Abstract
Human T-lymphotropic virus type 1 (HTLV-1) is a complex retrovirus encoding regulatory and accessory genes in four open reading frames (ORF I to IV) of the pX region. We have demonstrated an important role of pX ORF I expression, which encodes p12I, in establishment of HTLV-1 infection in a rabbit model and for optimal viral infectivity in quiescent primary lymphocytes. These data indicated that p12I may enhance lymphocyte activation and thereby promote virus infection. To further define the role of p12I in cell activation, we characterized the subcellular localization of p12I in transfected 293T cells and HeLa-Tat cells by multiple methods, including immunofluorescence confocal microscopy, electron microscopy, and subcellular fractionation. Herein, we demonstrate that p12I accumulates in the endoplasmic reticulum (ER) and cis-Golgi apparatus. The location of p12I was unchanged following treatments with both cycloheximide (blocking de novo protein synthesis) and brefeldin A (disrupting ER-to-Golgi protein transport), indicating that the protein is retained in the ER and cis-Golgi. Moreover, using coimmunoprecipitation assays, we identify the direct binding of p12I with both calreticulin and calnexin, resident ER proteins which regulate calcium storage. Our results indicate that p12I directly binds key regulatory proteins involved in calcium-mediated cell signaling and suggest a role of p12I in the establishment of HTLV-1 infection by activation of host cells.
Human T-lymphotropic virus type 1 (HTLV-1) is the etiologic agent of adult T-cell leukemia and lymphoma and appears to initiate a variety of immune-mediated disorders, including the chronic neural degenerative disease HTLV-1-associated myelopathy/tropical spastic paraparesis (16, 44). As a complex retrovirus, HTLV-1 contains the common retroviral genes gag, pol, and env, as well as several regulatory and accessory genes. These regulatory and accessory genes are present in four different open reading frames (ORFs) in the pX region between env and the 3′ long terminal repeat (LTR) (10, 15, 39, 40). ORFs IV and III encode the regulatory proteins Tax and Rex, respectively, which have been extensively characterized. Tax is a 40-kDa nuclear-localizing protein that increases viral transcription from the HTLV-1 LTR as well as a number of cellular genes involved in host cell proliferation (17, 30, 34). Rex is a 27-kDa nucleolar-localizing protein that acts at the posttranscriptional level by promoting the cytoplasmic accumulation of unspliced and singly spliced viral RNA (29).
Recent studies have provided important new data that indicate a role of the highly conserved ORF I-encoded protein p12I in HTLV-1 infection. ORF I mRNA has been detected in HTLV-1-infected cells derived from patients with both adult T-cell leukemia and lymphoma and HTLV-1-associated myelopathy/tropical spastic paraparesis and from asymptomatic carriers (3, 5, 9, 10). Moreover, recombinant p12I is recognized by sera from naturally infected humans and experimentally infected rabbits (14). Peptides derived from amino acid sequences unique to ORF I-encoded proteins are recognized by cytotoxic T lymphocytes isolated from infected subjects, indicating the chronic production of these proteins during HTLV-1 infection (37). Members of our group have demonstrated that the selective ablation of ORF I mRNA dramatically decreases the infectivity of ACH, an infectious molecular clone of HTLV-1, in a rabbit model of infection (12). Additionally, we also demonstrated that ORF I expression is required for optimal viral infectivity in quiescent primary lymphocytes, suggesting a role of p12I in T-lymphocyte activation (1).
These data imply a functional role of pX ORF I-encoded proteins in host cell activation. HTLV-1 p12I is a small hydrophobic protein and has distant homology with the bovine papillomavirus E5 protein (17). The viral protein contains four minimal proline-rich SH3 domain binding motifs (PXXP), which are commonly present in cellular proteins involved in regulating signal transduction. The protein associates with the 16-kDa subunit of the vacuolar H+ ATPase, enhances E5 transforming ability (18), and binds to the interleukin 2 receptor β and γ chain (33). Moreover, Koralnik et al. (25, 26) have observed by indirect immunofluorescence assays that p12I is predominantly found in the perinuclear region of HeLa-Tat cells and predicted that p12I is expressed in cellular endomembranes. These studies together with the predicted structure motifs of p12I imply a functional role for this protein in modulating cellular signals. Using subcellular fractionation, immunofluorescence assay, and confocal and electron microscopy, we demonstrate that the majority of p12I is enriched in the endoplasmic reticulum (ER) and cis-Golgi apparatus. Proteins that are expressed in the ER achieve their specific localization by direct retention or by retrieval from cellular compartments through recognition of specific cellular motifs. To test whether p12I maintained its localization by retention or retrieval, we used cycloheximide to block protein synthesis and tested for the expression of the protein over a 24-h period. The typical perinuclear localization of p12I did not change following either cycloheximide treatment alone or cycloheximide plus brefeldin A (BFA) cotreatment, indicating that p12I is retained in the ER and cis-Golgi compartment. The amino acid sequence of p12I does not contain typical ER retention motifs. Therefore, the retention of p12I in the ER is likely related to other structural features of the protein. Using serially deleted p12I expression vectors, we demonstrated that two regions containing predicted transmembrane domains (amino acids [aa] 1 to 47 and aa 47 to 99) are independently sufficient for the localization of p12I. Furthermore, p12I colocalized with and bound calreticulin and calnexin, resident ER proteins important for calcium storage and release from the ER. Our data suggest a potential mechanism for p12I in calcium-mediated cell activation to enhance HTLV-1 replication.
MATERIALS AND METHODS
Cell lines and plasmids.
The 293 cell line is a human kidney embryonic cell line (catalog number 1573, American Type Culture Collection). 293T is the 293 cell line which stably expresses the simian virus 40 (SV40) T antigen (obtained from G. Franchini, National Institutes of Health). HeLa-Tat is a human cervical carcinoma cell line, HeLa, which stably expresses human immunodeficiency virus type 1 Tat protein. This cell line was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institutes of Health, from Barbara Felber and George Pavlakis (HLtat, catalog number 1293). These three cell lines were maintained in Dulbecco's modified Eagle medium (supplemented with 10% fetal bovine serum, 100 μg of streptomycin plus penicillin/ml, and 2 mM l-glutamine [Gibco]). The R49 cell line (13) is a rabbit peripheral blood mononuclear cell line which was stably transformed with ACH, an infectious molecular clone of HTLV-1 (24). R49 cells were maintained in RPMI 1640 medium supplemented with 15% fetal bovine serum, 100 μg of streptomycin plus penicillin/ml, and 2 mM l-glutamine (Gibco). The pMEp12I plasmid contains influenza virus hemagglutinin (HA) epitope-tagged p12I sequence in the pME vector, which is driven by a hybrid promoter (SRα) containing the SV40 early promoter and the R region of the HTLV-1 LTR (33, 41). The pLEGFP-C1 plasmid is a retroviral green fluorescent protein expression vector (Clontech). The full-length p12I sequence was PCR amplified from the ACH plasmid and directionally subcloned into the pLEGFP-C1 plasmid to construct pLEGFPp12I. The pBCp12I plasmid was created by subcloning p12I-encoding sequence to the C terminus of a glutathione S-transferase (GST) sequence in the pBC vector (6), a mammalian GST expression vector driven by the SV40 promoter (obtained from C. Kredinger, Institute de Génétique et de Biologie Moléculaire et Cellulaire, CNRS/LGME-INSERM, Strasbourg, France).
Construction of serial deletion mutants of p12I.
Serial deletion mutants of p12I, C-terminally tagged with HA, were subcloned into the pME vector in two steps. Eight deletion mutants were first subcloned into the pME vector at EcoRI and NotI sites by PCR-mediated directional subcloning. The HA epitope was then synthesized as oligonucleotide and ligated at the C termini of all the deletion mutants using NotI and XbaI sites. The following primers were used in the PCRs to amplify the different regions of the p12I encoding sequence.
p12I 15-99 (5′, ATATGAATTCATGACGGCGCTCCTGCTC; 3′, ATATGCGGCCGCTTAGAAGAGGAAAGCCGC); p12I 32-99 (5′, AAATTTGAATTCATGCTCCGCCCGCCTCCT; 3′, ATATGCGGCCGCTTAGAAGAGGAAAGCCGC); p12I 48-99 (5′, ATAGAATTCATGATACTCAGCAATCTGCTT; 3′, ATATGCGGCCGCTTAGAAGAGGAAAGCCGC); p12I 1-86 (5′, ATATGAATTCATGCTGTTTCGCCTTCTC; 3′, ATTGCGGCCGCGGGGAGAAAGCGCCACCTCGC); p12I 1-47 (5′, ATATGAATTCATGCTGTTTCGCCTTCTC; 3′, ATTGCGGCCGCTTGAAAAGGAAGGAAGAGGAG); p12I 15-86 (5′, ATATGAATTCATGACGGCGCTCCTGCTC; 3′, ATTGCGGCCGCGGGGAGAAAGCGCCACCTCGC); p12I 15-69 (5′, ATATGAATTCATGACGGCGCTCCTGCTC; 3′, ATTGCGGCCGCGCTGAGGAGAAGAGGAAGCGA); p12I 15-47 (5′, ATATGAATTCATGACGGCGCTCCTGCTC; 3′, ATTGCGGCCGCTTGAAAAGGAAGGAAGAGGAG).
