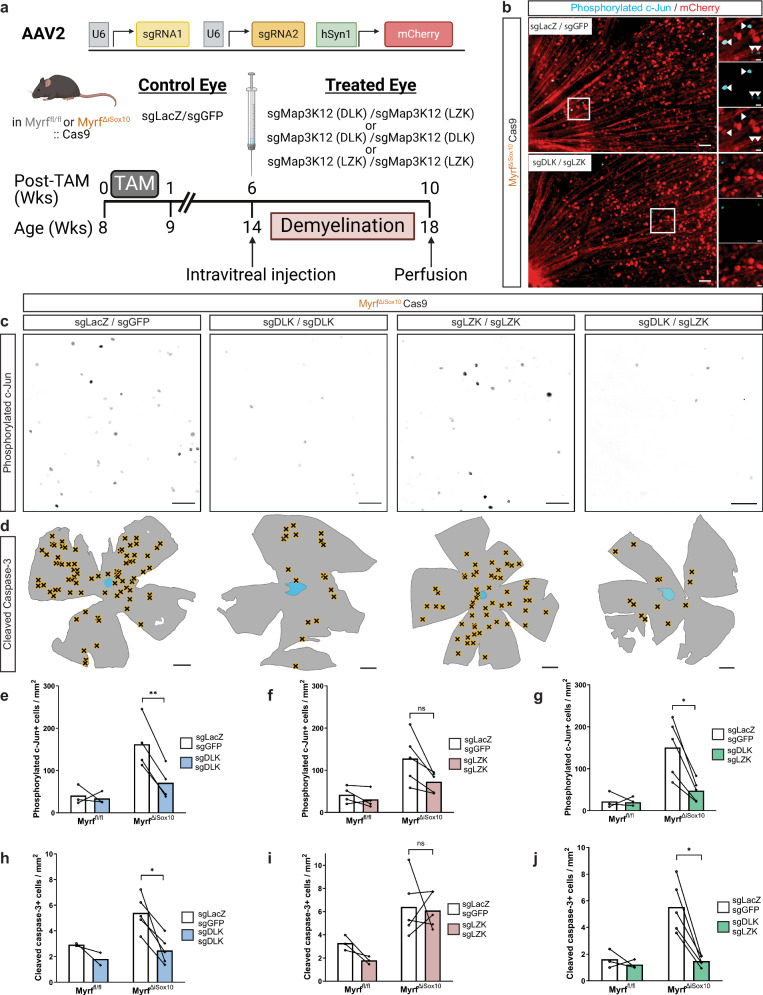
Genetic disruption of DLK inhibits apoptosis of demyelinated RGCs
We next used a CRISPR/Cas9 genetic disruption approach to determine the contributions of DLK, and its related kinase LZK, to apoptosis of chronically demyelinated RGCs. DLK and LZK can have partially redundant roles38, so we developed sgRNAs to disrupt one or both genes within RGCs (Fig. 7a). We injected adeno-associated viruses (AAVs) containing sgRNAs targeting either DLK (Map3k12), LZK (Map3k13) or both DLK and LZK intravitreally at six weeks post tamoxifen in MyrfΔiSox10 and Myrffl/fl mice crossed with mice constitutively-expressing Cas9 (MyrfΔiSox10 Cas9 or Myrffl/fl Cas9). Control AAVs with sgRNAs targeting eGFP and LacZ were injected into the contralateral eye. All AAVs effectively labeled RGCs throughout the retina (Fig. 7b). To determine whether Cas9-mediated disruption of DLK and/or LZK prevented the phosphorylation of c-Jun and apoptosis of RGCs, we stained retinal flatmounts with phosphorylated c-Jun (Fig. 7c) and cleaved caspase-3 (Fig. 7d). Retinae infected with either sgDLK alone or sgDLK/sgLZK have near-complete suppression of both c-Jun phosphorylation and RGC apoptosis (Fig. 7e, g, h, j). In contrast, knockout of LZK alone had a no significant effect on either phosphorylated c-Jun or cleaved caspase-3 cell density (Fig. 7f, i). Together, these data indicate DLK is the major MAP3K necessary for neuronal apoptosis following demyelination and subsequent remyelination failure.
Fig. 7. DLK is necessary for neuronal apoptosis following demyelination in MyrfΔiSox10 mice.
a Schematic of the viral CRISPR/Cas9 approach to disrupt DLK and/or LZK in retinal cells. b Retinae stained with phosphorylated c-Jun and mCherry at 10 weeks post tamoxifen following treatment with sgLacZ/sgGFP, or sgDLK/sgLZK on the opposite eye. Inlays are of boxed areas and scale bar is 5 µm in boxed areas. c Representative images of phosphorylated c-Jun immunostaining in retinal flatmounts following administration of sgLacZ/sgGFP, sgDLK/sgDLK, sgLZK/sgLZK, or sgDLK/sgLZK. d Overview of retinas with locations of cleaved caspase-3+ cells in the GCL indicated by black X in viral-treated eyes. e Density of phosphorylated c-Jun cells within the GCL following sgDLK/sgDLK administration. **p = 0.0080. Myrffl/fl n = 3, MyrfΔiSox10 n = 4 mice. f Density of phosphorylated c-Jun positive cells in the GCL following sgLZK/sgLZK administration. nsp = 0.0766. Myrffl/fl n = 4, MyrfΔiSox10 n = 5 mice. g Density of phosphorylated c-Jun cells within the GCL following sgDLK/sgLZK administration. *p = 0.0148. Myrffl/fl n = 3, MyrfΔiSox10 n = 5 mice. h Cleaved-caspase-3+ cells within the GCL following sgDLK/sgDLK administration. *p = 0.0254. Myrffl/fl n = 2, MyrfΔiSox10 n = 5 mice. i The density of cleaved-caspase-3+ cells within the GCL after sgLZK/sgLZK administration. nsP = 0.8530. Myrffl/fl n = 3, MyrfΔiSox10 n = 5 mice. j Cleaved-caspase-3+ cells within the GCL following sgDLK/sgLZK administration. *p = 0.0125. Myrffl/fl n = 3, MyrfΔiSox10 n = 5 mice. Scale bars are 50 µm in (b, c) and 500 µm in (d). Connected lines indicate retinae from the same mouse in (e–j). Paired Student’s t test with Holm-Šidák correction for multiple comparisons was used from (e–j). All statistical tests are two-sided. ns not statistically significant. Error bars are SEM. Source data for this Figure are provided as a Source Data file. a created in BioRender. Duncan (2023) BioRender.com/e99a533.
Discussion
By comparing our remyelination capable and deficient mouse lines, we find remyelination is associated with improved functional recovery and neuronal integrity. Mice with remyelination failure have elevated activation of MAPKs downstream of DLK and phosphorylation of the transcription factor c-Jun. We demonstrate that DLK is necessary for apoptosis of demyelinated RGCs using Cas9-mediated disruption of DLK from RGCs and pharmacological inhibition of DLK, both sufficient to suppress c-Jun phosphorylation and RGC apoptosis. We propose that neuroprotection in demyelinating disease can be achieved by targeting retrograde signaling mediated by DLK within demyelinated neurons or by promoting remyelination which is associated with suppression of the same DLK-mediated signaling cascade (Supplementary Fig. 11).
The two models of genetic demyelination induced by knockout of Myrf offer distinct advantages for subsequent studies. MyrfΔiPlp1 mice, which knock Myrf out from OLs, demonstrate highly reproducible, CNS-wide demyelination with clear behavioral and physiological readouts. The remyelination process begins rapidly in MyrfΔiPlp1 mice, even before all myelin destined to be lost has degenerated. We found the delay of conduction within the visual system correlates temporally with demyelination, whereas restoration of axonal conduction is associated with remyelination in MyrfΔiPlp1 mice. Likewise, motor function declines significantly with the onset of demyelination and slowly recovers during remyelination, as seen in other models of extensive demyelination47,48. In contrast to MyrfΔiPlp1 mice, MyrfΔiSox10 mice, do not effectively remyelinate and fail to recover, with the differentiation of remyelinating cells blocked at the COP stage. MyrfΔiSox10 mice are the first model that cell-selectively induces demyelination and impairs remyelination, allowing for investigation of how chronic demyelination impacts neurons and the efficacy of neuroprotective therapeutics.
