Summary
Insights into the role of ubiquitin-dependent signaling in the regulation of apoptosis have provided one of the most significant breakthroughs in recent years for cell death research. It has been revealed that all steps in the apoptotic cascade, including transcriptional regulation of apoptotic gene expression, outer mitochondrial membrane permeabilization and caspase activation, are under the control of the ubiquitin/proteasome system. This makes ubiquitin signaling one on the most critical life and death decision checkpoints in mammalian cells. Here we discuss the ubiquitylation-dependent regulation of the mitochondrial steps in apoptosis, with a focus on the role of regulated protein degradation in this process. The newly identified ubiquitylation-dependent processes in the Bcl-2 family-regulated outer mitochondrial membrane permeabilization, as well as the role of mitochondria-associated E3 ubiquitin ligases and other molecular components of the ubiquitin/proteasome system in the control of mitochondrial steps in apoptosis, are discussed.
Keywords: apoptosis, mitochondria, Bcl-2, ubiquitin, proteasome, E3 ubiquitin ligase
1. The ubiquitin/proteasome system (UPS): an overview
In the late 1970s and early 1980s Aaron Ciechanover, Avram Hershko and Irvin Rose made the groundbreaking observation that the highly conserved 76-amino acid peptide ubiquitin (Ub) serves as a tag in the process of lysosome-independent degradation of harmful or superfluous cellular proteins [1–3]. Following the initial findings, the cascade that mediates Ub conjugation to the target proteins was defined. Ubiquitylation occurs through a process involving Ub-activating (E1), Ub-conjugating (E2) and Ub-ligating (E3) proteins. E3 Ub ligases can be classified into two families with distinct modes of action: HECT domain (Homologous to the E6-AP Carboxyl Terminus; e.g. mammalian NEDD4/NEDD-like family of Ub ligases) E3 Ub ligases that transfer activated Ub from an E2 enzyme via a thioester intermediate to an acceptor lysine in the substrate protein that can also be another ubiquitin moiety, and RING (really interesting new gene)-type E3 Ub ligases which facilitate the direct transfer of activated Ub from the E2 enzyme onto the substrate. While K48-linked poly-Ub chains act as a signal for proteasomal degradation [2, 3], other types of ubiquitylation, K63-linked poly-Ub chains or monoubiquitylation in particular, are also involved in degradation-independent regulation of proteins. Moreover, in order to turn off the Ub signal, Ub can be removed from the substrate proteins by deubiquitinating enzymes (DUBs) [4–6]. Thus, Ub conjugation shares significant similarities with protein phosphorylation in regards to its reversibility. Poly- and monoubiquitylation are involved in organelle-specific regulation of a vast number of cellular processes ranging from autophagy and selective protein degradation to DNA repair and membrane transport [7–11]. Specificity in the process of ubiquitylation is achieved by the vast number of Ub ligases, many of which are anchored in different subcellular compartments and function in an organelle specific manner [11–13].
The accumulating evidence shows that Ub-conjugation and highly regulated proteasome-dependent degradation of many distinct proteins are key to apoptosis progression. Notably, all steps in the cascade leading to apoptosis including, (i) outer mitochondrial membrane (OMM) permeabilization, (ii) caspase activation, as well as (iii) transcriptional regulation of apoptotic gene expression, are under control of the Ub/proteasome system (UPS), making Ub-dependent signaling a critical component for life and death in mammalian cells.
This review examines information on the mechanisms and prevalence of Ub-dependent regulated protein degradation in the control of distinct steps in Bcl-2 protein family-mediated apoptosis. The recent evidence linking non-Bcl-2 family proteins and their Ub-dependent regulation to the progression of mitochondrial steps in apoptosis will be also discussed in detail. Other aspects of UPS-mediated regulation of apoptosis, including caspase activation and apoptotic signaling in the nucleus have been extensively reviewed by others [14–17], and therefore are not discussed here.
2. The ubiquitin/proteasome system (UPS) is an essential regulator of apoptosis
Apoptosis, a genetically driven form of cell death, results in an organized removal of dying cells (reviewed in [14, 18–20]). To achieve this, the apoptotic cell death program concludes in an organized dismantling of the dying cell that is facilitated by a family of evolutionarily conserved aspartate-specific cysteine-dependent proteases, called caspases [14, 20, 21].
Caspases are synthesized as pro-enzymes that require proteolytic activation at Asp-X sites to produce the mature, catalytically active forms. This activation occurs in an ordered manner, where initiator caspases (e.g. caspases-2, -8 and -9) after being activated by distinct pro-apoptotic stimuli, cleave inactive forms of effector caspases (e.g. caspases-3, -6 and -7), thereby activating them. Given the widespread expression of pro-caspases that have a potential to be activated in proteolytic cascades, various control mechanisms preventing inappropriate caspase activation have evolved. Ubiquitylation-dependent regulation of caspases is one of the mechanisms controlling unwanted caspase activation [14, 15, 22–24]. Until a certain threshold of an apoptosis-inducing signal is reached, inhibitor-of-apoptosis proteins (IAPs), suppress the activity of caspases and control potentially deleterious inadvertent activation of the caspase cascade [22–24]. IAPs carry N-terminal BIR (baculovirus IAP repeat) protein interaction domains and C-terminal RING domains that provide E3 Ub ligase activity. Through their BIR motifs, IAPs can bind to caspases and directly inactivate them [14, 22]. In some cases IAPs mediate caspase ubiquitylation and thereby target them for proteasomal degradation. The IAP-mediated ubiquitylation of caspases, as well as Ub-regulated activation of pro- and anti-survival genes and their roles in apoptosis regulation has been reviewed [14–17] and will therefore not be discussed here in detail.
A dynamic balance between anti- and pro-apoptotic proteins from the Bcl-2 family serves as another caspase activation checkpoint in many modes of stress-induced apoptosis. The main cellular targets of the Bcl-2 family are mitochondrial membranes, the OMM in particular. Bcl-2 proteins can either induce OMM permeabilization and thus promote apoptosis (e.g. Bax, Bak and Bok) or inhibit it thus promoting cell survival (e.g. Bcl-2, Bcl-xL, Mcl-1) [18–20] (see Figure 1). The ratio of pro-apoptotic versus anti-apoptotic Bcl-2 family members is a critical factor in regulating OMM permeabilization and subsequent caspase activation. As in the case of caspases, various mechanisms, including regulation of protein expression, as well as diverse posttranslational modifications regulate the balance of the activities of pro- and anti-apoptotic Bcl-2 family proteins in healthy cells, and upon induction of apoptosis [19, 20, 25]. Progression of Bcl-2 family-mediated mitochondrial steps in apoptosis also relies on polyubiquitylation-mediated regulated protein degradation (Figure 2).
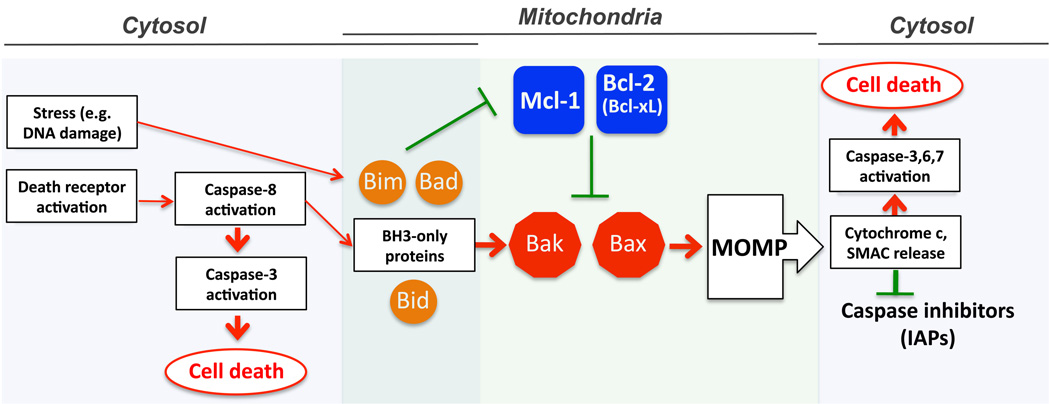
Figure 1. Apoptosis cascade: an overview.
