Summary
Cold stress is a major abiotic stress that threatens maize (Zea mays L.) production worldwide. Understanding the molecular mechanisms underlying cold tolerance is crucial for breeding resilient maize varieties. Tonoplast intrinsic proteins (TIPs) are a subfamily of aquaporins in plants. Here, we report that TIP family proteins are involved in maize cold tolerance. The expression of most TIP genes was responsive to cold stress. Overexpressing TIP2;1, TIP3;2 or TIP4;3 reduced the cold tolerance of maize seedlings, while loss‐of‐function mutants of TIP4;3 exhibited enhanced cold tolerance. Candidate gene‐based association analysis revealed that a 328‐bp transposon insertion in the promoter region of TIP4;3 was strongly associated with maize cold tolerance. This transposon insertion conferred cold tolerance by repressing TIP4;3 expression through increased methylation of its promoter region. Moreover, TIP4;3 was found to suppress stomatal closure and facilitate reactive oxygen species (ROS) accumulation under cold stress, thereby inhibiting the expression of cold‐responsive genes, including DEHYDRATION‐RESPONSIVE ELEMENT BINDING FACTOR 1 (DREB1) genes and a subset of peroxidase genes, ultimately attenuating maize cold tolerance. This study thus elucidates the mechanism underlying TIP‐mediated cold tolerance and identifies a favourable TIP4;3 allele as a potential genetic resource for breeding cold‐tolerant maize varieties.
Keywords: cold stress, maize, TIP4;3, natural variation, stomatal movement, reactive oxygen species
Introduction
Maize (Zea mays L.) is one of the most widely cultivated crops worldwide. Maize originated in the tropics and is especially sensitive to cold stress (yyGreaves, 1996). Cold stress negatively affects seed germination, seedling development and growth, culminating in lower grain yields (Verheul et al., 1996). Especially in regions with high‐latitude and altitude, cold stress during early spring presents a significant meteorological threat to maize production (Farooq et al., 2009). Therefore, enhancing the cold tolerance of maize during the germination and seedling stages can help mitigate the negative effects of temperature fluctuations.
The cold tolerance of maize is a complex trait governed by multiple quantitative trait loci (QTLs) throughout its genome. Various studies employing QTL mapping and genome‐wide association study (GWAS) methods have identified several genomic regions containing single nucleotide polymorphisms (SNPs) and insertions/deletions (InDels) associated with maize cold tolerance (Leipner et al., 2008; Zhang et al., 2020; Zhou et al., 2022). Metabolite GWAS has also played a significant role in elucidating the genetic basis of metabolic diversity during maize cold stress response (Pranneshraj et al., 2022; Zhu et al., 2023). For example, natural genetic variations in genes such as INDUCER OF CBF EXPRESSION 1 (ICE1), which encodes a basic helix–loop–helix (bHLH) transcription factor, have been associated with variations in nitrogen metabolism and cold tolerance in maize (Jiang et al., 2022). Recent lipidomic analyses have implicated a subset of key enzymes involved in lipid metabolism in maize cold tolerance, expanding the understanding of the metabolic mechanisms underlying cold response in maize (Gao et al., 2023). Additionally, advancements in transgenic technology have facilitated the identification of genes regulating maize cold tolerance through reverse genetic approaches. A subset of crucial genes involved in maize cold tolerance have been identified, including DEHYDRATION‐RESPONSIVE ELEMENT BINDING FACTOR1 (DREB1), also reported as C‐REPEAT BINDING FACTOR (CBF), type‐A response regulator 1 (ZmRR1), cellulose synthetase (CesA), mitogen‐activated protein kinases (MPK2 and MPK8) and basic leucine zipper 68 (bZIP68) (Li et al., 2022; Yang et al., 2023; Zeng et al., 2021).
The cold signalling pathway mediated by CBF/DREB1 key transcription factors have been extensively studied across plant species (Liu et al., 2018; Shi et al., 2018). In Arabidopsis thaliana, three CBF/DREB1 genes play a crucial role in regulating the expression of cold‐regulated (COR) genes. In maize, overexpression of DREB1 genes, such as DREB1A, DREB1.5, DREB1.7 and DREB1.10, enhances cold tolerance (Han et al., 2020; Li et al., 2022; Zeng et al., 2021), suggesting the conserved role of DREB1s in cold tolerance in different plant species (Yang et al., 2023). Natural variations in the ZmRR1 and bZIP68 were shown to be associated with cold tolerance through DREB1 pathway. For instance, a natural variant of ZmRR1 with a deletion containing 15‐amino acid residues in the coding region enhances cold tolerance by stabilizing ZmRR1 and promoting the expression of COR genes, e.g., DREB1 and CesA genes (Zeng et al., 2021). Conversely, bZIP68 acts as a negative regulator of maize cold tolerance by directly suppressing the expression of DREB1 genes. MPK8 phosphorylates bZIP68 to enhance its protein stability and DNA‐binding affinity (Li et al., 2022). Intriguingly, bZIP68 underwent selection during early domestication and the superior allele in teosinte was employed in maize (Li et al., 2022), suggesting that new gene resource for cold tolerance trait could be mined from the teosinte. Although numerous studies employing various strategies have identified potential candidate genes, validation of these candidates remains limited, thus constraining the comprehension of the molecular and genetic basis of maize cold tolerance.
Aquaporins (AQPs) belong to the main intrinsic protein family and regulate the water balance of plant cells and tissues by facilitating water transport across membranes (Johansson et al., 2000). Plant AQPs can transport small molecules such as CO2, hydrogen peroxide (H2O2) urea, ammonia, glycerol, silicon, boron, and arsenic (Sun et al., 2024). Emerging studies has shown that AQPs play a crucial role in mitigating damage caused by abiotic stresses, including salinity, alkali stress, drought and hypoxia stress, in many plant species (Afzal et al., 2016; Sudhakaran et al., 2021). For example, in Arabidopsis, NOD26‐LIKE INTRINSIC PROTEIN 2;1 (NIP2;1) acts as a lactic acid efflux channel to enhance plant survival during the hypoxia stress (Beamer et al., 2021). In rice (Oryza sativa), overexpressing RICE WATER CHANNEL 3 (RWC3), which encodes a plasma membrane intrinsic protein 1 (PIP1) type AQP, increased root hydraulic conductivity (Lpr) and improved water status under drought stress (Lian et al., 2004). In rose (Rosa sp.), drought‐induced phosphorylation of PIP2;1 promotes the nuclear translocation of the membrane‐tethered MYB transcription factor PHD TYPE TRANSCRIPTION FACTOR WITH TRANSMEMBRANE DOMAINS (PTM), thereby enhancing drought tolerance (Zhang et al., 2019). Similarly, tobacco (Nicotiana tabacum) AQP1 enhances water use efficiency, hydraulic conductivity and yield under salt stress (Sade et al., 2010). Conversely, an Ospip2;1 mutant was sensitive to alkali stress, with a reduced survival rate and poor growth (Zhang et al., 2023). Nevertheless, whether and how AQPs involve in cold tolerance in plants remains poorly understood.
In this study, we uncovered the negative role of tonoplast intrinsic protein (TIP) family proteins in maize cold tolerance by influencing stomatal movement and reactive oxygen species (ROS) accumulation. We identified a 328‐bp transposon insertion in the promoter region of TIP4;3. This insertion increased methylation levels, resulting in the repression of TIP4;3 transcription and consequently enhanced cold tolerance in maize. These findings highlight a crucial genetic target for development of cold‐tolerant maize varieties.
Results
Three TIPs have a negative effect on the cold tolerance of maize
To identify the components involved in cold tolerance in maize, we previously conducted a screening on transgenic maize plants overexpressing more than 700 maize genes in the background of the LH244 inbred line under cold treatment (Zeng et al., 2021). Among these genes, we found that three TIP genes encoding tonoplast aquaporin proteins showed a negative effect on the cold tolerance of maize seedlings (Chaumont et al., 2001) (Figure 1a–i). Specifically, maize seedlings overexpressing TIP2;1, TIP3;2, or TIP4;3 under the control of Ubi promoter exhibited impaired cold tolerance, with a higher area of leaf injury compared to wild‐type LH244 following cold treatment (Figure 1a–i). These findings suggest a negative role for these TIP genes in maize cold tolerance.
Figure 1.
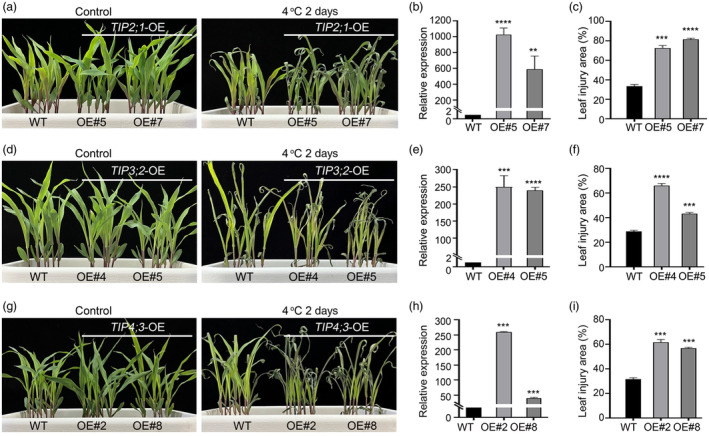
Analysis of cold tolerance in TIP2;1, TIP3;2 and TIP4;3‐overexpressing transgenic plants. (a–c) Cold‐tolerance phenotypes (a), relative TIP2;1 expression levels (b), and leaf injury area (c) of wild‐type (WT) and TIP2;1‐overexpressing transgenic plants (OE#5, OE#7) after cold treatment. (d–f) Cold‐tolerance phenotypes (d), relative TIP3;2 expression levels (e) and leaf injury area (f) of WT and TIP3;2‐overexpressing transgenic plants (OE#4, OE#5) after cold treatment. (g–i) Cold‐tolerance phenotypes (g), relative TIP4;3 expression levels (h), and leaf injury area (i) of WT and TIP4;3‐overexpressing transgenic plants (OE#2, OE#8) after cold treatment. In (a, d, g), 14‐day‐old seedlings grown at 25 °C were incubated at 4 °C for 2 days. Representative photographs were taken after 2 days of recovery at 25 °C. In (b, c, e, f, h, i), Error bars represent mean ± SD (standard deviation). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (two‐sided t‐test).