We use these models to determine the degree of neurodegeneration in mice without effective remyelination relative to those mice with efficient remyelination. An association between remyelination and neuroprotection has been difficult to establish in rodents, in part due to their rapid remyelination. Targeting the OL lineage with cell-specificity, we find greater RGC apoptosis in mice MyrfΔiSox10 mice which are unable to remyelinate. Apoptosis of RGCs is persistent at 10- and 12-weeks post tamoxifen in the MyrfΔiSox10 mice, supporting a notion of continuous neurodegeneration in the context of prolonged demyelination. Although the rate of apoptosis in RGCs is relatively modest (~0.06% of RGCs being apoptotic at any given time), the cumulative effect of such apoptosis on the population would be significant over time given the rapid rate of microglial clearance of apoptotic cells. Accelerating remyelination by deleting the M1 muscarinic receptor from OL lineage cells increases axon preservation and functional recovery in EAE49. Collectively, this lineage-specific gain of function experiment and our loss of function experiments provide compelling support that remyelination rate is a critical determinant of subsequent neurodegeneration. Additionally, both neuroimaging50–52 and histopathological14,53 studies measuring remyelination in MS lesions support a neuroprotective role. One limitation is our approach is incapable of dissociating the relative neuroprotective contributions of new OLs verses the deposition of new myelin. Future studies targeting specific mechanisms by which OLs may support neuronal health during remyelination will be crucial to disentangle the role of the oligodendrogenesis versus myelinogenesis on alleviating neurodegeneration.
Our experiments provide an important contrast to recent findings that myelinated axons are more at risk for degeneration during autoimmune-mediated demyelination30 and in Plp1 mutants54,55. Axonal damage in EAE is more prevalent in myelinated axons relative to their demyelinated counterparts, and hypomyelinated Mbp mutant mice incur less axonal damage during EAE30. Plp1 over-expressing mice, mimicking Plp1 duplication in humans, initiate progressive demyelination during early adulthood55. In these mice, axons are most vulnerable to damage during active demyelination and are relatively protected following the resolution of inflammation55. Likewise, Plp1 mutants with abnormal myelin are relatively protected from CD8+ T cell-mediated neurodegeneration when demyelinated54. Both in MS tissue14,53 and in experimental models30,55, chronically demyelinated neurons seem less vulnerable to damage relative to those undergoing active demyelination. Thus, axons are highly vulnerable during active demyelination or when deposited with dysfunctional myelin. Nevertheless, in MyrfΔiSox10 mice following chronic demyelination apoptotic RGCs are found at a more than four-fold increase relative to remyelinated and control mice, suggesting remyelination failure is associated with slow, but persistent neurodegeneration. Collectively, these animal studies indicate a substantial challenge for targeting myelin in MS; during the early stages of the disease damaged myelin is harmful to the axon and needs to be cleared and inflammation resolved, yet prolonged demyelination is also detrimental to both the function and viability of the neuron.
An important limitation of our studies is it remains unclear the contribution of inflammation to neurodegeneration following remyelination failure in MyrfΔiSox10 mice. Microglia have a crucial role in regulating the efficacy of remyelination56–58. Interestingly, we found the reciprocal is also true: preventing oligodendrogenesis and remyelination in MyrfΔiSox10 mice led to expansion of a microglial population, which could conceivably damage neurons. This microglial population is characterized by the elevated expression of lipid-binding and metabolism transcripts. Cholesterol is the major lipid constituent of myelin and both sterol synthesis and efflux in microglia are necessary for effective remyelination59,60, presumably by acting as a source of sterols for newly generated OLs. In MyrfΔiSox10 mice, it is intriguing to consider that in the absence of new remyelinating OLs, microglia lack their normal cellular sink for recycled cholesterol from damaged myelin. In support of impaired cholesterol export from microglia, we found an accumulation of neutral lipid droplets, which act to store cholesterol esters in microglia from MyrfΔiSox10 mice. Increased accumulation of cholesterol within microglia and poor efflux is associated with persistent inflammation60,61. With disease chronicity in MS, microglia become more diffusely activated62; it is conceivable that poor remyelination may contribute to persistent microglial activation and potentially neurodegeneration. Unfortunately, given the premature death of MyrfΔiSox10 mice, we cannot determine whether remyelination failure would continue to result in prolonged microglial inflammation and if that would subsequently damage neurons. Future work should determine if this this shift in microglia phenotype following remyelination failure directly contributes to neurodegeneration.
While remyelination failure has been associated with axonal damage in MS14,53, it has remained unclear how demyelination-induced damage to the axon may be discerned in the nucleus or if it drives loss of the soma. Periventricular lesions that fail to remyelinate in MS are associated with greater atrophy of brain regions they project from51, suggesting that demyelination of the axon may be perceived by the soma and culminate in neurodegeneration. Here, we find chronic demyelination of the axon is associated with elevated DLK-mediated phosphorylation of the transcription factor c-Jun in the nucleus, transcriptional changes and RGC apoptosis. Potentially, DLK could be activated in Myrf∆iSox10 mice either as a direct loss of OL support or due to other factors such as altered inflammation. However, DLK is localized within the axon and several stimuli known to drive DLK-mediated signaling including axonal cytoskeletal disruption63 as well as transport deficits64, were observed following demyelination. Mechanistically, injury to the axon results in palmitoylation of DLK and its localization to retrogradely trafficked vesicles; a process required for phosphorylation of downstream kinases including JNK45. JNK signaling is a well-known activator of c-Jun mediated transcription in response to axonal injury35,37, generally triggering apoptosis of CNS neurons65. c-Jun likely regulates apoptosis at least in part through modulating the expression of Bcl2 family members like Bcl2l2 and Bbc365,66. Taken together, we suggest DLK mediates a retrograde signal from the demyelinated axon that culminates in phosphorylation of c-Jun and subsequent apoptosis.
We found that DLK inhibitors protect neurons from apoptosis following chronic demyelination. Notably, we also see c-Jun phosphorylation and RGC apoptosis following inflammatory demyelination in EAE. This suggests the pathway may also be induced during acute inflammatory demyelination and that DLK inhibitors may also be protective in this context. Blocking DLK activity is therefore a strong candidate for neuroprotection in demyelinating disease. While a number of blood-brain barrier permeable DLK inhibitors have already been developed46,67, a recent phase I trial with a DLK inhibitor in ALS warrants caution as prolonged DLK inhibition was associated with considerable safety concerns68. It may be necessary to dissect and target the downstream responses induced by DLK that drive neurodegeneration in the context of remyelination failure to produce druggable therapeutic targets. However, our data strongly supports that remyelination is also capable of suppressing DLK-mediated signaling and neuronal apoptosis. Even the incomplete remyelination present within the optic nerve of MyrfΔiPlp1 mice at 10 weeks post tamoxifen is associated with suppressed MAPK signaling and apoptosis relative to the Myrf∆iSox10 line. Therefore, it is plausible that even promoting a relatively small degree of remyelination will reduce neuronal attrition and will be of value in treating demyelinating disease.
Methods
Mouse lines and husbandry
All animal procedures were performed in accordance with, and approved by, the Institutional Animal Care and Use Committee of OHSU (IP00001328 and TR01_IP00001328). All mice were housed and maintained in the Oregon Health & Science University animal facility in a pathogen-free temperature-controlled environment on a 12-h light/dark cycle from 6 a.m. to 6 p.m. Myrffl/fl mice were generated in the Barres laboratory15 (B6;129-Myrftm1Barr/J, JAX: 010607). After being crossed to C57BL/6 for over ten generations they were crossed to either Plp1-CreERT mice69 (B6.Cg-Tg[Plp1-cre/ERT]3Pop/J, JAX:005975) or Sox10-CreERT mice19 (CBA;B6-Tg[Sox10-icre/ERT2]388Wdr/J, JAX:026651). To allow for CRISPR/Cas9 mediated disruption of genes, MyrfΔiSox10 mice were crossed to constitutively-expressing Cas9 mice70 (Gt[ROSA]26Sortm1.1[CAG-cas9*,-EGFP]Fezh/J, JAX:024858) to produce MyrfΔiSox10 Cas9 and Myrffl/fl Cas9 mice. In all cases CreERT negative littermates served as non-demyelinated controls, and were matched by sex to knockout mice when possible. Plp1-CreERT mice and Sox10-CreERT mice were crossed with inducible Sun1-GFP mice71 (B6.129-Gt(ROSA)26Sortm5.1(CAG-Sun1/sfGFP)Nat/MmbeJ, JAX 030952) to assess which cells are recombined within the retina. Genotypes were determined by PCR analysis of ear clips, using established primers (Supplementary Table 1) for each line, and revalidated at experimental endpoint. All experiments were conducted in both sexes of eight week-old mice. Every week following tamoxifen administration mice were weighed and health assessments performed. When mice reached a motor score of three, indicated by ataxia and significant hindlimb weakness, they received wet food on the bottom of their cage to maintain hydration. Initial cohorts indicated that MyrfΔiSox10 mice developed seizures after 12 weeks post tamoxifen, so all analyses were limited to 12 weeks or earlier.