Critical steps in the cascade leading to apoptosis activation, (1) activation (2) outer mitochondrial membrane permeabilization and (3) caspase activation. The activation stages of intrinsic (stress-induced) and extrinsic (death ligand-induced) apoptotic pathways occur in extramitochondrial compartments of the cell (cytosol). Pro-apoptotic signals are then transferred through the activities of BH3-domain only Bcl-2 family proteins (e.g. Bid, Bad and Bim) to the mitochondria, where multi-BH domain Bcl-2 family proteins regulate the outer mitochondrial membrane permeability. Apoptosis is activated or enchanced upon permeabilization of the outer mitochondrial membrane and release of proteins, including cytchrome c, SMAC/Diablo (SMAC) from the intermembrane space to the cytosol. The subsequent steps of apoptosis signaling, including cytochrome c-dependent activation of caspase-9, and SMAC/Diablo-mediated inhibition of caspase inhibitors (IAPs) occur in the cytosol.
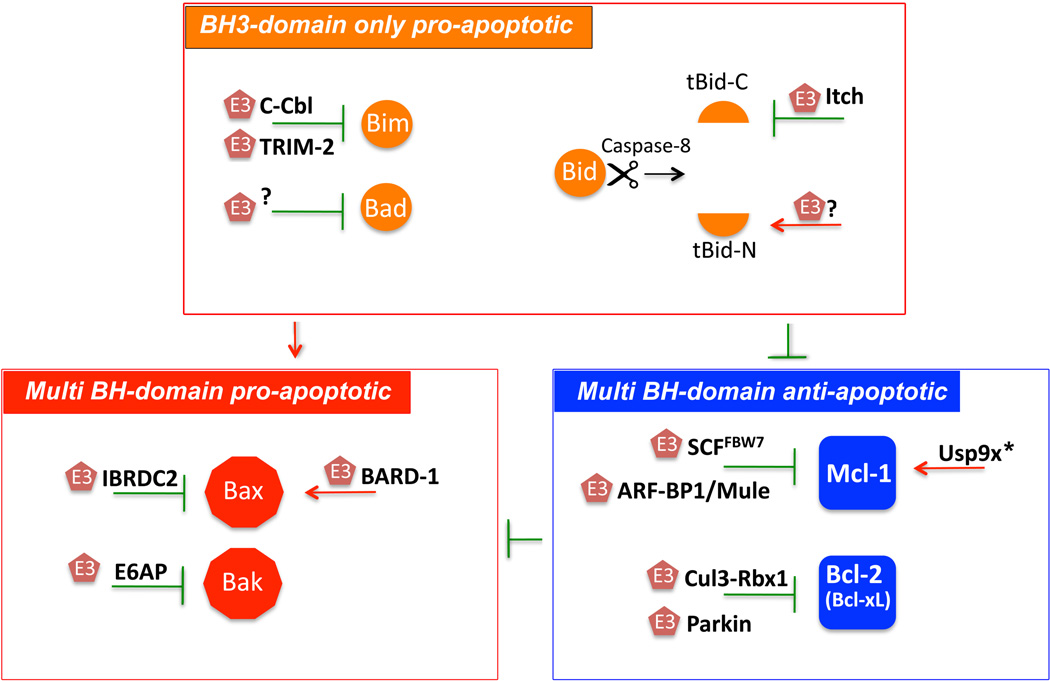
Figure 2. UPS-dependent regulation of Bcl-2 family protein levels.
The UPS regulates the ratio of pro- and anti-apoptotic proteins in Bcl-2 family. Distinct E3 Ub ligases (E3) are either activated or inhibited upon induction of apoptosis. This leads to a regulated degradation of anti-apoptotic Bcl-2 proteins (e.g. Mcl-1 or Bcl-2) or stabilization of pro-apoptotic Bcl-2 family proteins (e.g. Bim or Baxβ) and thereby activation of downstreamcell death program. Red arrows: depict activation of respective proteins by specific E3 Ub ligases (E3). Green “Ts” depict inhibition of respective proteins by specific E3 Ub Ligases (E3). *Usp9x is a deubiquitinase (DUB) that inhibits Mcl-1 degradation. For details see text and Table1.
3. Ubiquitylation-dependent control of mitochondrial steps in apoptosis
3.1 Overview of mitochondrial steps in apoptosis
As discussed above, mitochondria serve as a signaling platform critical for the synchronization and translation of pro-survival and pro-death signals initiated in various compartments of the cell. The so-called BH3 (Bcl-2-homology 3)-only proteins (e.g. Bad, Bid, Bim, Blk, Bmf, Noxa and Puma) are normally sequestered in different cellular compartments, but upon apoptotic stimuli, translocate to mitochondria. This translocation leads to changes in the balance of pro- and anti-apoptotic activities of multi-BH-domain Bcl-2 family proteins already present on the OMM. These include the anti-apoptotic proteins: Bcl-2, Bcl-xL, Bfl-1/A1, Bcl-w and Mcl-1 as well as the pro-apoptotic Bcl-2-family proteins: Bak and Bok. Bax, another pro-apoptotic multi-BH-domain protein traffics between the cytosol and the OMM in healthy cells, and in cells destined to die, accumulates on the OMM where it initiates permeabilization of the OMM. Although the mechanism of OMM permeabilization is still under evaluation, this event leads to the release of a number of proteins, including cytochrome c, AIF (apoptosis inducing factor) and SMAC/Diablo, from the mitochondrial intermembrane space (IMS) into the cytosol. Mitochondrial permeabilization and the release of aforementioned proteins are requisite for a subsequent activation of caspases in mammalian cells. While cytochrome c acts as a direct activator of caspase-9 as part of the apoptosome, SMAC/Diablo eliminates IAP-dependent caspase inhibition. The accumulating evidence indicates that the balance between pro- and anti-apoptotic Bcl-2 family proteins is controlled by the UPS (Figure 2, Table1). Importantly, this includes regulated degradation of different Bcl-2 family subclasses, BH3-only proteins and multi-BH domain pro- and anti-apoptotic factors, and therefore regulation of distinct steps of the process (Figure 2), making the UPS a critical checkpoint factor of mitochondrial apoptosis progression.
Table 1.
