Abstract
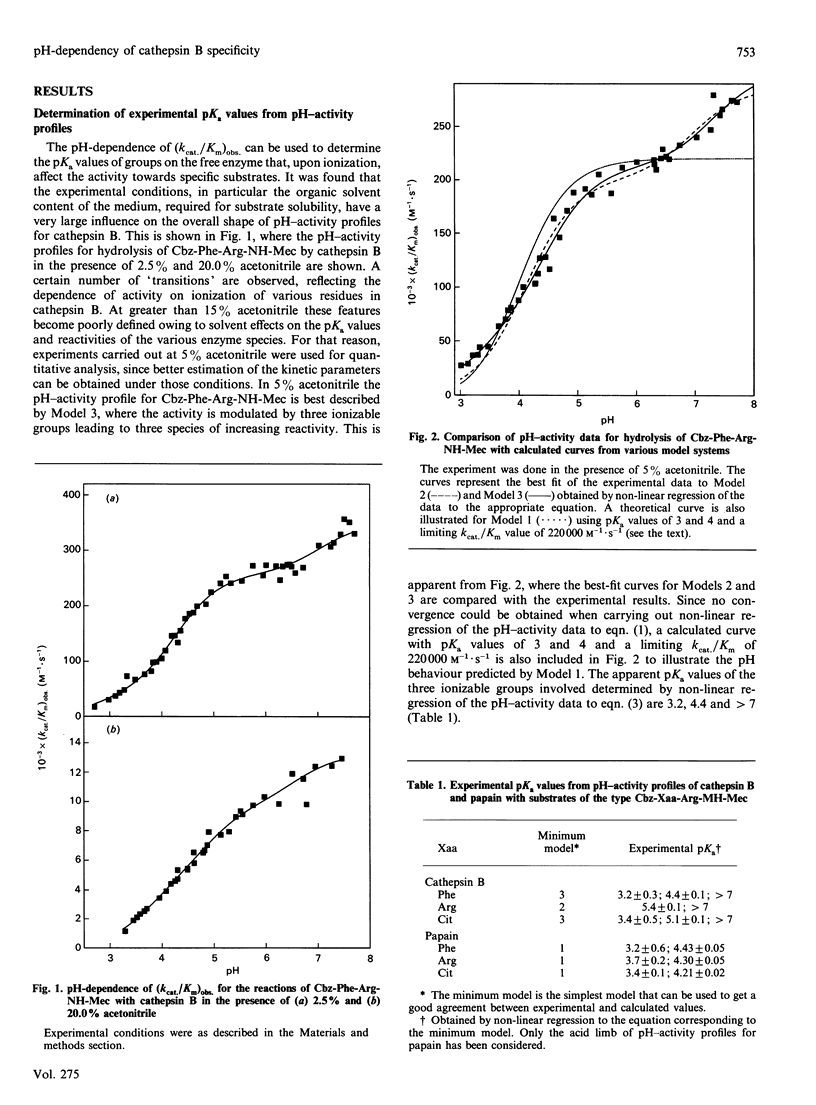
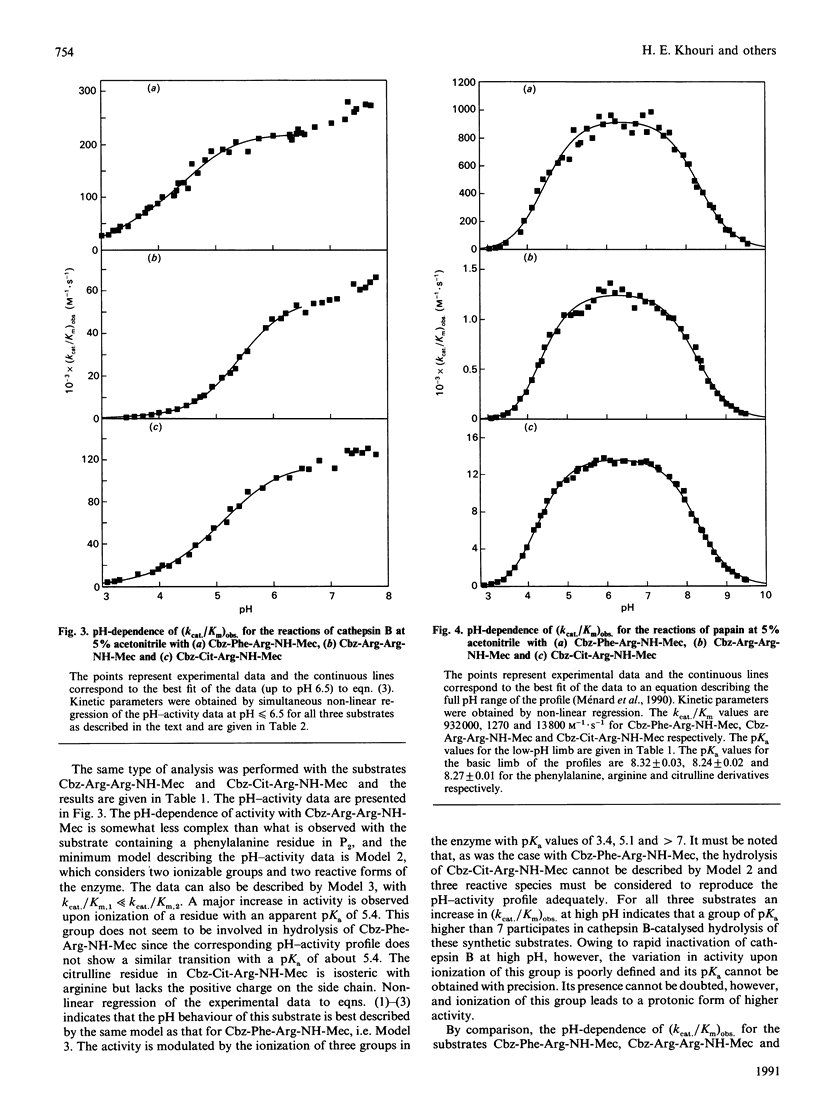
1. Three synthetic substrates of cathepsin B (EC 3.4.22.1) with various amino acid residues at the P2 position (Cbz-Phe-Arg-NH-Mec, Cbz-Arg-Arg-NH-Mec and Cbz-Cit-Arg-NH-Mec, where Cbz represents benzyloxycarbonyl and NH-Mec represents 4-methylcoumarin-7-ylamide) were used to investigate the pH-dependency of cathepsin B-catalysed hydrolyses and to obtain information on the nature of enzyme-substrate interactions. 2. Non-linear-regression analysis of pH-activity profiles for these substrates indicates that at least four ionizable groups on cathepsin B with pKa values of 3.3, 4.55, 5.46 and greater than 7.3 can affect the rate of substrate hydrolysis. 3. Ionization of the residue with a pKa of 5.46 has a strong effect on activity towards the substrate with an arginine in P2 (8.4-fold increase in activity) but has only a moderate effect on the rate of hydrolysis with Cbz-Cit-Arg-NH-Mec (2.3-fold increase in activity) and virtually no effect with Cbz-Phe-Arg-NH-Mec. The kinetic data are consistent with this group being an acid residue with a side chain able to interact with the side chains of an arginine or a citrulline in the P2 position of a substrate. Amino acid sequence alignment and model building with the related enzyme papain (EC 3.4.22.2) suggest that Glu-245 of cathepsin B is a likely candidate. The relative importance of electrostatic and hydrophobic interactions in the S2 subsite of cathepsin B is discussed. 4. For all three substrates, activity appears after ionization of a group with a pKa of 3.3, believed to be the active-site Cys-29 of cathepsin B. The identity of the groups with pKa values of 4.55 and greater than 7.3 remains unknown.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aronson N. N., Jr, Barrett A. J. The specificity of cathepsin B. Hydrolysis of glucagon at the C-terminus by a peptidyldipeptidase mechanism. Biochem J. 1978 Jun 1;171(3):759–765. doi: 10.1042/bj1710759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajkowski A. S., Frankfater A. The pH dependency of bovine spleen cathepsin B-catalyzed transfer of N alpha-benzyloxycarbonyl-L-lysine from p-nitrophenol to water and dipeptide nucleophiles. Comparisons with papain. J Biol Chem. 1983 Feb 10;258(3):1650–1655. [PubMed] [Google Scholar]
- Barrett A. J., Kirschke H. Cathepsin B, Cathepsin H, and cathepsin L. Methods Enzymol. 1981;80(Pt 100):535–561. doi: 10.1016/s0076-6879(81)80043-2. [DOI] [PubMed] [Google Scholar]