Tamoxifen administration
Tamoxifen (T5648, Sigma) was dissolved in corn oil (C8267, Sigma) at 20 mg/mL using heat (37 °C) and agitation. Mice received intraperitoneal injections at 100 mg/kg for five consecutive days at 8 weeks of age (P56–P62). Tamoxifen was prepared fresh prior to administration for each cohort of mice.
EdU administration
To examine proliferation of OPCs and differentiation of new OLs, we administered 5-ethynyl-2′-deoxyuridine (EdU) in the drinking water starting the week after tamoxifen injections until 10 weeks post-tamoxifen. EdU (NE08701, Carbosynth) was dissolved in water along with 0.2 mg/mL of dextrose (D16-500, Fisher) to encourage consumption. EdU water was changed every 2–3 days over the course of administration.
GNE-3511 administration
GNE-3511 (5331680001, Millipore-Sigma) was emulsified into 0.5% methylcellulose (M7140, Sigma) with 0.2% Tween 80, and vortexed prior to oral gavage36. At 10 weeks post tamoxifen administration, MyrfΔiSox10 mice and Myrffl/fl controls received two daily gavages with either GNE-3511 at 75 mg/kg or vehicle only. Gavages were at least 8 h apart for three consecutive days, for a total of six gavages.
EAE induction
Experimental autoimmune encephalomyelitis (EAE) was induced in eight week-old female C57BL/6 mice via immunization with myelin oligodendrocyte glycoprotein (MOG)35-55 (PolyPeptide Laboratories). A total of 200 µg MOG35-55 was dissolved in complete Freund’s adjuvant containing 400 µg of Mycobacterium tuberculosis (231141, Difco) and injected subcutaneously in 0.2 mL volume per mouse. Pertussis toxin (181, List Biological labs Inc) was administered via intraperitoneal injection after the MOG35-55 injection and 2 days later at 75 ng and 200 ng per mouse doses, respectively. Mice were perfused within 3 days of peak disease and all mice reached an EAE score of three. Following perfusion, retinae were prepared for flatmount staining and optic nerves frozen and subsequently cryosectioned.
Intravitreal injections
Intravitreal injections of viruses containing tandem sgRNAs were used to induce Cas9-mediated disruption of Map3k12 and Map3k13. Myrffl/fl Cas9 and MyrfΔiSox10 Cas9 mice were anesthetized with ketamine/xylazine (ketamine 100 mg/kg and xylazine 12 mg/kg). Proparacaine (NDC 13985-611-15, Akorn Inc) and tropicamide (NDC 17478-102-12, Akorn Inc) eye drops were applied to provide local analgesia and better visualization of the injection, respectively. One eye received AAV expressing small guide RNAs against LacZ and GFP under U6 promoters (AAV2-U6-sgLacZ-U6-sgGFP-hSyn1-mCherry) with the opposite eye receiving sgRNAs against Map3K12 (DLK), Map3K13 (LZK), or both genes. The concentration of viruses was adjusted to 2.8 × 1012 genome copies per mL immediately prior to injection in sterile 1x phosphate buffered saline (PBS). Under stereo microscopic control, the ora serata was incised with a 30 gauge needle without touching the lens. AAV vectors (1 µL/injection) were delivered into the vitreous through the incision using a 5 µL Hamilton microinjection syringe with a blunt 20 deg 33 gauge needle. After injection, paralube ophthalmic ointment (NDC 17033-211-38, Dechra) was applied and mice were placed on a heating pad until they awoke.
To determine if Myrf knockout of from RGCs induces apoptosis, AAV2-hSyn1-Cre along with AAV2-hSyn1-mCherry (Addgene 114472-AAV2) were mixed at 2.32 × 1012 genome copies per mL prior to injection and intravitreally injected into one eye of Myrffl/fl mice. The control eye received just AAV2-hSyn1-mCherry. These injections were conducted as above, and mice were perfused 10 weeks and retinae harvested for flatmount staining.
Optic nerve crush
Mice were prepared for surgery with the same ketamine/xylazine protocol for anesthesia as above. The right eye of each animal received an incision of the superior conjunctiva exposing the optic nerve, with care to avoid lesioning the orbital sinus. The nerve was crushed approximately 0.5 mm from the optic disk for 10 s using fine forceps (Dumont, #5). Following surgery paralube ophthalmic ointment was applied and 3.25 mg/kg of extended-release buprenorphine (Ethiqa XR formulation) was administered subcutaneously. The mouse was placed on a heating pad until it was awake.
Compound action potential recordings
Mice were deeply anaesthetized with ketamine and xylazine as above and an incision was made to expose the dorsal skull. The optic nerves were cut just behind the optic disk. The dorsal skull was then removed and olfactory bulbs were cut with fine surgical scissors, and the brain was gently lifted to expose the optic chiasm. The optic chiasm was then cut and optic nerves were carefully removed and placed in oxygenated artificial cerebrospinal fluid (aCSF) (124 mM NaCl, 1 mM NaH2PO4, 2.5 mM KCl, 26 mM NaHCO3, 10 mM glucose, 1 mM Na Ascorbate, 1 mM MgCl2, 2 mM CaCl2). Glass suction electrodes were prepared by heating the ends of glass capillaries over a flame until the tip was slightly constricted to match the nerve diameter, a few millimeters away the electrodes were bent at approximately 30 degrees to facilitate nerve holding. A silver wire was inserted into both recording and stimulating electrodes, and a reference wire was coiled around the electrodes to form a connection with the bath. Both electrodes were filled with aCSF. The optic nerve was transferred to a recording chamber continuously perfused with aCSF and held in place with wettened filter paper. Using gentle suction, the chiasmal side of the nerve was drawn into the recording electrode and the retinal side into the stimulating electrode. The nerve was then allowed to acclimate for 30 min to allow the tissue to form a seal with the constricted tip of the glass electrodes before recording began. The stimulating electrode was connected to a constant current isolated stimulator unit (Digitimer DS3) and nerves were stimulated at increasing amplitudes from 0.2 mA to 2 mA at 0.2 Hz for 100 µs until supramaximal threshold was found. Nerves were stimulated at 125% supramaximal threshold for recordings used in quantifications. Signals from the recording electrode were digitized via a Digidata 1440B, amplified using an Axon Instruments Multiclamp 700B amplifier and recorded using Clampex 10.7 software (Molecular Devices). CAP curves were subtracted from recordings following administration of TTX (1 µm, HB1035 HelloBio). Data was then analyzed using the Clampfit v10.7 software and to find both the latency to the highest peak and CAP area. CAP area is proportional to the number of stimulated axons72, and was measured as the area of the positive voltage deflection following stimulus. Each mouse was considered a biological replicate, and one nerve was measured from each mouse.
Visual evoked potential recordings
Low ambient light (13–18 Lux) was ensured in the room where VEPs were recorded. Mice were anesthetized via intraperitoneal injection using both xylazine (12.5 mg/kg) and ketamine (87.5 mg/kg) diluted in PBS. Pupils were dilated with tropicamide eye drops administered to each eye precisely 6 min after ketamine/xylazine administration. Mice were then placed in a sealed cardboard box (12 × 12 × 16 cm) for 5 min for dark adaptation. For the flash VEP one centimeter steel needle electrodes (Natus Neurology) were placed medially under the skin between the two eyes along the sagittal suture, with the needle inserted eight mm to optimize proximity to the visual cortex. A needle electrode inserted subcutaneously just above the tip of the nose served as reference electrode and an additional electrode inserted into the tail served as the ground electrode. A dome was next lowered to reduce ambient light and VEP stimulation and recording began precisely 13 min after administration of anesthesia. Flash based binocular visual electrophysiology was measured using an Espion Diagnosys system (Diagnosys LLC). Examinations consisted of three runs, with the following characteristics: pulse intensity 3 cd s/m2, frequency 1 Hz, on-time 4 ms, pulse color: white-6500K, and 100 sweeps per acquisition as per22. The standard VEP waveform with these parameters was characterized by a prominent negative deflection after approximately 70 ms which we identified as N1. N1 was defined as the first negative deflection after 50 ms. The two most representative/reproducible waves were used for analysis. The exam was performed by an operator blinded to mouse genotype.