Effectors of mitochondrial steps in apoptosis and the UPS-related factors regulating them
| Protein | Function/Category | E3 Ub ligases/ DUBs regulating mitochondrial actions of these proteins |
The UPS-dependent effect on mitochondrial steps in apoptosis |
References |
|---|---|---|---|---|
| Bim | Bcl-2-family BH3-only pro-apoptotic protein | cCbl &TRIM2/? | Pro-apoptotic conditions stabilize Bim and facilitate OMM permeabilization. | [58, 59, 61, 62] |
| tBid | Bcl-2-family BH3-only pro-apoptotic protein | Itch/? | tBid is degraded in apoptotic cells; the UPS-mediated tBid degradation has a potential role in fine-tuning of pro-apoptotic signal. | [54–56] |
| Bad | Bcl-2-family BH3-only pro-apoptotic protein | ?/? | c-RAF kinase-dependent phosphorylation of Bad induces its degradation; a mechanism limiting pro-apoptotic activity of Bad. | [57] |
| Bax | Bcl-2-family multi BH-domain pro-apoptotic protein; execution of OMM permeabilization | IBRDC2/? | IBRDC2 mediates removal of apoptotically active form of Baxα, a mechanism preventing inadvertent Baxα-induced OMM permeabilization. Also, Baxβ is kept at low levels in healthy cells via continuous UPS-dependent degradation. Baxβ degradation is inhibited upon induction of apoptosis. | [68–72] |
| Bak | Bcl-2-family multi BH-domain pro-apoptotic protein; execution of OMM permeabilization | E6AP/? | Human papiloviruses-encoded E6 oncogenes highjack E6AP to induce USP-mediated Bak degradation leading to inhibition of apoptosis enabling viral proliferation. | [84, 85] |
| Mcl-1 | Bcl-2-family anti-apoptotic protein | ARF-BP1/Mule and SCFFBW7/ Usp9x | A short half-life protein; rapidly degraded upon induction of apoptosis. Degradation of Mcl1 facilitates apoptosis. | [26–30]. |
| Bcl-2 | Bcl-2-family anti-apoptotic protein | Parkin&Cul3Rbx1/? | Accumulation and degradation of Bcl-2, mediated by the USP-dependent mechanism correlates with cell sensitivity to apoptosis. Parkin mono-ubiqutinates Bcl-2 and thereby stabilizes this protein. | [37–40, 48] |
| BARD1 | DNA repair factor in the nucleus; also associated with the OMM | BARD1 is an E3 Ub ligase | Mitochondia-associated subset of BARD1 stimulates Bax oligomerization and the OMM permeabilization. | [79] |
| MOAP-1 | Mitochondria associated; pro-apoptotic protein | ?/? | A short half-life protein; MOAP-1 is kept at low levels through the USP-dependent degradation. Apoptosis-induced accumulation of MOAP-1 induces Bax-dependent OMM permeabilization. | [81] |
| ARTS | Mitochondria associated; pro-apoptotic protein | ?/? | A short half-life protein; High levels of ARTS promotes apoptosis. Under normal growth conditions ARTS levels are kept low through the UPS-dependent degradation. | [43] |
| p97 | AAA-ATPase | ?/? | Required for the extraction of Mcl-1 from the OMM prior to the proteasomal degradation in the cytosol. | [31] |
3.2 UPS and activation of mitochondrial steps in apoptosis
3.2.1 Pro-apoptotic role of regulated degradation of Mcl-1
While little is known whether Bcl-2 and Bcl-xL turnover regulation serves as a checkpoint in apoptosis control, extensive evidence points to the proteasomal degradation of Mcl-1, another multi-BH domain anti-apoptotic protein, being an important pro-apoptotic event in many modes of cell death, allowing cells to rapidly switch from pro-survival to apoptosis mode [26–30]. The physiological importance of Mcl-1 turnover regulation is strongly supported by data showing that increased stability and abnormal cellular accumulation of Mcl-1 is linked to resistance to apoptosis and tumour development. For example, overexpression of Mcl-1 is linked to poor prognosis in lymphotic leukemias, multiple myeloma and breast cancer [26–28].
Unlike most Bcl-2 family proteins that are relatively stable, Mcl-1 has a short half-life of 40 to 60 min [29–31]. This unstable nature of Mcl-1 is utilized by various stress signals that can induce rapid downregulation of Mcl-1, occurring through both activation of proteasomal Mcl-1 degradation and inhibition of Mcl-1 synthesis. Distinct E3 Ub ligases regulate proteasomal degradation of Mcl1. Mule (Mcl-1 Ub ligase E3) [26], also known as LASU [32], a BH-3 domain-containing HECT E3 Ub ligase, was shown to control Mcl-1 degradation in DNA-damage-induced apoptosis [26]. ARF-BP1/Mule interacts with Mcl-1 through its BH3-domain [26, 32] and mediates Mcl-1 K48-linked polyubiquitylation, thereby tagging Mcl-1 for proteasomal degradation. Consistently, downregulation of ARF-BP1/Mule by RNA interference inhibited stress-induced Mcl-1 protein degradation and suppressed DNA-damage induced apoptosis [26]. SCFFBW7 (Skp1-Cullin1-F-box complex containing the F-box substrate recognition factor FBW7), another E3 Ub ligase, also interacts with and targets Mcl-1 for proteasomal degradation [27]. SCFFBW7-mediated degradation of Mcl-1 depends on the Mcl-1 phosphorylation level. Mcl-1 is phosphorylated by GSK3 (glycogen synthase kinase 3) mainly at S159 and T163 residues [27], and mutation of these sites not only inhibited Ub-dependent degradation of Mcl-1, but also blocked interaction of this protein with FBW7, indicating that Mcl-1 phosphorylation might serve as a signal for SCFFBW7 recruitment.
As in the case of ARF-BP1/Mule, depletion of distinct components of the SCFFBW7 complex, including FBW7, Cullin1 and Skp1, led to increases in the levels of Mcl-1 in cultured cells, thymus and thymic lymphomas [27] and increased resistance to apoptosis. Mcl-1 degradation in response to DNA-damaging agents was also hampered in FBW7−/− cells [27], confirming a physiological role for SCFFBW7 complex in promoting stress-induced Mcl-1 degradation.
The tissue specific and redundant roles for ARF-BP1/Mule and SCFFBW7 in Mcl-1 turnover regulation need to be established. However, the lack of correlation between ARF-BP1/Mule and Mcl-1 expression in T-cell acute lymphoblastic leukemia cells [27] suggests that this protein might regulate Mcl-1 degradation in other cell types. Alternatively, these two different E3 Ub ligases could respond to distinct pro-apoptotic triggers.
The efficiency of stress-induced degradation of Mcl-1 is also controlled by recently identified deubiquitinase, Usp9x (Ub-specific peptidase 9x). Usp9x removes the K48 poly-Ub chains from the Mcl-1 and as a consequence hampers proteasomal degradation of this protein in healthy cells [28]. Upon DNA damage, Usp9x activity is inhibited, allowing a more rapid Mcl-1 degradation thus facilitating the apoptotic program [28]. This occurs through the control of the Usp9x/Mcl-1 interaction and depends on Mcl-1 phosphorylation. Since mutations of Mcl-1 phosphorylation sites (S155A, S159A and T163A) stabilized or enhanced the interaction of Usp9x with Mcl-1, a dephosphorylated form of Mcl-1 appears to be preferentially recruiting Usp9x [28]. As discussed above, these conditions are antagonistically regulating the physical interaction between Mcl-1 and SCFFBW7 [27]. Thus upon stress-induced Mcl-1 dephosphorylation, SCFFBW7 is replaced by Usp9x providing a fast response to a changing pro-survival or pro-death cellular environment. As in a case of SCFFBW7 [27], Usp9x and Mcl-1 protein levels in human follicular lymphomas and in normal tissues are also highly correlated [28]. Whether in cells destined to die, Usp9x is also displaced by ARF-BP1/Mule is not known.
In addition to ARF-BP1/Mule-, SCFFBW7- and Usp9x-mediated direct regulation of Mcl-1 ubiquitylation, VCP/p97 (valosin-containing protein/p97; hereafter referred to as p97) is also required for homeostatic and stress-induced proteasomal degradation of Mcl-1 [31]. p97 is a ubiquitously expressed member of the AAA-ATPase (ATPases associated with diverse cellular activities) family of proteins. p97, as well as the yeast homologue CDC48, is an essential component of a wide range of Ub-associated biological pathways, including UPS-mediated protein degradation in the ER-associated degradation (ERAD) pathway [33–36]. p97 mediates the retrotranslocation of polypeptides from the ER and mitochondrial membranes to the cytosol prior to their proteasomal degradation [33, 34]. Removal of ubiquitylated Mcl-1 from the OMM and delivery to the proteasome is also mediated by p97 [31]. Consistent with this, p97 downregulation or expression of an ATPase hydrolysis-deficient mutant (p97QQ) stabilized OMM-associated Mcl-1 [31]. In addition, an in vitro reconstitution assay revealed that p97 is recruited to the OMM and required for extraction of Mcl-1 from this membrane [31].
Whether other Bcl-2 family proteins are also under the control of p97, as well as the role(s) for Mcl-1-specific E3 Ub ligases and Usp9x in mitochondrial recruitment of p97 remains to be established. However, since inhibition of p97 led to prominent stabilization of Mcl-1, but had a marginal effect on cellular sensitivity to apoptosis, (MK lab, unpublished observation) one might expect that p97 controls not only Mcl-1 but also other regulators of apoptosis.