We then performed a comprehensive analysis of the expression patterns of TIP genes in response to cold stress in maize. Heatmap analysis from a previous study revealed that the expression levels of nine genes out of 11 members of maize TIP family were detectable, and most of their transcriptions were responsive to cold stress (Alexandersson et al., 2005) (Figure S1a,b). Furthermore, examination of the expression of TIP2;1, TIP3;2 and TIP4;3 by RT‐qPCR showed that all these three genes were downregulated after cold treatment (Figure 2a). To explore natural variations at these three TIP loci, we examined their expression levels across cold‐sensitive and cold‐resistant inbred lines. Consistent with the above result, the transcript levels of these genes decreased under cold treatment (4 °C, 12 h) across all inbred lines (Figure 2b). Particularly, the cold‐mediated repression of TIP4;3 expression was stronger on average in the cold‐tolerant inbred lines compared to the cold‐sensitive inbred lines, while the expression levels of TIP2;1 or TIP3;2 were comparable between the two groups of inbred lines at the same cold treatment conditions (Figure 2b). These results suggest that while TIP2;1, TIP3;2 and TIP4;3 all involved in cold stress responses, the expression level of TIP4;3 is strongly associated with the variation in cold tolerance observed among maize inbred lines.
Figure 2.
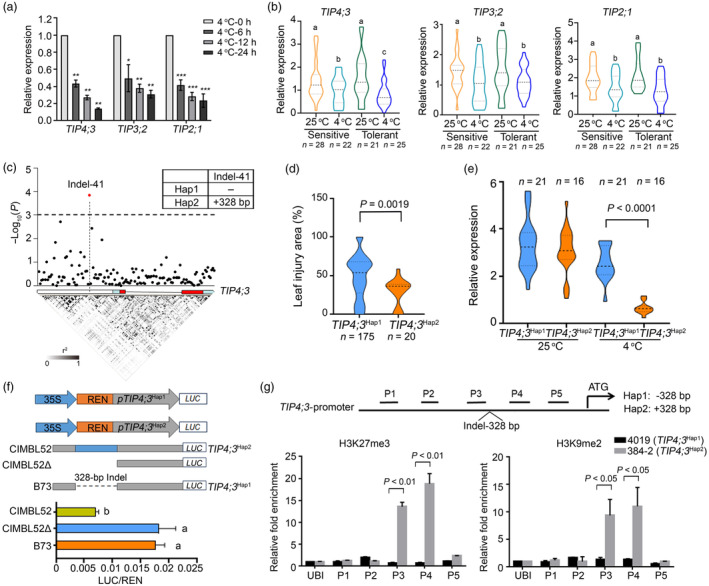
A 328‐bp insertion (InDel‐41) in TIP4;3 is significantly associated with maize chilling tolerance. (a) Relative expression levels of TIP2;1, TIP3;2 and TIP4;3 after cold treatment at 4 °C for 0, 6, 12 and 24 h. Samples from seedlings maintained at permissive conditions (25 °C) were collected as a 0‐h time point. (b) The gene expression of TIP4;3, TIP3;2 and TIP2;1 in hyper‐sensitive and hyper‐tolerant inbred lines at permissive condition (25 °C) or after cold treatment (4 °C) for 12 h. Different letters represent significant differences (P < 0.05, one‐way ANOVA). “n” represents the number of inbred lines. (c) Local Manhattan plot showing the association analysis of genetic variation at the TIP4;3 locus with chilling tolerance in maize and the pattern of pairwise linkage disequilibrium of DNA polymorphisms. A diagram of the TIP4;3 locus is shown. The white, light blue and red rectangles represent 1.3‐kb promoter, UTR and coding region, respectively. The most significant Indel (Indel‐41) in the promoter is connected to its location in the locus diagram by a dotted line. The transverse dotted line represents the threshold line. (d) Area of leaf injury (%) of inbred lines harbouring the Hap1 or Hap2 haplotypes of TIP4;3. (e) Relative TIP4;3 transcript levels in inbred lines harbouring the Hap1 or Hap2 haplotypes grown at 25 °C or exposed to 4 °C for 12 h. The 21 Hap1 and 16 Hap2 lines were randomly selected for gene expression analysis. (f) Dual luciferase (LUC) reporter assays, using firefly LUC controlled by the TIP4;3 B73, TIP4;3 CIMBL52 or TIP4;3 CIMBL52Δ promoter in maize protoplasts under 4 °C for 3 h. (g) Chromatin immunoprecipitation followed by quantitative PCR (ChIP‐qPCR) showing the chromatin state at the TIP4;3 4019 and TIP4;3 384‐2 promoters following cold stress treatment. Anti‐H3K27me3 (left) and anti‐H3K9me2 (right) antibodies were used for immunoprecipitation. Five regions (P1–P5) of the TIP4;3 promoter were tested; the maize Ubiquitin (Ubi) was used as a negative control.
Identification of a favourable TIP4;3 allele with enhanced cold tolerance
To identify a favourable natural allele of TIP4;3 that enhances maize cold tolerance, we re‐sequenced a 4.0‐kb genomic region containing TIP4;3 (comprising a 1.3‐kb promoter fragment and untranslated regions [UTRs]). This analysis revealed a total of 127 SNPs (causing nonsynonymous mutations) and 13 insertions/deletions (InDels) with a minor allele frequency >5%. Subsequently, candidate gene association analysis was performed using relative leaf injury area as a phenotype reflecting the cold tolerance by TASSEL (Bonferroni threshold P < 1.92 × 10−3). Among the identified variants, a 328‐bp insertion (InDel‐41) located 489 bp upstream of the translation start site of TIP4;3 showed the highest association with cold tolerance (P = 1.41 × 10−4) (Figure 2c). Based on this InDel, the 195 maize varieties were categorized into two haplotype groups: TIP4;3 Hap1 (175 inbred lines) and TIP4;3 Hap2 (20 inbred lines) respectively (Table S1). Notably, TIP4;3 Hap1 inbreds exhibited significantly higher leaf injury area than TIP4;3 Hap2 inbreds (P = 0.0019) (Figure 2d). Consistent with their cold tolerance, TIP4;3 expression was much lower in TIP4;3 Hap2 inbreds than in TIP4;3 Hap2 inbreds under cold treatment (Figure 2e). Therefore, we designated TIP4;3 Hap1 and TIP4;3 Hap2 as the sensitive and tolerant alleles respectively.
To investigate whether the natural variation in the TIP4;3 promoter contributes to the difference in expression levels, we performed a dual‐luciferase (LUC) reporter assay in maize protoplasts. Protoplasts were transfected with constructs containing the LUC reporter gene driven by the TIP4;3 promoter derived from either the B73 (Hap1) or CIMBL52 (Hap2) inbred line. Dual‐luciferase reporter assay revealed that the TIP4;3 CIMBL52 promoter exhibited lower LUC activity, achieving only 40% of the TIP4;3 B73 promoter (Figure 2f). These findings suggest that the sequence variation in the TIP4;3 promoter region contributes to differences in gene expression levels between the two haplotypes, thereby leading to the divergence in cold tolerance among maize inbred lines.
To investigate the effect of Indel‐41 in TIP4;3 expression, we conducted a BLAST search against the maize transposable element (TE) database using the 328‐bp sequence of Indel‐41 as a query (http://maizetedb.org/Bmaize/). We observed that this sequence matches the CACTA TE family. Sequence analysis of the 328‐bp fragment revealed the presence of (5′‐CACTA‐3′) terminal inverted repeats and 11‐bp sub‐terminal repeats (TTTGCCGAGTG) (Figure S2a,b). Transposon insertions have been known to influence the methylation levels of histones, subsequently affecting gene expression (Mao et al., 2015). Therefore, we examined the levels of di‐ and tri‐methylation of lysine 9 or lysine 27 of histone H3 (H3K9me2 and H3K27me3) along the promoters of TIP4;3 Hap1 inbred line (4019) and TIP4;3 Hap2 inbred line (384‐2) under cold stress. We observed a much higher histone methylation level in the TIP4;3 Hap2 promoter than the TIP4;3 Hap1 promoter (Figure 2g). These results suggest that the 328‐bp insertion leads to the elevated H3K9me2 and H3K27me3 levels at the TIP4;3 promoter region, thereby repressing TIP4;3 transcription.
The TIP4; 3 Hap2 allele improves maize cold tolerance
To validate the impact of the TIP4;3 Hap2 allele on maize cold tolerance, we introgressed TIP4;3 Hap2 allele from a cold‐tolerant inbred line (384‐2) into a cold‐sensitive inbred line (4019) carrying the TIP4;3 Hap1 allele, generating near‐isogenic lines (NILs) through three generations of successive backcrossing of the F1 plants (384‐2 × 4019) to 4019 as the recurring parent. In each generation, TIP4;3 Hap1/TIP4;3 Hap2 heterozygous plants were selected and backcrossed to 4019. After one generation of self‐pollination, BC3F2 plants were genotyped to identify plants homozygous for the cold‐tolerant TIP4;3 Hap2 or cold‐sensitive TIP4;3 Hap1 allele (Figure 3a; Figure S3a). Evaluation of cold tolerance in these plants revealed that the NIL‐TIP4;3 Hap2 plants exhibited greater cold tolerance than NIL‐TIP4;3 Hap1 plants. Furthermore, heterozygous TIP4;3 plants exhibited similar cold tolerance to NIL‐TIP4;3 Hap2, indicating that TIP4;3 Hap2 is a dominant allele (Figure 3a,b; Figure S3b). Moreover, lower TIP4;3 transcript levels were detected in NIL‐TIP4;3 Hap2 plants compared to in NIL‐TIP4;3 Hap1 plants, with a more pronounced effect after cold treatment (Figure 3c). These results indicate that the two natural alleles, TIP4;3 Hap1 and TIP4;3 Hap2, result in differential expression levels of TIP4;3, leading to diverse cold tolerance in maize.
Figure 3.