Tissue processing
Mice were deeply anaesthetized with ketamine (400 mg/kg) and xylazine (60 mg/kg) then transcardially perfused with 10 mL of PBS and 40 mL of freshly hydrolyzed 4% paraformaldehyde (19210, Electron Microscopy Sciences). Tissues were gently dissected and placed in 4% paraformaldehyde for post fixation. For optic nerves used for electron microscopy, nerves were post fixed in 2% paraformaldehyde (15710, Electron Microscopy Sciences) with 2% glutaraldehyde (16310, Electron Microscopy Sciences) instead of 4% paraformaldehyde. For immunohistochemistry, optic nerves were post fixed for 2 h, brains overnight, retinae for 1 h and spinal cords for 4 h. Optic nerves, brains, and spinal cords were then cryoprotected in 30% sucrose for at least 48 h. Tissues were embedded in OCT and frozen on dry ice and stored at −80 °C until sectioning on a cryostat (CM3050-S, Leica). Brain sections were mounted at 10 µm thickness on Superfrost Plus slides (1255015, Fisher Scientific) between 1.4 mm to −0.6 mm relative to bregma. Optic nerve sections were made between 2–4 mm retinal to the optic chiasm mounted at 10 µm thickness. All sections were stored at −80 °C until immunohistochemistry was performed. Eyes were washed with 1x PBS following post-fixation and the retinae were dissected and post-fixed in 4% paraformaldehyde for 30 min and washed three times in 1x PBS. Retinae for in situ hybridization were flash frozen on dry ice following intracardiac perfusion with 1x PBS.
Immunohistochemistry
Slides were thawed from the −80 °C until dry and then rehydrated in 1x PBS. For MBP staining, tissue delipidization was performed by immersing slides in ascending and descending ethanol solutions before being washed 3x in 1x PBS. Slides were blocked for 30 min at room temperature with 10% fetal calf serum (SH30910.03, Cytiva) with 0.2% Triton-X100 (10789704001, Sigma). Primary antibodies were applied overnight in 1x PBS with 0.2% Triton-X100 in a sealed container at room temperature. Primary antibodies included mouse anti-BCAS1 (1:200; NaBC1; sc-136342, Santa Cruz), chicken anti-MBP (1:200; MBP, Aves), mouse anti-CC1 (1:500; Clone CC1; OP80, Millipore), rabbit anti CD3 (1:500; Clone SP7, NB600-1441SS, Novus), rat anti CD4 (1:200; Clone GK1.5; MAB554, R and D Systems), rat anti CD8 (1:200; Clone 53-6.7; 550281, BD Biosciences), rabbit anti-OLIG2 (1:500; AB9610, Millipore), chicken anti OLIG2 (1:500; OLIG2-002, Aves), goat anti mouse-PDGFRα (1:200; AF1062, R&D Systems), rat anti-CD68 (1:500; Clone FA-11, MCA1957GA, Biorad), rabbit anti-Iba1 (1:1000; 019-19741, Wako), rabbit anti SYT4 (1:200; 105 143 Sysy Antibodies), mouse anti NKX2.2 (1:200; Clone 74.5A5; DSHB), rabbit anti-GFAP (1:1000; Z0334, Dako), mouse anti NFL-DegenoTag (1:1000; Clone MCA-6H63, Encor Biotechnology), mouse anti-E06 (Oxidized phospholipid, 1:500; 330001S, Avanti), chicken anti-mCherry (1:1000; mCherry-0100, Aves), chicken anti-GFP (1:1000; ab13970, Abcam), rabbit anti-cleaved caspase-3 (1:200; 559565, BD Pharminogen) and rabbit anti-cleaved caspase-3 (1:200; AF835, R and D Systems). Following incubation with primary antibodies, slides were washed three times in 1x PBS before appropriate Alexa Fluor 488, 555 or 647 secondary antibodies (Invitrogen) were applied for 2 h at room temperature. Slides were then again washed three times with 1x PBS before coverslipping with Fluoromount G (0100-01, Southern Biotech) for analysis.
For EdU labeling, slides with optic nerve sections were incubated at room temperature for 30 min protected from light in freshly-prepared Alexa-647 EdU Cell Proliferation Assay (C10340, Thermo Fisher Scientific) cocktail after immunohistochemistry. Slides were washed three times in 1x PBS and coverslipped in Fluoromount-G (01001-01, Southern Biotech). For BODIPY 493/503 (D3922, Invitrogen) staining, slides were incubated in BODIPY for 15 min in 0.5 µg/mL solution following application of secondary antibodies and then washed 3x in 1x PBS prior to coverslipping.
Retinal flatmounts were blocked in 10% fetal calf serum with 0.2% Triton-X100 for 1 h with agitation. Retinae were then incubated with primary antibodies at 4 °C with agitation in 1x PBS 0.2% Triton-X100. Primary antibodies included guinea pig anti-RBPMS (1:500; 1832, Phosphosolutions), rabbit anti-RBPMS (1:500; ABN1362, Millipore), rabbit anti-phosphorylated c-Jun (1:500; 9261 Cell Signaling), mouse anti AP-2α (1:500; Clone 3B5; sc-12726 Santa Cruz), mouse anti-AP-2β (1:25, Clone PCRP-TFAP2B-2E1; DSHB) and rabbit anti-cleaved caspase-3. Appropriate secondary antibodies were applied overnight with agitation at 4 °C. To prepare for mounting on slides, each retina was cut with 4–5 incisions along the radial axis from the edge to about 2/3rds the distance to the optic disk then mounted on slides. Prolong Glass Antifade Mountant (P36980, Thermo Fisher Scientific) was applied prior to coverslipping.
In situ hybridization
RNAscope in situ hybridization was used to detect Rbpms (527231, ACDBio) in combination with Ecel1 (1137301-C2, ACDBio) or Hrk (475331-C3, ACDBio). The assay was performed according to the manufacturer’s instructions (RNAscope Multiplex Fluorescent V2 Assay, ACDBio). Briefly, 20 µm thick sections were mounted on Superfrost slides (1255015, Fisher Scientific) and stored at −80 °C until in situ hybridization was performed. Slides were dehydrated in 50%, 70%, and 100% ethanol for 5 min before being placed into boiling target retrieval buffer for 5 min to unmask the target RNA. Slides were treated with H2O2 for 10 min at room temperature then Protease III was applied for 30 min at 40°C. Probes were hybridized for 2 h at 40 °C. Either Ecel 1 or Hrk was assigned to channel 2 or 3 and diluted 1:50 in Rbpms probes assigned to channel 1. To test RNA integrity within the tissue, probes against housekeeping genes Polr2a, Ppib and Ubc (320881, ACDBio) were applied to slides cut in parallel along with 3-Plex negative control probes on an additional slide (320871, ACDBio). Signal amplification was performed according to the instructions of the kit. Signal detection utilized Opal520 (OP-001001, Akoya) and Opal 690 (OP-00106, Akoya), which were diluted 1:1000 in TSA buffer (322809, ACDBio). Nuclei were detected by DAPI stain applied for 5 min prior to coverslipping with Prolong Gold (P36934, Thermo Fisher Scientific). Images were captured on a Zeiss LSM 980 microscope with Zen Blue Software (v3.7).
Electron microscopy tissue processing and analysis
Following postfixation, optic nerves were stored in a buffer of 1.5% paraformaldehyde, 1.5% glutaraldehyde, 50 mM sucrose, 22.5 mM CaCl2 2H2O in 0.1 M cacodylate buffer for at least 7 days before embedding. Optic nerves were trimmed to two millimeters from the optic chiasm prior to plastic embedding and sections obtained approximately 50 µm chiasmal of that to avoid dissection artifact. Optic nerves were post-fixed in 2% osmium tetroxide (19190, Electron Microscopy Sciences) with 1.5% potassium ferrocyanide (25154-20, Electron Microscopy Sciences) using a Biowave Pro+ microwave (Ted Pella). Contrast was enhanced by en bloc staining with 0.5% uranyl acetate (22400, Electron Microscopy Sciences) before dehydration in ethanol and embedding in Embed 812 (14120, Electron Microscopy Sciences). 0.5 µm sections were cut on an ultramicrotome and stained with 0.5% Toluidine Blue (22050, Electron Microscopy Sciences) with 0.5% sodium borate (21130, Electron Microscopy Sciences) to visualize the optic nerve for area measurements. 60 nm sections were mounted on copper grids (T400-Cu, Electron Microscopy Sciences) then counter stained with 5% Uranyl Acetate for 20 min followed by Reynold’s Lead Citrate (80 mM Pb(NO3)2 17900-25, Electron Microscopy Sciences and 120 mM Sodium Citrate, 21140, Electron Microscopy Sciences) for 6 min. Grids were imaged at 4800x on a FEI Tecnai T12 transmission electron microscope with a 16 Mpx camera (Advanced Microscopy Techniques Corp). The density of axons and those with organelle accumulations was measured within ten random images per nerve. Organelle accumulations were counted if more than half the area of the axon was occupied by two or more organelles. We then multiplied the average density of axons within the optic nerve by the area measured on the adjacent Toluidine Blue section to get the total number of axons per nerve. For g-ratio analyses, 5–6 images were analyzed per animal at ×4800 magnification. For quantifications at 10 and 20 weeks post tamoxifen, the axon and myelin were manually traced with the spline contour tool using the Zen 3.0 software (Zeiss) to determine the axon diameter relative to the axon diameter with myelin. For g-ratio measurements at eight weeks post tamoxifen, Myeltracer tool (v1.3 was used to calculate axon diameter relative to myelin diameter73. Demyelinated axons were not included in g-ratio analyses and analyses was conducted blinded to genotype.