3.2.2 UPS-mediated degradation of Bcl-2
Turnover of Bcl-2 (B-cell CLL/lymphoma 2), the founding member of the Bcl-2 family of proteins, is also regulated by the UPS [37–40]. Cytosolic inhibitor of Nrf2 (INRF2) functions as an adaptor for Cul3-Rbx1-mediated ubiquitylation of Bcl-2. Since ectopic expression of INRF2 in HEPA1 and HEK293 cells reduced Bcl-2 levels, while downregulation or mutation of INFR2 caused Bcl-2 accumulation [39], one might conclude that INRF2-regulated Bcl-2 degradation functions as an apoptosis-promoting factor. However, it is not clear whether, as in the case of Mcl-1, pro-apoptotic conditions accelerate INRF2- and UPS-dependent Bcl-2 degradation. The activity of INRF2 toward Bcl-2 correlated with cellular sensitivity to apoptosis and depended on the oxidative status of the cell [39], suggesting that INRF2 might specifically contribute to progression of oxidative stress-regulated modes of cell death. Furthermore, recent evidence suggests that under some conditions Bcl-2 degradation might serve as a dynamic apoptosis checkpoint mechanism.
For example, other apoptosis-related factors mediating UPS-dependent Bcl-2 turnover were identified, including a mitochondria-associated pro-apoptotic protein, ARTS. Expression of ARTS is lost in lymphoblasts of over 70% of childhood acute lymphoblastic leukemia patients [41], suggesting that ARTS can function as a tumor suppressor in leukemia with an important role in the pathogenesis of childhood acute lymphoblastic leukemia. On the other hand, overexpression of ARTS was reported in astrocyte tumors [42], indicating the lack of general correlation between tumor development and ARTS expression. One possible mechanism of ARTS-mediated apoptosis is its role in the degradation of anti-apoptotic Bcl-2 family proteins [43]. This notion is supported by data showing reduced mitochondrial levels of Bcl-xL and Bcl-2 in cells stably or transiently overexpressing ARTS [43]. Since caspase inhibition did not block this decrease, it has been proposed that ARTS might control Bcl-xL and Bcl-2 levels directly [43]. However, further studies are needed to verify this possibility. Interestingly, cellular levels of ARTS are also regulated through the UPS [43]. Like Mcl-1, ARTS has a short half-life of ~30 min in healthy cells and its levels are kept low through continuous Ub-mediated degradation [43]. Upon induction of apoptosis, ubiquitylation of ARTS is inhibited causing the stabilization of ARTS and subsequent activation of the mitochondrial cell death pathway. Given that ARTS resides in the IMS [44], the mechanism of how the UPS regulates this protein, including potential E3 Ub ligases, as well as how ARTS is exported from the IMS to the cytosol prior to proteasomal degradation needs to be established.
Of note, activation of ARTS leads to decreased XIAP (X-linked inhibitor of apoptosis protein) protein levels, caspase-3 activation and cell death [44], suggesting a role for ARTS in the regulation of the USP-dependent XIAP turnover. In mammalian cells, ARTS is released from mitochondria upon pro-apoptotic stimuli and then binds to XIAP [44], indicating that in addition to Bcl-2 (and Bcl-xL) degradation, ARTS binding to and inhibition of XIAP is related to its pro-apoptotic function. One possibility is that ARTS targets distinct E3 Ub ligases with specificity towards XIAP and Bcl-2 and therefore is implicated in different levels of apoptosis regulation.
Parkin, the mitochondria-acting IBR-domain E3 Ub ligase [10, 45], might also regulate Bcl-2 turnover and Bcl-2-linked apoptosis [46–48]. Expression of Parkin was found to slow-down ceramide-induced cell death in PC12 cells [46]. Furthermore, while excess Parkin prevented the release of cytochrome c from mitochondria isolated from rodent and human neuroblastoma cells, knock-down of Parkin by shRNAi caused increased cytochrome c release upon insult with BH3-only peptides [47]. Parkin was also identified as a E3 Ub ligase for the mono-ubiquitylation of Bcl-2 [48] causing its stabilization and increased interaction with Beclin 1 in the context of autophagy. However, Bcl-2 stabilization might also explain the observed anti-apoptotic function of Parkin. While the role of Parkin in mitochondria-specific autophagy is established [10, 45], the role of this protein in apoptosis is less well defined. Yet, due to the extensive crosstalk between apoptotic cell death and autophagy, one might anticipate the regulatory influence of Parkin on the execution of cell death.
3.2.3 Proteasomal degradation of tBid-N facilitates pro-apoptotic activity of tBid-C
In addition to degradation of multi-BH3 Bcl-2 family proteins, mitochondrial steps of apoptosis are also modulated by UPS-dependent BH3-only Bcl-2 family protein degradation, including Bid, and stabilization of Bim and Bad.
Upon induction of death receptor-mediated apoptosis, the 20-kDa protein Bid (BH3 interacting-domain death agonist) is activated through proteolytic cleavage in its unstructured loop by caspase-8. The Bid cleavage results in the formation of a ~6-kDa N-terminal part (tBid-N) and a ~14-kDa C-terminal part (truncated Bid or tBid-C) that contains the BH3 domain [49–51]. While full length Bid resides in the cytosol, cleaved Bid translocates to mitochondria and thus transduces apoptotic signals from the cell membrane to the mitochondria serving as a mechanism linking extrinsic (death receptor-mediated) mode of apoptosis with the mitochondria. Mitochondria-associated tBid-C interacts with OMM localized Bcl-2 family proteins and membrane lipids, thus facilitating OMM permeabilization [50–53].
Under basal growth conditions, full length Bid is a stable protein [54–56]. However, upon Bid cleavage, ubiquitylation and proteasomal degradation of N-terminal (tBid-N) and C-terminal (tBid-C) fragments have been shown to modulate the pro-apoptotic activity of cleaved Bid [54–56]. Consistent with this notion, caspase cleavage of full length Bid in TNFα-stimulated cells was not associated with accumulation of tBid fragments, indicating that tBid-N and tBid-C are unstable. Since proteasomal inhibition stabilized tBid-C, it was concluded that the UPS system controls rapid degradation of tBid-C in apoptotic cells [54]. Furthermore, expression of a stable mutant of tBid-C, sensitized HeLa cells to apoptosis [54], suggesting that Ub-dependent proteasomal degradation of tBid might serve as a fine-tuning mechanism regulating the strength of tBid-dependent signaling in apoptosis. The work by Azakir and co-workers [56] demonstrated that tBid-C is a substrate of the HECT-domain E3 Ub ligase Itch. Ectopic expression of Itch protected cells from tBid- and TRAIL-induced apoptosis, whereas downregulation of Itch by RNA interference had the opposite effects. Itch and tBid-C, but not full length Bid were found to form a molecular complex, likely facilitating Itch-induced increases in tBid-C ubiquitylation and proteasomal degradation [56]. Based on these data one might conclude that UPS, through rapid degradation of tBid-C, functions in inhibiting tBid-mediated apoptosis. However, the possibility that ubiquitylation and degradation of tBid-C serves as a molecular step in tBid-mediated apoptosis signaling also needs to be considered here.
Another mechanism of how UPS-dependent tBid ubiquitylation and degradation affects mitochondrial steps in apoptosis, has also been recently proposed [55]. It has been shown that inhibiting proteasomal degradation of tBid-N prevented Bid-dependent OMM permeabilization, without affecting the translocation of this protein to the OMM [55]. In contrast to tBid-C, Ub-conjugation of tBid-N does not require canonical lysine residues since no lysine residues are present in this fragment. Instead, glutamine (E14) and cysteine (C15) residues located in the α-helix 1 of Bid seem to be critical for ubiquitylation and destabilization of tBid-N [55]. Since in vitro studies have demonstrated that upon caspase cleavage of Bid, the tBid-C and tBid-N remain associated and the separation of these fragments appears to be critical for tBid-C activity (Chou et al., 1999; Zha et al., 2000; Kuwana et al., 2002), degradation of tBid-N is likely to facilitate pro-apoptotic activity of tBid-C. Consistent with this notion, overexpression of tBid-N blocked the pro-apoptotic activity of the tBid-C (Tan et al., 1999; Kudla et al., 2000; Renshaw et al., 2004).