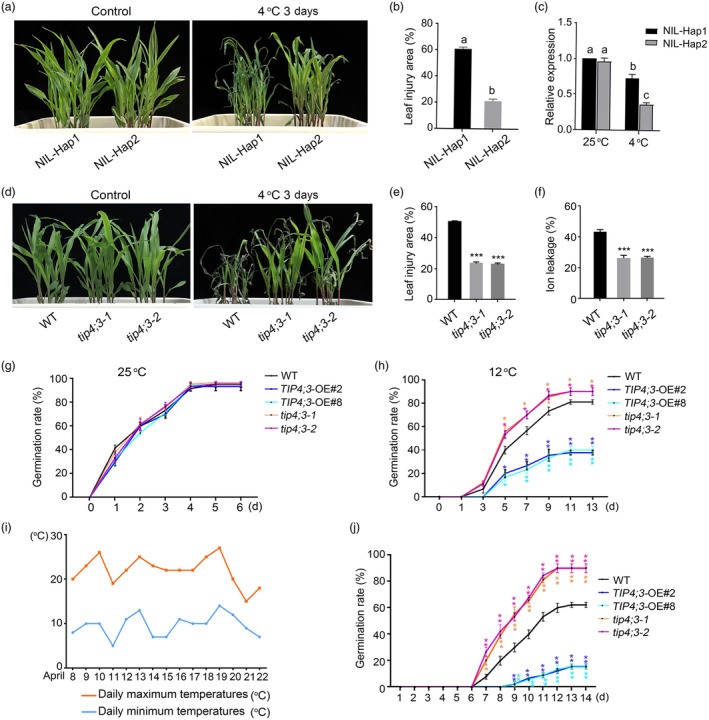
The TIP4;3 Hap2 allele improves cold tolerance in maize. (a) Chilling phenotypes of near‐isogenic lines (NILs) carrying the allele conferring cold tolerance (TIP4;3 Hap2) or the allele conferring cold sensitivity (TIP4;3 Hap1). (b) Leaf injury area of the NILs when exposed to cold stress. Bars represent 10 seedlings for each genotype in three independent experiments. Different letters represent significant differences (P < 0.05) determined by one‐way ANOVA with Tukey's multiple comparisons test. (c) Relative TIP4;3 expression levels in NIL‐Hap1 (TIP4;3 allele from 4019) and NIL‐Hap2 (TIP4;3 allele from 384‐2). Total RNA was extracted from three seedlings in each independent experiment. Different letters represent significant differences (P < 0.05) determined by one‐way ANOVA with Tukey's multiple comparisons test. (d–f) Chilling phenotypes (d), leaf injury area (e), and ion leakage (f) of WT and tip4;3 mutants. Fourteen‐day‐old seedlings grown at 25 °C were exposed to 4 °C for 3 days. Representative images were taken after 2 days of recovery at 25 °C. (g, h) Germination rates of WT, TIP4;3‐OE lines and tip4;3 mutants at 25 °C (g) or 12 °C For each assay, 30 seeds of WT, TIP4;3‐OE lines and tip4;3 mutants were placed in an incubator at 25 °C and 12 °C after soaking in water for 24 h. (i, j) Daily average minimum and maximum temperatures (i) and germination rates of WT, TIP4;3‐OE and tip4;3 mutants in natural field conditions (j). Seeds were sown on April 8, with 30 seeds per pot; the germination rate was scored over 14 consecutive days. The daily minimum temperature was 5 °C, and the maximum temperature was 27 °C. Error bars represent mean ± SD (standard deviation) from 3 biological replicates. *P < 0.05, **P < 0.01, *** P < 0.001, Student's two‐sided t‐test.
TIP4;3 compromises maize cold tolerance at the seedling and germination stages
To dissect the role of TIP4;3 in maize cold tolerance, we employed clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associated nuclease 9 (Cas9)‐mediated gene editing to generate knockout mutants in the LH244 inbred line which carries the Hap1 haplotype. The tip4;3 mutants harboured a 1‐bp deletion (tip4;3–1) or a 2‐bp insertion (tip4;3–2) in the first exon of the TIP4;3 gene, presumed to produce premature stop codon (Figure S3c). After cold treatment, the tip4;3 mutants displayed a reduced relative leaf injury area compared to the WT (Figure 3d–f). Additionally, we performed a cross between the tip4;3–1 and the tip4;3–2 mutant, and found that the F1 progeny displayed a cold tolerance phenotype similar to tip4;3–1 and tip4;3–2 (Figure S4). These results further support the notion that TIP4;3 acts as a negative regulator of cold tolerance of maize.
Cold stress usually reduces the early vigour of seeds after germination. To determine whether TIP4;3 affects the early vigour of maize seeds experiencing cold stress, we scored the germination rates of seeds at 25 °C or 12 °C in an incubator after seed hydration for 24 h using seeds from the WT, the tip4;3 mutants, and two TIP4;3‐overexpression lines (TIP4;3‐OE#2 and #8). The germination ratio showed no clear difference among WT, tip4;3 or TIP4;3‐OE plants at 25 °C (Figure 3g). When incubated at 12 °C, the germination rates of the tip4;3 mutant seeds were higher, whereas those of the TIP4;3‐OE lines were notably lower than the WT (Figure 3h). We repeated this assay in natural field conditions, when the day/night temperatures were around 20 °C/10 °C: the tip4;3 mutant seeds displayed higher germination rates than the WT, with the final rate reached 85% (tip4;3 mutants) versus 60% (WT). In contrast, the germination rates of TIP4;3‐OE seeds were severely delayed compared to the WT and final germination rates remained below 20% (Figure 3i,j). Taken together, these findings suggest that TIP4;3 negatively modulates both early vigour and seedling tolerance to cold stress in maize.
TIP4;3 acts as an aquaporin
TIP4;3 encodes a vacuolar aquaporin localizing to the tonoplast in maize protoplasts, as evidenced by the overlap between the green fluorescence signal from a TIP4;3‐green fluorescent protein (GFP) fusion protein and the vacuolar membrane marker AtTIP1;1 fused to the red fluorescent protein (Figure 4a). Furthermore, TIP4;3 was found to be expressed in various maize tissues, with particularly high expression levels observed in the leaf, tassel, silk and immature cob (Figure 4b). To assess the water transport activity of TIP4;3, the Xenopus oocyte system was employed. Oocytes injected with the cRNA of TIP4;3 or AtTIP1;1 (used as a positive control) exhibited rapid swelling upon switching from an isotonic to a hypotonic buffer, displaying an increase in osmotic water permeability (Po) (Figure 4c; Figure S5). This result indicates that like AtTIP1;1, TIP4;3 possesses water transport activity.
Figure 4.
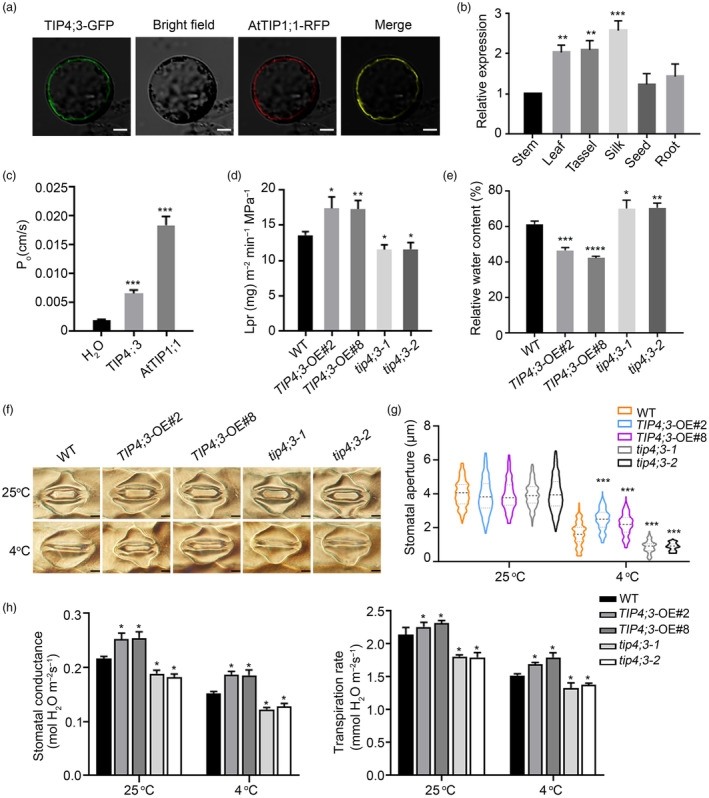
Biochemical characterization of TIP4;3 in response to cold stress. (a) Subcellular localization of TIP4;3. AtTIP1;1‐RFP (a fusion between Arabidopsis TIP1;1 and red fluorescent protein) was used as a vacuolar marker. pSuper:TIP4;3‐GFP and pSuper:AtTIP1;1‐RFP were co‐transfected into maize mesophyll protoplasts. GFP and RFP signals were visualized by confocal microscopy. Scale bars, 10 μm. (b) The relative expression of TIP4;3 expression in different tissues of maize. (c) Osmotic water permeability (Po) of TIP4;3 in Xenopus oocytes. Water transport was assayed at 18 °C for 3 days after cRNA injection. Water transport activity analysis was performed as described in materials and methods. Oocytes injected with water were used as a negative control. AtTIP1;1, an aquaporin, was used as a positive control for water permeability. (d) Root hydraulic conductivity (Lpr) in WT, TIP4;3‐OE and tip4;3 plants. Culture in hydroponic conditions was conducted for this experiment. Four‐week‐old plants were used to measure root hydraulic conductivity. (e) Relative water content of WT, TIP4;3‐OE and tip4;3 plants after 4 °C treatment for 24 h. (f) Cold‐induced stomatal closure was photographed from the leaves of WT, TIP4;3‐OE and tip4;3 plants before and after (4 °C). Scale bars, 5 μm. (g) Stomatal aperture in WT, TIP4;3‐OE and tip4;3. Bars represent 200 stomata in five replicates under 25 °C and 4 °C conditions. Eight‐day‐old seedlings were immersed in MES‐KOH buffer, exposed to light at 25 °C for 5 h to completely open stomata, and then treated at 4 °C for 20 min before being photographed. The dashed horizontal line represents the median, and the upper and lower dotted lines represent the third quartile and first quartile respectively. The bounds of the plot represent data density. ***P < 0.001, **P < 0.01, *P < 0.05 two‐sided t‐test. (h) Analysis of stomatal conductance and transpiration rate under permissive temperature and after cold treatment for 24 h. In b, c, d, e, g, h, bars represent mean ± SD (standard deviation) from three independent experiments. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05 (two‐sided t‐test).