Serum neurofilament light chain (NfL) detection
After deep anesthesia with ketamine and xylazine as above, 0.5 mL blood was acquired from mice immediately prior to perfusion via intracardiac puncture. Blood was allowed to clot for 1 h before spinning at 500 × g for 10 min. The serum was removed and snap frozen on dry ice and stored at −80 °C. NfL concentration was measured on the Simoa platform using the NF-light advantage kit V2 (Quanterix). To account for the high concentrations found in demyelinating mice that might go beyond the highest point of the calibrator, serum was bench-diluted to 1:4 or 1:8 (depending on available sample volume). On the Simoa, another 1:4 online dilution followed as part of the standard assay procedure, and final concentration was corrected for the applied dilution factor.
Immunofluorescence image analysis
Immunostained sections were captured with a Zeiss ApoTome2 at 20x using 0.8NA lens with Zen Blue software (v3.3). Optic nerve cross sections were imaged in their entirety with at least four sections 200 µm apart analyzed per mouse, per analysis. For cellular counts of OL lineage cells, the optic nerve was manually outlined using the spline contour tool in Zen 3.0 (Zeiss) and OLIG2-positive nuclei were counted first. Each OLIG2-positive cell was examined to see if it expresses CC1, PDGFRα, SYT4, NKX2.2 or EdU. For microglial and T-cell counts, only Iba1-positive or CD3-positive cells with DAPI nuclear staining were considered to be positive. For analysis of BCAS1, MBP, E06 and CD68 images were manually thresholded by an observer blinded to genotype and timepoint in Fiji ImageJ 1.53 K (NIH) to determine the area occupied relative to the size of the optic nerve. For analysis of BODIPY immunofluorescence in microglia, a microglia/macrophage were first identified by IBA1+ staining then BODIPY fluorescence was measured within IBA1+ cells using ImageJ. Likewise, for E06, an IBA1+ mask was placed over the image and the area of E06 staining outside and inside microglia were quantified. To quantify phosphorylated c-Jun positive neurons in the spinal cord, whole spinal cords were imaged and the gray matter outlined using the NEUN channel and double positive cells were counted and examined to ensure there was DAPI present. At least four images of the lumbar spinal cord were quantified per animal.
Retinae were imaged in their entirety for analysis at 20x with 0.8NA lens using a Zeiss ApoTome2. To quantify RBPMS or phosphorylated c-Jun, cells were manually counted in the GCL within a 200 µm × 200 µm box placed at 500 µm, 1000 µm, 1500 µm and 2000 µm from the optic disk in each quadrant for a total of 16 regions. For quantifications of whether phosphorylated c-Jun+ cells were AP-2α/β or RBPMS-positive, each phosphorylated c-Jun positive cell was counted as above then determined if the cells were AP-2α/β or RBPMS positive. Cleaved caspase-3-positive cells were counted over the extent of the retina. For analysis of cleaved caspase-3 expression in AP2-α/β or RBPMS positive cells, each caspase-3 positive cell was individually evaluated for these markers per retina. The retina was manually outlined using the spline contour tool (Zen 3.0, Zeiss) to determine the area for density measurements. All analyses were conducted blind to genotype or treatment.
Western blot
Mice were deeply anesthetized as above and perfused with 1x PBS before the optic nerve was removed and flash frozen on dry ice and stored at −80 °C until protein was extracted. Thawed optic nerves were dounce homogenized in RIPA buffer along with complete protease inhibitors (11836153001, Roche) and phosphatase inhibitors (04906837001, Roche). Following homogenization, samples were spun at 13,000 × g for 15 min and the protein lysate was removed and frozen. Lysates from four nerves (two mice) were combined and protein was run on Bis-Tris-gel (NP0335BOX, Invitrogen). To transfer to a PVDF membrane (IPVH00010, Thermo Scientific), the gel blotting sandwich cassette (A25977, Thermo Fisher Scientific) was placed in transfer buffer (NP0006-01, Thermo Scientific) under 25 V for 90 min. Following transfer, blots were rinsed in 1x TBS with 0.1% Tween-20 (TBST) before blocking in 1x TBST with 5% milk powder for 1 h. Blots were probed with antibodies against DLK (GTX124127, Genetex), pMKK4 (9156, Cell Signaling), MKK4 (9152, Cell Signaling), pJNK (9251, Cell Signaling), JNK (9252, Cell Signaling), MOG (supernatant from clone 8-18C5, kind gift of R. Reynolds, Imperial College, London, UK), MBP (MAB386, Millipore) diluted in TBST with 2% BSA (BP9706-100, Fisher Scientific) overnight to 1:1000. After overnight incubation, blots were washed in 1x TBST and incubated with appropriate HRP-conjugated secondary (Goat anti-rat 7077, Cell Signaling, Goat anti-mouse 7076, Cell Signaling, Goat anti-rabbit 7074, Cell Signaling) for 2 h with 2% milk powder in TBST. Immunoreactivity was visualized using chemiluminescence (34080, Thermo Fisher Scientific) and imaged on a Syngene GBox iChemiXT using GeneSys software (v1.2.5.0). Blots were subsequently re-probed with β-actin-HRP as a loading control (Clone AC-15; A3854, Sigma). Densitometric analysis was performed in ImageJ v1.53 by quantifying the intensity of bands relative to loading control and then normalized relative to the mean of the Myrffl/fl control group. All unprocessed full blots used for quantification are supplied in the Source Data file attached to the manuscript.
Laser capture microscopy
Eyes were dissected and snap frozen in OCT, then sectioned on a cryostat at 20 µm thickness and mounted onto Poly-L-Lysine (P1524, Sigma) coated membrane slides (414190-9041-000, Zeiss). Sections were fixed in 70% ethanol for 2 min before staining in Harris Modified Hematoxylin (HHS32, Sigma) with 0.2% glacial acetic acid for 30 s. Sections were then immersed in 70% ethanol twice and 100% ethanol twice for 30 s each and stored at −80 °C in a sealed container until LCM was performed. A Zeiss Palm Microbeam microscope was used to conduct LCM with cut segments extracted onto the lid of adhesive cap tubes (415190-9181-000, Zeiss). The GCL was identified and sectioned—at least 20 sections per animal. For sampling of the whole retina, laser incisions were made through each retinal layer. Samples were treated with RLT lysis buffer and RNA isolated using the MicroRNAeasy kit (74004, Qiagen) as per the manufacturer’s instructions and frozen −80 °C until sequencing.
Bulk RNAseq
Following RNA isolation, RNA quantity and quality were evaluated on an Agilent 2100 Bioanalyzer using the Eukaryote Total RNA Pico. cDNA libraries were produced by loading 15 ng of RNA for use with the Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero Plus kit and sequenced with a NovaSeq 6000 at 50 million reads per sample. Raw reads were sorted based on barcodes and FASTQC files were produced. Reads were aligned to Mus musculus (GRCm38/Mm10) and expression counts were performed using STAR. DeSeq2 was run using Basepair software to determine differentially expressed genes between whole retina and the GCL or between the GCL of MyrfΔiSox10 and Myrffl/fl mice. A total of six Myrffl/fl and six MyrfΔiSox10 were compared for statistical analyses.