Taken together, these data indicate that cleavage of Bid by caspase-8 results in two products that are degraded by the proteasome in an ubiquitylation-dependent manner, and this degradation controls the downstream steps mediated by tBid. However, whether or not the degradation rate of tBid-C is lower than that of tBid-N, which would be necessary to enable the pro-apoptotic activity of tBid-C, needs to be elucidated.
3.3 UPS and inhibition of mitochondrial steps in apoptosis
Turnover of other than Bid BH3-only proteins, including Bad [57], Bim [58, 59] and Bik [60], is also under control of the UPS. However, unlike degradation of Bid fragments, proteasome-dependent homeostatic degradation of these BH3-only proteins promotes cell survival. The data also show crosstalk between the phosphorylation status of these proteins and Ub-conjugation that appears to be critical for Bad (Bcl-2-associated death promoter) and BimEL (Bcl-2-interacting mediator of cell death, extra long) proteasome-dependent turnover regulation.
3.3.1 Degradation of Bad
Reversible phosphorylation of the Bcl-2 family proteins serves as an important regulatory mechanism vital for cell survival. For example, multiple kinases neutralize the pro-apoptotic activity of Bad by phosphorylation of serine residues (S75, S99, and S118 in human Bad) thus facilitating Bad sequestration in the cytosol by 14-3-3 proteins. Phosphorylation has been also shown to regulate UPS-dependent Bad turnover [57]. In NIH3T3 cells, c-Raf kinase pathway activation led to a rapid phosphorylation of Bad, associated with ubiquitylation and destabilization of this protein, resulting in a 2-fold increase in the rate of Bad turnover [57]. Thus, in addition to cytosolic sequestration, a rapid UPS-dependent degradation of Bad might be another mechanism for limiting the pro-apoptotic activity of this protein. Whether Bad phosphorylation serves as an E3 Ub ligase-recruiting signal or induces a degradation-prone conformation needs to be elucidated, as well as the consequences of this mechanism for the cell sensitivity to apoptosis.
3.3.2 Degradation of Bim
Phosphorylation of BimEL has been also linked to turnover of this protein. BimEL is dephosphorylated and thus stabilized in response to many distinct stress signals [58, 59, 61, 62]. In the cell detachment (anoikis) model of cell death, MEK-ERK (mitogen-activated protein kinase/ extracellular signal regulated kinase) inhibitors induced BimEL-dependent apoptosis [58]. MEK inhibitors suppressed BimEL S69 phosphorylation and consequently induced accumulation of this protein in both attached and detached cells through inhibition of its ubiquitylation and proteasomal degradation. Notably, pro-apoptotic activity of MEK inhibitors does not depend on increased levels of BimEL, per se, but rather mitochondrial translocation of BimEL is critical for cell death induction, apparent in detached but not in attached cells [58]. Thus, accumulation of BimEL due to inhibition of proteasomal degradation of this protein, while likely increasing the strength of the pro-apoptotic signal, might require additional triggers specific to detached cells for cell death induction.
The pro-survival role of UPS-dependent degradation of Bim is further supported in other models of cell death. Upon growth factor deprivation osteoclasts undergo apoptosis associated with a rapid and sustained increase in Bim protein levels [61]. In this case reduced ubiquitylation and proteasomal degradation promote Bim stabilization [61]. In osteoclasts the RING domain E3 Ub ligase c-Cbl might be involved in Bim ubiquitylation [61]. c-Cbl was found to co-immunoprecipitate with Bim in the presence of the proteasome inhibitor, MG132. This physical association was enhanced following treatment with pro-survival cytokines. Furthermore, proteasome inhibition increased Bim expression in c-Cbl overexpressing cells [61]. Consistent with the role for c-Cbl in Bim ubiquitylation, ectopic expression of c-Cbl enhanced Bim ubiquitylation while c-Cbl mutant expression suppressed it, even in the presence of proteasome inhibitor [61]. In summary, these data suggest that c-Cbl plays a role in the ubiquitylation and proteasome-dependent degradation of Bim and serves as a critical pro-survival regulator of cytokine withdrawal-induced apoptosis in osteoclasts.
Bim ubiquitylation and degradation is also the basis for the pro-survival effect of ischemic preconditioning in cultured cortical neurons. In contrast to c-Cbl-mediated degradation of Bim in osteoclasts, degradation of Bim in the neuronal model of ischemic preconditioning appears to be mediated by another RING domain E3 Ub ligase, TRIM2 (tripartite motif-containing protein 2) [59]. TRIM2 binds to Bim following phosphorylation at the S69 residue by p42/p44 MAPK, but does not interact with a phospho-site mutant of Bim [59]. Importantly, shRNAi-induced downregulation of TRIM2 resulted in the stabilization of Bim, as well as the suppression of ischemic tolerance [59], suggesting a critical role for Bim stability regulation in ischemia/reperfusion-induced neuronal apoptosis.
Of note, based on the fact that lysine-deficient Bim mutant could be degraded by the proteasome without prior polyubiquitylation, it has been proposed that proteasomal degradation of Bim might not require ubiquitylation [63]. Bim, as well as other BH3-only proteins Bad and Bmf, are considered an intrinsically disordered protein (IDP) [63, 64]. IDPs can be degraded by uncapped proteasomes [65, 66], and at least under some conditions, a similar mechanism might govern Bim (and Bad) turnover. However, the extent and role in apoptosis progression of ubiquitylation-independent proteasomal degradation of BH-3 only proteins are currently not clear.
3.3.3 UPS-dependent degradation of apoptotically active Bax
Regulation of multi BH-domain pro-apoptotic proteins [19], including Bax (Bcl-2-associated X protein), Bak (Bcl-2 homologous antagonist/killer) and Bok (BCL-2-related ovarian killer), is considered the main task of other Bcl-2 family proteins. While Bok expression is restricted to the reproductive tissues [67], Bax and Bak are more ubiquitously expressed, and function as mitochondrial executioners of pro-apoptotic signals.
Evidence indicates that UPS-dependent regulated degradation of Bax serve as a widespread mechanisms promoting cell survival. [68–72]. While the most common isoform of Bax, 21-kD Baxα is a stable protein, its turnover might vary depending on the cellular context. For example, Baxα is a short-lived protein in malignant B-cell lines [69]. Supporting the importance of stringent regulation of Baxα turnover, increased proteasomal degradation of this protein is a common feature of poor prognosis in chronic lymphocytic leukemia [69]. Furthermore, the abnormal proteasome-dependent degradation of Baxα also appears to be an important factor in the high resistance of malignant B-cells to TRAIL-induced apoptosis [68].
In healthy cells Bax resides mostly in the cytosol or loosely associated with the OMM as a monomer in an apoptotically dormant conformation [73, 74]. Only after disengagement of αhelix 9 from the hydrophobic pocket does Bax accumulate on the mitochondria, oligomerize and thereby induce OMM permeabilization [74–76]. Inadvertent change in Bax conformation is likely to represent a strong pro-apoptotic stimulus. Indeed, supporting the possibility that self-activation of Bcl-2 family proteins plays a role in apoptosis activation, it has been shown that an activated Bak mutant induced a conformation change in, and oligomerization of, non-activated Bak [77]. Truncated Bax (H2–H3) could also activate full length Bax [78]. Thus, as H2–H3 mimics the part of Bax that under physiological conditions is exposed after Bax is activated by BH3-only proteins, it has been proposed that active Bax-induced Bax activation occurs during apoptosis [78]. Therefore, although anti-apoptotic Bcl-2 family proteins normally control Bax conformation, elimination of accidentally formed active Bax, or when pro-survival mechanisms overcome stress, might be central for cell survival. Recent data indicate that IBRDC2, an IBR (in between RING)-type RING finger E3 Ub ligase, exclusively facilitates removal of apoptotically active Baxα [72]. While overexpression of IBRDC2 increased Baxα ubiquitylation, shRNAi-mediated downregulation of this protein led to cellular accumulation of active Baxα, spontaneous apoptosis and increased cell sensitivity to actinomycin D- and staurosporin-induced cell death [72]. Notably, in healthy cells IBRDC2 resides mainly in the cytosol and accumulates on the mitochondria upon induction of apoptosis, in synchrony with Baxα. It has also been shown that mitochondrial accumulation of IBRDC2 requires activated Baxα [72], suggesting that the apoptotically active, but not the dormant form of Baxα is targeted for proteasomal degradation by IBRDC2-dependent ubiquitylation. The extent to which IBRDC2 expression corresponds to the cell sensitivity to apoptosis, especially in malignant B-cell lines, and its role in development of poor prognosis chronic lymphocytic leukemia, as well as other Bax inhibition-linked tumors, remains to be established.
