Abstract
Metal oxide nanoparticles synthesized with the aid of medicinal plant extracts are showing potential as treatment options for inflammatory diseases. Two key benefits of this synthesis method is: the synthesis process is environmentally benign and the utilization of medicinal plant-derived extracts adds to the medicinal value of the synthesized nanoparticle. Earlier, sesquiterpene lactone zaluzanin-D (ZD) has been isolated from leaves of Vernonia arborea. ZD showed ability to reduce inflammation in activated monocytes. Copper oxide nanoparticles (bCuO-NPs) were synthesized using ZD-enriched leaf extract of V. arborea and characterized by UV-vis spectroscopy, FT-IR, XRD, particle size analyzer, and TEM. Synthesized bCuO-NPs did not show significant toxicity to human monocytic cell lines (THP-1) at the tested concentrations. The bCuO-NPs showed radical scavenging ability indicating anti-oxidant properties. Flow cytometry experiments proved the capability of bCuO-NPs to reduce intracellular ROS in peroxide-activated THP-1 cells. The NPs also showed a significant ability to reduce inflammatory adhesion in PMA-activated THP-1 cells. In the DNA methylation studies, bCuO-NPs behaved similarly to ZD and prevented DNA hypomethylation at the MMP-9 promoter region. These properties strongly indicate the ability of bCuO-NPs to reduce inflammation in the activated monocytes. Furthermore, in zebrafish (Danio rerio) embryos, the developmental toxicity of bCuO-NPs was assessed. The studies indicated the reduced toxicity and compatibility of the NPs with biological organisms. Based on the results, it can be concluded that the bCuO-NPs produced from ZD-enriched leaf extract have significant anti-oxidant capabilities and the ability to reduce inflammation in monocytic cell lines. Overall, reduced in vitro and in vivo toxicity, along with its antioxidant and anti-inflammatory properties, makes bCuO-NPs a potential candidate for anti-inflammatory drugs.
Copper oxide nanoparticles successfully synthesized using Zaluzanin-D enriched leaf extract of Vernonia arborea, showed antioxidant properties, anti-inflammatory activity in activated monocytes (THP-1) and reduced toxicity in zebra fish model.
Introduction
Application of nanomaterials in the fields of bioimaging, targeted drug delivery, biosensing, and other biomedical applications is widely being investigated. This is mainly due to their favorable properties like reduced toxicity and enhanced bioactivity of the nanomaterial.1 These properties are directly attributed to the changes in the physiochemical nature of the material due to its nanoform.2 One of the key challenges in generating nanoparticles with biological applications is to avoid or reduce the use of harmful chemical methods that may affect downstream biological applications. This clean and eco-friendly green synthesis approach arose as a feasible alternative to synthesis of nano-sized particles.3
An efficient substitute for chemical and physical processes is the biosynthesis of metal and metal oxide nanoparticles (NPs), such as those containing iron, titanium, palladium, copper, gold, zinc oxide, and silver.4 Synthesis of metal oxide NPs using a biological approach has gained popularity due to its environmental friendliness, simplicity, and economic benefits. This approach involves utilizing natural materials like plants, bacteria, and algae for reducing and stabilizing metals to their corresponding NPs in a single step. Alkaloids, flavonoids, saponins, steroids, terpenoids, and tannins are examples of natural organic phytoconstituent biomolecules found in bioactive extracts that function as reducing and stabilizing agents because of their complex and varied compositions.5 An added advantage of nanomaterials synthesized using plant-based compounds is their therapeutic biocompatibility since the use of toxic and harmful reagents and hazardous conditions have been eliminated from the NPs synthesis process.6
Studies show metal oxide NPs possess a broad spectrum of excellent therapeutic qualities. Copper oxide nanoparticles (CuO-NPs) have been reported to possess a wide range of therapeutic applications. The most common application is as an antimicrobial agent. Apart from this, the effect of CuO-NPs on cellular oxidant and apoptotic pathways has also been studied. These studies correlate the physiochemical properties of the CuO-NPs, such as their surface charge and size, to cellular toxicity and microbial activity. As mentioned earlier generating these CuO-NPs by utilizing green methods of synthesis could be advantageous. Earlier studies indicate a wide range of materials including extracts from algae, and plant materials rich in polyphenols have been used for synthesizing CuO-NPs.6 Such bio-synthesized NPs showed promising bioactivity including broad-spectrum antimicrobial activity. These types of novel plant-based CuO-NPs also showed higher antioxidant potential, reduced cytotoxicity, and other bioactivity with potential commercial benefits.7–11
Zaluzanin-D (ZD), a sesquiterpene lactone with antifungal activity was isolated from the leaf extract of the Vernonia arborea plant.7,12 Very few studies are available based on this compound, especially in terms of its application in nanomaterial synthesis for bioactivity applications. Earlier work has identified the anti-inflammatory and epigenetic-modulating ability of ZD in human blood samples and in vitro studies. Studies were also carried out to synthetically modify ZD to enhance its anticancer activity.13 Despite these promising results not much work has been done to further explore the potential of ZD. The current studies, aim to utilize the ZD-enriched extract of Vernonia arborea leaf extract to bio-synthesize CuO-NPs (bCuO-NPs) and study its ability to modulate inflammation in human monocytic cell lines. This includes studying its ability to alter intracellular ROS generation and cell adhesion properties of activated human monocytic cell lines (THP-1). To the best of our knowledge, this is the first report on the anti-inflammatory effect of bCuO-NPs synthesized using ZD-enriched Vernonia arborea leaf extract.
Materials and methods
Plant collection and extraction
The leaves of Vernonia arborea were collected from Kumili, Idukki district, Kerala, India. It was authenticated by a plant taxonomist (Dr K. N. Sunil Kumar Siddha Central Research Institute, Chennai-600106) with a voucher ID – V2304220A. The leaves (5 kg) were shade-dried, powdered, and extracted with methanol (2.5 L). The methanol extract was dried and sequentially partitioned with water (250 ml) and varying combinations of 250 ml ethyl acetate/hexane (0, 5, 25, 50, 75 and 100%).14 The organic layer was collected and dried under reduced pressure to remove solvent. TLC screening of the separated fraction indicated the presence of ZD in the fraction extracted with 25% EtOAc/hexane (ZDE). The presence of ZD in the ZDE was confirmed by isolation and characterization using earlier reported methods (mentioned in the ESI data Fig. 1 & 2†).
Qualitative phytochemical analysis
The dried ZDE extract was dissolved in MeOH and analyzed for the presence of phytochemical compounds like alkaloids, tannins, terpenoids, flavonoids, saponin, steroids, phenolics, etc., using earlier standardized protocols.15
Alkaloids
1 ml, 2 N HCL was added to ZDE shaken vigorously followed by a few drops of freshly prepared Mayer's reagent. A cream-coloured precipitate indicated the presence of alkaloids in the ZDE.
Flavonoids
1 ml, 20% NaOH solution was added to ZDE. The solution turns yellow before becoming colorless with the addition of a few drops of diluted HCL indicating the presence of flavonoids.
Phenols
5 ml of distilled water was added to ZDE. Along with this 5% neutral ferric chloride was also added. The solution turned deep green indicating the presence of phenolic compounds.
Saponins
Frothing tests were conducted to determine the presence of saponins. The extract was shaken thoroughly with H2O for 10 min and allowed to rest for 10 min. A fairly stable frothing after 10 min rest for the test solution indicated the presence of saponins.
Steroids
The presence of steroids was determined using the Libermann–Burchard reaction. The presence of steroids in the extract was indicated by the formation of a green and blue ring upon the addition of the test solution.
Tannins
Formation of a yellowish-brown precipitate upon adding FeCl3 solution to the extract solution. The color disappearance through the addition of dil. H2SO4 indicated the presence of tannins.
Terpenoids
The addition of H2SO4 to the extract dissolved in CHCl3 resulted in the formation of a reddish-brown precipitate at the junction. This indicated the presence of terpenoids.
Biosynthesis of copper oxide nanoparticles
10 mM copper sulfate pentahydrate (CuSO4·5H2O) (purum p.a., crystallized, ≥99.0% RT Merck KGaA, Darmstadt, Germany) was added into 80 ml of Milli-Q water in a 500 ml glass beaker. 20 ml of the ZDE was added into the solution and stirred at 300 rpm at 70 °C for 4 h.9 The color change of the solution from green to brown indicated the formation of nanoparticles (bCuO-NPs).