The HA oligonucleotides included the sense oligonucleotide GGCCGCGTACCCATACGATGTTCCAGATTACGCTAGCTTGGCCGC GGCTTTCCTCTTCTAAT and the antisense oligonucleotide TAGATTAGAAGAGGAAAGCCGCGGCCAAGCTAGCGTAATCTGGAACATCGTATGGGTACGC. Sanger sequencing was used to confirm all of the inserted p12I nucleotide sequences and to ensure that the sequences were in-frame.
Immunoblot assay.
293T cells were seeded at approximately 40% confluence in 10-cm-diameter tissue culture dishes 1 day before transfection. Cells were transfected with 10 μg of pMEp12I, pME, or deletion mutants of p12I using Lipofectamine Plus (Gibco). Transfected 293T cells were lysed at 48 h after transfection in Triton X-100 buffer [1% deoxycholic acid, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), with protease inhibitor 20-μg/ml leupeptin, 20-μg/ml aprotinin, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF), and 1 mM sodium orthovanadate]. The protein concentration from all lysates was measured using a Micro BCA protein assay kit (Pierce). Forty micrograms of each lysate was separated by SDS–10% polyacrylamide gel electrophoresis (SDS-PAGE). After transfer to nitrocellulose membranes (Millipore), the expression of pMEp12I and that of the deletion mutants of p12I were detected with a mouse monoclonal anti-HA antibody (1:1,000; Babco), followed by incubation with a secondary antibody conjugated to horseradish peroxidase (1:1,000; New England Biolabs). Bands were visualized by using a chemiluminescence detection system (New England Biolabs).
Immunocytochemistry.
For indirect immunofluorescence assays, HeLa-Tat cells were seeded into chamber slides (Fisher Scientific) and were transfected with 4 μg of pMEp12I or deletion mutants of pMEp12I using Lipofectamine Plus. Two days posttransfection, cells were fixed for 15 min with 4% paraformaldehyde, followed by incubation with primary antibodies: mouse monoclonal anti-HA (1:200; Babco), rabbit polyclonal anticalreticulin (1:500; Affinity Bioreagents), or mouse monoclonal anti-adaptin γ (1:200; Sigma) for 1 h at room temperature in antibody dilution buffer (0.01 M sodium phosphate, 0.5 M NaCl, 0.5% Triton X-100, 2% bovine serum albumin). Cells were washed three times with phosphate-buffered saline (PBS) (Gibco) and were incubated with either fluorescein isothiocyanate-labeled anti-mouse antibody plus indocarbocyanine (Cy3)-labeled anti-rabbit or Cy3 labeled anti-mouse antibody (Jackson Immunogen) at room temperature for 1 h. To identify subcellular organelles, specific markers, including the Golgi marker, BODIPY TR ceramide, the mitochondrion marker, MitoTracker Orange, and the F-actin marker, rhodamine phalloidin, were used to costain with p12I expression according to the manufacturer's protocol (Molecular Probes). Briefly, 5 μM BODIPY TR ceramide was added to the secondary antibody dilution buffer, and cells were stained at room temperature for 1 h. MitoTracker Orange (100 nM) was added to Dulbecco's modified Eagle medium, and live cells were stained for 30 min, followed by the indirect immunofluorescence detection of p12I as described above. Rhodamine phalloidin (1:100) was added to the secondary antibody dilution buffer, and cells were costained for both F-actin and p12I. The images were collected using the Lasersharp software program with a Bio-Rad MRC1024 confocal microscope.
BFA and cycloheximide treatments.
To test the effect of BFA treatment on p12I localization, pLEGFPp12I-transfected HeLa-Tat cells were treated with 10 μg of BFA/ml for 30 min at 2 days posttransfection. The localization of GFPp12I and that of adapter complex protein 1 (AP-1) were detected by direct and indirect immunofluorescence assays, respectively, as described above. For cycloheximide studies, 10 μg of cycloheximide (Sigma)/ml was added into pMEp12I-transfected HeLa-Tat cell medium at 24 h posttransfection, and cells were treated for 0.5, 2, 4, 8, 12, or 24 h, followed by detection of p12I accumulation by indirect immunofluorescence assay. For cycloheximide and BFA cotreatment, cells were incubated with 10 μg of BFA/ml for 30 min before cell fixation at different time periods of cycloheximide treatments, followed by indirect immunofluorescence assay to examine p12I expression.
Electron microscopy.
Electron microscopy was used to further identify the subcellular organelles expressing p12I. HeLa-Tat cells were transfected with 15 μg of pMEp12I. Cells were fixed with 2.5% paraformaldehyde and 0.5% glutaraldehyde at 48 h posttransfection, followed by dehydration and embedding in a commercial resin (LR White, Electron Microscopy Science). Embedded samples were sectioned with an LKB Ultratome and were deposited on nickel grids. The grids were incubated with mouse monoclonal anti-HA antibody (1:50; Babco) at room temperature for 1 h, followed by incubation with the 10-nm colloidal gold-conjugated anti-mouse antibody (1:100; Amersham Pharmacia Biotech) for 30 min at room temperature. The samples were examined by electron microscopy (Phillips 300) at ×45,000 or ×53,000 magnification, and images were processed using standard photographic methods.
Membrane and cytosol fractionation.
The membrane and cytosol fractions of pMEp12I-transfected cells were separated to test the association of p12I with the membrane. Transfected 293T cells were resuspended in 1 ml of homogenization buffer (200 mM HEPES [pH 7.5], 5 mM sodium pyrophosphate, 5 mM EGTA, 1 mM MgCl2, 1 mM AEBSF, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 1 mM sodium orthovanadate), followed by sonication and centrifugation at 11,000 rpm for 1 min. The supernatant was ultracentrifuged at 100,000 × g for 1 h at 4°C, and the resulting cytosol fraction (the supernatant portion) was collected. The pellet was washed twice with PBS and resuspended in 1 ml of extraction buffer (20 mM Tris-HCl [pH 7.5], 1% Triton X-100, 100 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 5 mM NaF, 1 mM AEBSF, 1 mM sodium orthovanadate), followed by shaking for 1 h at 4°C and centrifugation at 100,000 × g for 45 min. The supernatant was collected as the membrane fraction. The protein concentrations from both membrane and cytosol fractions were measured using the Micro BCA protein assay kit. The expression of p12I, Ras-related GTP binding protein 1B (Rab1B), and Fas-associated death domain protein (FADD) in both membrane and cytosol fractions were tested by immunoblot analysis using mouse monoclonal anti-HA antibody, rabbit polyclonal anti-Rab1B antibody (Zymed), and mouse monoclonal anti-FADD antibody (Transduction Lab), respectively, as described previously (7, 27, 45).
Subcellular membrane fractionation.
To identify the subcellular membranes that contain p12I, gradient ultracentrifugation and fractionation were performed. Approximately 108 pMEp12I-transfected 293T cells at 48 h posttransfection were resuspended in 3 ml of homogenization buffer (HB) (containing 10 mM HEPES [pH 7.4], 1 mM EDTA, 0.25 M sucrose, 20-μg/ml leupeptin, 20-μg/ml aprotinin, 1 mM AEBSF, and 1 mM phenylmethylsulfonyl fluoride). Cells were disrupted by 10 strokes with a Dounce homogenizer followed by 5 to 10 passages through a 27-gauge needle to obtain about 95% broken cells. Nuclei and unbroken cells were pelleted by centrifugation at 1,500 rpm for 10 min in an Eppendorf 5714R centrifuge. The postnuclear supernatant was centrifuged in an SW55 rotor (Beckman) at 65,000 × g for 1 h. The resulting pellet was resuspended in 0.8 ml of HB buffer. Discontinuous iodixanol (OptiPrep, 60% wt/vol; Gibco) gradients were prepared in an SW41 centrifugation tube as described previously (46). Briefly, iodixanol was diluted to 50% in HB buffer as the stock solution, which was then used to make various concentrations of iodixanol used in gradient experiments. Gradients were set up in a 13-ml centrifuge tube by underlaying solutions with defined percentages of iodixanol with a syringe and metal needle to create a gradient consisting from top to bottom of 2.5, 5, 7.5, 10, 12.5, 15, 17.5, 20, and 30% iodixanol. The resuspended pellet was loaded on top of the gradients and centrifuged in an SW41 rotor at 41,000 rpm for 2.5 h. The resulting gradient was collected as 1-ml fractions (total, 13 fractions). The fractions were solubilized by adding Triton X-100 to a final concentration of 1%, followed by immunoblot analysis to test for the expression of p12I, the ER marker, calnexin, and the Golgi 58-kDa protein as described previously (4, 19, 38, 47).