Given the strong pro-apoptotic consequences of Bax activation, UPS-dependent apoptosis-linked degradation of several non-Bcl-2 family proteins also regulate Bax activity. These include, the RING domain E3 Ub ligase BARD1 (BRCA1-associated RING domain 1) [79]. In the nucleaus, BARD1 forms heterodimers with BRCA1 and participates in nuclear DNA repair. In contrast, the mitochondrial function of BARD1 does not require BRCA1 but has been associated with stimulation of Bax oligomerization at the mitochondria [79]. The mechanistic link between Bax oligomerization and E3 Ub ligase activity of BARD1 is currently unknown. Mitochondrial targets for this protein, as well as the mechanism regulating mitochondrial localization of BARD1, also need to be established in more detail. The BRCA1/BARD1 RING complex functions as an Ub ligase with a substantially higher activity than the BRCA1 or BARD1 subunits by themselves [80], suggesting a distinct activity and substrate specificity for BARD1 on mitochondria.
Activation of Bax on mitochondria is also linked to proteasomal degradation of MOAP-1 (modulator of apoptosis-1) [81]. MOAP-1 contains a BH3-like motif and was initially identified by a yeast two-hybrid screen as a binding partner of Bax [82]. Ectopic expression of this protein is able to trigger apoptosis in mammalian cells, and vice versa, reduced levels of MOAP-1 confer resistance to apoptosis induction by a variety of triggers [81]. Consistent with the role of MOAP-1 in Bax activation, mitochondria isolated from MOAP-1 knock down cells were resistant to recombinant Bax-induced OMM permeabilization, as evidenced by the cytochrome c release assay [81]. Under normal growth conditions, MOAP-1 is a low abundance, unstable protein with an estimated turnover rate of ~25min. However, upon induction of apoptosis, MOAP-1 accumulates in a time frame similar to the accumulation of apoptotically-active Bax and the release of cytochrome c from the mitochondria to the cytosol [81]. This suggests that MOAP-1 might be important for OMM permeabilization, perhaps through the regulation of Bax conformation. Notably, pro-apoptotic stimuli prevent MOAP-1 ubiquitylation and thereby inhibit proteasomal degradation of this protein [81]. The stability of MOAP-1 is also regulated by TRIM39 (tripartite motif 39), a RING and B-box domain-containing protein [83]. Whether TRIM39 is the E3 ubiquitin ligase regulating the MOAP-1 mediated pathway needs to be established. However, since ectopic expression of TRIM39 stabilized MOAP-1 [83] it is unlikely that TRIM39 is responsible for K48 polyubiquitylation and proteasomal degradation of this protein.
In contrast to the stable nature of Baxα, the turnover of Baxβ, a 24 KD splice variant of Bax, is much faster [71]. It appears that Baxβ has a stronger pro-apoptotic potential then Baxα due to spontaneous accumulation on mitochondria and induction of OMM permeabilization [71]. A high rate of Baxβ turnover facilitates maintenance of low levels of this protein in healthy cells. The work by Fu et al. [71] also demonstrates that proteasomal control of Baxβ stability is important for apoptosis regulation. Upon stress-induced apoptosis, Baxβ accumulates in cells likely due to inhibition of its degradation by the proteasome [71]. The mechanism and molecular components vital for Baxβ ubiquitylation and/or regulation of proteasomal degradation of this protein remain unknown.
3.3.4 Inhibition of apoptosis by destabilization of Bak
In contrast to Bax, there is no information available on the regulation of Bak turnover in cells under normal growth conditions. However, certain viral proteins are able to induce the proteasome-dependent degradation of Bak thereby suppressing cell death following viral infection [84, 85]. E6 proteins of various human papillomaviruses (HPVs) are oncogenes linked to cervical cancer development [84–86]. In addition to destablizing the tumor suppressor p53 [87], E6 proteins can also induce degradation of Bak [84, 85]. E6-dependent Bak destabilization involves proteasomal degradation and the activity of the HECT domain E3 Ub ligase, E6AP (E6 associated protein; the prototypical HECT domain E3 Ub ligase). In addition, expression of distinct E6 proteins triggers the proteasome-dependent degradation of Bak following UV irradiation. This mechanism appears to be specific for Bak, since other Bcl-2 family proteins were not affected by E6 protein expression. Importantly, E6-dependent degradation of Bak was sufficient to prevent cytochrome c release and apoptosis induction, indicating that upon viral infection the molecular machinery that controls Bak turnover under normal conditions might be hijacked by viral proteins to prevent premature cell death, thus facilitating cell survival and thereby viral proliferation [84, 85].
4. Concluding remarks
As summarized above, accumulating evidence indicates that UPS-dependent regulated protein degradation controls the mitochondrial steps in apoptosis. However, in contrast to other molecular steps in apoptosis, including UPS-dependent caspase regulation in the cytosol or p53-dependent regulation of apoptotic genes in the nucleus, an understanding of the molecular mechanisms of how ubiquitylation influences apoptotic activities of mitochondria-associated proteins is, in most cases, preliminary. Further research is needed to solidify and add mechanistic understanding to these findings. For example, since several of the reports discussed here utilize only protein overexpression and distinct cell culture models, genetic analyses of how depletion of certain E3 Ub ligases affects apoptotic signaling at mitochondria will likely provide further progress in this important and exciting research area. Although we have focused on the Bcl-2 family mediated regulation of mitochondrial steps in apoptosis, several non-Bcl-2 family factors, including mitochondrial fusion and fission regulators [88–92] are also implicated in this process. Since these proteins are also regulated by the UPS [10, 31, 93–97] the connection between UPS and regulated degradation of mitochondrial fusion and fission factors is likely. We are currently investigating these problems.
The connection between Ub-dependent Bcl-2 family protein degradation and therapeutic potential of proteasome inhibitors such as bortezomib for the treatment of certain cancers has been already suggested. Proteasomal inhibition in chronic lymphatic leukemia cells triggers the induction of apoptosis [98], probably through the stabilization of pro-apoptotic proteins such as Bax and subsequent release of cytochrome c [68, 99]. A similar mechanism might mediate the anti-proliferative effect of the naturally occurring proteasome inhibitor curcumin (diferuloylmethane) [100–102].
The coming years of research will likely uncover currently unknown features of Ub-dependent regulation of mitochondrial homeostasis and thus, create several new questions further stimulating this growing area of research.
Highlights.
Insights into the role of degradative and non-degradative ubiquitin-dependent signaling in the regulation of apoptosis have provided one of the most significant breakthroughs in the recent years of cell death research. It has been revealed that all major steps in apoptosis cascade signaling, including transcriptional regulation of apoptotic gene expression, outer mitochondrial membrane permeabilization and caspase activation, are under the control of the ubiquitin/proteasome system. This makes ubiquitin signaling one on the most critical life and death decision checkpoints in mammalian cells. The focus of this review is the ubiquitination-dependent regulation of the mitochondrial steps in apoptosis. The newly identified ubiquitination-dependent processes in the Bcl-2 family-regulated outer mitochondrial membrane permeabilization, as well as the overall role of mitochondria-associated E3 ubiquitin ligases and other molecular components of the ubiquitin/proteasome system during mitochondria-governed apoptosis are discussed.