Chemical and physical characterization of bCuO-NPs
The bCuO-NPs were investigated using a UV-vis spectrophotometer (LAB INDIA, UV-3200 twin beam spectrophotometer) in the 200–700 spectrum range for surface plasmon resonance detection (SPR). Fourier transform infrared spectroscopy (FT-IR) (Shimadzu IR tracer 100) was used to examine the functional groups of metal oxide and phyto components attached to the NPs. The crystalline structure and average crystal size of the bCuO-NPs were investigated using an X-ray diffractometer (XRD-BRUKER USA D8 Advance, Davinci). The particle size distribution and charge of bCuO-NPs were measured using a particle size analyzer (Horiba, SZ-100). The morphological examination and elemental composition of the bCuO-NPs were assessed using a Hi-Resolution Scanning Electron Microscope (HR-SEM) and a high-resolution-transmission electron microscope (HR-TEM) (JEOL Japan, JEM-2100 plus) coupled with Energy Dispersive X-ray spectroscopy (EDX). A drop of bCuO-NPs solution was applied to the TEM grid and allowed to cure before being mounted on a specimen holder and imaged.
Biological studies
Fetal Bovine Serum (FBS), Phosphate Buffered Saline (PBS), RPMI-1640 and trypsin was obtained from Sigma Aldrich Co, St Louis, USA. EDTA, glucose and antibiotics from Hi-Media Laboratories Ltd, Mumbai.
Cell culture and determination of IC50 values of bCuO-NPs by MTT assay
Human monocytic leukaemia cell line (THP-1) (RRID: CVCL0006), was procured from National Centre for Cell Sciences (NCCS), Pune, India. The cell line was cultured in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum (FBS), 100 U ml−1 penicillin and 0.1 mg ml−1 streptomycin in a humidified atmosphere with 5% CO2 at 37 °C. At 70–80% confluency, a suspension of 5 × 104 cells per ml of viable cells was seeded in a 96-well microtiter plate and incubated for the assays.12
The bCuO-NPs were tested on THP-1, to identify its effect on cell viability. MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) assay was carried out as mentioned earlier, using 20 000 cells per ml treated for 24 h with different concentrations of bCuO-NPs (1.25–10 mg ml−1). Doxorubicin (0.1 mg ml−1) was used as a positive control.16 After suitably processing the samples, the final absorbance of the MTT assay was read at 570 nm using a Thermo Multiskango 96-well microplate reader. Cell viability was determined as the relative percentage of treated cells to the untreated cells and plotted.
CLSM imaging of cell viability of bCuO-NPs by CLSM
Imaging experiments were carried out using a confocal laser scanning microscope, LSM-700, Carl Zeiss, Germany. These cells were then treated with PBS containing 50 μl of both, acridine orange (AO) and ethidium bromide (EB). The stained cells were visualized under CLSM to image the live (green fluorescence, 526 nm Em) and dead cells (red fluorescence, 650 nm Em).
Anti-oxidant evaluation of bCuO-NPs
DPPH (2,2-diphenyl-1-picrylhydrazyl) and ABTS ((2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid))) assays were carried out to identify the antioxidant potential of bCuO-NPs. All these assays were carried out as triplicates with the bCuO-NPs evaluated within the concentration range of (0.135–5 mg ml−1). Ascorbic acid was used as positive control. The following formula was used to calculate the antioxidant activity of bCuO-NPs:Radical scavenging activity (%) = [(Acontrol − Asample)/Acontrol] × 100%
For the DPPH assay, the reaction mixture consisting of DPPH (0.1 mM) and bCuO-NPs was incubated for 30 min at room temperature and its absorbance (A) was measured at 517 nm using a Thermo Multiskan go 96-well Microplate reader.17 ABTS radical scavenging activity was done as earlier reported with slight modifications.18 7 mM ABTS solution was mixed with 2.45 mM potassium persulfate to produce ABTS radical cation (ABTS+). The reaction mixture was diluted to an absorbance of 0.70 ± 0.02 at 734 nm using 0.2 M PBS (pH 7.4) at 30 °C. 20 μl of bCuO-NPs was added to 180 μl of diluted ABTS+ solution and incubated at 30 °C for 6 min. A decrease in absorbance at 420 nm was monitored using a spectrophotometer (UV1800, SHIMADZU, Kyoto, Japan). The antioxidant effect of bCuO-NPs against ABTS+ was measured and calculated using the formula mentioned above.
Intracellular ROS scavenging activity of bCuO-NPs
Effect of bCuO-NPs on Intracellular Reactive Oxygen Species (ROS) was measured in THP-1 cells using an earlier standardized method with minor modifications.19 The cells were cultured in a 6-well plate at a density of 7 × 104 cells per well per ml. Cells were pretreated with H2O2 followed by the treatment with bCuO-NPs (10 mg ml−1). At the end of incubation with compounds and activators, the cells were treated with 10 μM of DCFH-DA (2′,7′-dichlorodihydrofluorescein diacetate) for 30 min. Cells were washed, resuspended, and analysed for change in intracellular ROS levels, using BDFACS Calibur (Ex 488 nm/Em 535 nm).
Effect of bCuO-NPs on PMA-activated THP-1 cell adhesion
THP-1 cells were pre-treated with bCuO-NPs for 24 hours to investigate the effect of bCuO-NPs on monocyte cell adhesion. Pre-treated cells were washed and treated with PMA (Phorbol 12-myristate 13-acetate) (50 mg ml−1) for 48 h. After completion of incubation, the cells were gently washed with PBS containing 2% FBS, and the non-adhered cells were collected to determine the percentage adherence.
Toxicity of biosynthesized bCuO-NPs in a non-target organism
Breeding of zebrafish and production of embryos
The adult male and female zebrafish of the AB strain were obtained from the Tharun Aquaculture fish farm in Padappai, Chennai, 601301. The zebrafish were kept at constant pH (7.2–7.6), conductivity (130–100 S cm−1), temperature (26 ± 1.00 °C), dissolved oxygen (6.90 mg L−1), total hardness (100 mg L−1 as CaCO3) and salinity (0.5 ppt). The fish were maintained in 25 liter glass water tanks. In addition to live brine shrimp (Artemia nauplii; CIBA, Chennai), micro pellet feed was supplied three times a day. The breeding ratio (2 : 1) male and female was introduced in the hatching chambers after suitable acclimatization. Fertile eggs were recovered one to two hours after the light cycle began (14 h Lt. & 10 h dark pp). To ensure that exposure to bio-synthesized bCuO NPs began after the late cleavage or early blastula stages (between 2 and 3 hours after fertilization, or hpf), permitting the least amount of growth variation, each embryos were examined under a dissecting microscope following a specific protocol, as described by Kamaraj et al.20 and Priyadharsan et al.21
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of SRM Institute of Science and Technology and approved by the Institutional Animal Ethics Committee of SRM Institute of Science and Technology (Clearance No. SAF/IAEC/290/2022).
Toxicity test on zebrafish embryos
The bCuO-NPs stock solution was ultrasonically agitated for five minutes. Following dilution and filtration via Whatman® membrane filters (diameter 47 mm, pore size 0.45 μm), the samples were sterilized using dechlorinated tap water (DTW) without the use of dispersants. A total of 25 zebrafish embryos per well were placed into six-well plates following three DTW rinses. The embryos were subsequently subjected, for a total of 120 hours, to suspensions of bCuO-NPs at different concentrations (1.25, 2.5, 5, and 10 mg L−1), with a control of 25 ml of DTW applied. Before each test suspension of NPs was added to the well, it underwent a 20 second vortex agitation. The plates were incubated at a constant temperature of 24 ± 2.00 °C for the acute assessments; no replacements or refreshments of suspensions were made. Dead embryos and larvae were discovered in each well upon inspection and analysis at 24, 48, 72, 96, and 120 hours post-fertilization (hpf), as indicated by a white precipitate inside the chorion.22 The analysis only included trials in which the survival rate of the control well exceeded 85%. The results of the acute toxicity experiments showed that, although not substantially differentiating from the untreated control (p > 0.05), exposure to 10 mg L−1 bCuO-NPs caused the mortality rate (42.6%) and the fewest observable morphological abnormalities. The hatchability percentage was measured 48 hours following treatment exposure. It was determined that the animals were dead 120 hours after exposure, according to Priyadharsan et al.21
Effect of bCuO-NPs on DNA methylation at MMP-9 promotor region
Earlier studies from the lab have proven the ability of ZD to reduce DNA methylation at MMP-9 gene promoter region in activated THP-1 cells. The experiment was repeated with bCuO-NPs treated THP-1 cells to investigate the ability of the NPs to alter DNA methylation at the MMP9 promoter region. The experimental method followed the same protocol as reported by Cheeran and Munuswamy-Ramanujam, 2020.12 DNA was isolated from the bCuO-NPs treated and control THP-1 samples and evaluated for change in methylation pattern. Isolated DNA samples were subject to restriction digestion as per earlier protocol. PCR was carried out for the digested and control samples and the PCR products were analyzed for appropriate band patterns and results were recorded.