Immunoprecipitation assay for calreticulin and calnexin binding.
To investigate the association of p12I with calreticulin and calnexin, 293T cells were transfected with 10 μg of pMEp12I. Cells were lysed with Triton X-100 buffer. Cell lysates were precleared with both 30 μl of protein A Sepharose beads (Sigma) and 6 μl of normal rabbit serum for 6 h, followed by incubation with either 1:150 diluted rabbit polyclonal anticalreticulin (Affinity Bioreagents) or 1:150 diluted rabbit polyclonal anticalnexin (StressGene) overnight. The immune complex mixture was incubated with 30 μl of protein A Sepharose beads for 2 h. Beads were washed twice in Triton X-100 lysis buffer and boiled in SDS sample buffer, and the supernatants were separated by SDS-PAGE. The proteins were examined for p12I expression by immunoblot assay using a mouse monoclonal anti-HA antibody (1:1,000; Babco). The endogenous expression of calreticulin and that of calnexin were tested by immunoblot assay using chicken polyclonal anticalreticulin (Affinity Bioreagents) and the same rabbit polyclonal anticalnexin described above. As an additional control, 293T cells were transfected with 10 μg of pHCMV-16-kDa-AU1 (18) and were lysed with Triton X-100 buffer. Cell lysates were precleared and immunoprecipitated with either anticalreticulin or anticalnexin as described above. The 16-kDa subunit of the vacuolar H+ ATPase was examined by immunoblot assay using mouse monoclonal anti-AU (1:1,000; Babco).
GST binding assay.
To verify the interaction of p12I with calreticulin and calnexin, pBCp12I- and pBC-transfected 293T cells were lysed with Triton X-100 buffer. One milligram of total lysate was used to bind 40 μl of a 50% slurry of glutathione-agarose beads previously blocked by 5% nonfat dry milk for 2 h. Beads were washed three times with lysis buffer and boiled in SDS sample buffer, followed by SDS-PAGE and immunoblot analysis with chicken polyclonal anticalreticulin or rabbit polyclonal anticalnexin as described above. The same membranes were stripped and tested for the expression of GSTp12I or GST by immunoblot assay using rabbit polyclonal anti-GST (1:7,000).
Glycoprotein analysis.
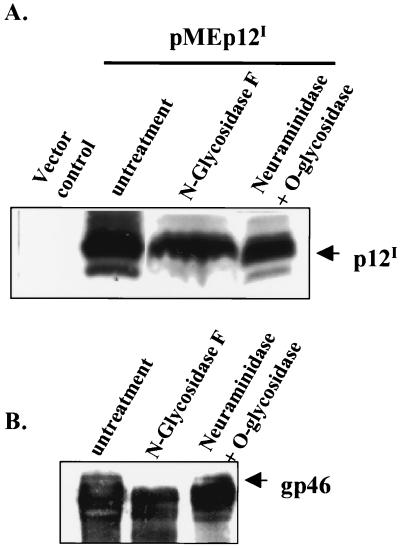
To test whether p12I is expressed as a glycoprotein, pMEp12I-transfected 293T cells were lysed with RIPA buffer (1% NP-40, 1% deoxycholic acid, 10% glycerol, 150 mM NaCl, 50 mM Tris-HCl [pH 7.5] with protease inhibitor 20-μg/ml leupeptin, 20-μg/ml aprotinin, 1 mM AEBSF, and 1 mM sodium orthovanadate). One milligram of total lysate was immunoprecipitated with rabbit polyclonal anti-HA antibody (1:150; Babco) as described above. The immune-complex-bound beads were washed three times with PBS. For the release of N-linked glycans from the protein, the beads were boiled for 5 min in 45 μl of 1× glycosidase incubation buffer (Glyco Systems) containing 0.1% SDS and 50 mM β-mercaptoethanol, followed by addition of 0.75% NP-40. The supernatant was collected and enzymatically treated overnight at 37°C using 1 mU of N-glycosidase F (Glyco Systems). For the release of O-linked glycans from the protein, the beads were boiled for 5 min in 45 μl of 1× O-glycanase reaction buffer (Sigma). The supernatant was collected and enzymatically treated at 37°C for 1 h using 10 mU of neuraminidase (Sigma), followed by overnight digestion with 1.5 mU of O-glycanase at 37°C. Samples were then analyzed by SDS-PAGE and p12I expression, tested using an immunoblot assay as described above. The HTLV-1 surface envelope protein gp46 was used as a positive control for N-glycosidase F treatment. R49 cells were lysed with RIPA buffer. One milligram of total lysate was immunoprecipitated by 1:150 diluted mouse monoclonal anti-gp46 IC-11 (36). The immune-complex-bound beads were treated with N-glycosidase F or neuraminidase plus O-glycanase to release N-glycans or O-glycans by the same method described above. The expression of gp46 was tested using an immunoblot assay with IC-11.
RESULTS
p12I is enriched in ER membranes.
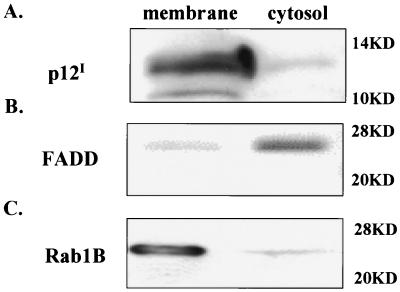
HTLV-1 p12I has been demonstrated to localize in the perinuclear region and in fine reticular patterns consistent with cellular endomembranes in transfected cells (25, 26). To define the subcellular localization of p12I in plasma and cellular membranes, we transiently expressed p12I in 293T cells and performed membrane and cytosol fractionation. The majority of p12I was detected in membrane fractions (Fig. 1A), indicating that p12I is a membrane-associated protein. FADD is a cytosolic adapter protein critical for Fas signaling (27). Rab1B associates with intracellular membrane organelles and participates in the transport of glycoproteins between the ER and Golgi compartments (21). These two proteins were used as markers for cytosol fractions (Fig. 1B) and membrane fractions (Fig. 1C), respectively.
FIG. 1.
p12I is a membrane-associated protein. Total proteins were prepared from p12I-transfected 293T cells and separated into membrane and cytosol fractions. Equal amounts of proteins from both fractions were analyzed by immunoblot assay. (A) The majority of p12I was detected in the membrane fraction. FADD (B) and Rab1B (C) were used to verify the cytosol and membrane fractions, respectively. Molecular mass markers are given at right.