- Berger A., Schechter I. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):249–264. doi: 10.1098/rstb.1970.0024. [DOI] [PubMed] [Google Scholar]
- Bond J. S., Barrett A. J. Degradation of fructose-1,6-bisphosphate aldolase by cathepsin B. Biochem J. 1980 Jul 1;189(1):17–25. doi: 10.1042/bj1890017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst K., Willenbrock S. J., Salih E. Effects of conformational selectivity and of overlapping kinetically influential ionizations on the characteristics of pH-dependent enzyme kinetics. Implications of free-enzyme pKa variability in reactions of papain for its catalytic mechanism. Biochem J. 1983 Jun 1;211(3):701–708. doi: 10.1042/bj2110701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenth J., Kalk K. H., Swen H. M. Binding of chloromethyl ketone substrate analogues to crystalline papain. Biochemistry. 1976 Aug 24;15(17):3731–3738. doi: 10.1021/bi00662a014. [DOI] [PubMed] [Google Scholar]
- Dufour E. Sequence homologies, hydrophobic profiles and secondary structures of cathepsins B, H and L: comparison with papain and actinidin. Biochimie. 1988 Oct;70(10):1335–1342. doi: 10.1016/0300-9084(88)90004-1. [DOI] [PubMed] [Google Scholar]
- ELLMAN G. L. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959 May;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Gray P. J., Duggleby R. G. Analysis of kinetic data for irreversible enzyme inhibition. Biochem J. 1989 Jan 15;257(2):419–424. doi: 10.1042/bj2570419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao T., Hara K., Takahashi K. Purification and characterization of cathepsin B from monkey skeletal muscle. J Biochem. 1984 Mar;95(3):871–879. doi: 10.1093/oxfordjournals.jbchem.a134680. [DOI] [PubMed] [Google Scholar]
- Kamphuis I. G., Drenth J., Baker E. N. Thiol proteases. Comparative studies based on the high-resolution structures of papain and actinidin, and on amino acid sequence information for cathepsins B and H, and stem bromelain. J Mol Biol. 1985 Mar 20;182(2):317–329. doi: 10.1016/0022-2836(85)90348-1. [DOI] [PubMed] [Google Scholar]
- Katunuma N., Towatari T., Tamai M., Hanada K. Use of new synthetic substrates for assays of cathepsin L and cathepsin B. J Biochem. 1983 Apr;93(4):1129–1135. doi: 10.1093/oxfordjournals.jbchem.a134238. [DOI] [PubMed] [Google Scholar]