Statistical analysis
The results are presented as the mean ± SD, for at least three independent experiments statistical analyses were performed between the groups using one-way ANOVA, followed by a post hoc test. Differences between groups were considered to be significant at a P value of <0.05. Statistical analyses were performed with GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA).
Results and discussion
Extraction and biosynthesis of bCuO-NPs
The dried methanolic extracts were subjected to fractionation as mentioned earlier and each fraction was tested for its ability to synthesize bCuO-NPs. Based on the UV results it was observed the fraction extracted using 25% EtOAc in hexane provided a better yield of NPs under the given conditions. The other fractions either did not show color change or did not show the expected absorbance when subjected to UV spectrometry. It is to be noted that the TLC of the 25% fraction showed a significant presence of sesquiterpene lactone ZD (compared to the standard). The presence of ZD in the extract was confirmed by isolating the compound from the extract using standard column chromatography technique and analyzing using spectroscopy techniques (ESI Fig. 1 & 2†).23
Phytochemical analysis of zaluzanin-D enriched EtOAc extract
The 25% EtOAc/hexane fraction that was significantly effective in generating bCuO-NPs was subjected to phytochemical analysis. The presence of alkaloids, tannins, terpenoids, flavonoids, steroids, phenolics, and steroids were all examined in the selected fraction. The fraction showed the presence of tannins, flavonoids, and terpenoids (Table 1).
Phytochemical analysis of zaluzanin-D enriched leaf VA extract.
| Chemical compounds | Presence (+)/absence (−) |
|---|---|
| Tannins | + |
| Flavonoids | + |
| Phenolics | − |
| Alkaloids | − |
| Steroids | − |
| Terpenoids | + |
| Saponins | − |
UV-vis spectroscopy analysis
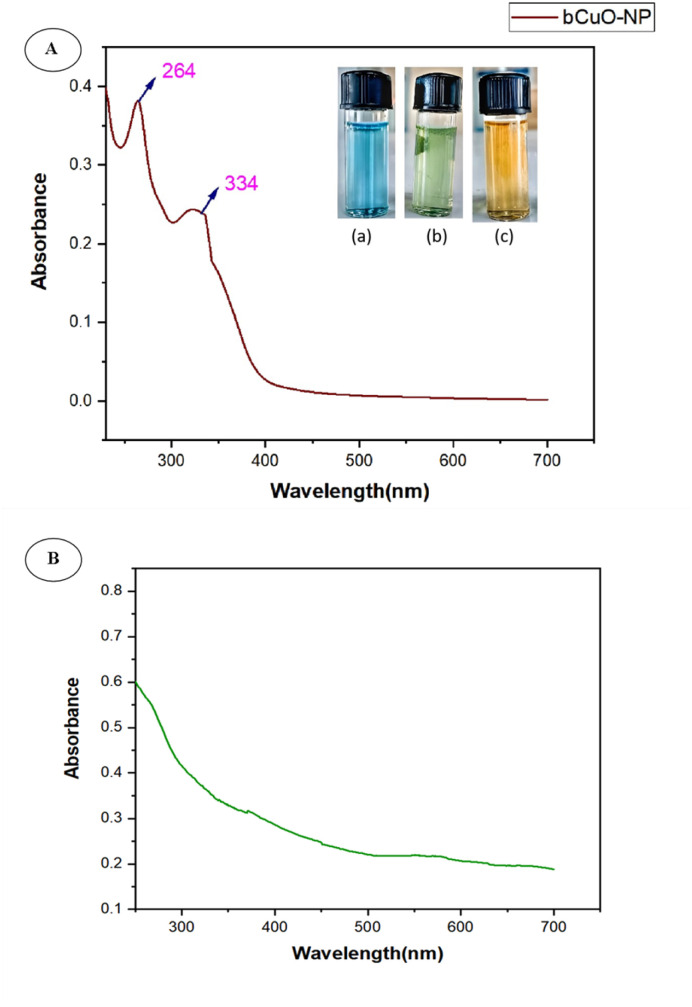
UV-vis spectroscopy investigation of the bCuO-NPs revealed two peaks, indicating that the ZD enriched fraction of Vernonia arborea promoted bio synthesis of copper oxide NPs. The first peak was observed at 264 nm and the second was at 334 nm (ref. 24) (Fig. 1A). The absorption peaks at 264 and 334 nm corresponded to the inter-band transition of the copper metal's core electrons, which is what makes CuO NPs.8,25 The broad size distribution of bCuO-NPs could explain the broad peaks in UV-vis spectra.
Fig. 1. (A) UV-vis spectrum of bCuO-NPs, (B) UV-vis spectrum of plant extract.
We predicted the second peak to be the Surface Plasmon Resonance (SPR), indicating the presence of CuNPs. In earlier investigations, CuNPs demonstrated SPR in a region between 562 and 573 nm. However, our work detected SPR in the vicinity of 334 nm. The location of the plasmon absorption peak, which depends on various factors such as the size, shape, and type of solvent used, most likely caused it.26 These variables correlated with the refractive index of the nanoparticle medium. The large particle surfaces maintain sufficient separation from each other. Electrons require less energy to move from their equilibrium positions compared to smaller particles, thereby reducing the adequate restoring force of ions on electrons. The same understanding also applies to forms related to the surface. Furthermore, some variations in the SPR and λmax are also caused by structural changes in phytochemicals adsorbed on the nanosurface. Numerous studies in the literature on the synthesis of copper/CuNPs often produce a mixture of CuO and Cu2O nanoparticles using various methods. The color shift was linked to the produced NPs surface plasmon resonance (SPR), indicating that the metabolites in the fraction are effective at reducing metal Cu to the nanoscale27,28 And there is no significant peak observed in the UV spectra of the plant extract (Fig. 1B) in the same wavelength compared with that of bCuO-NPs.28,29
FT-IR analysis
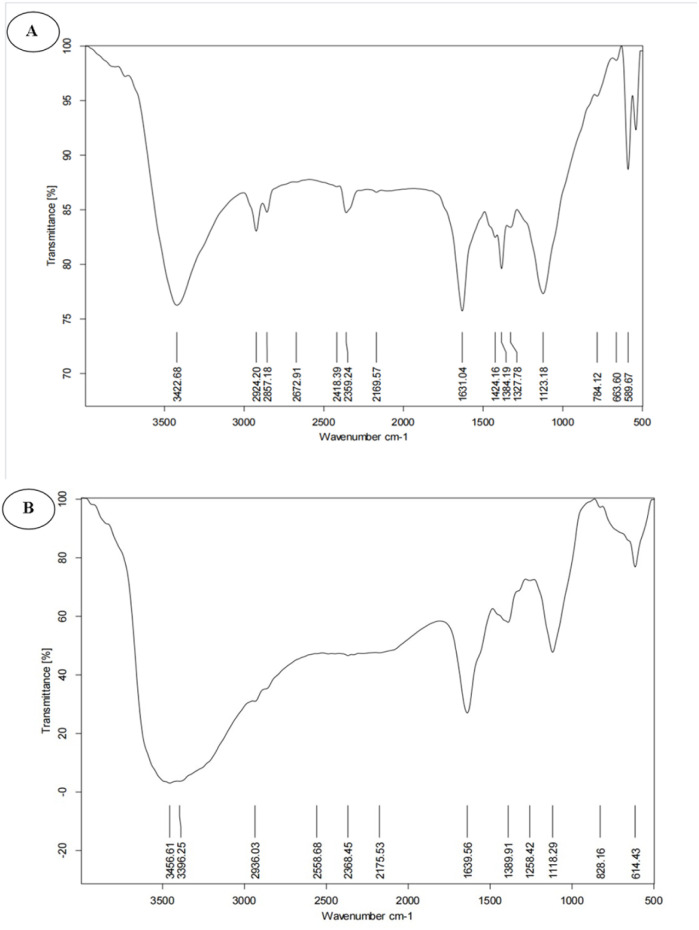
FT-IR analysis was used to characterize the functional groups present in the bCuO-NPs or to investigate the capping on the surface of copper nanoparticles. As shown in Fig. 2(A), ZD enriched ZDE showed peaks at 614, 828, 1118, 389, 1639, 2368, and 3396 cm−1. O–H vibration peak and stretching C C–H conjugated alkene had large absorption peaks at 3396 and 1639 cm−1 respectively.30,31 The C C–H bending of alkene was represented by a peak at 614 cm−1, while the strong stretching of hetero-bonded hydrogen was represented by a peak at 2368 cm−1. FT-IR spectrum of bCuO-NPs revealed a similar peak pattern at 3422 cm−1 (O–H stretching), 2924 cm−1, 2857 cm−1 (medium C–H stretching, alkane), 2359 (hetero bonded hydrogen), 1631 cm−1 (strong C C, alkene), 1424 cm−1 (C–H medium, bending), 1384 cm−1 (medium, C–H bending, alkene) 1327 cm−1 (O–H bending), 1123 cm−1 (strong C–O stretching, secondary alcohol), 784 cm−1 (strong C–H bending). The bands between 663 and 589 cm−1 correspond to metal–oxygen, indicating the presence of bCuO-NPs.29 This spectroscopy has long been used as an effective tool for obtaining additional information about the nature of copper oxides. These bands at 589 cm−1 and 663 cm−1 correspond to CuO vibrations, confirming the formation of CuO/Cu2O NPs and aligning with literature values.26,27 A similarity between the functional group profile of the ZD enriched ZDE and bCuO-NPs indicates the presence of extract material in the NPs. Earlier studies have shown that this type of biosynthesized NPs is a result of the extract phyto components that may play a role in the reduction, capping, and stabilizing of the NPs.32 In a previous study (Babu Nagati et al., 2012), the surface of silver NPs synthesized with Cajanus cajan leaf extract revealed the presence of alkaloids and terpenoids.33
Fig. 2. (A) FT-IR spectrum ofbCuO-NPs, and (B) zaluzanin-D enriched Vernonia arborea ethyl acetate extract.