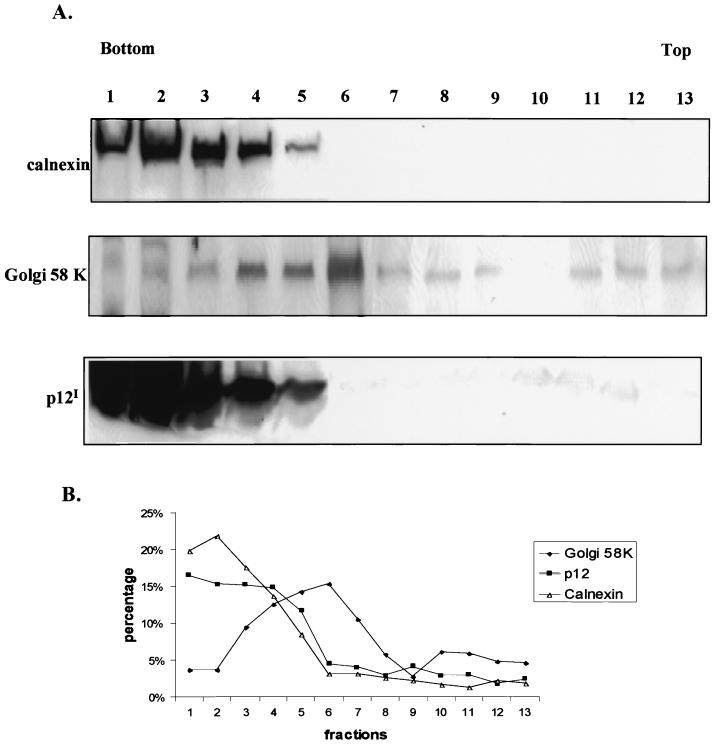
To further identify the p12I-containing membrane pool, we performed gradient ultracentrifugation. Calnexin, an ER integral membrane protein, was used to identify the ER-containing fractions (Fig. 2A, fractions 1 to 5). The Golgi 58-kDa protein, a Golgi membrane resident protein, was used to identify Golgi apparatus-containing fractions (Fig. 2A, fractions 4 to 8). Fractions 4 and 5 contained both calnexin and the Golgi 58-kDa protein, consistent with the ER-to-Golgi intermediate compartment and cis-Golgi. The peak level of expression of p12I was detected in fractions 1 to 5, which corresponded to the calnexin-reactive ER fractions and cis-Golgi-containing fractions (Fig. 2).
FIG. 2.
p12I is enriched in ER membrane fractions. (A) The subcellular distribution of calnexin, the Golgi 58-kDa protein (Golgi 58 K), and p12I was analyzed with 293T cells transiently transfected with the pMEp12I plasmid. Following fractionation and SDS-PAGE separation, the calnexin, Golgi 58-kDa protein, and p12I were detected in an immunoblot assay by incubation with rabbit polyclonal anticalnexin, mouse monoclonal anti-Golgi 58-kDa protein, and mouse monoclonal anti-HA followed by secondary antibody incubation. The p12I and calnexin were both enriched in the ER-rich fractions 1 to 4. Numbers above the panels correspond to the fractions analyzed in panel B. (B) Graphical representation of results from subcellular fractionation of p12I-transfected 293T cells. Distribution of calnexin, the Golgi 58-kDa protein, and p12I was determined by immunoblot assay followed by densitometry image analysis. The results are expressed as percentages of the respective signals in each fraction to the sum of signals in all fractions. The ER marker calnexin was located primarily in fractions 1 to 4, and the Golgi marker Golgi 58-kDa protein was detected in fractions 4 to 12, among which fractions 4 to 7 showed enhanced expression.
p12I localizes in the ER and Golgi apparatus.
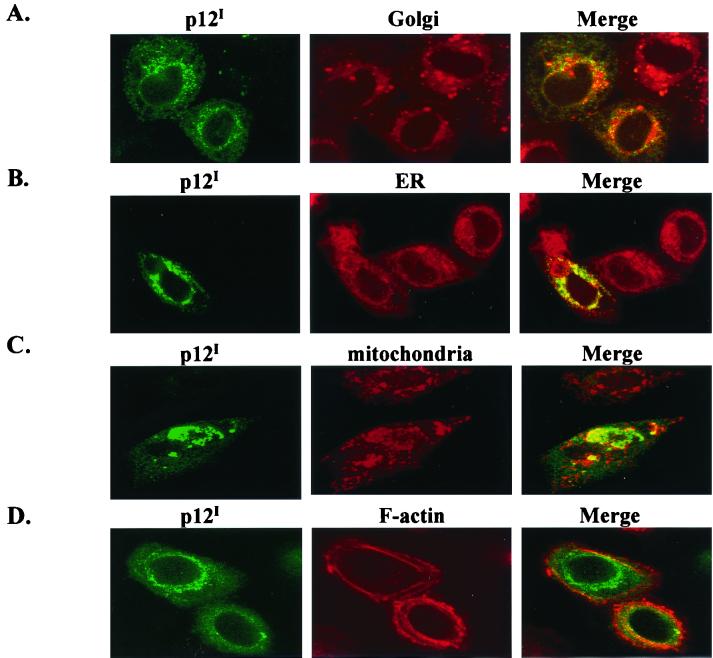
The two major subcellular compartments in the perinuclear region are the ER and the Golgi apparatus. To further identify the subcellular compartment of p12I accumulation, multiple markers were used, including a Golgi marker (BODIPY TR ceramide), an ER marker (calreticulin), a mitochondrion marker (MitoTracker Orange), and an F-actin marker (rhodamine phalloidin). p12I accumulation colocalized with the Golgi marker (Fig. 3A) and calreticulin (Fig. 3B), an ER luminal resident protein. However, p12I accumulation did not appear to localize directly with the mitochondrial marker (Fig. 3C) or colocalize with F-actin staining (Fig. 3D). These results suggested that the accumulation of p12I was distributed within the ER and the Golgi apparatus.
FIG. 3.
p12I colocalizes with the ER and Golgi apparatus. HeLa-Tat cell were transfected with pMEp12I, the localization of p12I was examined by indirect immunofluorescence assay, and images were collected by confocal microscopy as described in Materials and Methods. The images in the first column display the staining for p12I expression. The images in the second column are the subcellular markers in the same field. The images in the third column are the merged images for p12I expression and subcellular marker costaining. (A) p12I colocalizes with BODIPY Ceramide (Golgi marker). (B) p12I colocalizes with calreticulin (ER marker). (C) p12I does not colocalize with MitoTracker Orange (mitochondria marker). (D) p12I does not colocalize with rhodamine phalloidin (F-actin marker).
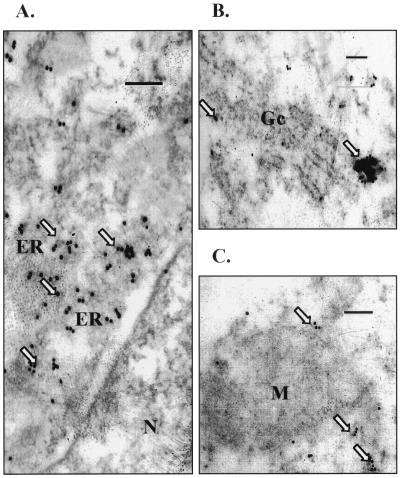
Electron microscopy using immunogold labeling was performed to verify the subcellular localization of p12I. Gold particles corresponding to p12I accumulation were localized in the membrane structures outside the nucleus, including the ER and Golgi apparatus (Fig. 4A and B) but not mitochondria (Fig. 4C). Indirect immunofluorescence and electron microscopy studies were also performed using 293 cells with similar results (data not shown).
FIG. 4.
p12I expression identified by immunogold particles in cytoplasmic membranes. p12I-transfected 293T cells were fixed, embedded, and stained as described in Materials and Methods. (A) Portion of a cell showing a nucleus (N); immunogold particles are concentrated in membrane structures outside the nucleus, consistent with the ER. (B) Portion of a cell containing Golgi stacked membranes (Gc) with accumulated gold particles. (C) Gold particles external to mitochondria (M). Magnification, ×40,500. Bar = 0.2 μm.
p12I localization is not sensitive to treatment with BFA.
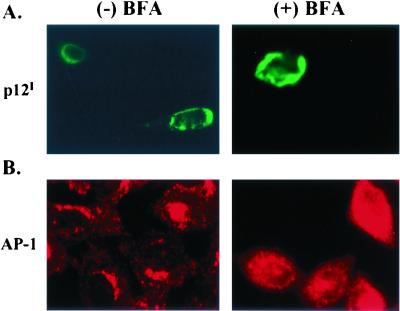
BFA is a fungal product which blocks ADP ribosylation factor activation, resulting in interference with ER-to-Golgi anterograde protein trafficking. In addition, BFA leads to the disruption of the trans-Golgi apparatus in treated cells (32, 43). To test whether p12I localization is affected by BFA treatment, transfected HeLa-Tat cells were treated with 10 μg of BFA/ml for 30 min. The localization of p12I was unchanged by BFA treatment (Fig. 5A). However, the localization of AP-1, a major component of the adapter protein complex, which is localized in the trans-Golgi apparatus, changed from perinuclear staining to a dispersed staining pattern following BFA treatment (Fig. 5B). These results indicate that p12I accumulates in the ER or cis-Golgi apparatus and the localization of the protein is not ADP ribosylation factor dependent.