Nuclei isolation and snRNAseq
Blood was removed from deeply anesthetized mice via intracardiac perfusion with 10 mL of 1x PBS at 4-7 PM to reduce circadian fluctuations. Optic nerves were dissected out and immediately snap-frozen on dry ice. Frozen tissue was stored at −80 °C for up to 6 months until subsequent processing. The nuclei isolation buffer (NIB, 146 mM NaCl, 5 mM Tris-HCl, 1 mM CaCl2, 21 mM MgCl2, 0.03% Tween-20, 0.01% BSA, 1 µg/mL actinomycin D, pH 7.5) was prepared with one tablet of protein inhibitor cocktail (cOMPLETE Mini lacking EDTA, 11873580001, Roche) along with 15 uL of RNAsin (N2615, Promega) per 10 mL NIB. When optic nerves were removed from the freezer, they were immediately placed in cooled 2 mL NIB solution in a 7 mL Dounce grinder and ground 20 times with a loose pestle. Then, the homogenate was passed through a 200 µm strainer (43-50200-03, Pluriselect). The homogenate was ground 10 additional times with a tight pestle, then 2 mL NIB was added and the homogenate was passed through a 40 µm strainer (43-50040-03, Pluriselect). The homogenate was ground five times with a tight pestle (B), then passed through a 20 µm filter (43-50020-03, Pluriselect). The sample was then centrifuged at 500 × g for 5 min at 4 °C three times with the supernatant discarded and new NIB added each time. Following the last centrifugation step, the pellet was resuspended in 0.5 mL NIB with 5 µL SuperaseIN (AM2696, Thermo Fisher Scientific) along with 1% BSA and mixed 1:200 with RedDot (40060, Biotium). The samples were then isolated from debris by fluorescence-activated nuclei sorting (FANS). Two main gates were used: a 638 emission for the RedDot stain and a low trigger pulse width as singlet discriminator using BD FACS software (v1.2.0.142) (Supplementary Fig. 12). A total of 100,000 nuclei were aimed to be sorted. Following sorting, samples were centrifuged at 300 × g for 1 min at 4 °C, held on ice for 1 min then spun for 1 min at 300 × g at 4 °C. The top supernatant was carefully removed and the nuclei were then prepared for snRNAseq using a Chromium Next GEM Single Cell 3′ Reagent Kit v3.1 (10x Genomics). Single nuclei were partitioned in droplets with single gel beads, which contained primers with cell-tagging indexes. Single nucleus suspensions were targeted to 10,000 nuclei per sample with 500 million reads per sample. The resulting cDNA was used as a template for library preparation. Samples were sequenced using a NovaSeq 6000 and FASTQ files were prepared using bcl2fastq (Illumina) and then aligned to the mouse GRCm38/mm10 reference genome using Cellranger (v7.0.0, 10x Genomics). Reads were mapped to both exonic and intronic regions.
snRNA-seq analyses
Data was analyzed using R (v4.2.3) and the Seurat package (v4.3.0.1)74. First, quality control of each sample was performed. Ambient RNA was removed using SoupX75 (v1.6.2). To remove doublets and debris, nuclei were filtered based on 1000 < nFeature_RNA < 4000 as well as 1250 < nCount_RNA < 10,000. Mitochondrial genes were removed by manually excluding all features starting with “mt-“. One sample (a Myrffl/fl sample) failed quality control and was not included in further analysis. Following quality control, the seven samples were normalized with “SCTransform”76 (v0.3.5) and integrated using “FindIntegrationAnchors” function in Seurat. The integrated dataset was visualized by UMAP plotting techniques. Nuclei were first clustered using 35 principal component dimensions. Differentially expressed genes (DEGs) were identified using Seurat’s “FindMarkers” with the following analysis criteria: Log2 fold change >0.322, the minimum percentage of nuclei expressing the gene = 0.25, and adjusted p value (calculated with the Wilcox significance test) <0.05. Top genes were used to identify each cluster. Tables with marker genes for all clusters (Supplementary Data 1), comparisons of DEGs in MyrfΔiPlp1 and MyrfΔiSox10 nuclei as well as microglia nuclei are provided in Supplementary Data 2–5.
Several samples had neuronal nuclear contamination from the dissection indicated by nuclei with high Rbfox3, Syt7, and Snap25 and these clusters were manually excluded. After the neuronal contamination was removed, we re-clustered using 33 principal component dimensions to account for the removal of a highly diverse neuronal population that may have affected the original UMAP clustering. We re-ran the DEG analysis for the final UMAP clustering using the analysis criteria stated above and annotated the clusters using established markers. Next, we compared the DEGs between the control mice derived from different lines (Myrffl/fl; Plp1-CreERT-negative and Myrffl/fl; Sox10CreERT-negative) using the analysis criteria stated. Because both control lines had less than 10 DEGs in total, for the ease of analysis between genotypes, we combined the three control samples (hereby referred to as Myrffl/fl). After combining the controls, DEGs between Myrffl/fl and MyrfΔiPlp1 or MyrfΔiSox10 were identified with the analysis criteria. Likewise, DEGs between the two knock-out lines (MyrfΔiPlp1 or MyrfΔiSox10) were identified using the analysis criteria above. DEGs for KOOLs were then displayed using a volcano plot (EnhancedVolcano v1.16 and ggplot2 3.42). and transcripts with log2 fold change >0.50 were highlighted as enriched in MyrfΔiPlp1 or MyrfΔiSox10 nuclei.
For microglia re-clustering, we subdivided these cells and reclustered using eight principal component dimensions based off the elbowplot. DEGs for microglia subclusters were identified with the same analysis criteria as above. DIM3 population DEGs were calculated with the following analysis criteria: log2 fold change >0.50 and visualized with a volcano plot (ggplot2 3.42). Gene set enrichment analysis (GSEA) for molecular function pathways was performed on the DIM3 cluster differentially expressed genes (p adjusted <0.05) using ClusterProfiler (v4.6.2). Trajectory pathway analysis in psuedotime was performed on the OL lineage using Monocle3 (v1.3.1).
Production of viral constructs
SgRNAs were designed using CRISPROn77 and compared to previously published sgRNAs78. Five sgRNAs for Map3K12 and Map3K13 were tested in total for indel formation in cultured Neuro2a cells expressing Cas9. Px333 (Addgene # 64073) was modified by restriction enzymes to insert oligonucleotides with the MluI site and ApaI site at the XhoI and KpnI sites, respectively. This allows the removal of tandem U6 promoters with sgRNAs and insertion into AAV-U6-sgRNA-hSyn-mCherry (Addgene #87916) in place of the single U6-sgRNA when digested with MluI and ApaI. SgRNAs were tested in the modified px333 plasmid and validated by TIDE (tracking of indels by decomposition)79. The two best sgRNAs were chosen based on the degree of decomposition after the PAM site. While all sgRNAs tested demonstrated decomposition after the PAM site, the sgRNAs which were used in the study to target Map3K12 at exons 4 and 9 had the highest decomposition and have the following sequences AGGGTGTTCGGGTTTCATGG and TGTAGAGAGCACATCAGCGG. The guides utilized against Map3K13 had the sequences TCTGGGGAACAGCAACACTG and GGTCACGGTGTATAATCTTG78 targeting exons 3 and 5 of Map3K13, respectively. SgRNAs against LacZ and GFP for the control AAV were taken from previously validated sgRNAs80,81. AAV2-hSyn1-Cre was generated by replacing the mCherry open reading frame from AAV-U6-sgRNA-hSyn-mCherry (Addgene #87916) with the Cre open reading frame from AAV:ITR-U6-sgRNA-pCBh-Cre-WPRE-hGHpA-ITR (Addgene #60229). Large-scale packaging into AAV2 of viral vectors for intravitreal injection was completed by Vector Biolabs (AAVs with sgRNAs against Map3K12, Map3K13 or Map3K12/Map3K13) or Vector Biosystems (sgLacZ/sgGFP) or using the OHSU Molecular Virology Core (AAV2-hSyn1-Cre).
Cas9-expressing Neuro2a cell culture
Mouse Neuro2a cells (CCL-131, ATCC) expressing Cas9 (cells were transduced with Cas9 in the pQXCIH plasmid) were grown in Dulbeco’s modified Eagle medium (11960-044, Gibco) (DMEM) with 10% FBS supplemented with glutamine (25030-081, Gibco), Penicillin-Streptomycin (100 U/mL penicillin, 100 mg/mL streptomycin; 15140-122, Gibco); and Sodium Pyruvate (11360-070, Gibco). Neuro2a cells were incubated at 37 °C and 5% CO2 and were passaged every 3 days. To transfect Neuro2a cells with sgRNA expressing plasmids, we used Lipofectamine 2000 (52758, Invitrogen) and the cells were collected 24 h later and DNA extracted using DNeasy Blood and Tissue Kit as per the manufacturer’s instructions (69504, Qiagen).
