Abstract
CtBP has been shown to be a highly conserved corepressor of transcription. E1A and all the various transcription factors to which CtBP binds contain a conserved PLDLS CtBP-interacting domain, and EBNA3C includes a PLDLS motif (amino acids [aa] 728 to 732). Here we show that EBNA3C binds to CtBP both in vitro and in vivo and that the interaction requires an intact PLDLS. The C terminus of EBNA3C (aa 580 to 992) has modest trans-repressor activity when it is fused to the DNA-binding domain of Gal4, and deletion or mutation of the PLDLS sequence ablates this and unmasks a transactivation function within the fragment. However, loss of the CtBP interaction motif had little effect on the ability of full-length EBNA3C to repress transcription. A striking correlation between CtBP binding and the capacity of EBNA3C to cooperate with (Ha-)Ras in the immortalization and transformation of primary rat embryo fibroblasts was also revealed.
CtBP (E1A C-terminal binding protein) was initially identified as a cellular factor interacting with the COOH terminus (amino acids [aa] 225 to 238) of adenovirus E1A oncoproteins. Although the precise significance of this interaction remains unknown, it is essential for the immortalization of primary rodent cells by E1A and has also been reported to negatively modulate E1A-mediated transformation, tumorigenicity, and metastasis (2, 23, 24, 27, 29). More recently it has been shown that this E1A-binding protein is one of a highly conserved family of (co)repressors of transcription. The Drosophila melanogaster homologue dCtBP mediates transcriptional repression by at least six different transcription factors, including Knirps, Kruppel, Snail, Zfh-1, Polycomb, and Hairy (16, 17, 20, 22, 28). CtBP also interacts with Xenopus and human Polycomb proteins (28), and human hCtBP acts as a corepressor for the ZEB transcription factor that is involved in the regulation of lymphocyte and muscle differentiation (23). The mouse homologue mCtBP is a corepressor for the NET member of the Ets family of transcription factors and oncogenes (3). mCtBP also participates in the Ikaros repression complex (10). It has been reported that in some situations CtBP can recruit chromatin-modifying histone deacetylase (HDAC) enzymes 1, 4, 5, and 7 and that it can also bind Sin3A. However, the precise molecular mechanism by which CtBP inhibits transcription is unknown and may turn out to be different in different situations (3, 10, 15, 30, 36). All these various cellular transcription factors and also the E1A proteins contain a conserved Pro-X-Asp-Leu-Ser (PLDLS) CtBP interaction domain that is necessary and probably sufficient for the interaction. A second mammalian CtBP was recently described, and the two family members—which are referred to as CtBP1 and CtBP2—are largely homologous, although they may have distinct tissue distributions (5, 32). The protein described in this report is human CtBP1 and hereafter will be referred to as CtBP.
The PLDLS amino acid motif is also found in a human cellular protein called CtIP (CtBP interacting protein), which also has (co)repressor activity and was recently shown to bridge an interaction between the retinoblastoma tumor suppressor protein (pRb) and CtBP, forming a complex which can repress E2F-regulated genes and thus participate in regulation of the cell proliferation cycle; CtIP probably bridges p130 and CtBP in a similar manner (6, 15). CtIP has also been shown to bind to the carboxyl-terminal region of the breast cancer-associated tumor suppressor and transcription factor BRCA1 and may be involved in regulation of the p21WAF1 and GADD45 genes by BRCA1 (11, 12, 34, 35).
Using recombinant viruses, EBNA3C has been shown to be one of the five viral genes which are absolutely essential for the activation and immortalization of human B cells by Epstein-Barr virus (EBV) (8, 9, 31). The large (992 aa) nuclear protein that it encodes is first expressed in EBV-infected resting human B cells during activation into their first proliferation cycle; thereafter, the steady-state level of EBNA3C in lymphoblastoid cell lines (LCLs) is remarkably constant. To date, detailed genetic analysis of EBNA3C function in this immortalization process has not been possible. The only recombinant EBV in which EBNA3C has been modified are unable to express a functional protein, and these viruses fail to immortalize B cells (31). The little we know about the activities of EBNA3C in the immortalization process has been extrapolated from evidence gained from in vitro biochemical studies and the transfection of EBNA3C expression plasmids. These approaches have revealed that EBNA3C can act as a potent repressor of transcription when it is targeted to DNA as a fusion with the DNA-binding domain (DBD) of Gal4 (1, 33). Moreover, the unfused wild-type protein can specifically repress reporter plasmids containing the EBV Cp latency-associated promoter. EBNA3C binds a transcriptional repression complex which includes the chromatin-modifying enzyme HDAC1, and the data are consistent with this being targeted to Cp by the cellular DNA-binding protein CBF1/RBP-Jκ (25, 26). Since Cp is the main promoter for EBNA mRNA initiation, EBNA3C might contribute to a negative autoregulatory control loop. Although the full-length EBNA3C represses transcription when it is targeted to DNA, a cryptic (recessive) transactivation domain located in the C terminus (aa 724 to 826) has also been described (14), and in certain circumstances EBNA3C can transactivate the EBV LMP1 promoter (14, 37).