Aknowlegements
We thank Pamela Wright for insightful comments on the manuscript. We also gratefully acknowledge financial support from the National Institute of General Medical Science (grant RO1 GM083131 to MK).
Abbreviations
- Ub
ubiquitin
- OMM
outer mitochondrial membrane
- IMM
inner mitochondrial membrane
- IMS
mitochondria inter membrane space
- UPS
ubiquitin/proteasome system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciechanover A, Elias S, Heller H, Ferber S, Hershko A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J Biol Chem. 1980;255:7525–7528. [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Millard SM, Wood SA. Riding the DUBway: regulation of protein trafficking by deubiquitylating enzymes. J Cell Biol. 2006;173:463–468. doi: 10.1083/jcb.200602082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frappier L, Verrijzer CP. Gene expression control by protein deubiquitinases. Curr Opin Genet Dev. 2011;21:207–213. doi: 10.1016/j.gde.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 8.Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- 9.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pines J, Lindon C. Proteolysis: anytime, any place, anywhere? Nat Cell Biol. 2005;7:731–735. doi: 10.1038/ncb0805-731. [DOI] [PubMed] [Google Scholar]
- 12.Neutzner A, Neutzner M, Benischke AS, Ryu SW, Frank S, Youle RJ, et al. A systematic search for ER membrane-associated RING finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J Biol Chem. 2011 doi: 10.1074/jbc.M110.197459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karbowski M, Youle RJ. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr Opin Cell Biol. 2011;23:476–482. doi: 10.1016/j.ceb.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- 15.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 16.Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol. 2009;19:130–140. doi: 10.1016/j.tcb.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Zhang HG, Wang J, Yang X, Hsu HC, Mountz JD. Regulation of apoptosis proteins in cancer cells by ubiquitin. Oncogene. 2004;23:2009–2015. doi: 10.1038/sj.onc.1207373. [DOI] [PubMed] [Google Scholar]
- 18.Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 20.Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 22.Morizane Y, Honda R, Fukami K, Yasuda H. X-linked inhibitor of apoptosis functions as ubiquitin ligase toward mature caspase-9 and cytosolic Smac/DIABLO. J Biochem. 2005;137:125–132. doi: 10.1093/jb/mvi029. [DOI] [PubMed] [Google Scholar]
- 23.Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, et al. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- 24.Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 29.Cuconati A, Mukherjee C, Perez D, White E. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 2003;17:2922–2932. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, et al. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warr MR, Acoca S, Liu Z, Germain M, Watson M, Blanchette M, et al. BH3-ligand regulates access of MCL-1 to its E3 ligase. FEBS Lett. 2005;579:5603–5608. doi: 10.1016/j.febslet.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 33.Ye Y, Meyer H, Rapoport T. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 34.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 35.Jentsch S, Rumpf S. Cdc48 (p97): a "molecular gearbox" in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Halawani D, Latterich M. p97: The cell's molecular purgatory? Mol Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Azad N, Vallyathan V, Wang L, Tantishaiyakul V, Stehlik C, Leonard SS, et al. S-nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J Biol Chem. 2006;281:34124–34134. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- 38.Breitschopf K, Haendeler J, Malchow P, Zeiher AM, Dimmeler S. Posttranslational modification of Bcl-2 facilitates its proteasome-dependent degradation: molecular characterization of the involved signaling pathway. Mol Cell Biol. 2000;20:1886–1896. doi: 10.1128/mcb.20.5.1886-1896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niture SK, Jaiswal AK. INrf2 (Keap1) targets Bcl-2 degradation and controls cellular apoptosis. Cell Death Differ. 2011;18:439–451. doi: 10.1038/cdd.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Zhang J, Yan H, Wu YP, Li C, Zhang GY. Activation of GluR6-containing kainate receptors induces ubiquitin-dependent Bcl-2 degradation via denitrosylation in the rat hippocampus after kainate treatment. J Biol Chem. 2011;286:7669–7680. doi: 10.1074/jbc.M110.156299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elhasid R, Sahar D, Merling A, Zivony Y, Rotem A, Ben-Arush M, et al. Mitochondrial pro-apoptotic ARTS protein is lost in the majority of acute lymphoblastic leukemia patients. Oncogene. 2004;23:5468–5475. doi: 10.1038/sj.onc.1207725. [DOI] [PubMed] [Google Scholar]
- 42.Gottfried Y, Voldavsky E, Yodko L, Sabo E, Ben-Itzhak O, Larisch S. Expression of the pro-apoptotic protein ARTS in astrocytic tumors: correlation with malignancy grade and survival rate. Cancer. 2004;101:2614–2621. doi: 10.1002/cncr.20675. [DOI] [PubMed] [Google Scholar]
- 43.Lotan R, Rotem A, Gonen H, Finberg JP, Kemeny S, Steller H, et al. Regulation of the proapoptotic ARTS protein by ubiquitin-mediated degradation. J Biol Chem. 2005;280:25802–25810. doi: 10.1074/jbc.M501955200. [DOI] [PubMed] [Google Scholar]
- 44.Gottfried Y, Rotem A, Lotan R, Steller H, Larisch S. The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. EMBO J. 2004;23:1627–1635. doi: 10.1038/sj.emboj.7600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darios F, Corti O, Lucking CB, Hampe C, Muriel MP, Abbas N, et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12:517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- 47.Berger AK, Cortese GP, Amodeo KD, Weihofen A, Letai A, LaVoie MJ. Parkin selectively alters the intrinsic threshold for mitochondrial cytochrome c release. Hum Mol Genet. 2009;18:4317–4328. doi: 10.1093/hmg/ddp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen D, Gao F, Li B, Wang H, Xu Y, Zhu C, et al. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem. 2010;285:38214–38223. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 50.Willis SN, Adams JM. Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol. 2005;17:617–625. doi: 10.1016/j.ceb.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ott M, Norberg E, Zhivotovsky B, Orrenius S. Mitochondrial targeting of tBid/Bax: a role for the TOM complex? Cell Death Differ. 2009;16:1075–1082. doi: 10.1038/cdd.2009.61. [DOI] [PubMed] [Google Scholar]
- 52.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Goonesinghe A, Mundy ES, Smith M, Khosravi-Far R, Martinou JC, Esposti MD. Pro-apoptotic Bid induces membrane perturbation by inserting selected lysolipids into the bilayer. Biochem J. 2005;387:109–118. doi: 10.1042/BJ20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breitschopf K, Zeiher AM, Dimmeler S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J Biol Chem. 2000;275:21648–21652. doi: 10.1074/jbc.M001083200. [DOI] [PubMed] [Google Scholar]
- 55.Tait SW, de Vries E, Maas C, Keller AM, D'Santos CS, Borst J. Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J Cell Biol. 2007;179:1453–1466. doi: 10.1083/jcb.200707063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azakir BA, Desrochers G, Angers A. The ubiquitin ligase Itch mediates the antiapoptotic activity of epidermal growth factor by promoting the ubiquitylation and degradation of the truncated C-terminal portion of Bid. FEBS J. 2010;277:1319–1330. doi: 10.1111/j.1742-4658.2010.07562.x. [DOI] [PubMed] [Google Scholar]
- 57.Fueller J, Becker M, Sienerth AR, Fischer A, Hotz C, Galmiche A. C-RAF activation promotes BAD poly-ubiquitylation and turn-over by the proteasome. Biochem Biophys Res Commun. 2008;370:552–556. doi: 10.1016/j.bbrc.2008.03.141. [DOI] [PubMed] [Google Scholar]
- 58.Fukazawa H, Noguchi K, Masumi A, Murakami Y, Uehara Y. BimEL is an important determinant for induction of anoikis sensitivity by mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitors. Mol Cancer Ther. 2004;3:1281–1288. [PubMed] [Google Scholar]