FIG. 5.
p12I localization is not sensitive to BFA treatment. GFPp12I-transfected HeLa-Tat cells were treated with BFA (10 μg/ml) (right column) for 30 min or left untreated (left column). Cells were fixed and stained using a monoclonal antibody against AP-1, followed by Cy3-labeled anti-mouse antibody staining. (A) p12 localization did not change following BFA treatment. (B) AP-1 staining became diffuse following BFA treatment.
p12I is retained in the ER.
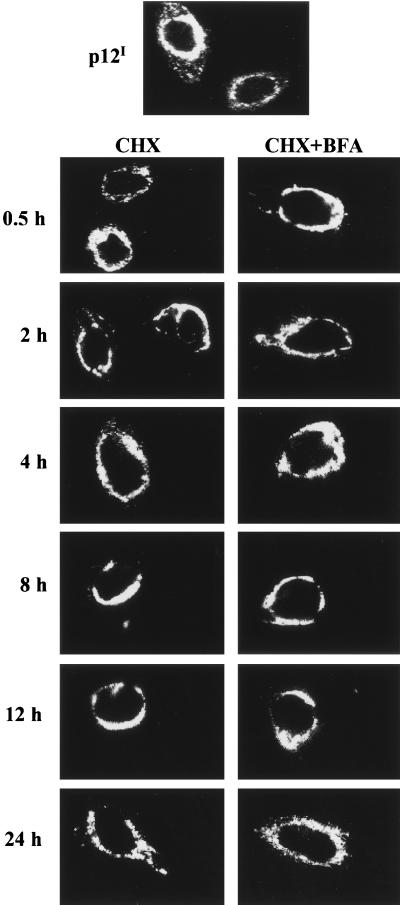
ER resident proteins maintain their localization by one of two mechanisms, retention or retrieval. To test which of these mechanisms was used by p12I, we treated p12I-transfected HeLa-Tat cells with cycloheximide to block new protein synthesis and then tested for p12I expression. If a protein translocates from ER-Golgi compartments to the plasma membrane and then is retrieved to the ER, the protein should be transiently observed at these locations following the cycloheximide treatment. p12I, however, was detected in the same perinuclear region, consistent with ER and cis-Golgi retention, at all time periods tested following cycloheximide treatment (Fig. 6). These data indicate that p12I is retained in the ER and cis-Golgi compartment. Moreover, incubation with both cycloheximide and BFA did not change the p12I localization, further demonstrating the retention of p12I in the ER and cis-Golgi compartments. Since the half-life of p12I has been determined to be 16 to 24 h (42), we stopped the cycloheximide treatment at 24 h postincubation. As expected, after 24 h of treatment we observed the number of p12I-expressing cells to be decreased following continuous cycloheximide treatment (data not shown).
FIG. 6.
p12I is retained in the ER and cis-Golgi compartments. HeLa-Tat cells were transfected with pMEp12I and were treated with cycloheximide (CHX) at 24 h posttransfection. The localization of p12I was examined at various time points following cycloheximide treatment by indirect immunofluorescence assay. The perinuclear staining pattern did not change following continuous cycloheximide treatment and BFA treatment (CHX+BFA).
Two regions of p12I (aa 1 to 47 and 48 to 99) independently determine cellular localization.
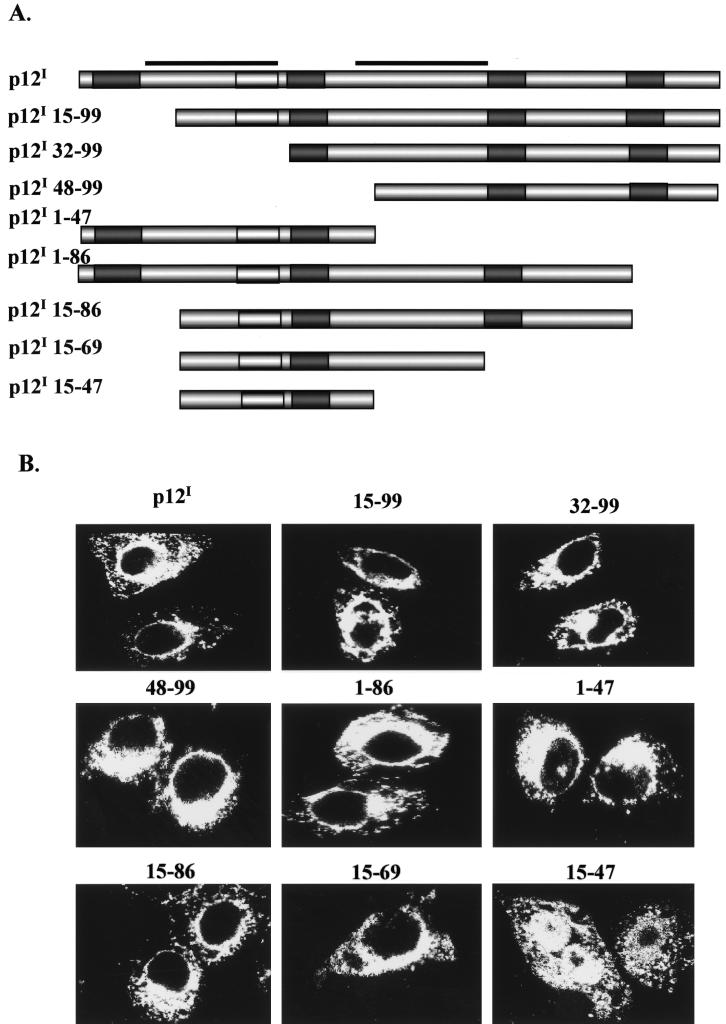
The amino acid sequence of p12I suggests the presence of two transmembrane domains (aa 12 to 32 and 48 to 68) within the two regions of p12I that independently determine cellular localization (18). To identify regions in p12I required for protein localization, eight serial deletion mutants were created (Fig. 7A). Figure 7 illustrates the serial p12I deletion mutants and highlights dileucine and PXXP motifs as well as predicted transmembrane domains. The expression and correct molecular weights of wild-type p12I and HA-tagged deletion mutants were confirmed by immunoblot assay (data not shown). The localization of the mutants was then examined by indirect immunofluorescence assay with transfected HeLa-Tat cells (Fig. 7B). Mutants p12I 1-86 and p12I 1-47 maintained the perinuclear staining typical of wild-type p12I, implying that the presence of the domain in the N-terminal half of p12I (aa 1 to 47) is sufficient for ER and cis-Golgi localization. Mutants p12I 15-99, p12I 32-99, and p12I 48-99 also maintained the wild-type protein localization, suggesting that the C-terminal half of p12I (aa 48 to 99) contained a second domain sufficient for ER and cis-Golgi localization. Mutants p12I 15-86, p12I 15-69, and p12I 15-47 represented both an N-terminal 14-aa deletion and sequential deletions in the C terminus of the protein. The mutated proteins 12I 15-86 and p12I 15-69 maintained the wild-type p12I pattern of localization. However, p12I 15-47 was diffusely expressed throughout the cell. Taken together, these data suggest that either of the two protein regions (aa 1 to 47 or 48 to 99) previously predicted to contain putative transmembrane domains (aa 12 to 32 and 48 to 68, respectively) (18) is sufficient for the viral protein to maintain ER and cis-Golgi localization. In addition, our data indicate that the N-terminal 14 aa of the protein are critical for the first transmembrane region to maintain the wild-type protein accumulation pattern.
FIG. 7.
Schematic representation and localization of p12I serial deletion mutants. (A) Schematic representation of serial p12I deletion mutants. Open box, dileucine motif; shaded box, PXXP motif; bar, transmembrane domain. (B) Localization of wild-type p12I and serial deletion p12I mutants. The images were examined at 48 h posttransfection by indirect immunofluorescence assay. The N-terminal deletion mutants p12I 15-99, p12I 32-99, and p12I 48-99 maintained the perinuclear staining typical of wild-type p12I. C-terminal deletion mutants p12I 1-86, p12I 1-47, and dual deletion mutants p12I 15-86 and p12I 15-69 also displayed perinuclear staining. The last mutant, p12I 15-47, lost the perinuclear staining pattern and was diffusely expressed.
p12I is associated with calreticulin and calnexin.