In addition to modulating transcription, EBNA3C can substitute for papillomavirus type 16 E7 and adenovirus E1A in oncogenic transformation assays; like these other viral oncoproteins, EBNA3C also enables activated (Ha-)Ras to transform primary rodent embryo fibroblasts (REFs). Also like E7 and E1A, it can overcome the repressive effect of p16INK4 in REF assays (18). These results indicated that EBNA3C might override normal signals for growth arrest at the restriction point (R-point) in G1 of the cell cycle, when pRb (the operational target of p16INK4) is primarily active. This was subsequently confirmed when it was shown that EBNA3C over expression can direct cell cycle progression in serum-deprived cells and suppress the accumulation of the cyclin-dependent kinase inhibitor p27KIP1 that is normally associated with exit from the cell cycle. Overexpression of EBNA3C also leads to polyploidy and the emergence of cells with multiple nuclei, suggesting that it might deregulate additional cell cycle checkpoints (19).
Inspection of the predicted amino acid sequence of EBNA3C revealed a motif of five residues (aa 728 to 732) that matched perfectly the CtBP-binding site in E1A. Since CtBP plays a role in transcriptional repression and EBNA3C can repress transcription, the ability of in vitro-translated EBNA3C to bind to a glutathione S-transferase (GST) fusion with CtBP was tested. As predicted by the presence of a PLDLS sequence, EBNA3C was precipitated by GST-CtBP bound to Sepharose beads (Fig. 1A). The GST pulldown assays were performed essentially as described previously (25, 26).
FIG. 1.
(A) PLDLS motif in EBNA-3C is essential for CtBP binding in vitro, but flanking residues affect the affinity of the interaction. Bacterially expressed GST or GST-CtBP fusion protein (CtBP) was incubated with equal amounts of [35S]methionine-labeled wild-type EBNA-3C (WT) or EBNA-3C deleted for the PLDLS motif (ΔPLDLS), point mutated (Pro728 into Ala728 and Leu731 into Ala731) (ALDAS), deleted for the PLDLS motif and point mutated (Gln901 to Asp901 and Asp902 to Leu902), creating a substitute 899PLDLS903 (PLDLS−/+). Each assay was done in triplicate and consistently produced similar results; all these mutants are illustrated in panel B. The results of the binding experiments shown in panel A were quantified using a Storm 860 (Molecular Dynamics), and the average values are shown in panel C. The degree of binding to wild-type EBNA3C was given an arbitrary value of 100.
PLDLS motif in EBNA3C is essential and sufficient for interaction with CtBP.
Mutants of EBNA3C were generated by recombinant PCR or site-directed mutagenesis (Quickchange; Stratagene) in order to test whether the PLDLS motif was necessary for this binding. In one mutant, the PLDLS encoding nucleotides were deleted (ΔPLDLS), and in the other the critical proline and leucine residues (13) were replaced with alanine (ALDAS). These proteins were translated in vitro and tested in GST pulldown assays, and the results, illustrated in Fig. 1A, showed that both mutants had lost the ability to bind to CtBP. These data are entirely consistent with PLDLS being essential for the interaction between EBNA3C and CtBP.