Root hydraulic conductivity (Lpr) reflects the ability of roots to take up water (Cabrera et al., 2024). The Lpr of wild‐type, TIP4;3‐OE and tip4;3 mutant plants was determined using the pressure‐chamber approach. Compared to the WT, TIP4;3‐OE plants exhibited higher Lpr (Figure 4d), indicating that overexpression of TIP4;3 enhances root hydraulic conductivity. Conversely, tip4;3 mutant plants displayed lower Lpr compared to the WT plants (Figure 4d), suggesting that the absence of TIP4;3 leads to a reduction in root hydraulic conductivity. These results suggest that TIP4;3 plays a role in promoting Lpr and water uptake by roots.
TIP4;3 regulates stomatal movement
Considering that TIP4;3 has a negative impact on cold tolerance, but plays a positive role in water uptake, we next measured the water content in WT, TIP4;3‐OE and tip4;3 mutant plants in response to cold stress. We observed that the relative leaf water content was lower in TIP4;3‐OE lines, but higher in tip4;3 mutants compared to the WT after cold treatment (Figure 4e). These findings prompted us to test whether TIP4;3 affects stomatal movement by measuring stomatal aperture on WT, TIP4;3‐OE and tip4;3 leaves under cold treatment. Accordingly, we immersed plants in MES‐KOH buffer in the light to ensure that stomata were completely open, followed by cold treatment. While the stomatal apertures were comparable among the three genotypes under permissive (25 °C) conditions, cold stress induced different degrees of stomatal closure in WT, TIP4;3‐OE and tip4;3 plants. Compared to the WT, the stomatal apertures of TIP4;3‐OE plants were larger, while those of the tip4;3 mutants were smaller after cold treatment (Figure 4f,g). Furthermore, we determined the stomatal conductance and transpiration rate of 21‐day‐old WT, TIP4;3‐OE and tip4;3 mutant plants grown at 25 °C and treated at 4 °C for 24 h. We observed that stomatal conductance and transpiration rate were higher in TIP4;3‐OE plants but lower in tip4;3 mutants compared to the WT with or without cold treatment (Figure 4h,i). Based on these findings, we conclude that TIP4;3 attenuates cold tolerance by suppressing stomatal closure and promoting transpiration upon cold stress.
Transcriptome profiling of TIP4;3
To further investigate the role of TIP4;3 in cold tolerance of maize, we conducted transcriptome deep sequencing (RNA‐seq) using 2‐week‐old seedlings of WT and TIP4;3‐OE plants grown at 25 °C treated or not at 4 °C for 12 h in three independent experiments. We identified differentially expressed genes (DEGs) based on the criteria of a significant difference (P < 0.05) with an absolute fold‐change ≥2, yielding 2620 DEGs in the WT seedlings after exposure to cold (4 °C) compared to the WT seedlings without cold treatment (25 °C) (Table S2). We defined TIP4;3‐affected genes as those DEGs between TIP4;3‐OE and WT plants based on the criteria fold change ≥2 and P < 0.05 under permissive conditions (25 °C) or cold treatment (4 °C). We identified a total of 2192 genes affected by TIP4;3, 992 genes (255 up and 737 down) at 25 °C (Figure 5a; Table S3) and 1200 (759 up and 441 down) after cold treatment (Figure 5a; Table S4). Based on the defined cold‐responsive transcriptome (Zhang et al., 2017), 536 out of these 2192 genes showed significant changes in response to cold treatment; we refer to these genes as COR genes affected by TIP4;3 (Figure 5b).
Figure 5.
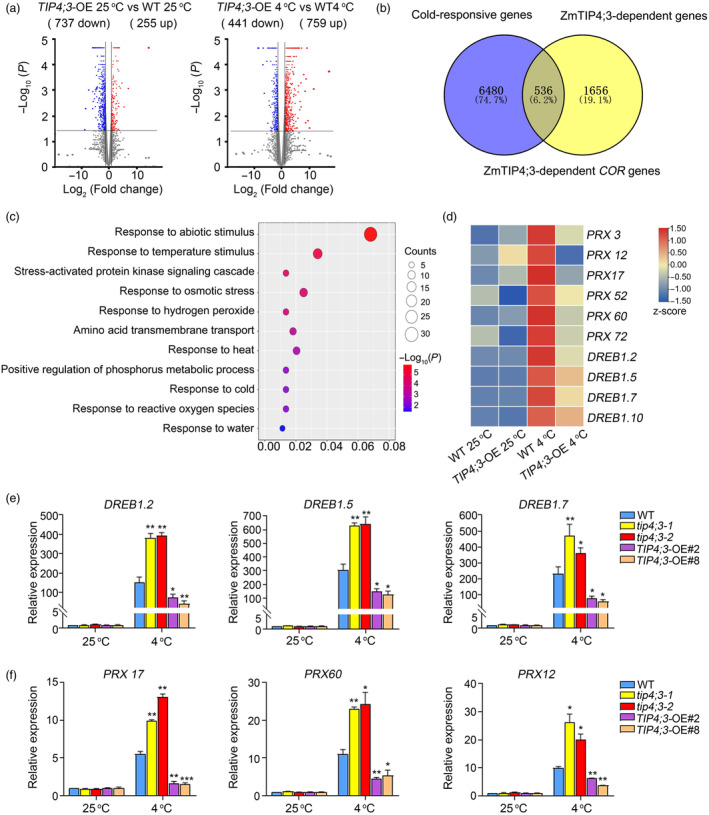
Transcriptome analysis of TIP4;3‐dependent genes. (a) Volcano plots showing the number of differentially expressed genes (DEGs) between WT and TIP4;3‐OE plants under control conditions (25 °C) or following cold treatment (25 °C). DEGs were identified using a P‐value <0.05 and absolute log2 (fold‐change) value ≥1 as criteria. (b) Venn diagram showing the extent of overlap between TIP4;3‐dependent genes and COLD‐REGULATED (COR) genes. (c) Gene ontology term enrichment analysis of TIP4;3‐dependent COR genes. (d) Heatmap representation of expression levels for some DEGs modulated by TIP4;3. FPKM values were Z‐score normalized. (e, f) Relative expression levels of DREB1.2, DREB1.5, DREB1.7, PRX12, PRX17 and PRX60 in WT, TIP4;3‐OE lines and tip4;3 mutants under cold stress. Error bars represent mean ± SD (standard deviation) (n = 3; two‐sided t‐test). **P < 0.01, *P < 0.05.
Gene ontology (GO) term analysis indicated that the genes affected by TIP4;3 are involved in various biological processes, including response to temperature stimuli, osmotic stress, hydrogen peroxide and ROS (Figure 5c). We noticed that the expression of several DREB1s, key genes promoting maize cold tolerance, was affected by TIP4;3 (Figure 5d). Indeed, the expression of DREB1.2, DREB1.5 and DREB1.7 was remarkably lower in the TIP4;3‐OE lines, but higher in the tip4;3 mutants compared to the WT (Figure 5e). Notably, we also observed that a subset of genes encoding peroxidases responsible for ROS scavenging, including peroxidase 12 (PRX12), PRX17 and PRX60, were downregulated in TIP4;3‐OE lines and upregulated in tip4;3 mutants compared to the WT (Figure 5f). These results suggest that the various TIP4;3 genotypes have distinct sensitivities to cold stress, which result in distinct transcriptional responses of COR genes, probably through an indirect mechanism.
TIP4; 3 facilitates ROS accumulation under cold stress
Given that TIP4;3 impacts on the expression of ROS‐scavenging genes, we speculated that TIP4;3 may influence the accumulation of ROS in response to cold stress. Indeed, overexpressing TIP4;3 led to a substantial induction of ROS accumulation in maize, particularly under cold conditions, as evidenced by 3′3′‐diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining (Figure 6a,b). Furthermore, cytoplasmic ROS levels were evaluated using the ROS probe 2′,7′‐dichlorodihydro‐dichlorofluorescin diacetate (H2DCF‐DA). We observed little difference in ROS signals among all genotypes tested in the absence of cold treatment (Figure 6c,d). However, after cold treatment, TIP4;3‐overexpressing plants exhibited dramatically increased cytosolic ROS accumulation, while tip4;3 mutants showed lower cytosolic ROS accumulation compared to the WT (Figure 6c,d). Additionally, the application of glutathione (GSH), a ROS‐scavenging reagent, partially alleviated the cold injury symptoms induced by cold stress (Figure 6e,f), indicating the involvement of ROS in cold‐induced injury. These results suggest that TIP4;3 facilitates over‐accumulation of ROS under cold conditions.
Figure 6.
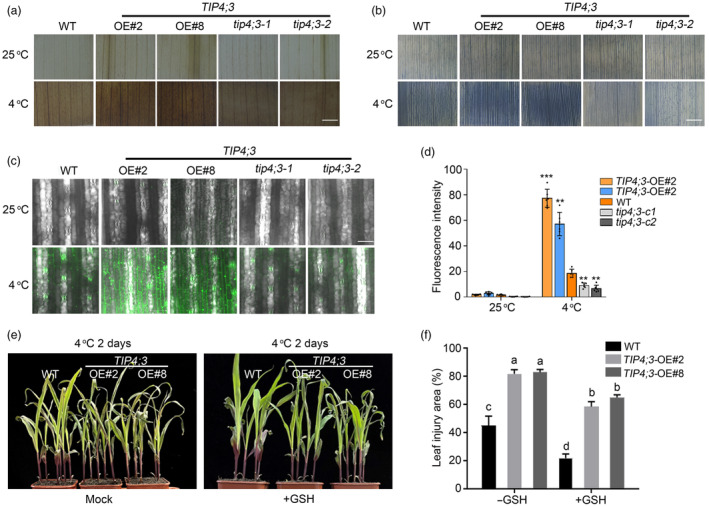
TIP4;3 affects ROS accumulation under cold stress. (a, b) Diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) histochemical staining of WT, TIP4;3‐OE and tip4;3 leaves under cold stress. The experiments were independently repeated three times. Representative photographs of leaf histochemical staining are shown. Scale bars, 1 mm. (c) Imaging of ROS levels in WT, TIP4;3‐OE and tip4;3 leaves under 25 °C and 4 °C conditions detected with H2DCFDA. (d) Quantification of ROS levels in (c). Error bars represent mean ± SD (standard deviation) from three independent experiments (n = 10). ***P < 0.001, **P < 0.01, *P < 0.05 (two‐sided t‐test). (e) Exogenous application of glutathione (GSH) partially rescues the compromised cold sensitivity of tip4;3 mutants. Maize seedlings were treated with 10 mM GSH under 4 °C for 2 days. Representative photographs were taken after 2 days of recovery. (f) Leaf injury area for the seedlings in (e). Error bars represent mean ± SD (standard deviation) from 3 biological replicates (n = 5 plants for each replicate). Different letters represent significant differences (P < 0.05) determined by one‐way ANOVA with Tukey's multiple comparisons test.