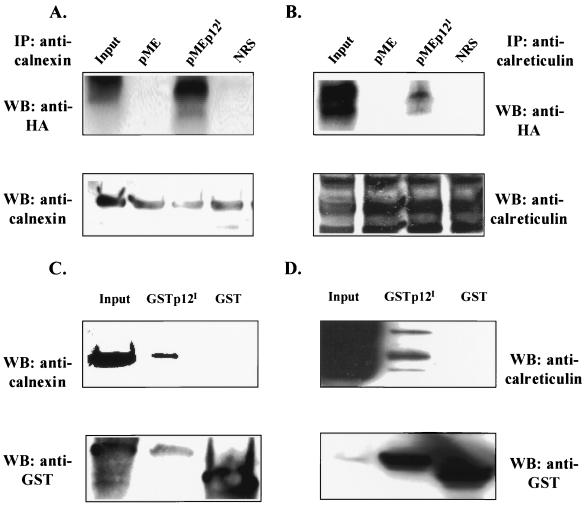
Calreticulin and calnexin, ER resident proteins, bind to a variety of viral glycoproteins, including hepatitis C virus (HCV) E1 and E2 (8), human immunodeficiency virus type 1 envelope gp160 (35), and human cytomegalovirus glycoprotein B (48) to promote protein folding. Among these, the HCV E1 and E2 proteins are retained in the ER. The fact that p12I colocalizes with calreticulin and calnexin in the ER implied that calreticulin and calnexin, as molecular chaperones, may bind to p12I. To address this question, we performed both coimmunoprecipitation and GST binding assays. Our data indicated that polyclonal anticalnexin and anticalreticulin coprecipitated p12I from transfected cells (Fig. 8A and B). Additionally, GSTp12I was able to pull down calnexin and calreticulin in the GST binding assay (Fig. 8C and 8D). The expressions of calnexin, calreticulin, GSTp12I, and GST were tested in Western blots (lower panels in Fig. 8). Normal rabbit serum did not precipitate p12I (Fig. 8A and B). The GST vector alone did not bind to either calnexin or calreticulin (Fig. 8C and D). In addition, we tested the possible association between another membrane-associated protein, the 16-kDa subunit of the vacuolar H+ ATPase, and calreticulin or calnexin. The 16-kDa subunit protein exhibited moderate binding to calnexin but failed to bind calreticulin (data not shown).
FIG. 8.
Association of p12I with calreticulin and calnexin. (A and B) Coimmunoprecipitation of p12I with calreticulin or calnexin. p12I- and vector-transfected 293T cell lysates were immunoprecipitated with rabbit polyclonal anticalnexin (A) or anticalreticulin (B), followed by SDS-PAGE separation and Western blot analysis using mouse monoclonal anti-HA. NRS, normal rabbit serum. The expression of calreticulin or calnexin in each lysate was concurrently tested by immunoblot assay using rabbit polyclonal anticalnexin (A, lower panel) or chicken polyclonal anticalreticulin (B, lower panel). (C and D) GSTp12I binds to calreticulin and calnexin. GSTp12I- and GST-transfected 293T cell lysates were incubated with glutathione-agarose beads, followed by SDS-PAGE and Western blot analysis using rabbit polyclonal anticalnexin or chicken polyclonal anticalreticulin. The membrane was stripped and was tested for the expression of GSTp12I and GST.
Deglycosylation analysis of p12I.
Both calreticulin and calnexin serve as ER chaperones and predominantly bind to N-linked glycoproteins to promote their folding. Therefore, we performed deglycosylation analysis of p12I to test whether p12I is a glycoprotein. N-glycosidase F or neuraminidase plus O-glycanase were used to enzymatically digest immunoprecipitated p12I to release possible N-linked glycans from asparagine or possible Galβ1-3GalNAc from serine or threonine residues in the protein. The electrophoretic mobility of p12I did not change following treatment with either N-glycoside F or O-glycanase (Fig. 9A), indicating that p12I is not a glycosylated protein. The N-linked glycoprotein HTLV-1 gp46, which migrated faster following treatment with N-glycosidase (Fig. 9B), served as a positive control for N-glycosidase F digestion. Taken together, our data are the first to show that a nonglycosylated viral protein colocalizes with and specifically binds to calreticulin and calnexin in the ER. Calreticulin and calnexin also function to modulate calcium storage in the ER. Taken together, our data suggest a role for p12I in cell activation through calcium-mediated cell signaling initiated from the ER.
FIG. 9.
p12I is not a glycoprotein. (A) p12I-transfected 293T cell lysates were immunoprecipitated by rabbit polyclonal anti-HA. The immune complex-bound beads were treated with either N-glycosidase F or neuraminidase plus O-glycanase overnight. The beads were boiled in SDS sample buffer, and the proteins were separated by SDS-PAGE. Expression of p12I was tested by mouse monoclonal anti-HA. (B) R49 cells, rabbit peripheral blood mononuclear cells transformed by an HTLV-1 molecular clone (ACH), were lysed and immunoprecipitated by IC11 antibody (mouse monoclonal anti-gp46). The immune-complex-bound beads were treated with either N-glycosidase F or neuraminidase plus O-glycanase overnight. The beads were boiled in SDS sample buffer, and the proteins were separated by SDS-PAGE. The expressions of HTLV-1 envelope (gp46) were tested by IC11.
DISCUSSION
In this study, we determined the subcellular accumulation of HTLV-1 p12I to further define the possible function for this protein in viral infection. We demonstrate that p12I is retained as a membrane-associated protein and is expressed predominantly in the ER and cis-Golgi apparatus. Two regions of the protein containing predicted transmembrane domains are independently responsible for this pattern of localization. Importantly, we are the first to identify the interaction of p12I with both calreticulin and calnexin in the ER, suggesting a possible function of the viral protein in calcium-mediated cell signaling leading to host cell activation. These findings are consistent with our previous studies that demonstrated a requirement for HTLV-1 p12I in viral infectivity both in a rabbit model of infection (12), in primary lymphocytes in vitro (1), and in calcium-dependent nuclear factor of activated T cells-mediated transcription (B. Albrecht et al., unpublished data).
HTLV-1 p12I is highly hydrophobic and has been shown to localize to the perinuclear region (25, 26). Consistent with these previous reports, we biochemically confirmed, using cell fractionation, that p12I was associated with cellular membranes. Since the ER and the Golgi apparatus are two major organelles present in the perinuclear region, it was likely that p12I was localized to these two compartments. We costained p12I with various subcellular markers to more specifically identify the accumulation pattern of p12I in these compartments. p12I colocalized with both calreticulin, an ER resident protein, and the Golgi apparatus. These findings were further verified by immunoelectron microscopy. Thus, our data indicate that p12I accumulation is confined principally to the ER and cis-Golgi apparatus. This tenet was further confirmed by our studies using BFA, which acts to block ADP ribosylation factor-GTP formation and therefore disrupts ER-to-Golgi protein transport. The localization of a trans-Golgi protein, AP-1, was disrupted by BFA treatment. In contrast, the accumulation pattern of p12I was unchanged by BFA treatment, suggesting that the ER and cis-Golgi apparatus are the major compartments in which the viral protein accumulates. These data were confirmed by our subcellular fractionation study. Importantly, we used three different expression plasmids, including pMEp12I, GFPp12I, and GSTp12I, to investigate the localization of p12I. Although these plasmids contain different promoters and vary in their level of expression of the tagged protein (25), they all exhibited identical patterns of p12I accumulation, indicating the strong tendency of the protein to be retained in the ER and cis-Golgi compartments.
In general, proteins that are expressed in the ER achieve their specific localization in two different but potential overlapping mechanisms: by direct retention or by retrieval from cellular compartments through recognition of specific cellular motifs. To test whether p12I maintained its localization by retention or retrieval, we used cycloheximide to block protein synthesis and tested for the expression of the protein over a 24-h period. The typical perinuclear localization of p12I did not change following either cycloheximide treatment alone or cotreatment with cycloheximide plus BFA, indicating that p12I is retained in the ER and cis-Golgi compartment. Among the most extensively studied ER localization signals is a C-terminal KDEL amino acid sequence (20), which is responsible for the localization of calreticulin to the ER (28). Two well-characterized motifs, the C-terminal KKXX sequence and the N-terminal double arginine (RR) motif, are sufficient to retain type I integral membrane protein (amino terminus in the lumen) and type II integral membrane protein (C terminus in the lumen) to the ER, respectively (20). The amino acid sequence of p12I does not contain a KDEL, KKXX, or RR motif. Therefore, the ER retention of p12I is likely related to other structural features of the protein.