In order to show that PLDLS is not only necessary but also sufficient for CtBP binding by EBNA3C, another mutant was generated. The ΔPLDLS mutant was further mutated at the codons for residues 901Q and 902D to create the substitute 899PLDLS903 (EBNA3C.PLDLS−/+). GST pulldown experiments were performed and consistently showed that ΔPLDLS−/+ was able to bind GST-CtBP, but with reduced efficiency relative to wild-type EBNA3C (Fig. 1A). These results showed that PLDLS is sufficient for the interaction with CtBP but that the binding affinity could be affected by the flanking residues and/or the position of the motif in the polypeptide (summarized in Fig. 1B and C).
CtBP and EBNA3C interact in vivo.
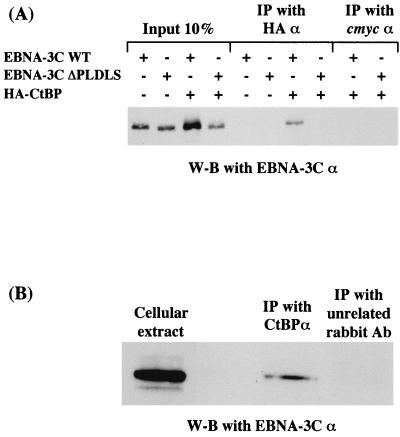
Further experiments were performed in order to determine whether EBNA3C and CtBP interact in vivo. Initially, a series of cotransfections were performed using a plasmid encoding a hemagglutinin (HA)-tagged CtBP protein. This was expressed from pSG5-HA-CtBP. DG75 Burkitt's lymphoma-derived B cells were transfected with plasmids encoding wild type EBNA3C (WT) or EBNA3C.ΔPLDLS with or without the plasmid encoding the tagged CtBP. Protein extracts from the transfected cells were subjected to immunoprecipitation (IP) using monoclonal antibodies recognizing the HA epitope or an irrelevant antibody against the c-Myc epitope. Approximately 107 DG75 cells were transiently transfected, by electroporation, with vector DNA encoding HA-tagged CtBP, EBNA3C, or EBNA3C.ΔPLDLS in the combinations indicated. At 48 h after transfection, DG75 cells were harvested and lysed in 600 μl of lysis buffer (50 mM Tris [pH 8], 150 mM NaCl, 10% glycerol, 0.5% Triton X-100, 2 mM phenylmethylsulfonyl fluoride, 2 mM proteinase inhibitor cocktail [Boehringer Mannheim]). Lysates were then centrifuged at 14,000 rpm at 4°C for 10 min, and 200 μl of the supernatant was incubated with protein G-Sepharose beads (Amersham Pharmacia Biotech) for 1 h at 4°C. The mixture was then centrifuged for 1 min at 1,500 rpm at 4°C, and the supernatant was transferred to a fresh tube. The supernatant was incubated with the appropriate antibody for 2 h at 4°C before 30 μl of protein G-Sepharose beads was added, and the mixture was incubated for a further hour at 4°C. Next the mixture was centrifuged for 1 min at 1,500 rpm at 4°C, the supernatant was removed, and the precipitate was washed with 1 ml of ice-cold lysis buffer. The mixture was then centrifuged for 1 min at 1,500 rpm at 4°C. This washing procedure was repeated four times. After boiling in sodium dodecyl sulfate (SDS) sample buffer and removal of the protein G-Sepharose beads by centrigugation, the proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose. IP products were then Western blotted using the A.10 monoclonal antibody directed against EBNA3C as described previously (18, 19, 25, 26). Representative results (Fig. 2A) show that EBNA3C (WT) is precipitated by anti-HA only in the presence of HA-CtBP. In contrast, EBNA3C.ΔPLDLS was not precipitated by anti-HA. Similar results were obtained with EBNA3C.ALDAS (data not shown).
FIG. 2.
(A) EBNA-3C WT but not EBNA-3C.ΔPLDLS can interact with HA-CtBP in vivo. DG75 cells were transfected with 10 μg of pSG5-EBNA-3C WT or 10 μg of pSG5-EBNA-3C ΔPLDLS with or without 10 μg of pSG5-HA-CtBP. Forty-eight hours after transfection, cell extracts were subjected to IP with mouse monoclonal antibodies against HA or c-Myc as indicated. After resolution on an SDS–7.5% polyacrylamide gel, the proteins were Western blotted (W-B) and probed with anti-EBNA-3C A.10. (B) EBV-encoded EBNA-3C interacts with cellular protein CtBP in vivo. Cell extracts from an LCL immortalized by EBV were subjected to IP with rabbit polyclonal antibodies against CtBP or a rabbit anti-mouse immunoglobulin serum (Dako) as indicated. After resolution on an SDS–7.5% polyacrylamide gel, the proteins were Western blotted and probed with anti-EBNA-3C A.10.