Discussion
Plants contain the largest number and maximum diversity of aquaporin homologues with diverse subcellular localization patterns, solute specificity and gating properties (Afzal et al., 2016). As primary water transporter proteins, TIPs are important in maintaining water balance, regulating stomatal movement, and responding to environmental stresses (Afzal et al., 2016). The physiological roles of AQPs throughout plant growth and development have been intensively studied during the last few decades (Alexandersson et al., 2005; Maurel, 2007; Maurel et al., 2002, 2015; Maurel and Chrispeels, 2001). However, the contribution and specific roles of TIPs in cold stress remain unclear. In this study, we provide physiological and genetic evidence showing that maize TIP4;3 compromises cold tolerance by suppressing stomatal closure and increasing water loss through transpiration, potentially leading to dehydration stress under cold conditions. The identification of genetic variation associated with AQPs may provide targets for improving cold tolerance in maize.
Previous studies have reported that the proper expression and abundance of AQPs are crucial for plant adaptation to low temperatures (Lee et al., 2005; Tyerman et al., 2002; Verdoucq et al., 2008). To explore the natural variation of AQP genes, we employed gene‐based association analysis and identify a causative variation for cold tolerance at the TIP4;3 locus, where a 328‐bp CACTA‐like element insertion in the promoter region was found to increase histone methylation, reduced the expression of TIP4;3. Previous study showed that TEs are abundant in plant genomes and can be transcriptionally activated by cold stress (Chang et al., 2020; Liang et al., 2021; Makarevitch et al., 2015). Given that TIP4;3 expression was notably lower in TIP4;3 Hap2 compare to TIP4;3 Hap2 under cold treatment, we propose that the CACTA‐like element may serve as a cold stress‐responsive TEs that leading to the difference in AQP gene expression through epigenetic regulation.
Under cold conditions, plants undergo water deficit and develop a survival strategy to prevent water loss through reduced root hydraulic conductivity, and stomatal closure (Maurel et al., 2008; Muraoka et al., 2023). In rice, the down‐regulation of AQP gene expression is associated with the decrease in Lpr under cold stress (Murai‐Hatano et al., 2008). Similarly, in cucumber and maize, the reduction in Lpr due to low temperatures might be attributed to aquaporin dysfunction (Aroca et al., 2005; Lee et al., 2004, 2005). These findings collectively establish a connection between the cold‐induced decrease in Lpr and AQP activity in various plant species. In our study, we observed a continuous decrease in the expression of several TIP genes, TIP2;1, TIP3;2 and TIP4;3, in maize leaves following cold treatment (Figure 2b). Also, we found the overexpression of TIP4;3 in plants resulted in an elevated Lpr, whereas tip4;3 mutation showed a decreased Lpr. This suggests that the downregulation of TIP genes could confer a cold‐conferred decrease in Lpr, which is crucial for modulating the water permeability of the tonoplast and thereby minimizing water loss of maize seedlings exposed to cold stress.
Maintaining optimal stomatal function is essential to prevent excessive water loss while ensuring supply of carbon dioxide for photosynthesis. AQPs are likely to have impacts on crop physiological performance under stress conditions through their effects on water transport, and ultimately stomatal conductance (Moshelion et al., 2015). In this study, we noticed that stomatal conductance and transpiration rate were higher in TIP4;3‐OE plants but lower in tip4;3 mutants compared to the WT with or without cold treatment (Figure 4h,i). Similarly, overexpression of aquaporin genes such as TIP2;1 in grapevine, NtAQP1 in tobacco and HvPIP2;1 in barley increased stomatal conductance and transpiration rates (Hanba et al., 2004; Pou et al., 2013; Uehlein et al., 2003). All these findings support the notion that AQPs‐mediated adjustment in tissue hydraulics can have an indirect impact on stomatal conductance and transpiration (Maurel et al., 2016).
Many studies have reported increased accumulation of ROS under cold stress. AQPs are known to be involved in the transport of H2O2, one of the best characterized ROS (Cruz de Carvalho, 2008). Generally, it is considered that TIPs are involved in the import of H2O2 into the vacuoles and improve antioxidative system to protect plants (Shivaraj et al., 2021). Meanwhile, ROS promotes the internalization of AQPs, thereby reducing their density at the cell surface and hydraulic conductivity (Boursiac et al., 2008; Wudick et al., 2015). This mechanism could provide feedback regulation where AQPs contribute to the initial steps of ROS signalling (Maurel et al., 2021). In this study, we showed that plants overexpressing TIP4;3 had an elevated accumulation of cytosolic ROS, while tip4;3 mutants exhibited reduced levels compared to the wild type under cold stress. Given the importance to ROS in maize hydraulic conductance and cold tolerance (Aroca et al., 2001), it is speculated that in cold‐sensitive genotypes with constitutive high Lpr, such as TIP4;3‐overexpressors and TIP4;3 Hap1, displayed an increased sensitivity to cold‐induced dehydration, leading to ROS burst and possibly membrane damage. In contrast, in cold‐tolerant genotypes, such as tip4;3 and TIP4;3 Hap2, the reduced Lpr helps preserve water permeability and maintain ROS homeostasis, thus enhancing the cold tolerance of maize (Figure 7).
Figure 7.
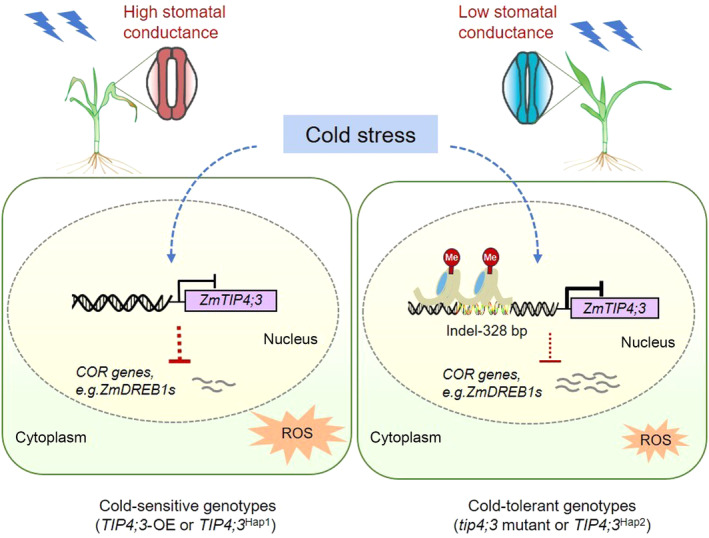
Model of TIP4;3‐mediated cold tolerance in maize. Under cold stress conditions, TIP4;3, a tonoplast aquaporin, suppresses stomatal closure and facilitates reactive oxygen species (ROS) accumulation, as well as downregulates the expression of cold‐responsive genes, thereby negatively regulating maize cold tolerance. Hap2 maize inbred lines harbour a 328‐bp transposon insertion in the TIP4;3 Hap2 promoter. This insertion increases the level of histone modifications H3K9me2 and H3K27me3, contributing to the repression of TIP4;3 expression. Thus, TIP4;3 Hap2 inbred lines show lower TIP4;3 expression and better cold tolerance than TIP4;3 Hap1 inbred lines.
TIPs generally play roles in the water‐stress response during drought, salinity and osmotic stresses (Afzal et al., 2016). In this study, we observed that tip4;3 mutants exhibited enhanced cold tolerance during the germination and seedling stages (Figure 3). Intriguingly, these mutants also showed increased drought tolerance, with higher survival rates than wild‐type plants under drought conditions (Figure S6). These enhanced stress tolerances did not compromise yield‐related traits, such as hundred‐grain weight, ear row number, or ear length, in field tests (Figure S7). This study thus provides valuable genetic resources for developing new maize varieties with both cold and drought tolerance without yield penalties.
Methods
Plant materials and growth conditions
The maize (Zea mays L.) transgenic plants were obtained from the Center for Crop Functional Genomics and Molecular Breeding, China Agricultural University. All constructs were transformed into inbred line LH244 (wild type) using Agrobacterium‐mediated transformation (Lee and Zhang, 2014). For association analysis, we used 195 maize natural variation panel, which consists of inbred lines from tropical and temperate backgrounds (Li et al., 2013). Maize seeds were planted in pots (30 × 20 × 15 cm) containing plant ash, vermiculite and Pindstrup soil mix (Denmark) (3:1:1) and grown at 25 °C under a 16 h light/8 h dark photoperiod with 200 μmol/m2/s white light and 60% relative humidity. For cold treatment, maize plants were grown for 14 days with irrigation and exposed to 4 °C under a 16‐h light/8‐h dark photoperiod for 2–4 days using a cold chamber (Conviron CMP6010). After cold treatment, the seedlings were recovered at 25 °C for 2 days prior to photography. For germination assays, dry seeds were hydrated at 25 °C for 24 h and then transferred to a glass dish containing a wet filter paper for germination in an incubator at 25 °C or 12 °C for the indicated period.
Physiological analyses
The relative injured area was measured as described previously (Zeng et al., 2021). The water content was measured as follows: the seedlings were recovered at 25 °C for 2 days after cold treatment, all the above‐ground parts of the seedlings were collected and weighed to obtain the fresh weight (Wf). Subsequently, the collected samples were dried in a 70 °C oven and weighed to obtain the dry weight (Wd). The water content percentage was calculated using the formula: (Wf − Wd)/Wf × 100%.