XRD analysis
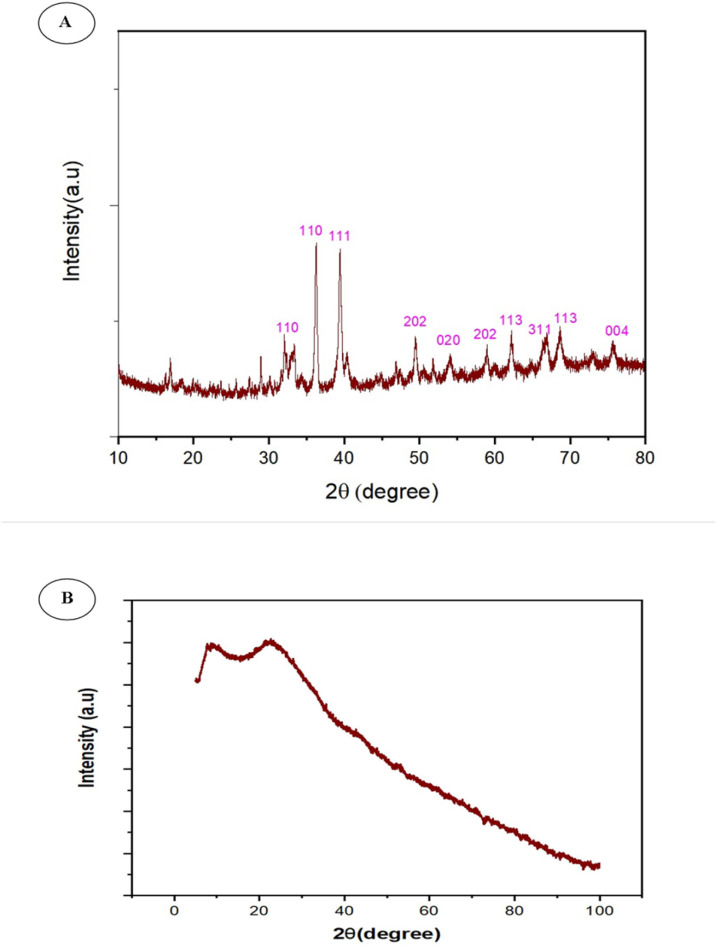
It is standard practice to use XRD patterns to confirm the crystalline nature of bCuO-NPs. XRD of bCuO-NPs revealed ten peaks at 32.41°, 35.59°, 39.13°, 49.13°, 53.88°, 58.35°, 61.79°, 66.76°, 68.42°, and 75.37°, at 2 theta values (Fig. 3A). These peaks are designated to (110), (110), (111), (202), (020), (202), (113), (311), (113), and (004) respectively.34 The diffraction peaks are well synchronized with the JCPDS Card (Joint Committee on Powder Diffraction Standards, 89-2529), indicating that the green synthesized bCuONPs are crystalline, and the additional peaks represent the organic materials present in the secondary metabolites from ZDE as a capping/stabilizing agent in the redox reaction. The peaks confirmed the presence of bCuO-NPs.34 Additional peaks observed in the XRD pattern could be attributable to precipitation of plant metabolites. The XRD analysis of ZDE in Fig. 3B lacks distinct sharp peaks due to its plant origin and amorphous solid state upon drying. Instead of sharp XRD peaks, it exhibits broad halos at 2θ = 20° and 100°.
Fig. 3. (A) XRD pattern of bCuO-NPs showed intense peaks at specific 2 theta values and (B) plant extract.
Particle size analysis of bCuO-NPs
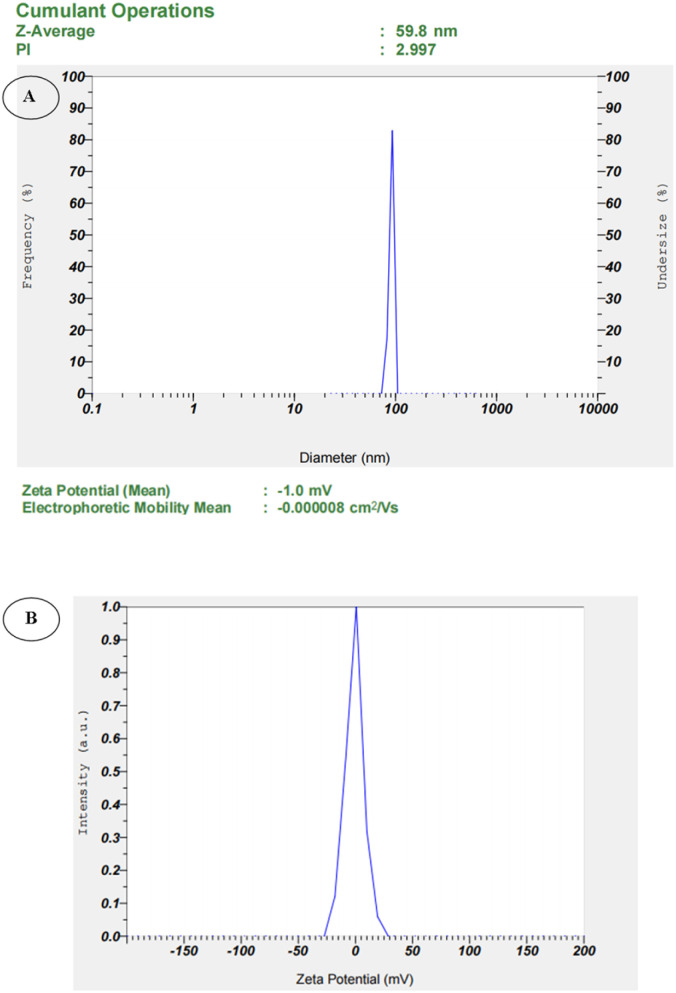
The average size and charge of the bCuO-NPs were determined using particle size analysis. The NPs were discovered to have an average size of 59.8 nm (Fig. 4A) and a charge of −1.0 mV (Fig. 4B). The movement of the NPs under the influence of the electric field was used to determine the surface charge of the NPs.35 The influence of the solvent, pH, and ionic strength on the surface charge of NPs is significant. As a result of its amphipathic character, we chose Milli-Q water as a solvent, which does not affect surface charge during analysis. The negative charge suggested that the bCuO-NPs are stable and maintain strong repulsion between them, preventing the bCuO-NPs from aggregating. The magnitude of the charge on the bCuO-NPs has a direct relationship with its stability.36
Fig. 4. (A) Particle size analysis of bCuO-NPs, (B) zeta potential of bCuO-NPs.