Coimmunoprecipitation experiments were performed on extracts from LCL cells using an anti-CtBP rabbit serum. The LCLs express the complete range of EBV latent proteins expressed from the episomal genome. The products of the immunoprecipitation were Western blotted with the anti-EBNA3C monoclonal antibody A.10. These experiments confirmed that in vivo, some fraction of CtBP and of physiologically normal levels of EBNA3C interact and coprecipitate (Fig. 2B shows a representative experiment). A Western blot of the LCL extract probed with the anti-CtBP serum confirmed the specificity of this reagent and showed that there was no cross-reactivity with EBNA3C (data not shown).
Mutation of the CtBP binding site in EBNA3C unmasks an activation domain located in the C terminus of EBNA3C.
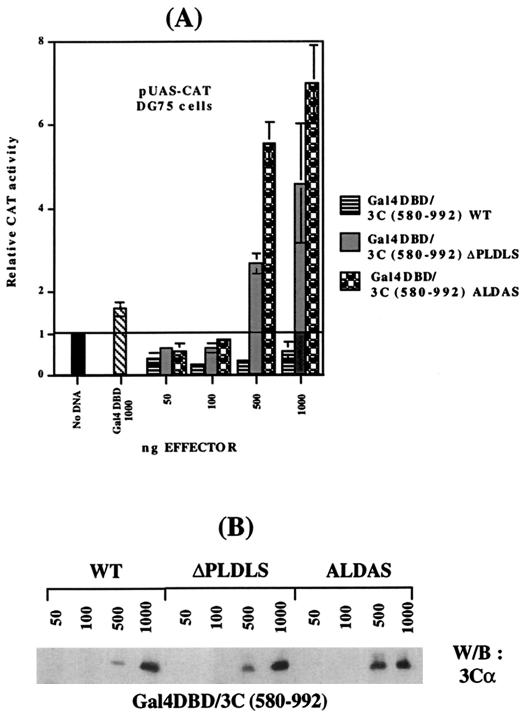
Previous studies showed that the C-terminal 412 aa of EBNA3C fused to the DNA-binding domain of Gal4 have modest repressor activity on the pUAS-CAT reporter (1). This fragment of EBNA3C includes the PLDLS motif. In order to determine the effect of deleting or changing the amino acid composition of the CtBP interaction site on this repressor activity, DG75 cells were transfected with pUAS-CAT (a Gal4-responsive reporter) together with pCDNA3-Gal4/3C(580–992).WT, pCDNA3-Gal4/3C(580–992).ΔPLDLS, or pCDNA3-Gal4/3C(580–992).ALDAS plasmid DNA. The electroporation procedure and activity assays were performed as described previously (25, 26). The results showed that while wild-type DNA produced modest repression (four- to fivefold) with up to 500 ng of transfected DNA, in similar experiments ΔPLDLS and ALDAS both had little or no effect at similar concentrations of input DNA (50 to 100 ng). However, at 500 ng and above, the mutants actually activated the pUAS-CAT reporter by up to eightfold (Fig. 3). These data indicate that EBNA3C and CtBP functionally interact in vivo and suggest that an activation domain located within this fragment of EBNA3C is occluded by CtBP binding to the PLDLS motif.
FIG. 3.
(A) Mutation of the CtBP binding site in EBNA-3C unmasks an activation domain located in the C terminus of EBNA-3C. DG75 cells were transfected with 5 μg of the pUAS-CAT reporter plasmid and with the amounts of pCDNA-3/Gal4DBD-HA (Gal4DBD), pCDNA-3/Gal4DBD-HA-EBNA-3C(580–992). WT, pCDNA-3/Gal4DBD-HA-EBNA-3C(580–992).ΔPLDLS, or pCDNA-3/Gal4DBD-HA-EBNA-3C(580–992).ALDAS effector plasmid DNA as indicated. Cell extracts were prepared 48 h after transfection, and chloramphenicol acetyctransferase (CAT) activity was determined. After normalization to β-galactosidase activity (2 μg of pSV-βgal per transfection was used), the data were expressed as CAT activity relative to the activity from pUAS-CAT with empty control vectors, which was given an arbitrary value of 1. Mean values and standard deviations from three independent experiments are shown. (B) Western -blot analysis of the cell extracts pooled from three independent experiments. The protein extract from transfected cells was resolved by SDS-PAGE (7.5%), blotted, and probed with anti-EBNA-3C A.10.