Statistics
Statistical analyses were conducted with Prism 10.2.2 (Graphpad) or SPSS v13.0 and all data is presented as mean ± standard error. Sample sizes are indicated by the number of dots in the figures or are otherwise explicitly stated. All tests are two-sided. To test for normality, the Shapiro-Wilk test was used. To test for homogeneity of variance, Levene’s test was used for comparison of two groups and Brown-Forsythe test for three or more. If data was not normally distributed appropriate non-parametric tests were run. If a time point had less groups than others in the analysis because of premature euthanasia of MyrfΔiSox10 mice, this time point was treated as an independent experiment for statistics. For all statistical analyses n represents a single animal, except for the optic nerve snRNAseq and western blots, where two animals worth of nerves were combined per sample (n). Degree of significance was indicated in figures by *p < 0.05, **p < 0.01, ***p < 0.001 or ****p < 0.0001 unless stated otherwise. Animals were assigned to group based on genotype or to treatment by random selection. The number of mice used in experiments was based on previous publications with similar methodology. Statistical comparisons are outlined in Supplementary Table 2 and t, F, H statistics, p values and degrees of freedom reported when appropriate.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank Brett Hilton for critically reading and providing feedback on this manuscript. We thank Pamela Canaday and Christina Metea for their assistance and guidance with flow cytometry. Alex Klug and Amy Carlos provided 10x microfluidics and library production support. Brian Jenkins, Hannah Bronstein and Stefanie Kaech Petrie provided guidance for microscopy image analysis. Paul Meraner kindly provided Cas9-expressing Neuro2a cells for sgRNA validation. This work received support from the Flow Cytometry, Gene Profiling/RNA and DNA Services Shared Resource, OHSU Molecular Virology Core, Massively Parallel Sequencing Shared Resource, Multiscale Microscopy Core (MMC) and Advanced Light Microscopy Cores at OHSU. This work was supported by grants from Race to Erase MS, the NINDS (R01NS120981), NINDS P30 (NS061800), Collins Medical Trust (1016373), the National Multiple Sclerosis Society (RG-2001-35775), American Heart Association (20CDA35320169) and National Institute of Diabetes and Digestive and Kidney Diseases (K01DK121737). G.J.D. was supported by a postdoctoral fellowship (FG-1808-32238) and a career transition award (TA-2105-37636) from the National Multiple Sclerosis Society. C.C. was supported by FISM—Fondazione Italiana Sclerosi Multipla—cod. 2019/BC/002 and financed or co-financed with the “5 per mille” public funding. B.E. was supported by an endowment from the Warren family. G.J.D. and B.E. thank Caron and Larry Ogg for their generous financial support to support G.J.D. and helping to make this work possible.
Author contributions
G.J.D. and B.E. designed and conceived the experiments. G.J.D., S.D.I., J.H., M.M., J.E.J., C.C., A.A., N.J., K.A., S.J.F., B. Stedelin, B. Sivyer and B.E. performed experiments and acquired data. G.J.D., K.E. and J.N. curated data and provided code for analysis of the snRNAseq data. G.J.D., K.E., C.C., A.A., J.N. and B.E. analyzed data. J.H. and S.A.A. provided expertise and collected data for electron microscopy analysis. A.M. trained and supervised G.J.D. in acquisition of CAPs from the optic nerve. J.N. trained and supervised G.J.D. and K.E. in snRNAseq analysis. T.A.W. provided expertise with MAPK reagents and biology. T.S.S. provided expertise for EAE experiments. A.J.G. provided expertise for and supervised experiments analyzing serum neurofilament levels and visual evoked potentials. G.J.D. and B.E. wrote the manuscript with inputs from all authors. B.E. supervised the project.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Data availability
The SnRNAseq data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database under accession code GSE243788. The bulk RNAseq data generated in this study have been deposited in the NCBI GEO database under accession code GSE245362. All raw data generated and analyzed in this study are available from the corresponding authors on request. Source data are provided with this paper.
Materials availability
Plasmids generated in this study for DLK or LZK knockout have been deposited at Addgene (Plasmid #208834, 208835, 208836, 208837) and AAV2-hSyn1-Cre is available upon request. Transgenic mouse lines are available at Jackson laboratory (Sox10-CreERT JAX #027651, Plp1-CreERT JAX #005975, Myrffl/fl JAX #010607, Rosa26-Cas9 mice JAX #024858, Sun1-GFP mice JAX #030952) and the Myrffl/fl line on a C57BL/6 background is available from the Emery lab upon request. All other reagents were from commercial sources.
Code availability
All original code has been deposited at (Github: https://github.com/EmeryLab/Myrf_iCKO_OpticNerve)82. SnRNA-Seq data can be viewed in our interactive browser at https://emerylab.shinyapps.io/Myrf_iCKO_OpticNerve/.
Competing interests
T.S.S. and B.E. are co-founders of Autobahn Therapeutics, B.E. has received consulting fees from Autobahn Therapeutics and T.S.S. is a Senior Advisor to Autobahn Therapeutics. B.E. and G.J.D. have received licensing fees for the use of Myrf inducible conditional knockout mice. These potential conflicts of interest have been reviewed and managed by OHSU. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Greg J. Duncan, Email: duncangr@ohsu.edu
Ben Emery, Email: emeryb@ohsu.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53429-5.
References
- 1.Zawadzka, M. et al. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell6, 578–590 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tripathi, R. B., Rivers, L. E., Young, K. M., Jamen, F. & Richardson, W. D. NG2 glia generate new oligodendrocytes but few astrocytes in a murine experimental autoimmune encephalomyelitis model of demyelinating disease. J. Neurosci.30, 16383–16390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duncan, I. D. et al. The adult oligodendrocyte can participate in remyelination. Proc. Natl Acad. Sci. USA115, E11807–E11816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mezydlo, A. et al. Remyelination by surviving oligodendrocytes is inefficient in the inflamed mammalian cortex. Neuron111, 1748–1759.e1748 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Neely, S. A. et al. New oligodendrocytes exhibit more abundant and accurate myelin regeneration than those that survive demyelination. Nat. Neurosci.25, 415–420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldschmidt, T., Antel, J., Konig, F. B., Bruck, W. & Kuhlmann, T. Remyelination capacity of the MS brain decreases with disease chronicity. Neurology72, 1914–1921 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Frischer, J. M. et al. Clinical and pathological insights into the dynamic nature of the white matter multiple sclerosis plaque. Ann. Neurol.78, 710–721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trapp, B. D. & Stys, P. K. Virtual hypoxia and chronic necrosis of demyelinated axons in multiple sclerosis. Lancet Neurol.8, 280–291 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Nave, K. A. Myelination and the trophic support of long axons. Nat. Rev. Neurosci.11, 275–283 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Franklin, R. J., ffrench-Constant, C., Edgar, J. M. & Smith, K. J. Neuroprotection and repair in multiple sclerosis. Nat. Rev. Neurol.8, 624–634 (2012). [DOI] [PubMed] [Google Scholar]
- 11.De Stefano, N. et al. Axonal damage correlates with disability in patients with relapsing-remitting multiple sclerosis. Results of a longitudinal magnetic resonance spectroscopy study. Brain121, 1469–1477 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Singh, S. et al. Relationship of acute axonal damage, Wallerian degeneration, and clinical disability in multiple sclerosis. J. Neuroinflammation14, 57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodini, B. et al. Dynamic imaging of individual remyelination profiles in multiple sclerosis. Ann. Neurol.79, 726–738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornek, B. et al. Multiple sclerosis and chronic autoimmune encephalomyelitis: a comparative quantitative study of axonal injury in active, inactive, and remyelinated lesions. Am. J. Pathol.157, 267–276 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emery, B. et al. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell138, 172–185 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bujalka, H. et al. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol.11, e1001625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koenning, M. et al. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J. Neurosci.32, 12528–12542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duncan, G. J. et al. Myelin regulatory factor drives remyelination in multiple sclerosis. Acta Neuropathol.134, 403–422 (2017). [DOI] [PubMed] [Google Scholar]
- 19.McKenzie, I. A. et al. Motor skill learning requires active central myelination. Science346, 318–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartley, M. D. et al. Myelin repair stimulated by CNS-selective thyroid hormone action. JCI Insight4, e126329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fard, M. K. et al. BCAS1 expression defines a population of early myelinating oligodendrocytes in multiple sclerosis lesions. Sci. Transl. Med.9, eaam7816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cordano, C. et al. Validating visual evoked potentials as a preclinical, quantitative biomarker for remyelination efficacy. Brain145, 3943–3952 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou, J. et al. Transcriptomic atlas and interaction networks of brain cells in mouse CNS demyelination and remyelination. Cell Rep.42, 112293 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey, S. et al. Disease-associated oligodendrocyte responses across neurodegenerative diseases. Cell Rep.40, 111189 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Park, H. et al. Single-cell RNA-sequencing identifies disease-associated oligodendrocytes in male APP NL-G-F and 5XFAD mice. Nat. Commun.14, 802 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connor, J. R. & Menzies, S. L. Relationship of iron to oligodendrocytes and myelination. Glia17, 83–93 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity50, 253–271.e256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walther, T. C., Chung, J. & Farese, R. V. Jr. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol.33, 491–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu, B. & Simon, M. C. BODIPY 493/503 staining of neutral lipid droplets for microscopy and quantification by flow cytometry. Bio Protoc.6, e1912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaffner, E. et al. Myelin insulation as a risk factor for axonal degeneration in autoimmune demyelinating disease. Nat. Neurosci.26, 1218–1228 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar, J. M. et al. Oligodendroglial modulation of fast axonal transport in a mouse model of hereditary spastic paraplegia. J. Cell Biol.166, 121–131 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdelhak, A. et al. Plasma neurofilament light chain levels suggest neuroaxonal stability following therapeutic remyelination in people with multiple sclerosis. J. Neurol. Neurosurg. Psychiatry93, 972–977 (2022). [DOI] [PMC free article] [PubMed]