Plasmid construction and plant transformation
The transgenic plants (TIP4;3‐OE#2, ‐OE#8) used in this study were derived from maize inbred line LH244. These constructs were generated by cloning TIP4;3 coding sequences into pBCXUN (Li et al., 2022). To generate mutants of tip4;3 using CRISPR‐Cas9 genome editing, fragments of the first intron were selected as guide RNA targets and cloned into pBUE411 vector (Xing et al., 2014). Transgenic plants were obtained by Agrobacterium‐mediated transformation (Lee and Zhang, 2014). The promoter sequences of 1.0‐kb CIMBL52 (Hap2), 672 bp B73 (Hap1, deleting 328 bp), and 489 bp CIMBL52Δ were amplified and cloned into pGreenII0800‐LUC vectors using in‐fusion PCR cloning systems (C112‐02; Vazyme, Nanjing, China) to obtain the pTIP4;3 B73 :LUC, pTIP4;3 CIMBL52 :LUC and pTIP4;3 CIMBL52Δ :LUC, respectively. These three plasmids were purified by Plasmid maxiprep kit (EM123‐01; Transgen, Beijing, China) and transformed separately into maize protoplast to measure the transcriptional activation assay (Li et al., 2022).
RT‐qPCR and RNA‐seq
RT‐qPCR and RNA‐seq were conducted following established protocols (Zeng et al., 2021). Total RNAs were extracted from the leaves of 14‐day‐old maize plants using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNA with M‐MLV reverse transcriptase (Promega, Shanghai, China). The expression levels target genes were detected using SYBR Green reagent (Mei5bio, MF013‐01) in a StepOnePlus Real‐Time PCR System (Applied Biosystem, Waltham, MA, USA). The maize Ubi gene was used as an internal control. Primer sequences for qPCR are listed in Table S5. Each experiment was independently repeated three times (three biological replicates).
For RNA‐seq assays, 14‐day‐old seedlings of wild type and TIP4;3‐OE grown at 25 °C were subjected to treatment before and after exposure to 4 °C for 12 h. Total RNAs were extracted from pooled wild‐type or TIP4;3‐OE leaves using Trizol reagent. For each sample, leaves from three individual plants were harvested. Two independent biological replicates were performed. The RNA samples were then subjected to Illumina HiSeq deep sequencing (Illumina HiSeq 6000; Illumina, San Diego, CA, USA). The generated reads were initially processed to remove adapter sequences and low‐quality reads (Q score < 20). Subsequently, the reads were mapped to the Maize genome (B73 RefGen_v4, AGPv4) using HISAT2 (v.2.2.0) with default parameters. The read counts for each gene were obtained using Feature Counts (v.2.0.1). GO enrichment analysis of differentially expressed gene (DEG) clusters was performed using the program Phyper (http://www.geneontology.org/). The significance of the GO terms was corrected using false discovery rate (FDR) < 0.05. A heatmap of the expression levels of the DEGs was constructed using TBtools software (Chen et al., 2020).
Subcellular localization
Subcellular localization of TIP4;3 protein was performed as described previously (Zeng et al., 2021). Briefly, pSuper:TIP4;3‐GFP and AtTIP1;1‐RFP were transformed into maize protoplasts and incubated at 25 °C for 16 h. GFP and RFP fluorescence were observed by confocal microscopy (ZEISS710; Carl Zeiss, Oberkochen, Germany) with exciting light 488 and 561.
Transient expression assays in maize protoplasts
Dual‐LUC assays was performed as described previously (Li et al., 2022). pTIP4;3 B73 :LUC, pTIP4;3 CIMBL52 :LUC and pTIP4;3 CIMBL52Δ :LUC were used as reporter genes, REN gene driven by 35S promoter in pGreenII0800‐LUC was used as an internal control. Vectors were co‐transformed into maize protoplasts and incubated at 25 °C for 16 h. The luciferase signal was measured via the Dual‐Luciferase Reporter Assay system (Promega) on a GLOMAX 20/20 luminometer (Promega, Madison, WI, USA). Relative LUC activity was calculated by normalizing LUC activity to REN activity.
Histochemical detection of H2O2 , O2 −· and ROS
Two‐week‐old maize seedlings were grown in mixed soil in a greenhouse and then treated with 4 °C for 12 h. H2O2 and O2−· was stained with 3′3′‐diaminobenzidine (DAB, Sigma‐Aldrich, St. Louis, MO, USA) and nitroblue tetrazolium (NBT, Beyotime, Shanghai, China), respectively, as described previously (Jiang et al., 2022). For DAB staining, leaf pieces were incubated in 1 mg/ml DAB for 10 h, decolorized with 80% alcohol until the decolorizing solution was colourless and then observed under an Olympus microscope (SZX16). For NBT staining, leaf pieces were immersed in 10 mL NBT staining solution (6 mM diluted with potassium phosphate) for 10 h, decolorized with 80% ethanol and then observed under an Olympus microscope (SZX16). The ROS level in guard cells was detected using H2DCFDA staining as described (Gao et al., 2022). Portions of leaves were immersed in staining buffer (10 mM Tris–HCl, 50 mM KCl, 50 μM H2DCFDA, 0.02% Tween‐20 at pH 7.2) and subjected to vacuum filtration for 20 min at room temperature in the dark. The leaves were then washed with distilled water to remove excess dye. Fluorescence was examined using a confocal laser‐scanning microscope (ZEISS710).
TIP4;3‐based association analysis
According to the reference sequences of B73, the genomic regions of TIP4;3 from 195 maize genotypes were amplified and sequenced (Yang et al., 2010). The sequences were aligned by MEGA 7.0, and DNA variations among inbred lines were identified. 195 maize inbred lines were analysed for the association between the genetic variations in TIP4;3 and the relative injured area. The standard mixed linear model was applied (Yu et al., 2006), in which the population structure (Q) and kinship (K) were estimated for TIP4;3‐based association analysis. P value was calculated by the mixed linear model (MLM). SNP loci association with the phenotype and pairwise linkage disequilibrium was calculated by TASSEL software (Bradbury et al., 2007; Zhang et al., 2010).
Stomatal aperture analysis
The stomatal aperture assay was conducted as described (Guo et al., 2023). In brief, a 1‐cm piece of the first fully expanded leaf from 8‐day‐old maize seedlings was excised in the middle of leaf and immersed in stomatal opening solution MES buffer (10 mM MES–KOH, pH 5.7, 10 mM KCl, and 50 μM CaCl2) in the light (150 μmol/m2/s2) for 3 h at 25 °C to open the stomata. Then, 12 μM ABA was added into the MES buffer for another 2 h. The abaxial epidermis of leaf was evenly coated with colourless nail polish. After drying, the nail polish was peeled off with transparent scotch tape. Stomata sticking to the tape were photographed using an Olympus BX53 microscope (Olympus, Tokyo, Japan). The stomata were observed with a 40‐fold objective lens. Stomatal apertures were measured using the ImageJ software. Approximately 200 stomatal apertures from each seedling were measured in each experiment.
Osmotic water permeability assay
The osmotic water permeability assay was conducted as described (Zhang and Verkman, 1991). Oocytes were transferred 2–3 days after mRNA injection from Barth's solution (200 mosM/kg) at room temperature to the same solution diluted to 40 mosM/kg with distilled water. Changes in cell volume were monitored using a microscope (Nikon Ti‐U), and photographs were taken at 30‐ to 45‐s intervals. Oocyte diameters were measured four times along two sets of perpendicular axes. The volume V was estimated as the mean of two ellipsoid volumes. The osmotic permeability coefficient (Pf) was calculated using the formula: Pf = Vo[d(V/V O)/dt]/[S × Vw (Osmin‐Osmout)], with initial oocyte volume V O = 9 × 10−4 cm3, initial oocyte surface area S = 0.045 cm2 and molar volume of water Vw = 18 cm3/mol, respectively.
Root hydraulic conductivity (Lpr) assay
Root hydraulic conductivity (Lpr) was evaluated using the pressure chamber method, which was calculated by the slope of the root water flow rate curve at different pressures as described (He et al., 2023). Four‐week‐old maize plants grown hydroponically were used for the assay. The first leaf base was cut from maize plants under hydroponic conditions. The entire root system was placed in a sealed pressure chamber, with the incision exposed outdoors through a gasketed hole. Pressure was gradually increased from 0.1 to 1.2 MPa, with measurements taken at intervals of 0.1 MPa. A 1.5‐mL centrifuge tube (EP tube) filled with absorbent paper was used to collect the effluent juice at least three times at each pressure for 1 min each time. The mass of EP tube was weighed before and after water absorption, and the xylem fluid flow per unit time (Q, mg/min) was calculated when the stable flow rate was reached under each pressure. Roots were then cleaned, dried, and weighed. The amount of effluent juice collected per unit root weight and per unit outflow time (water flow rate, Jv, mg/(g·min)) was calculated. The slope of the curve relating water flow rate to pressure represents the root water conductivity (Lpr).
GSH treatment assays for cold tolerance
Fourteen‐day‐old seedlings were grown in pots containing equal‐weight soil for the GSH treatment assays as described (Jiang et al., 2022). Young maize seedlings were deprived of water for 3 days before the treatment. The equal amounts of 10 mM GSH solution were applied to soil‐grown plants. After the 12‐h treatment, plants were exposed to cold treatment at 4 °C for 2 days. At least five individual plants were compared with distilled water‐treated (mock) plants for each test. The statistical data based on obtained data from three independent experiments were collected.
Stomatal conductance and transpiration rate assays
The third leaf of the 21‐day‐old seedlings of wild type (WT), TIP4;3‐OE and tip4;3 mutant was used to measure stomatal conductance and transpiration rate by LI‐6400XT portable photosynthesis measurement system as described (He et al., 2023). The greenhouse humidity was between 60% and 65%; CO2 concentration was stabilized using CO2 cylinders. Four plants were tested for each genotype and three biological replicates were carried out.
Construction of phylogenetic tree
Amino acid sequences of TIP4;3 homologues were downloaded from Gramene protein database (https://www.gramene.org/). MEGA7.0 was used to build the phylogenetic tree. The numbers at each branch of the tree mean the bootstrap value.