Morphological analysis (HR-SEM) of bCuO-NPs
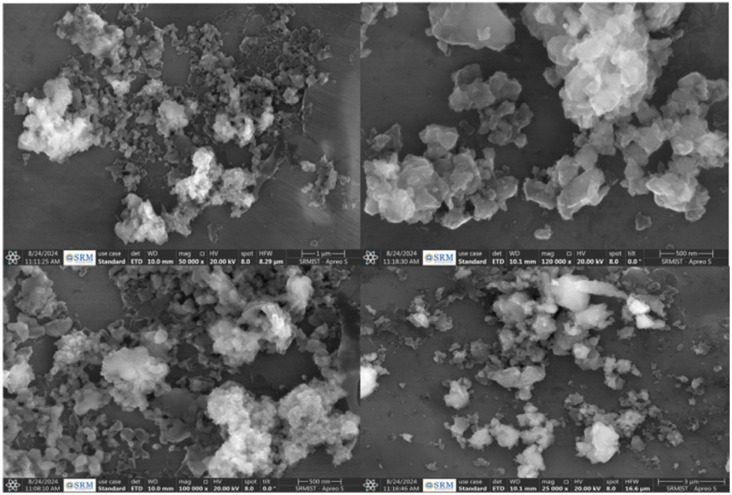
The morphology of the bCuO-NPs was examined using High Resolution Scanning Electron Microscopy (HR-SEM) (Fig. 5). HR-SEM images exhibited bCuONPs to be aggregated and with spherical shapes.37,38 This can be due to the coating of different surface functional groups from the prepared extracts.
Fig. 5. HR-SEM images of synthesized bCuO-NPs.
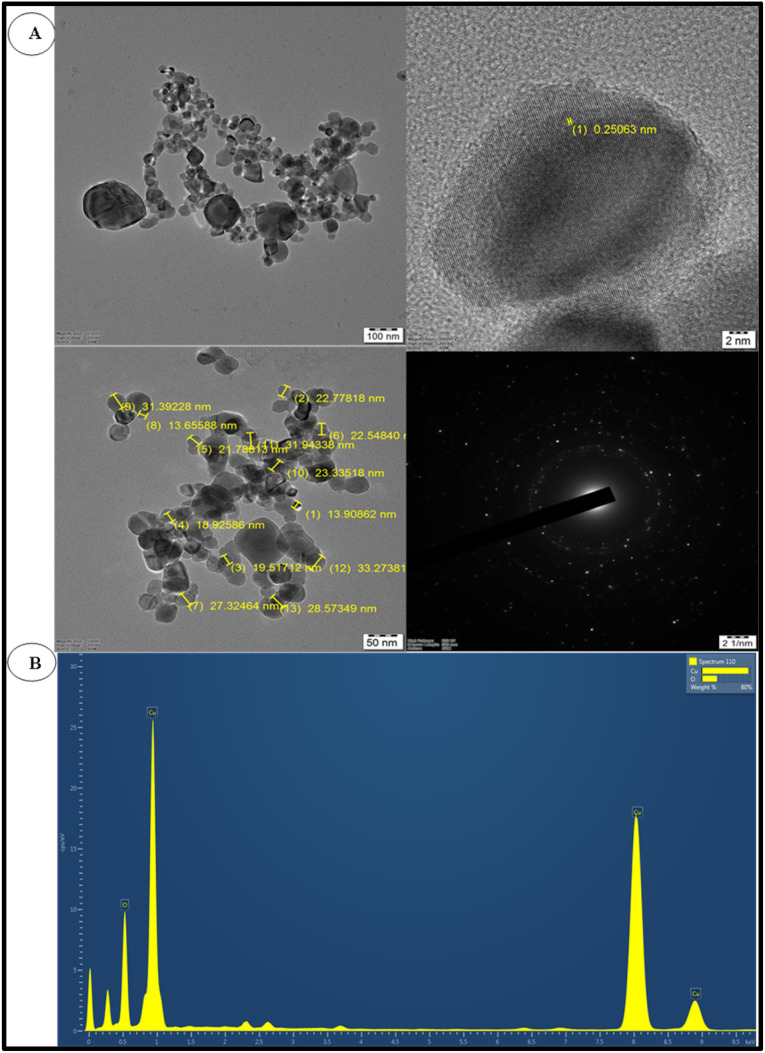
TEM and EDS profile analysis of bCuO-NPs
The shape and size of the bCuO-NPs were investigated using TEM (Fig. 6A) and EDS profile analysis (Fig. 6B). Fig. 6A confirms the effective synthesis of spherical bCuO-NPs from ZD enriched ZDE. The NPs were spherical in shape and mono-dispersed as per the XRD and TEM examination.39–41 This phenomenon could be advantageous in biotechnological and medicinal applications where the size of the NPs must be consistent. The potential application of NPs grows as their size shrinks. The findings are consistent with the Aglaia elaeagnoidea flower extract mediated synthesis of spherical CuO-NPs of 26–54 nm.42 The elemental makeup of the bCuO-NPs was validated by the EDS analysis. The bCuO-NPs were identified in the EDS profile (Fig. 6B), and it was determined that their composition was 75.26% Cu and 24.74% O. According to the EDS profile, the creation of bCuO-NPs was revealed by the prominent absorption peaks of Cu and O38 (Table 2).
Fig. 6. (A) TEM images of bCuO-NPs and SAED pattern and (B) EDS profile of bCuO-NPs.
Percentage of elemental composition in bCuO-NPs.
| Element | Wt% |
|---|---|
| O | 24.74 |
| Cu | 75.26 |
| Total | 100.00 |
Evaluating the cytotoxicity of bCuO-NPs
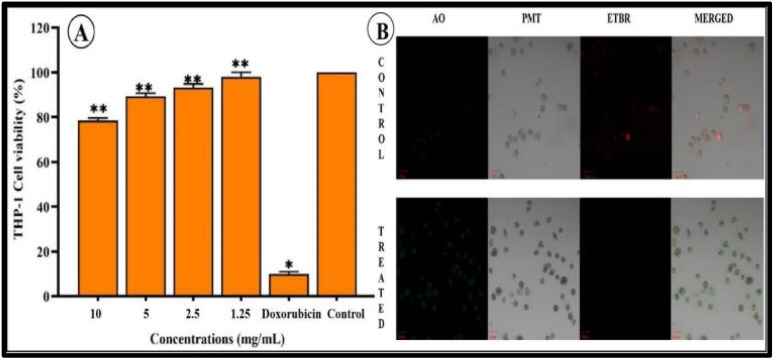
To establish the effect of bCuO-NPs on cell viability, MTT assay was carried out. THP-1 cells were used for the assay. These are human leukemic monocyte cells and used as an in vitro representation of monocytes present in the human blood.43 THP-1 cells were treated with bCuO-NPs in escalating doses of 1.25–10 mg ml−1 (Fig. 7A) with doxorubicin as positive control. The anticancer drug doxorubicin showed a THP-1 cell viability of 10% which was along the expected line. The bCuO-NPs showed relatively less toxicity (21.51%) even at the highest tested dose of 10 mg ml−1. At the lowest dose of 1.25 mg ml−1 the bCuO-NPs did not show any significant cytotoxicity compared to the control untreated group. The results of the MTT assay were confirmed by the CLSM images of the treated and control THP-1 cells. The cells treated with AO/EtBr after incubating with bCuO-NPs for 18 h showed sparse uptake of EtBr (red fluorescence) (Fig. 7B). THP-1 cells also did not show any significant change in their morphology between the treatment and the untreated control group. This confirmed that the bCuO-NPs do not have appreciable toxicity against the cells at the tested concentrations. The MTT assay thus indicated a very low toxicity for the bCuO-NPs. This enhances the possibility of NP's therapeutic as a treatment option.
Fig. 7. (A) Percentage of THP-1 cell viability treated with different concentrations (1.25–10 mg ml−1) of bCuO-NPs, and (B) CLSM images of THP-1 cells treated with control doxorubicin and bCuO-NPs (*, ** denote significant differences between the treated and control).
DPPH and ABTS based anti-oxidant evaluation of bCuO-NPs
To further investigate the potential of bCuO-NPs as an anti-inflammatory agent, anti-oxidant assays were carried out. The antioxidant assays with DPPH and ABTS are a useful measure to evaluate the antioxidant capacity of the bCuO-NPs. These colorimetric assays provide reliable information about the antioxidant scavenging potential of the test material. This in turn could help understand the ability of the NP to be used as anti-inflammatory agent.