- Knight C. G. Human cathepsin B. Application of the substrate N-benzyloxycarbonyl-L-arginyl-L-arginine 2-naphthylamide to a study of the inhibition by leupeptin. Biochem J. 1980 Sep 1;189(3):447–453. doi: 10.1042/bj1890447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R. W. Interaction of lysosomal cysteine proteinases with alpha 2-macroglobulin: conclusive evidence for the endopeptidase activities of cathepsins B and H. Arch Biochem Biophys. 1989 Sep;273(2):367–374. doi: 10.1016/0003-9861(89)90495-5. [DOI] [PubMed] [Google Scholar]
- McKay M. J., Offermann M. K., Barrett A. J., Bond J. S. Action of human liver cathepsin B on the oxidized insulin B chain. Biochem J. 1983 Aug 1;213(2):467–471. doi: 10.1042/bj2130467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R., Khouri H. E., Plouffe C., Dupras R., Ripoll D., Vernet T., Tessier D. C., Lalberté F., Thomas D. Y., Storer A. C. A protein engineering study of the role of aspartate 158 in the catalytic mechanism of papain. Biochemistry. 1990 Jul 17;29(28):6706–6713. doi: 10.1021/bi00480a021. [DOI] [PubMed] [Google Scholar]
- Polgár L., Csoma C. Dissociation of ionizing groups in the binding cleft inversely controls the endo- and exopeptidase activities of cathepsin B. J Biol Chem. 1987 Oct 25;262(30):14448–14453. [PubMed] [Google Scholar]
- Rich D. H., Brown M. A., Barrett A. J. Purification of cathepsin B by a new form of affinity chromatography. Biochem J. 1986 May 1;235(3):731–734. doi: 10.1042/bj2350731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock F., Brocklehurst K. A general framework of cysteine-proteinase mechanism deduced from studies on enzymes with structurally different analogous catalytic-site residues Asp-158 and -161 (papain and actinidin), Gly-196 (cathepsin B) and Asn-165 (cathepsin H). Kinetic studies up to pH 8 of the hydrolysis of N-alpha-benzyloxycarbonyl-L-arginyl-L-arginine 2-naphthylamide catalysed by cathepsin B and of L-arginine 2-naphthylamide catalysed by cathepsin H. Biochem J. 1985 Apr 15;227(2):521–528. doi: 10.1042/bj2270521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock F., Brocklehurst K. Natural structural variation in enzymes as a tool in the study of mechanism exemplified by a comparison of the catalytic-site structure and characteristics of cathepsin B and papain. pH-dependent kinetics of the reactions of cathepsin B from bovine spleen and from rat liver with a thiol-specific two-protonic-state probe (2,2'-dipyridyl disulphide) and with a specific synthetic substrate (N-alpha-benzyloxycarbonyl-L-arginyl-L-arginine 2-naphthylamide). Biochem J. 1984 Sep 15;222(3):805–814. doi: 10.1042/bj2220805. [DOI] [PMC free article] [PubMed] [Google Scholar]