Data significant test
Significant differences were analysed by Student's t‐test or Tukey's test.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
Y.S., conceived, designed, and directed the project. R.Z., and X.Z., performed most of the experiments. S.Z., performed the maize transformation. G.S., and Q.L., performed yield measurement. R.Z., M.L., Z.Z., and D.F., performed association and RNA‐seq analyses. R.Z., X.Z., L.G., F.T., S.Y., and S.Y., analysed the data. R.Z., S.Y., and Y.S., wrote the manuscript with comments from all authors.
Supporting information
Figure S1 Expression of TIP family genes in maize response to cold stress.
Figure S2 Schematic diagram of the 328 bp sequence.
Figure S3 Identification of NIL lines of TIP4;3 and TIP4;3 mutants.
Figure S4 Phenotypic testing of two alleles of tip4;3 mutants.
Figure S5 Representative photographs of oocytes that were injected with the cRNA of TIP4;3, AtTIP1;1 (a positive control), H2O (a negative control) after switching from isotonic to hypotonic buffer.
Figure S6 TIP4;3 negatively regulates drought tolerance in maize.
Figure S7 Yield‐related traits of tip4;3 mutant lines.
Table S1 Leaf injury area and haplotype of 195 maize inbred lines.
Table S2 A total of 2,620 differentially expressed genes (DEGs) based on the criteria of a significant difference (P < 0.05) with an absolute fold‐change ≥ 2 identified in WT seedlings after exposure to cold tratment.
Table S3 255 upregulated genes and 737 downregulated genes were identified under permissive conditions (25°C) in TIP4;3‐OE lines compared to the wild type.
Table S4 759 TIP4;3‐induced genes and 441 TIP4;3‐repressed genes were identified under cold treatment in TIP4;3‐OE compared to the wild type.
Table S5 Primers used in this study.
Acknowledgements
The transgenic seeds of maize were created by Center for Crop Functional Genomics and Molecular Breeding of China Agricultural University. This work was supported by STI 2030‐Major Projects (2023ZD0407104), the State Key Project of Research and Development Plan (2022YFF1001603), the National Natural Science Foundation of China (32201728, 32272025, and 31921001), and the Chinese Universities Scientific Fund (2022TC137 and 2023TC019).
Data availability statement
The raw RNA‐seq data in this paper have been deposited at NCBI under Bioproject with the accession number PRJNA1054219.
References
- Afzal, Z. , Howton, T.C. , Sun, Y. and Mukhtar, M.S. (2016) The roles of aquaporins in plant stress responses. J. Dev. Biol. 4, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersson, E. , Fraysse, L. , Sjövall‐Larsen, S. , Gustavsson, S. , Fellert, M. , Karlsson, M. , Johanson, U. et al. (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59, 469–484. [DOI] [PubMed] [Google Scholar]
- Aroca, R. , Amodeo, G. , Fernández‐Illescas, S. , Herman, E.M. , Chaumont, F. and Chrispeels, M.J. (2005) The role of aquaporins and membrane damage in chilling and hydrogen peroxide induced changes in the hydraulic conductance of maize roots. Plant Physiol. 137, 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca, R. , Tognoni, F. , Irigoyen, J.J. , Sánchez‐Díaz, M. and Pardossi, A. (2001) Different root low temperature response of two maize genotypes differing in chilling sensitivity. Plant Physiol. Biochem. 39, 1067–1073. [Google Scholar]
- Beamer, Z.G. , Routray, P. , Choi, W.G. , Spangler, M.K. , Lokdarshi, A. and Roberts, D.M. (2021) Aquaporin family lactic acid channel NIP2;1 promotes plant survival under low oxygen stress in Arabidopsis . Plant Physiol. 187, 2262–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac, Y. , Boudet, J. , Postaire, O. , Luu, D.T. , Tournaire‐Roux, C. and Maurel, C. (2008) Stimulus‐induced downregulation of root water transport involves reactive oxygen species‐activated cell signalling and plasma membrane intrinsic protein internalization. Plant J. 56, 207–218. [DOI] [PubMed] [Google Scholar]
- Bradbury, P.J. , Zhang, Z. , Kroon, D.E. , Casstevens, T.M. , Ramdoss, Y. and Buckler, E.S. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. [DOI] [PubMed] [Google Scholar]
- Cabrera, J.C.B. , Vanderborght, J. , Couvreur, V. , Behrend, D. , Gaiser, T. , Nguyen, T.H. and Lobet, G. (2024) Root hydraulic properties: An exploration of their variability across scales. Plant Direct. 8, e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.N. , Zhu, C. , Jiang, J. , Zhang, H.M. , Zhu, J.K. and Duan, C.G. (2020) Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 62, 563–580. [DOI] [PubMed] [Google Scholar]
- Chaumont, F. , Barrieu, F. , Wojcik, E. , Chrispeels, M.J. and Jung, R. (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.J. , Chen, H. , Zhang, Y. , Thomas, H.R. , Frank, M.H. , He, Y.H. and Xia, R. (2020) TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. [DOI] [PubMed] [Google Scholar]
- Cruz de Carvalho, M.H. (2008) Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 3, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq, M. , Aziz, T. , Wahid, A. , Lee, D.J. and Siddique, K.H.M. (2009) Chilling tolerance in maize: agronomic and physiological approaches. Crop Pasture Sci. 60, 501–516. [Google Scholar]
- Gao, H.J. , Cui, J.J. , Liu, S.X. , Wang, S.H. , Lian, Y.Y. , Bai, Y.T. , Zhu, T.F. et al. (2022) Natural variations of modulate the trade‐off between drought resistance and yield by affecting ZmRBOHC‐mediated stomatal ROS production in maize. Mol. Plant. 15, 1558–1574. [DOI] [PubMed] [Google Scholar]
- Gao, L. , Jiang, H. , Li, M. , Wang, D. , Xiang, H. , Zeng, R. , Chen, L. et al. (2023) Genetic and lipidomic analyses reveal the key role of lipid metabolism for cold tolerance in maize. J. Genet. Genomics 51, 326–337. [DOI] [PubMed] [Google Scholar]
- Guo, Y.Z. , Shi, Y.B. , Wang, Y.L. , Liu, F. , Li, Z. , Qi, J.S. , Wang, Y. et al. (2023) The clade F PP2C phosphatase ZmPP84 negatively regulates drought tolerance by repressing stomatal closure in maize. New Phytol. 237, 1728–1744. [DOI] [PubMed] [Google Scholar]
- Han, Q. , Qi, J. , Hao, G. , Zhang, C. , Wang, C. , Dirk, L.M. , Downie, A.B. et al. (2020) ZmDREB1A regulates RAFFINOSE SYNTHASE controlling raffinose accumulation and plant chilling stress tolerance in maize. Plant Cell Physiol. 61, 331–341. [DOI] [PubMed] [Google Scholar]
- Hanba, Y.T. , Shibasaka, M. , Hayashi, Y. , Hayakawa, T. , Kasamo, K. , Terashima, I. and Katsuhara, M. (2004) Overexpression of the barley aquaporin HvPIP2;1 increases internal CO2 conductance and CO2 assimillation in the leaves of transgenic rice plants. Plant Cell Physiol. 45, 521–529. [DOI] [PubMed] [Google Scholar]
- He, R. , Su, H.Q. , Wang, X. , Ren, Z.J. , Zhang, K. , Feng, T.Y. , Zhang, M.C. et al. (2023) Coronatine promotes maize water uptake by directly binding to the aquaporin ZmPIP2;5 and enhancing its activity. J. Integr. Plant Biol. 65, 703–720. [DOI] [PubMed] [Google Scholar]
- Jiang, H.F. , Shi, Y.T. , Liu, J.Y. , Li, Z. , Fu, D.Y. , Wu, S.F. , Li, M.Z. et al. (2022) Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat. Plants. 8, 1176–1190. [DOI] [PubMed] [Google Scholar]
- Johansson, I. , Karlsson, M. , Johanson, U. , Larsson, C. and Kjellbom, P. (2000) The role of aquaporins in cellular and whole plant water balance. Biochim. Biophys. Acta 1465, 324–342. [DOI] [PubMed] [Google Scholar]
- Lee, H. and Zhang, Z.Y.J. (2014) Agrobacterium‐mediated transformation of maize (Zea mays) immature embryos. Methods Mol. Biol. 1099, 273–280. [DOI] [PubMed] [Google Scholar]
- Lee, S.H. , Chung, G.C. and Steudle, E. (2005) Gating of aquaporins by low temperature in roots of chilling‐sensitive cucumber and chilling‐tolerant figleaf gourd. J. Exp. Bot. 56, 985–995. [DOI] [PubMed] [Google Scholar]
- Lee, S.H. , Singh, A.P. , Chung, G.C. , Ahn, S.J. , Noh, E.K. and Steudle, E. (2004) Exposure of roots of cucumber (Cucumis sativus) to low temperature severely reduces root pressure, hydraulic conductivity and active transport of nutrients. Physiol. Plant. 120, 413–420. [DOI] [PubMed] [Google Scholar]
- Leipner, J. , Jompuk, C. , Camp, K.H. , Stamp, P. and Fracheboud, Y. (2008) QTL studies reveal little relevance of chilling‐related seedling traits for yield in maize. Theor. Appl. Genet. 116, 555–562. [DOI] [PubMed] [Google Scholar]
- Li, H. , Peng, Z. , Yang, X. , Wang, W. , Fu, J. , Wang, J. , Han, Y. et al. (2013) Genome‐wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 45, 43–50. [DOI] [PubMed] [Google Scholar]
- Li, Z.Y. , Fu, D.Y. , Wang, X. , Zeng, R. , Zhang, X. , Tian, J.G. , Zhang, S.S. et al. (2022) The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell. 34, 2833–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, H.L. , Yu, X. , Ye, Q. , Ding, X.S. , Kitagawa, Y. , Kwak, S.S. , Su, W.A. et al. (2004) The role of aquaporin RWC3 in drought avoidance in rice. Plant Cell Physiol. 45, 481–489. [DOI] [PubMed] [Google Scholar]
- Liang, Z.K. , Anderson, S.N. , Noshay, J.M. , Crisp, P.A. , Enders, T.A. and Springer, N.M. (2021) Genetic and epigenetic variation in transposable element expression responses to abiotic stress in maize. Plant Physiol. 186, 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.Y. , Shi, Y.T. and Yang, S.H. (2018) Insights into the regulation of C‐repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60, 780–795. [DOI] [PubMed] [Google Scholar]