DPPH radical scavenging activity assay
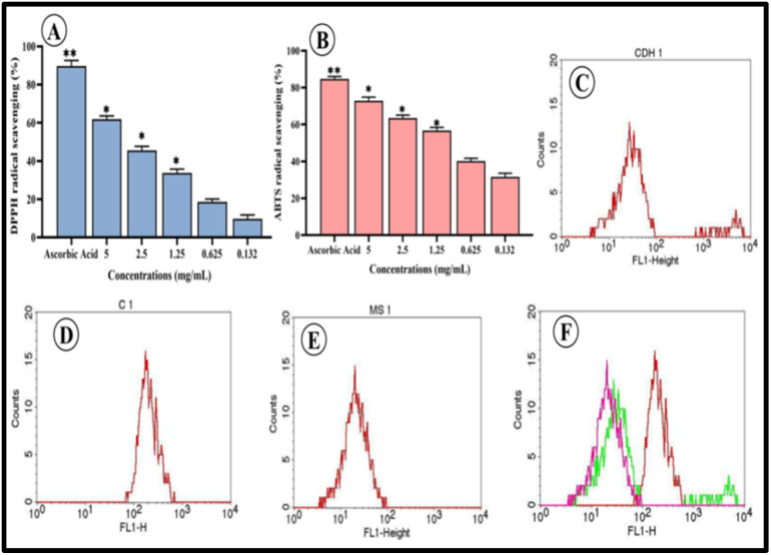
The stable free radical DPPH, which has nitrogen at its core, turns yellow when reduced. The antioxidant bCuO-NPs ability to scavenge free radicals as it lowers the DPPH radical by donating hydrogen is what causes the shift in colour.44 In the present result, strong radical scavenging capabilities were demonstrated by bCuO-NPs and the concentration of 0.132–5 mg ml−1, with higher concentrations exhibiting a high radical scavenging capacity (9.78–61.91%) (Fig. 8A).
Fig. 8. (A) DPPH, (B) ABTS radical scavenging activities of various concentrations of bCuO-NPs, (C) FACS data of CDH1 – DCFDA treated THP-1 cells (D) C1 – DCFDA + H2O2 treated THP-1 cells (E) MS1 – DCFDA + H2O2 + bCuO-NPs treated THP-1 cells (F) overlay diagram of the FACS analysis (*, ** denote significant differences between the treated and control).
ABTS radical scavenging assay
The ABTS radical, which is used to assess the antioxidant potential of compounds or extracts, is both a lipid-soluble and a water-soluble radical.17 At all investigated concentrations (0.132–5 mg ml−1), the ABTS free radical was effectively scavenged by the bCuO-NPs. Concentration-dependent anti-oxidant activity (31.64–72.94%) was demonstrated by the bCuO-NPs (Fig. 8B). The ABTS scavenging activity of the positive control, ascorbic acid, ranged from 62% to 92%.
Intracellular ROS scavenging activity of bCuO-NPs
The promising results from the DPPH assay and the ABTS assay indicated the potential of bCuO-NPs as an antioxidant agent. Hence, bCuO-NPs ability to regulate intracellular reactive oxygen species (ROS) was tested. ROS assay was carried out by DCFDA staining of H2O2 activated THP-1 cells. The H2O2 activated DCFDA stained cells show increased fluorescence (green) in FACS. An effective anti-oxidant agent will reduce the intracellular ROS levels in the cells thereby decreasing the fluorescent intensity of the cells.18 The bCuO-NPs at 1.25 mg ml−1 concentration considerably decreased the intracellular ROS in comparison to cells treated with peroxide alone. This was observed in FACS histogram wherein THP-1 cells treated with bCuO-NPs and peroxide showed a considerable drop in the fluorescence intensity in the FL1 detector (Fig. 8C–F) compared to the cells treated with peroxide alone. A key factor in controlling the cellular inflammatory process is the prevention of ROS production in cells triggered by peroxide.11 Therefore, the compounds' capacity to neutralize intracellular ROS in activated THP-1 cells acts as a marker indicating the potential of bCuO-NPs as an anti-inflammatory agent.
bCuO-NPs reduce PMA induced adhesion of THP-1 monocytic cells
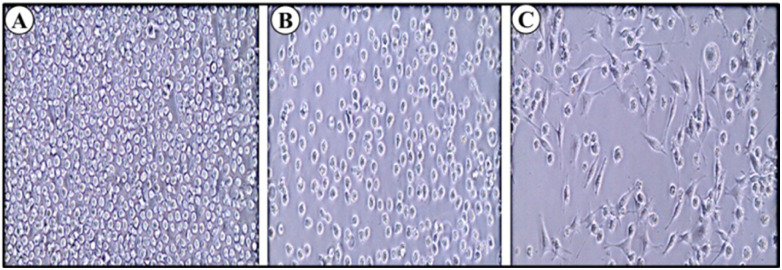
Adhesion of monocytes to endothelium is one of the preliminary inflammatory responses in the arterial walls; this is followed by diapedesis or invasion of monocytes into inflamed tissues. PMA (phorbol 12-myristate 13-acetate) is a well-known protein kinase C stimulator and other kinases, which subsequently leads to increased adherence and expression of markers associated with macrophage-like cells. PMA is used in in vitro assays to mimic inflammatory response and differentiation of THP-1 monocytes leading to activated monocytes.11 In this study, a previously standardized PMA concentration of 50 mg ml−1 was used to activate the THP-1 cells for 48 h. After incubating with PMA for 48 h the cells showed stable adhesion with minimal effect on cell survival. THP-1 cells were pre-treated (24 h) with bCuO-NPs followed by activation with PMA as mentioned above. THP-1 cells treated with the NPs showed significantly reduced adhesion (∼70%) (Fig. 9–C) compare to the control.45 This ability of the bCuO-NPs to reduce adhesion indicates the potential of bCuO-NPs to act as anti-inflammatory material.
Fig. 9. (A) Control: THP-1 cells – non-adhered (shows circular shaped cells) (B) THP-1 cells + PMA + bCuO-NPs (C) THP-1 cells + PMA (shows elongated adhered cells). The microscopic image shows the ability of bCuO-NPs to inhibit PMA activated adhesion in monocytes.
Embryo mortality and hatching rate induced by CuO NPs
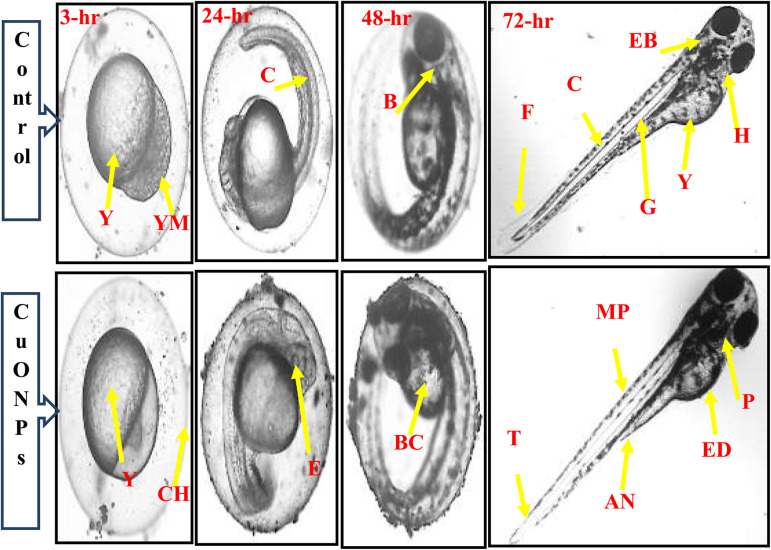
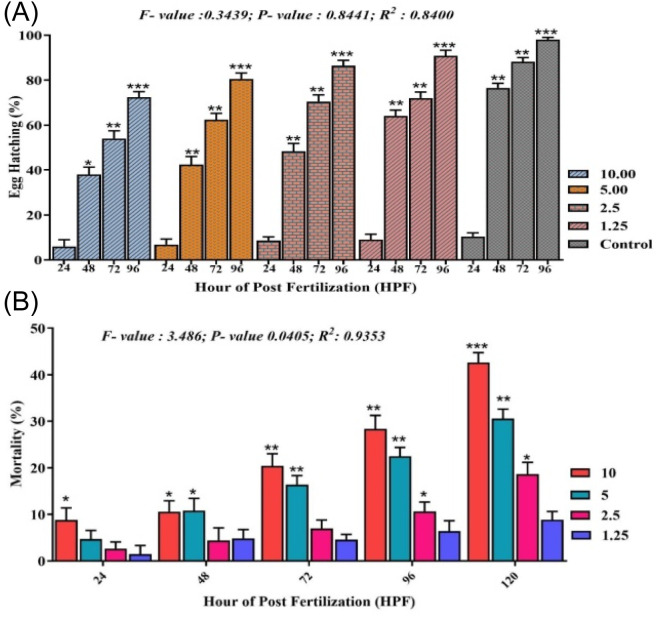
Reduced toxicity is one of the crucial features of a potential drug to be used in disease treatment. In vitro experiments with bCuO-NPs repeatedly proved its potential as an anti-inflammatory drug. The MTT assay also proved the lack of toxicity to the NP at 1.25 mg ml−1. To further confirm the safety of bCuO-NPs, testing in sensitive models could provide important information before proceeding to other in vivo assays. In this scenario, the zebrafish toxicity model is trusted for its ability to provide accurate cytotoxicity data. To determine the potential toxicity of subjecting zebrafish embryos to biogenic bCuO-NPs (1.25–10 mg L−1), we examined the rate of egg hatching and death during the duration of the current observation period (Fig. 10). Most embryos hatch 48 to 96 hours following fertilization. The egg hatching rate was less inhibited in the embryos exposed to bCuO-NPs, as seen in Fig. 11A. The test groups treated with 10 mg L−1 bCuO-NPs had a significantly high hatching rate (78.4%) at 96 hpf compared to the control group (94.28%). The mortality at the lower concentration did not differ noticeably, as shown in Fig. 11B. The mortality of the 5 and 10 mg L−1 treatment groups increased considerably to the control group as the dosages were raised. Our findings indicate that the degree and duration of exposure to bCuO-NPs influenced the rise in embryonic toxicity and at the lower concentrations there was no significant difference in toxicity.