To map the region required for the localization of p12I to the ER, we sequentially deleted both the amino terminus and the carboxyl terminus of p12I. Interestingly, either N-terminal mutants (p12I 15-99, p12I 32-99, p12I 48-99) or C-terminal mutants (p12I 1-86, p12I 1-47) alone maintained patterns of staining typical of the full-length protein. A computer analysis of the amino acid sequence of p12I predicted the presence of two transmembrane domains (aa 12 to 32 and 48 to 68) within these two regions (18). Similar to the findings of Koralnik et al. (26), our data indicated that two regions in p12I (aa 1 to 47 and 48 to 99) are independently sufficient for the localization. In this previous study, however, deletion of the first 12 aa from the N-terminal region of the protein did not influence the perinuclear accumulation of the protein. Our results indicate that deletion of the first N-terminal 14 aa (mutant p12I 15-47) resulted in a loss of perinuclear localization. Taken together, these data indicate the importance of aa 12 to 14 in the function of the first transmembrane domain of p12I. Ongoing work in our laboratory seeks to further define critical motifs of the protein in both localization and functional studies (1). Our data are consistent with studies which indicate the importance of transmembrane domains in the ER localization for both HCV E1 (11) and rubella virus E1 (22).
Calreticulin and calnexin each have been demonstrated to modulate calcium storage and control protein folding, including that of several viral glycoproteins, in the ER (28, 31). Our data indicate that p12I binds to each of these ER resident proteins. Within the ER, p12I may serve to modulate calcium-mediated signals involved in cell activation. Our parallel studies indicate that expression of p12I in Jurkat T cells enhances reporter gene activity mediated by the nuclear factor of activated T cells in a calcium-dependent manner (Albrecht et al., unpublished). Alternatively, these proteins may serve as molecular chaperones to regulate the folding of p12I. As molecular chaperones, both calreticulin and calnexin have been predominantly shown to bind N-linked glycoproteins. Asparagine at aa 51 of p12I is a possible N-linked glycosylation site for the viral protein. However, our deglycosylation analysis revealed neither N-linked glycosylation nor O-linked glycosylation in p12I. Further studies will be required to determine the possible role of p12I in calcium storage and release from the ER. Interestingly, Johnson et al. (23) have reported that p12I binds to the heavy chain of major histocompatibility complex (MHC) class I and prevents its association with β2-microglobulin, impairing the traffic of the protein complex. Calreticulin also acts as a chaperone in the assembly and expression of MHC class I molecules in activated human T lymphocytes (2). One potential mechanism to explain the ability of p12I to interfere with MHC class I complex transport is binding and retaining of calreticulin-MHC class I complexes in the ER or cis-Golgi.
In summary, our data illustrate that p12I is accumulated and retained in the ER and cis-Golgi apparatus. We have determined that two regions in p12I are independently sufficient for p12I localization, and we are the first to identify the association between a nonglycosylated viral protein, p12I, and resident ER proteins, suggesting a role of p12I in calcium-mediated signals during cell activation. Our data support emerging evidence for the role of p12I in early events of T-cell signaling to enhance the replicative ability of HTLV-1.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants RR-14324 from the National Center for Research Resources and CA-70529 from the National Cancer Institute, awarded through the Ohio State University Comprehensive Cancer Center. M. Lairmore is supported by an Independent Scientist Career Award from the National Institutes of Health (K02 AI01474). B. Albrecht is supported by a Boehringer Ingelheim Fonds predoctoral fellowship.
We thank Y. Rikihisa, E. Handly, J. Nisbet, and D. Dangel for technical assistance. We also thank G. Franchini, K. M. Coggeshall, C. Kredinger, P. Green, and K. Boris-Lawrie for providing valuable reagents and discussions.
REFERENCES
- 1.Albrecht B, Collins N D, Burniston M T, Nisbet J W, Ratner L, Green P L, Lairmore M D. Human T-lymphotropic virus type 1 open reading frame I p12(I) is required for efficient viral infectivity in primary lymphocytes. J Virol. 2000;74:9828–9835. doi: 10.1128/jvi.74.21.9828-9835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arosa F A, de Jesus O, Porto G, Carmo A M, de Sousa M. Calreticulin is expressed on the cell surface of activated human peripheral blood T lymphocytes in association with major histocompatibility complex class I molecules. J Biol Chem. 1999;274:16917–16922. doi: 10.1074/jbc.274.24.16917. [DOI] [PubMed] [Google Scholar]
- 3.Berneman Z N, Gartenhaus R B, Reitz M S, Blattner W A, Manns A, Hanchard B, Ikehara O, Gallo R C, Klotman M E. Expression of alternatively spliced human T-lymphotropic virus type 1 pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc Natl Acad Sci USA. 1992;89:3005–3009. doi: 10.1073/pnas.89.7.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom G S, Brashear T A. A novel 58-kDa protein associates with the Golgi apparatus and microtubules. J Biol Chem. 1989;264:16083–16092. [PubMed] [Google Scholar]
- 5.Cereseto A, Berneman Z, Koralnik I, Vaughn J, Franchini G, Klotman M E. Differential expression of alternatively spliced pX mRNAs in HTLV-I-infected cell lines. Leukemia. 1997;11:866–870. doi: 10.1038/sj.leu.2400665. [DOI] [PubMed] [Google Scholar]
- 6.Chatton B, Bahr A, Acker J, Kedinger C. Eukaryotic GST fusion vector for the study of protein-protein associations in vivo: application to interaction of ATFa with Jun and Fos. BioTechniques. 1995;18:142–145. [PubMed] [Google Scholar]
- 7.Chou J H, Jahn R. Binding of Rab3A to synaptic vesicles. J Biol Chem. 2000;275:9433–9440. doi: 10.1074/jbc.275.13.9433. [DOI] [PubMed] [Google Scholar]
- 8.Choukhi A, Ung S, Wychowski C, Dubuisson J. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J Virol. 1998;72:3851–3858. doi: 10.1128/jvi.72.5.3851-3858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciminale V, D'Agostino D, Zotti L, Franchini G, Felber B K, Chieco-Bianchi L. Expression and characterization of proteins produced by mRNAs spliced into the X region of the human T-cell leukemia/lymphotropic virus type II. Virology. 1995;209:445–456. doi: 10.1006/viro.1995.1277. [DOI] [PubMed] [Google Scholar]
- 10.Ciminale V, Pavlakis G N, Derse D, Cunningham C P, Felber B K. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J Virol. 1992;66:1737–1745. doi: 10.1128/jvi.66.3.1737-1745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocquerel L, Duvet S, Meunier J C, Pillez A, Cacan R, Wychowski C, Dubuisson J. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J Virol. 1999;73:2641–2649. doi: 10.1128/jvi.73.4.2641-2649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins N D, Newbound G C, Albrecht B, Beard J L, Ratner L, Lairmore M D. Selective ablation of human T-cell lymphotropic virus type 1 p12(I) reduces viral infectivity in vivo. Blood. 1998;91:4701–4707. [PubMed] [Google Scholar]
- 13.Collins N D, Newbound G C, Ratner L, Lairmore M D. In vitro CD4+ lymphocyte transformation and infection in a rabbit model with a molecular clone of human T-cell lymphotropic virus type 1. J Virol. 1996;70:7241–7246. doi: 10.1128/jvi.70.10.7241-7246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekaban G A, Peters A A, Mulloy J C, Johnson J M, Trovato R, Rivadeneira E, Franchini G. The HTLV-I orfI protein is recognized by serum antibodies from naturally infected humans and experimentally infected rabbits. Virology. 2000;274:86–93. doi: 10.1006/viro.2000.0406. [DOI] [PubMed] [Google Scholar]
- 15.Felber B K, Paskalis H, Kleinmanewing C. The pX protein of HTLV-1 is a transcriptional activator of its long terminal repeats. Science. 1985;229:675–679. doi: 10.1126/science.2992082. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira O C, Jr, Planelles V, Rosenblatt J D. Human T-cell leukemia viruses: epidemiology, biology, and pathogenesis. Blood Rev. 1997;11:91–104. doi: 10.1016/s0268-960x(97)90015-1. [DOI] [PubMed] [Google Scholar]