- Makarevitch, I. , Waters, A.J. , West, P.T. , Stitzer, M. , Hirsch, C.N. , Ross‐Ibarra, J. and Springer, N.M. (2015) Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 11, e1004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H. , Wang, H. , Liu, S. , Li, Z. , Yang, X. , Yan, J. , Li, J. et al. (2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 6, 8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel, C. (2007) Plant aquaporins: Novel functions and regulation properties. FEBS Lett. 581, 2227–2236. [DOI] [PubMed] [Google Scholar]
- Maurel, C. , Boursiac, Y. , Luu, D.T. , Santoni, V. , Shahzad, Z. and Verdoucq, L. (2015) Aquaporins in Plants. Physiol. Rev. 95, 1321–1358. [DOI] [PubMed] [Google Scholar]
- Maurel, C. and Chrispeels, M.J. (2001) Aquaporins. A molecular entry into plant water relations. Plant Physiol. 125, 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel, C. , Javot, H. , Lauvergeat, V. , Gerbeau, P. , Tournaire, C. , Santoni, V. and Heyes, J. (2002) Molecular physiology of aquaporins in plants. Int. Rev. Cytol. 215, 105–148. [DOI] [PubMed] [Google Scholar]
- Maurel, C. , Tournaire‐Roux, C. , Verdoucq, L. and Santoni, V. (2021) Hormonal and environmental signaling pathways target membrane water transport. Plant Physiol. 187, 2056–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel, C. , Verdoucq, L. , Luu, D.T. and Santoni, V. (2008) Plant aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624. [DOI] [PubMed] [Google Scholar]
- Maurel, C. , Verdoucq, L. and Rodrigues, O. (2016) Aquaporins and plant transpiration. Plant Cell Environ. 39, 2580–2587. [DOI] [PubMed] [Google Scholar]
- Moshelion, M. , Halperin, O. , Wallach, R. , Oren, R. and Way, D.A. (2015) Role of aquaporins in determining transpiration and photosynthesis in water‐stressed plants: crop water‐use efficiency, growth and yield. Plant Cell Environ. 38, 1785–1793. [DOI] [PubMed] [Google Scholar]
- Murai‐Hatano, M. , Kuwagata, T. , Sakurai, J. , Nonami, H. , Ahamed, A. , Nagasuga, K. , Matsunami, T. et al. (2008) Effect of low root temperature on hydraulic conductivity of rice plants and the possible role of aquaporins. Plant Cell Physiol. 49, 1294–1305. [DOI] [PubMed] [Google Scholar]
- Muraoka, Y. , Yang, G. , Munemasa, S. , Takeuchi, Y. , Ishimaru, Y. , Murata, Y. , Uozumi, N. et al. (2023) An outward‐rectifying plant K+ channel SPORK2 exhibits temperature‐sensitive ion‐transport activity. Curr. Biol. 33, 5488–5494. [DOI] [PubMed] [Google Scholar]
- Pou, A. , Medrano, H. , Flexas, J. and Tyerman, S.D. (2013) A putative role for TIP and PIP aquaporins in dynamics of leaf hydraulic and stomatal conductances in grapevine under water stress and re‐watering. Plant Cell Environ. 36, 828–843. [DOI] [PubMed] [Google Scholar]
- Pranneshraj, V. , Sangha, M.K. , Djalovic, I. , Miladinovic, J. and Djanaguiraman, M. (2022) Lipidomics‐assisted GWAS (lGWAS) approach for improving high‐temperature stress tolerance of crops. Int. J. Mol. Sci. 23, 9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade, N. , Gebretsadik, M. , Seligmann, R. , Schwartz, A. , Wallach, R. and Moshelion, M.J.P.P. (2010) The role of tobacco aquaporin1 in improving water use efficiency, hydraulic conductivity, and yield production under salt stress. Plant Physiol. 152, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Ding, Y. and Yang, S. (2018) Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637. [DOI] [PubMed] [Google Scholar]
- Shivaraj, S.M. , Sharma, Y. , Chaudhary, J. , Rajora, N. , Sharma, S. , Thakral, V. , Ram, H. et al. (2021) Dynamic role of aquaporin transport system under drought stress in plants. Environ. Exp. Bot. 184, 104367. [Google Scholar]
- Sudhakaran, S. , Thakral, V. , Padalkar, G. , Rajora, N. , Dhiman, P. , Raturi, G. , Sharma, Y. et al. (2021) Significance of solute specificity, expression, and gating mechanism of tonoplast intrinsic protein during development and stress response in plants. Physiol. Plant. 172, 258–274. [DOI] [PubMed] [Google Scholar]
- Sun, Q. , Liu, X. , Kitagawa, Y. , Calamita, G. and Ding, X. (2024) Plant aquaporins: Their roles beyond water transport. Crop J. 12, 641–655. [Google Scholar]
- Tyerman, S.D. , Niemietz, C.M. and Bramley, H. (2002) Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant Cell Environ. 25, 173–194. [DOI] [PubMed] [Google Scholar]
- Uehlein, N. , Lovisolo, C. , Siefritz, F. and Kaldenhoff, R. (2003) The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425, 734–737. [DOI] [PubMed] [Google Scholar]
- Verdoucq, L. , Grondin, A. and Maurel, C. (2008) Structure‐function analysis of plant aquaporin PIP2;1 gating by divalent cations and protons. Biochem. J. 415, 409–416. [DOI] [PubMed] [Google Scholar]
- Verheul, M.J. , Picatto, C. and Stamp, P. (1996) Growth and development of maize (Zea mays L) seedlings under chilling conditions in the field. Eur. J. Agron. 5, 31–43. [Google Scholar]
- Wudick, M.M. , Li, X. , Valentini, V. , Geldner, N. , Chory, J. , Lin, J. , Maurel, C. et al. (2015) Subcellular redistribution of root aquaporins induced by hydrogen peroxide. Mol. Plant. 8, 1103–1114. [DOI] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.H. , Yan, J.B. , Shah, T. , Warburton, M.L. , Li, Q. , Li, L. , Gao, Y.F. et al. (2010) Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor. Appl. Genet. 121, 417–431. [DOI] [PubMed] [Google Scholar]
- Yang, Z.R. , Cao, Y.B. , Shi, Y.T. , Qin, F. , Jiang, C.F. and Yang, S.H. (2023) Genetic and molecular exploration of maize environmental stress resilience: Toward sustainable agriculture. Mol. Plant. 16, 1496–1517. [DOI] [PubMed] [Google Scholar]
- Yu, J.M. , Pressoir, G. , Briggs, W.H. , Bi, I.V. , Yamasaki, M. , Doebley, J.F. , McMullen, M.D. et al. (2006) A unified mixed‐model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. [DOI] [PubMed] [Google Scholar]
- yyGreaves, J. (1996) Improving suboptimal temperature tolerance in maize‐the search for variation. J. Exp. Bot. 47, 307–323. [Google Scholar]
- Zeng, R. , Li, Z.Y. , Shi, Y.T. , Fu, D.Y. , Yin, P. , Cheng, J.K. , Jiang, C.F. et al. (2021) Natural variation in a type‐A response regulator confers maize chilling tolerance. Nat. Commun. 12, 4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Yu, F. , Xie, P. , Sun, S. , Qiao, X. , Tang, S. , Chen, C. et al. (2023) A Gγ protein regulates alkaline sensitivity in crops. Science 379, eade8416. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Xu, Q. , Wang, D. , Di, H. , Huang, J. , Yang, X. et al. (2020) Identification of candidate tolerance genes to low‐temperature during maize germination by GWAS and RNA‐seqapproaches. BMC Plant Biol. 20, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R.B. and Verkman, A.S. (1991) Water and urea permeability properties of Xenopus oocytes: expression of mRNA from toad urinary bladder. Am. J. Phys. 260, C26–C34. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Feng, M. , Chen, W. , Zhou, X. , Lu, J. , Wang, Y. , Li, Y. et al. (2019) In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat Plants. 5, 290–299. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Ngu, D.W. , Carvalho, D. , Liang, Z.K. , Qiu, Y.M. , Roston, R.L. and Schnable, J.C. (2017) Differentially regulated orthologs in sorghum and the subgenomes of maize. Plant Cell 29, 1938–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z.W. , Ersoz, E. , Lai, C.Q. , Todhunter, R.J. , Tiwari, H.K. , Gore, M.A. , Bradbury, P.J. et al. (2010) Mixed linear model approach adapted for genome‐wide association studies. Nat. Genet. 42, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.M. , Muhammad, I. , Lan, H. and Xia, C. (2022) Recent advances in the analysis of cold tolerance in maize. Front. Plant Sci. 13, 866034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, F. , Bulut, M. , Cheng, Y. , Alseekh, S. and Fernie, A.R. (2023) Metabolite‐based genome‐wide association studies of large‐scale metabolome analysis to illustrate alterations in the metabolite landscape of plants upon responses to stresses. Methods Mol. Biol. 2642, 241–255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Expression of TIP family genes in maize response to cold stress.
Figure S2 Schematic diagram of the 328 bp sequence.
Figure S3 Identification of NIL lines of TIP4;3 and TIP4;3 mutants.
Figure S4 Phenotypic testing of two alleles of tip4;3 mutants.
Figure S5 Representative photographs of oocytes that were injected with the cRNA of TIP4;3, AtTIP1;1 (a positive control), H2O (a negative control) after switching from isotonic to hypotonic buffer.
Figure S6 TIP4;3 negatively regulates drought tolerance in maize.
Figure S7 Yield‐related traits of tip4;3 mutant lines.
Table S1 Leaf injury area and haplotype of 195 maize inbred lines.
Table S2 A total of 2,620 differentially expressed genes (DEGs) based on the criteria of a significant difference (P < 0.05) with an absolute fold‐change ≥ 2 identified in WT seedlings after exposure to cold tratment.
Table S3 255 upregulated genes and 737 downregulated genes were identified under permissive conditions (25°C) in TIP4;3‐OE lines compared to the wild type.
Table S4 759 TIP4;3‐induced genes and 441 TIP4;3‐repressed genes were identified under cold treatment in TIP4;3‐OE compared to the wild type.
Table S5 Primers used in this study.
Data Availability Statement
The raw RNA‐seq data in this paper have been deposited at NCBI under Bioproject with the accession number PRJNA1054219.