Fig. 10. Effect of bCuO-NPs and control (1.25–10 mg ml−1) showing normal zebrafish embryogenesis at different HPF development. The inverted microscope was used to produce the images. There were 4 periods depicted: blastula (3 HPF), segmentation (12 HPF), and pharyngula (24 HPF), and hatching (48 HPF). MP—melanophores; G—gut; Y—yolk sac; Ed—edema; Ch—chorion; F—fin; YM—yolk membrane; B—brain; E—eye anlage; T—tail P—pericardium; EB—ear bud; H—heart; C—chorda; BC—blood cells; AN—anus; (scale bar = 0.5 mm).
Fig. 11. Egg hatching rate of zebrafish embryos induced by (A) bCuO-NPs at 24–96 hpf, and (B) mortality of embryos increased in dose- and time-dependent manner. Data are expressed as means ± S. E. from three independent experiments (p > 0.05).
Many researches have revealed a decline in the survival and hatching rates of zebrafish embryos in different nanomaterials.21,45–47 The embryo's developmental delay was directly caused by the suppression of the egg hatching rate. In this investigation, we found that bCuO-NPs (1.25–10 mg L−1) caused anomalies in the head and tail of the embryos in addition to edema inside the yolk sac. The most prevalent abnormalities produced by embryos exposed to NPs were edema and a tail deformity, according to the results displayed in Fig. 10. Xu et al. 2012 (ref. 48) reported, that the embryos treated to titanium dioxide NPs exhibited severe abnormalities associated with the pericardial edema and yolk sac.48 Apart from this, the most frequently seen malformations in the embryos treated with silver NPs were fin fold and tail deformity regions.49 Consequently, we were also able to confirm that zebrafish embryos exposed to various NPs exhibited a range of abnormalities at high concentrations.
MMP-9 DNA promotor methylation studies
Earlier studies have shown the ability of pure ZD to reduce inflammation in THP-1 cells. The compound also altered the gene and protein expression in an anti-inflammatory manner. This included changes in levels of MMP-9 and other inflammatory cytokines. One key finding of the study was the ability of ZD to alter MMP-9 gene promoter DNA methylation.12 In the current study, the bCuO-NPs showed an anti-inflammatory profile similar to ZD. Hence, investigating the NP's effect on the DNA methylation pattern of the MMP-9 gene promotor region would be of interest. RFLP experiments carried out with bCuO-NPs treated THP-1 cells showed similar epigenetic modification capability for the NPs. Hypomethylation was observed in the samples of PMA-activated THP-1 cells treated with NPs (ESI Fig. 3†). The similarity in activity between pure ZD and the NPs could be attributed to the ZD-enriched extract present in the NP. Once again, the studies indicate the ability of bCuO-NPs to modulate inflammation similar to the active compound ZD found in the leaves of VA.
Conclusions
In the present study, CuSO4·5H2O was used to synthesize NPs via the biosynthesis route. A method standardized to generate ZD-enriched Vernonia arborea was developed and the enriched extract was successfully used to synthesize CuO-NPs from CuSO4·5H2O. The bCuO-NPs synthesized using enriched extract showed a spherical structural form, a mono-dispersed size distribution with nanoscale dimensions, and a monoclinic crystal structure. bCuO-NPs showed an effective ability to inhibit antioxidants. The NPs showed reduced toxicity both in in vitro human monocytic cell lines as well as in vivo zebrafish models. The antioxidant ability of the biosynthesized NPs extended into in vitro model, wherein they reduced intracellular ROS levels in THP-1 cell lines. Further, the NPs prevented PMA activation of the monocytes, indicating the ability to reduce inflammation in monocytes. Overall the studies identified the ability of zaluzanin D enriched extract to generate CuO NPs with the ability to modulate inflammation.
Data availability
Data will be made available on request.
Author contributions
Muhammad Sadiq: writing – original draft, validation, methodology, formal analysis, data curation, conceptualization. Arvind Sivasubramanian: writing – review & editing, validation. Aswathy Karanath-Anilkumar: methodology, formal analysis. Shazia Anjum-Musthafa: methodology, formal analysis. Ganesh Munuswamy-Ramanujam: writing – original draft, formal analysis, project administration, validation. Chinnaperumal Kamaraj: methodology, writing – review & editing, formal analysis.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary Material
Acknowledgments
The authors are grateful to the Department of Chemistry, SRM Institute of Science and Technology, Kattankulathur-603 203, Tamil Nadu, India and Interdisciplinary Institute of Indian System of Medicine (IIISM), SRM Institute of Science and Technology, Kattankulathur-603 203, Tamil Nadu, India, for providing the facilities required to finish this work.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04032e
References
- Bansal V. Bharde A. Ramanathan R. Bhargava S. K. Adv. Colloid Interface Sci. 2012;179:150–168. doi: 10.1016/j.cis.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Sankar R. Maheswari R. Karthik S. Shivashangari K. S. Ravikumar V. Mater. Sci. Eng., C. 2014;44:234–239. doi: 10.1016/j.msec.2014.08.030. [DOI] [PubMed] [Google Scholar]
- Sankar R. Karthik A. Prabu A. Karthik S. Shivashangari K. S. Ravikumar V. Colloids Surf., B. 2013;108:80–84. doi: 10.1016/j.colsurfb.2013.02.033. [DOI] [PubMed] [Google Scholar]
- Ishak N. A. I. M. Kamarudin S. K. Timmiati S. N. Mater. Res. Express. 2019;6:112004. [Google Scholar]
- Das R. K. Brar S. K. Nanoscale. 2013;5:10155–10162. doi: 10.1039/c3nr02548a. [DOI] [PubMed] [Google Scholar]
- Jadoun S. Arif R. Jangid N. K. Meena R. K. Environ. Chem. Lett. 2021;19:355–374. [Google Scholar]
- Kumari G. N. K. Masilamani S. Ganesh M. R. Aravind S. Sridhar S. R. Fitoterapia. 2003;74:479–482. doi: 10.1016/s0367-326x(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Abboud Y. Saffaj T. Chagraoui A. El Bouari A. Brouzi K. Tanane O. Ihssane B. Appl. Nanosci. 2014;4:571–576. [Google Scholar]
- Karuppannan S. K. Ramalingam R. Khalith S. B. M. Dowlath M. J. H. Raiyaan G. I. D. Arunachalam K. D. Biocatal. Agric. Biotechnol. 2021;31:101904. [Google Scholar]