- 17.Franchini G. Molecular mechanisms of human T-cell leukemia/lymphotropic virus type I infection. Blood. 1995;86:3619–3639. [PubMed] [Google Scholar]
- 18.Franchini G, Mulloy J C, Koralnik I J, Lo Monico A, Sparkowski J J, Andresson T, Goldstein D J, Schlegel R. The human T-cell leukemia/lymphotropic virus type I p12I protein cooperates with the E5 oncoprotein of bovine papillomavirus in cell transformation and binds the 16-kilodalton subunit of the vacuolar H+ ATPase. J Virol. 1993;67:7701–7704. doi: 10.1128/jvi.67.12.7701-7704.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y S, Alvarez C, Nelson D S, Sztul E. Molecular cloning, characterization, and dynamics of rat formiminotransferase cyclodeaminase, a Golgi-associated 58-kDa protein. J Biol Chem. 1998;273:33825–33834. doi: 10.1074/jbc.273.50.33825. [DOI] [PubMed] [Google Scholar]
- 20.Gomord V, Wee E, Faye L. Protein retention and localization in the endoplasmic reticulum and the Golgi apparatus. Biochimie. 1999;81:607–618. doi: 10.1016/s0300-9084(99)80118-7. [DOI] [PubMed] [Google Scholar]
- 21.Goud B, McCaffrey M. Small GTP-binding proteins and their role in transport. Curr Opin Cell Biol. 1991;3:626–633. doi: 10.1016/0955-0674(91)90033-u. [DOI] [PubMed] [Google Scholar]
- 22.Hobman T C, Lemon H F, Jewell K. Characterization of an endoplasmic reticulum retention signal in the rubella virus E1 glycoprotein. J Virol. 1997;71:7670–7680. doi: 10.1128/jvi.71.10.7670-7680.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson J M, Mulloy J C, Ciminale V, Fullen J, Nicot C, Franchini G. The MHC class I heavy chain is a common target of the small proteins encoded by the 3′ end of HTLV type 1 and HTLV type 2. AIDS Res Hum Retrovir. 2000;16:1777–1781. doi: 10.1089/08892220050193308. [DOI] [PubMed] [Google Scholar]
- 24.Kimata J T, Wong F, Wang J, Ratner L. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology. 1994;204:656–664. doi: 10.1006/viro.1994.1581. [DOI] [PubMed] [Google Scholar]
- 25.Koralnik I J, Fullen J, Franchini G. The p12I, p13II, and p30II proteins encoded by human T-cell leukemia/lymphotropic virus type 1 open reading frames I and II are localized in three different cellular compartments. J Virol. 1993;67:2360–2366. doi: 10.1128/jvi.67.4.2360-2366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koralnik I J, Mulloy J C, Andresson T, Fullen J, Franchini G. Mapping of the intermolecular association of the human T cell leukaemia/lymphotropic virus type I p12(I) and the vacuolar H+- ATPase 16 kDa subunit protein. J Gen Virol. 1995;76:1909–1916. doi: 10.1099/0022-1317-76-8-1909. [DOI] [PubMed] [Google Scholar]
- 27.Koya R C, Fujita H, Shimizu S, Ohtsu M, Takimoto M, Tsujimoto Y, Kuzumaki N. Gelsolin inhibits apoptosis by blocking mitochondrial membrane potential loss and cytochrome c release. J Biol Chem. 2000;275:15343–15349. doi: 10.1074/jbc.275.20.15343. [DOI] [PubMed] [Google Scholar]
- 28.Krause K H, Michalak M. Calreticulin. Cell. 1997;88:439–443. doi: 10.1016/s0092-8674(00)81884-x. [DOI] [PubMed] [Google Scholar]
- 29.Kusuhara K, Anderson M, Pettiford S M, Green P L. Human T-cell leukemia virus type 2 Rex protein increases stability and promotes nuclear to cytoplasmic transport of gag/pol and env RNAs. J Virol. 1999;73:8112–8119. doi: 10.1128/jvi.73.10.8112-8119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mesnard J M, Devaux C. Multiple control levels of cell proliferation by human T-cell leukemia virus type 1 tax protein. Virology. 1999;257:277–284. doi: 10.1006/viro.1999.9685. [DOI] [PubMed] [Google Scholar]
- 31.Michalak M, Corbett E F, Mesaeli N, Nakamura K, Opas M. Calreticulin: one protein, one gene, many functions. Biochem J. 1999;344:281–292. [PMC free article] [PubMed] [Google Scholar]
- 32.Moss J, Vaughan M. Activation of toxin ADP-ribosyltransferases by eukaryotic ADP-ribosylation factors. Mol Cell Biochem. 1999;193:153–157. [PubMed] [Google Scholar]
- 33.Mulloy J C, Crowley R W, Fullen J, Leonard W J, Franchini G. The human T-cell leukemia/lymphotropic virus type 1 p12I protein binds the interleukin-2 receptor β and γc chains and affects their expression on the cell surface. J Virol. 1996;70:3599–3605. doi: 10.1128/jvi.70.6.3599-3605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuveut C, Jeang K T. HTLV-I Tax and cell cycle progression. Prog Cell Cycle Res. 2000;4:157–162. doi: 10.1007/978-1-4615-4253-7_14. [DOI] [PubMed] [Google Scholar]
- 35.Otteken A, Moss B. Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperone function similar to that of calnexin. J Biol Chem. 1996;271:97–103. doi: 10.1074/jbc.271.1.97. [DOI] [PubMed] [Google Scholar]
- 36.Palker T, Tanner M, Scearce R, Streilen R, Clark M, Haynes B. Mapping of immunogenic regions of human T-cell leukemia virus type 1 (HTLV-I) gp46 and gp21 envelope glycoproteins with Env-encoded synthetic peptides and a monoclonal antibody to gp46. J Immunol. 1989;142:971–978. [PubMed] [Google Scholar]
- 37.Pique C, Ureta-Vidal A, Gessain A, Chancerel B, Gout O, Tamouza R, Agis F, Dokhelar M C. Evidence for the chronic in vivo production of human T cell leukemia virus type I Rof and Tof proteins from cytotoxic T lymphocytes directed against viral peptides. J Exp Med. 2000;191:567–572. doi: 10.1084/jem.191.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puertollano R, Alonso M A. Substitution of the two carboxyl-terminal serines by alanine causes retention of MAL, a component of the apical sorting machinery, in the endoplasmic reticulum. Biochem Biophys Res Commun. 1999;260:188–192. doi: 10.1006/bbrc.1999.0876. [DOI] [PubMed] [Google Scholar]
- 39.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodroski J G, Rosen C A, Haseltine W A. Trans-acting transcriptional activation of the long terminal repeat of human T-lymphotropic viruses in infected cells. Science. 1984;223:381–385. doi: 10.1126/science.6330891. [DOI] [PubMed] [Google Scholar]
- 41.Takebe Y, Seiki M, Fujisawa J I, Hoy P, Yokota K, Arai K I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trovato R, Mulloy J C, Johnson J M, Takemoto S, Deoliveira M P, Franchini G. A lysine-to-arginine change found in natural alleles of the human T-cell lymphotropic leukemia virus type 1 p12 protein greatly influences its stability. J Virol. 1999;73:6460–6467. doi: 10.1128/jvi.73.8.6460-6467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai S C, Adamik R, Haun R S, Moss J, Vaughan M. Effects of brefeldin A and accessory proteins on association of ADP-ribosylation factors 1, 3, and 5 with Golgi. J Biol Chem. 1993;268:10820–10825. [PubMed] [Google Scholar]
- 44.Uchiyama T. Human T cell leukemia virus type I (HTLV-I) and human diseases. Annu Rev Immunol. 1997;15:15–37. doi: 10.1146/annurev.immunol.15.1.15. [DOI] [PubMed] [Google Scholar]
- 45.Wilson A L, Erdman R A, Maltese W A. Association of Rab1B with GDP-dissociation inhibitor (GDI) is required for recycling but not initial membrane targeting of the Rab protein. J Biol Chem. 1996;271:10932–10940. doi: 10.1074/jbc.271.18.10932. [DOI] [PubMed] [Google Scholar]
- 46.Xia W, Zhang J, Ostaszewski B L, Kimberly W T, Seubert P, Koo E H, Shen J, Selkoe D J. Presenilin 1 regulates the processing of beta-amyloid precursor protein C-terminal fragments and the generation of amyloid beta-protein in endoplasmic reticulum and Golgi. Biochemistry. 1998;37:16465–16471. doi: 10.1021/bi9816195. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Kang D E, Xia W, Okochi M, Mori H, Selkoe D J, Koo E H. Subcellular distribution and turnover of presenilins in transfected cells. J Biol Chem. 1998;273:12436–12442. doi: 10.1074/jbc.273.20.12436. [DOI] [PubMed] [Google Scholar]
- 48.Zheng Z, Maidji E, Tugizov S, Pereira L. Mutations in the carboxyl-terminal hydrophobic sequence of human cytomegalovirus glycoprotein B alter transport and protein chaperone binding. J Virol. 1996;70:8029–8040. doi: 10.1128/jvi.70.11.8029-8040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]