- 59.Thompson S, Pearson AN, Ashley MD, Jessick V, Murphy BM, Gafken P, et al. Identification of a novel Bcl-2-interacting mediator of cell death (Bim) E3 ligase, tripartite motif-containing protein 2 (TRIM2), and its role in rapid ischemic tolerance-induced neuroprotection. J Biol Chem. 2011;286:19331–19339. doi: 10.1074/jbc.M110.197707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chinnadurai G, Vijayalingam S, Rashmi R. BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. 2008;27(Suppl 1):S20–S29. doi: 10.1038/onc.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI, et al. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 2003;22:6653–6664. doi: 10.1093/emboj/cdg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Zhou JY, Wu GS. Bim protein degradation contributes to cisplatin resistance. J Biol Chem. 2011;286:22384–22392. doi: 10.1074/jbc.M111.239566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiggins CM, Tsvetkov P, Johnson M, Joyce CL, Lamb CA, Bryant NJ, et al. BIM(EL), an intrinsically disordered protein, is degraded by 20S proteasomes in the absence of polyubiquitylation. J Cell Sci. 2011;124:969–977. doi: 10.1242/jcs.058438. [DOI] [PubMed] [Google Scholar]
- 64.Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DC, et al. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007;14:128–136. doi: 10.1038/sj.cdd.4401934. [DOI] [PubMed] [Google Scholar]
- 65.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 66.Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- 67.Hsu SY, Kaipia A, McGee E, Lomeli M, Hsueh AJ. Bok is a pro-apoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti-apoptotic Bcl-2 family members. Proc Natl Acad Sci U S A. 1997;94:12401–12406. doi: 10.1073/pnas.94.23.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu FT, Agrawal SG, Gribben JG, Ye H, Du MQ, Newland AC, et al. Bortezomib blocks Bax degradation in malignant B cells during treatment with TRAIL. Blood. 2008;111:2797–2805. doi: 10.1182/blood-2007-08-110445. [DOI] [PubMed] [Google Scholar]
- 69.Agrawal SG, Liu FT, Wiseman C, Shirali S, Liu H, Lillington D, et al. Increased proteasomal degradation of Bax is a common feature of poor prognosis chronic lymphocytic leukemia. Blood. 2008;111:2790–2796. doi: 10.1182/blood-2007-10-110460. [DOI] [PubMed] [Google Scholar]
- 70.Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci U S A. 2000;97:3850–3855. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu NY, Sukumaran SK, Kerk SY, Yu VC. Baxbeta: a constitutively active human Bax isoform that is under tight regulatory control by the proteasomal degradation mechanism. Mol Cell. 2009;33:15–29. doi: 10.1016/j.molcel.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 72.Benard G, Neutzner A, Peng G, Wang C, Livak F, Youle RJ, et al. IBRDC2, an IBR-type E3 ubiquitin ligase, is a regulatory factor for Bax and apoptosis activation. EMBO J. 2010;29:1458–1471. doi: 10.1038/emboj.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsu YT, Youle RJ. Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J Biol Chem. 1998;273:10777–10783. doi: 10.1074/jbc.273.17.10777. [DOI] [PubMed] [Google Scholar]
- 75.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 76.Antonsson B, Montessuit S, Sanchez B, Martinou JC. Bax is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J Biol Chem. 2001;276:11615–11623. doi: 10.1074/jbc.M010810200. [DOI] [PubMed] [Google Scholar]
- 77.Ruffolo SC, Shore GC. BCL-2 selectively interacts with the BID-induced open conformer of BAK, inhibiting BAK auto-oligomerization. J Biol Chem. 2003;278:25039–25045. doi: 10.1074/jbc.M302930200. [DOI] [PubMed] [Google Scholar]
- 78.Tan C, Dlugosz PJ, Peng J, Zhang Z, Lapolla SM, Plafker SM, et al. Auto-activation of the apoptosis protein Bax increases mitochondrial membrane permeability and is inhibited by Bcl-2. J Biol Chem. 2006;281:14764–14775. doi: 10.1074/jbc.M602374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tembe V, Henderson BR. BARD1 translocation to mitochondria correlates with Bax oligomerization, loss of mitochondrial membrane potential, and apoptosis. J Biol Chem. 2007;282:20513–20522. doi: 10.1074/jbc.M702627200. [DOI] [PubMed] [Google Scholar]
- 80.Brodie KM, Henderson BR. Differential modulation of BRCA1 and BARD1 nuclear localisation and foci assembly by DNA damage. Cell Signal. 2010;22:291–302. doi: 10.1016/j.cellsig.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 81.Fu NY, Sukumaran SK, Yu VC. Inhibition of ubiquitin-mediated degradation of MOAP-1 by apoptotic stimuli promotes Bax function in mitochondria. Proc Natl Acad Sci U S A. 2007;104:10051–10056. doi: 10.1073/pnas.0700007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan KO, Tan KM, Chan SL, Yee KS, Bevort M, Ang KC, et al. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J Biol Chem. 2001;276:2802–2807. doi: 10.1074/jbc.M008955200. [DOI] [PubMed] [Google Scholar]
- 83.Lee SS, Fu NY, Sukumaran SK, Wan KF, Wan Q, Yu VC. TRIM39 is a MOAP-1-binding protein that stabilizes MOAP-1 through inhibition of its poly-ubiquitination process. Exp Cell Res. 2009;315:1313–1325. doi: 10.1016/j.yexcr.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 84.Underbrink MP, Howie HL, Bedard KM, Koop JI, Galloway DA. E6 proteins from multiple human betapapillomavirus types degrade Bak and protect keratinocytes from apoptosis after UVB irradiation. J Virol. 2008;82:10408–10417. doi: 10.1128/JVI.00902-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simmonds M, Storey A. Identification of the regions of the HPV 5 E6 protein involved in Bak degradation and inhibition of apoptosis. Int J Cancer. 2008;123:2260–2266. doi: 10.1002/ijc.23815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McLaughlin-Drubin ME, Munger K. Oncogenic activities of human papillomaviruses. Virus Res. 2009;143:195–208. doi: 10.1016/j.virusres.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 88.Karbowski M. Mitochondria on guard: role of mitochondrial fusion and fission in the regulation of apoptosis. Adv Exp Med Biol. 2010;687:131–142. doi: 10.1007/978-1-4419-6706-0_8. [DOI] [PubMed] [Google Scholar]
- 89.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 90.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 91.Karbowski M, Youle RJ. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 2003;10:870–880. doi: 10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- 92.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. Embo J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang B, Huang J, Li HL, Liu T, Wang YY, Waterman P, et al. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 2008;18:900–910. doi: 10.1038/cr.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pahler JC, Ruiz S, Niemer I, Calvert LR, Andreeff M, Keating M, et al. Effects of the proteasome inhibitor, bortezomib, on apoptosis in isolated lymphocytes obtained from patients with chronic lymphocytic leukemia. Clin Cancer Res. 2003;9:4570–4577. [PubMed] [Google Scholar]
- 99.Premkumar DR, Jane EP, Agostino NR, Didomenico JD, Pollack IF. Bortezomib-induced sensitization of malignant human glioma cells to vorinostat-induced apoptosis depends on reactive oxygen species production, mitochondrial dysfunction, Noxa upregulation, Mcl-1 cleavage, and DNA damage. Mol Carcinog. 2011 doi: 10.1002/mc.21835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jana NR, Dikshit P, Goswami A, Nukina N. Inhibition of proteasomal function by curcumin induces apoptosis through mitochondrial pathway. J Biol Chem. 2004;279:11680–11685. doi: 10.1074/jbc.M310369200. [DOI] [PubMed] [Google Scholar]
- 101.Milacic V, Banerjee S, Landis-Piwowar KR, Sarkar FH, Majumdar AP, Dou QP. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 2008;68:7283–7292. doi: 10.1158/0008-5472.CAN-07-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chiu TL, Su CC. Curcumin inhibits proliferation and migration by increasing the Bax to Bcl-2 ratio and decreasing NF-kappaBp65 expression in breast cancer MDA-MB-231 cells. Int J Mol Med. 2009;23:469–475. doi: 10.3892/ijmm_00000153. [DOI] [PubMed] [Google Scholar]