- Moulton M. C. Braydich-Stolle L. K. Nadagouda M. N. Kunzelman S. Hussain S. M. Varma R. S. Nanoscale. 2010;2:763–770. doi: 10.1039/c0nr00046a. [DOI] [PubMed] [Google Scholar]
- Kumar P. P. N. V. Shameem U. Kollu P. Kalyani R. L. Pammi S. V. N. Bionanoscience. 2015;5:135–139. [Google Scholar]
- Cheeran V. Munuswamy-Ramanujam G. Int. Immunopharmacol. 2020;87:106803. doi: 10.1016/j.intimp.2020.106803. [DOI] [PubMed] [Google Scholar]
- Hibasami H. Yamada Y. Moteki H. Katsuzaki H. Imai K. Yoshioka K. Komiya T. Int. J. Mol. Med. 2003;12:147–151. [PubMed] [Google Scholar]
- Nagaraj B. Musthafa S. A. Muhammad S. Munuswamy-Ramanujam G. Chung W. J. Alodaini H. A. Hatamleh A. A. Al-Dosary M. A. Ranganathan V. J. King Saud Univ., Sci. 2022;34:101989. [Google Scholar]
- Gul R. Jan S. U. Faridullah S. Sherani S. Jahan N. Sci. World J. 2017:5873648. doi: 10.1155/2017/5873648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musthafa S. A. Kasinathan T. Bhattacharyya R. Muthu K. Kumar S. Munuswamy-Ramanujam G. Mater. Today: Proc. 2021;40:S216–S223. [Google Scholar]
- Sannasimuthu A. Kumaresan V. Pasupuleti M. Paray B. A. Al-Sadoon M. K. Arockiaraj J. Algal Res. 2018;35:519–529. [Google Scholar]
- Sannasimuthu A. Kumaresan V. Anilkumar S. Pasupuleti M. Ganesh M.-R. Mala K. Paray B. A. Al-Sadoon M. K. Albeshr M. F. Arockiaraj J. Free Radical Biol. Med. 2019;135:198–209. doi: 10.1016/j.freeradbiomed.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Ramachandran S. Loganathan S. Cheeran V. Charles S. Munuswamy-Ramanujan G. Ramasamy M. Raj V. Mala K. Leuk. Res. Rep. 2018;9:28–35. doi: 10.1016/j.lrr.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaraj C. Deepak P. Balasubramani G. Karthi S. Arul D. Aiswarya D. Amutha V. Vimalkumar E. Mathivanan D. Suseem S. R. Ecotoxicol. Environ. Saf. 2018;161:221–230. doi: 10.1016/j.ecoenv.2018.05.062. [DOI] [PubMed] [Google Scholar]
- Priyadharsan A. Ragavendran C. Kamaraj C. Kadaikunnan S. Raja R. K. Thiruvengadam M. J. Environ. Chem. Eng. 2023;11:110541. [Google Scholar]
- Badawy A. A. Abdelfattah N. A. H. Salem S. S. Awad M. F. Fouda A. Biology. 2021;10:233. doi: 10.3390/biology10030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari G. N. K. Masilamani S. Ganesh M. R. Aravind S. Phytochemistry. 2003;62:1101–1104. doi: 10.1016/s0031-9422(02)00667-2. [DOI] [PubMed] [Google Scholar]
- Rabiee N. Bagherzadeh M. Kiani M. Ghadiri A. M. Etessamifar F. Jaberizadeh A. H. Shakeri A. Int. J. Nanomed. 2020:3983–3999. doi: 10.2147/IJN.S255398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafique M. Shaikh A. J. Rasheed R. Tahir M. B. Gillani S. S. A. Usman A. Imran M. Zakir A. Khan Z. U. H. Rabbani F. J. Inorg. Organomet. Polym. Mater. 2018;28:2455–2462. [Google Scholar]
- Zayyoun N. Bahmad L. Laânab L. Jaber B. Appl. Phys. A. 2016;122:488. [Google Scholar]
- Djamila B. Eddine L. S. Abderrhmane B. Nassiba A. Barhoum A. Biomass Convers. Biorefin. 2024;14:6567–6580. [Google Scholar]
- Atarod M. Nasrollahzadeh M. Sajadi S. M. RSC Adv. 2015;5:91532–91543. [Google Scholar]
- Krithiga N. Jayachitra A. Rajalakshmi A. Indian J. Nanosci. 2013:6–15. [Google Scholar]
- Saravanakumar K. Shanmugam S. Varukattu N. B. MubarakAli D. Kathiresan K. Wang M.-H. J. Photochem. Photobiol., B. 2019;190:103–109. doi: 10.1016/j.jphotobiol.2018.11.017. [DOI] [PubMed] [Google Scholar]
- Mohamed A. A. Abu-Elghait M. Ahmed N. E. Salem S. S. Biol. Trace Elem. Res. 2021;199:2788–2799. doi: 10.1007/s12011-020-02369-4. [DOI] [PubMed] [Google Scholar]
- Sundararajan B. Mahendran G. Thamaraiselvi R. Ranjitha Kumari B. D. Bull. Mater. Sci. 2016;39:423–431. [Google Scholar]
- Banala R. R. Nagati V. B. Karnati P. R. Saudi J. Biol. Sci. 2015;22:637–644. doi: 10.1016/j.sjbs.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy P. Ilavenil S. Srigopalram S. Maniam G. P. Yusoff M. M. Govindan N. Choi K. C. J. Cleaner Prod. 2017;141:1023–1029. [Google Scholar]
- Pugazhendhi A. Prabhu R. Muruganantham K. Shanmuganathan R. Natarajan S. J. Photochem. Photobiol., B. 2019;190:86–97. doi: 10.1016/j.jphotobiol.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Clogston J. D. and Patri A. K., Charact. nanoparticles Intend. drug Deliv., 2011, pp. 63–70 [Google Scholar]
- Priya M. Venkatesan R. Deepa S. Sana S. S. Arumugam S. Karami A. M. Vetcher A. A. Kim S.-C. Sci. Rep. 2023;13:18838. doi: 10.1038/s41598-023-46002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punniyakotti P. Panneerselvam P. Perumal D. Aruliah R. Angaiah S. Bioprocess Biosyst. Eng. 2020;43:1649–1657. doi: 10.1007/s00449-020-02357-x. [DOI] [PubMed] [Google Scholar]
- Fouda A. Hassan S. E.-D. Abdo A. M. El-Gamal M. S. Biol. Trace Elem. Res. 2020;195:707–724. doi: 10.1007/s12011-019-01883-4. [DOI] [PubMed] [Google Scholar]
- Manjari G. Saran S. Arun T. Rao A. V. B. Devipriya S. P. J. Saudi Chem. Soc. 2017;21:610–618. [Google Scholar]
- Duan X. Liao Y. Liu T. Yang H. Liu Y. Chen Y. Ullah R. Wu T. J. Photochem. Photobiol., B. 2020;202:111718. doi: 10.1016/j.jphotobiol.2019.111718. [DOI] [PubMed] [Google Scholar]
- Shaheen T. I. Fouda A. Salem S. S. Ind. Eng. Chem. Res. 2021;60:1553–1563. [Google Scholar]
- Schwende H. Fitzke E. Ambs P. Dieter P. J. Leukocyte Biol. 1996;59:555–561. [PubMed] [Google Scholar]
- Batool R. Khan M. R. Sajid M. Ali S. Zahra Z. BMC Chem. 2019;13:1–15. doi: 10.1186/s13065-019-0549-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ilan O. Louis K. M. Yang S. P. Pedersen J. A. Hamers R. J. Peterson R. E. Heideman W. Nanotoxicology. 2012;6:670–679. doi: 10.3109/17435390.2011.604438. [DOI] [PubMed] [Google Scholar]
- de Assis Valadares L. P. Lima L. C. O. de Saboia-Morais S. M. T. Arantes T. M. Cristovan F. H. da Silva N. M. Andrade A. B. Ribeiro S. A. B. Alves B. G. Virote B. do C. R. Aquat. Toxicol. 2023;258:106454. doi: 10.1016/j.aquatox.2023.106454. [DOI] [PubMed] [Google Scholar]
- Yan Z. Zhou Y. Zhu P. Bao X. Su P. Front. Mar. Sci. 2023;10:1195125. [Google Scholar]
- Xu Z. Zhang Y.-L. Song C. Wu L.-L. Gao H.-W. PLoS One. 2012;7:e32818. doi: 10.1371/journal.pone.0032818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J. Browning L. M. Nallathamby P. D. Desai T. Cherukuri P. K. Xu X.-H. N. Chem. Res. Toxicol. 2012;25:1029–1046. doi: 10.1021/tx300021u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.