- 33.Kwong, J. M., Caprioli, J. & Piri, N. RNA binding protein with multiple splicing: a new marker for retinal ganglion cells. Invest. Ophthalmol. Vis. Sci.51, 1052–1058 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez, A. R., de Sevilla Muller, L. P. & Brecha, N. C. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J. Comp. Neurol.522, 1411–1443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins, T. A. et al. DLK initiates a transcriptional program that couples apoptotic and regenerative responses to axonal injury. Proc. Natl Acad. Sci. USA110, 4039–4044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Pichon, C. E. et al. Loss of dual leucine zipper kinase signaling is protective in animal models of neurodegenerative disease. Sci. Transl. Med.9, eaag0394 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Welsbie, D. S. et al. Functional genomic screening identifies dual leucine zipper kinase as a key mediator of retinal ganglion cell death. Proc. Natl Acad. Sci. USA110, 4045–4050 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welsbie, D. S. et al. Targeted disruption of dual leucine zipper kinase and leucine zipper kinase promotes neuronal survival in a model of diffuse traumatic brain injury. Mol. Neurodegener.14, 44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, Z., Maroney, A. C., Dobrzanski, P., Kukekov, N. V. & Greene, L. A. The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Mol. Cell Biol.21, 4713–4724 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirai, S., Izawa, M., Osada, S., Spyrou, G. & Ohno, S. Activation of the JNK pathway by distantly related protein kinases, MEKK and MUK. Oncogene12, 641–650 (1996). [PubMed] [Google Scholar]
- 41.Ghosh, A. S. et al. DLK induces developmental neuronal degeneration via selective regulation of proapoptotic JNK activity. J. Cell Biol.194, 751–764 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syc-Mazurek, S. B., Rausch, R. L., Fernandes, K. A., Wilson, M. P. & Libby, R. T. Mkk4 and Mkk7 are important for retinal development and axonal injury-induced retinal ganglion cell death. Cell Death Dis.9, 1095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itoh, T. et al. ZPK/DLK and MKK4 form the critical gateway to axotomy-induced motoneuron death in neonates. J. Neurosci.34, 10729–10742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu, J. et al. Palmitoylation couples the kinases DLK and JNK3 to facilitate prodegenerative axon-to-soma signaling. Sci. Signal.15, eabh2674 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holland, S. M. et al. Palmitoylation controls DLK localization, interactions and activity to ensure effective axonal injury signaling. Proc. Natl Acad. Sci. USA113, 763–768 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel, S. et al. Scaffold-hopping and structure-based discovery of potent, selective, and brain penetrant N-(1H-pyrazol-3-yl)pyridin-2-amine inhibitors of dual leucine zipper kinase (DLK, MAP3K12). J. Med. Chem.58, 8182–8199 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Duncan, I. D., Brower, A., Kondo, Y., Curlee, J. F. Jr & Schultz, R. D. Extensive remyelination of the CNS leads to functional recovery. Proc. Natl Acad. Sci. USA106, 6832–6836 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Traka, M. et al. A genetic mouse model of adult-onset, pervasive central nervous system demyelination with robust remyelination. Brain133, 3017–3029 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mei, F. et al. Accelerated remyelination during inflammatory demyelination prevents axonal loss and improves functional recovery. Elife5, e18246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ricigliano, V. A. G. et al. Spontaneous remyelination in lesions protects the integrity of surrounding tissues over time in multiple sclerosis. Eur. J. Neurol.29, 1719–1729 (2022). [DOI] [PubMed] [Google Scholar]
- 51.Tonietto, M. et al. Periventricular remyelination failure in multiple sclerosis: a substrate for neurodegeneration. Brain146, 182–194 (2023). [DOI] [PubMed] [Google Scholar]
- 52.Lazzarotto, A. et al. Time is myelin: early cortical myelin repair prevents atrophy and clinical progression in multiple sclerosis. Brain147, 1331–1343 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapp, B. D. et al. Axonal transection in the lesions of multiple sclerosis. N. Engl. J. Med.338, 278–285 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Groh, J. et al. Microglia-mediated demyelination protects against CD8(+) T cell-driven axon degeneration in mice carrying PLP defects. Nat. Commun.14, 6911 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar, J. M. et al. Demyelination and axonal preservation in a transgenic mouse model of Pelizaeus-Merzbacher disease. EMBO Mol. Med.2, 42–50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotter, M. R., Li, W. W., Zhao, C. & Franklin, R. J. Myelin impairs CNS remyelination by inhibiting oligodendrocyte precursor cell differentiation. J. Neurosci.26, 328–332 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miron, V. E. et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat. Neurosci.16, 1211–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruckh, J. M. et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell10, 96–103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cantuti-Castelvetri, L. et al. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science359, 684–688 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Berghoff, S. A. et al. Microglia facilitate repair of demyelinated lesions via post-squalene sterol synthesis. Nat. Neurosci.24, 47–60 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marschallinger, J. et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci.23, 194–208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kutzelnigg, A. et al. Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain128, 2705–2712 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Valakh, V., Walker, L. J., Skeath, J. B. & DiAntonio, A. Loss of the spectraplakin short stop activates the DLK injury response pathway in Drosophila. J. Neurosci.33, 17863–17873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller, B. R. et al. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat. Neurosci.12, 387–389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandes, K. A., Harder, J. M., Kim, J. & Libby, R. T. JUN regulates early transcriptional responses to axonal injury in retinal ganglion cells. Exp. Eye Res.112, 106–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harris, C. A. & Johnson, E. M. Jr. BH3-only Bcl-2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J. Biol. Chem.276, 37754–37760 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Craig, R. A. et al. Discovery of potent and selective dual leucine zipper kinase/leucine zipper-bearing kinase inhibitors with neuroprotective properties in in vitro and in vivo models of amyotrophic lateral sclerosis. J. Med. Chem.65, 16290–16312 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Katz, J. S. et al. A phase 1 study of GDC-0134, a dual leucine zipper kinase inhibitor, in ALS. Ann. Clin. Transl. Neurol.9, 50–66 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doerflinger, N. H., Macklin, W. B. & Popko, B. Inducible site-specific recombination in myelinating cells. Genesis35, 63–72 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron86, 1369–1384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stys, P. K., Ransom, B. R. & Waxman, S. G. Compound action potential of nerve recorded by suction electrode: a theoretical and experimental analysis. Brain Res.546, 18–32 (1991). [DOI] [PubMed] [Google Scholar]
- 73.Kaiser, T. et al. MyelTracer: a semi-automated software for myelin g-ratio quantification. eNeuro8, ENEURO.0558–20.2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol.33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience9, giaa151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol.20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiang, X. et al. Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat. Commun.12, 3238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Summers, D. W., Milbrandt, J. & DiAntonio, A. Palmitoylation enables MAPK-dependent proteostasis of axon survival factors. Proc. Natl Acad. Sci. USA115, E8746–E8754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res.42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Swiech, L. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol.33, 102–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang, J. P. et al. Different effects of sgRNA length on CRISPR-mediated gene knockout efficiency. Sci. Rep.6, 28566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duncan, G. J. et al. Remyelination protects neurons from DLK-mediated neurodegeneration. Zenodo10.5281/zenodo.13755371 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The SnRNAseq data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database under accession code GSE243788. The bulk RNAseq data generated in this study have been deposited in the NCBI GEO database under accession code GSE245362. All raw data generated and analyzed in this study are available from the corresponding authors on request. Source data are provided with this paper.
Plasmids generated in this study for DLK or LZK knockout have been deposited at Addgene (Plasmid #208834, 208835, 208836, 208837) and AAV2-hSyn1-Cre is available upon request. Transgenic mouse lines are available at Jackson laboratory (Sox10-CreERT JAX #027651, Plp1-CreERT JAX #005975, Myrffl/fl JAX #010607, Rosa26-Cas9 mice JAX #024858, Sun1-GFP mice JAX #030952) and the Myrffl/fl line on a C57BL/6 background is available from the Emery lab upon request. All other reagents were from commercial sources.
All original code has been deposited at (Github: https://github.com/EmeryLab/Myrf_iCKO_OpticNerve)82. SnRNA-Seq data can be viewed in our interactive browser at https://emerylab.shinyapps.io/Myrf_iCKO_OpticNerve/.