CtBP binding makes little contribution to repression by Gal4-EBNA3C and no apparent contribution to repression of Cp by EBNA3C.
Further experiments were performed in order to determine what contribution CtBP might make to the repression of transcription by Gal4 fusions with the full-length EBNA3C. A series of transfections in DG75 cells showed that a full-length EBNA3C fusion with the Gal4 DNA-binding domain was able to repress pUAS-CAT activity by up to approximately 10-fold. However, both the ΔPLDLS and the ALDAS mutant proteins consistently exhibited only very slightly reduced capacity to repress in similar assays (Fig. 4A). The reduction in activity was consistent but very modest, suggesting that binding to CtBP makes only a minor contribution to the ability of Gal4-EBNA3C to repress transcription by complementing the activity of other factors. This is consistent with our previous demonstration that the N-terminal half of EBNA3C also has repressor activity and that this is associated partly with binding to HDAC1 and partly with an unidentified factor(s) (1, 26). It seems that transcriptional repression by EBNA3C involves multiple repression domains and protein-protein interactions.
FIG. 4.
(A) CtBP makes little contribution to the repression by a full-length EBNA-3C/Gal4DBD fusion. DG75 cells were transfected with 5 μg of the pUAS-CAT reporter plasmid and with the amounts of pCDNA-3/Gal4DBD-HA (Gal4DBD), pCDNA-3/Gal4DBD-HA-EBNA-3C.WT, pCDNA-3/Gal4DBD-HA-EBNA-3C.ΔPLDLS, or pCDNA-3/Gal4DBD-HA-EBNA-3C.ALDAS effector plasmid DNA as indicated. Cell extracts were prepared 48 h after transfection, and CAT activity was determined. After normalization to β-galactosidase activity (2 μg of pSV-βgal per transfection was used), the data were expressed as fold repression relative to the activity from pUAS-CAT with empty control vectors, which was given an arbitrary value of 1. Mean values and standard deviations from three independent experiments are shown. (B) Western blot analysis of the cell extracts pooled from three independent experiments. The protein extract from transfected cells was resolved by SDS-PAGE (7.5%), blotted, and probed with anti-EBNA-3C A.10. (C) CtBP makes no significant contribution to the repression of Cp by a full-length EBNA-3C. DG75 cells were transfected with 5 μg of the 4XCp-TK-CAT reporter plasmid and with the amounts of pSG5-EBNA-3CWT, pSG5-EBNA-3C.ΔPLDLS, or pSG5-EBNA-3C.ALDAS effector plasmid DNA as indicated. Cell extracts were prepared 48 h after transfection, and CAT activity was determined. After normalization to β-galactosidase activity (2 μg of pSV-βgal per transfection was used), the data were expressed as fold repression relative to the activity from pUAS-CAT with empty control vectors, which was given an arbitrary value of 1. Mean values and standard deviations from three independent experiments are shown. (D) Western blot analysis of the cell extracts pooled from three independent experiments. The protein extract from transfected cells was resolved by SDS-PAGE (7.5%), blotted, and probed with anti-EBNA-3C A.10.
When the abilities of unfused EBNA3C.ΔPLDLS and EBNA3C.ALDAS to repress the EBV Cp promoter were compared with the wild-type protein, no significant differences were detected (Fig. 4C). Although this repression involves HDAC activity (26), it appears to be independent of CtBP.
CtBP-binding mutants of EBNA3C are impaired in their capacity to cooperate with (Ha-)Ras in the immortalization and transformation of REFs.
CtBP was originally defined as a protein that bound to the C terminus of the E1A oncoprotein of adenovirus. Furthermore, CtBP binds to at least one pRb-interacting protein, CtIP, and this interaction is probably central to the repression of transcription and regulation of cell proliferation by pRb (7, 15).
Since EBNA3C can bind CtBP, also cooperates with activated Ras to immortalize and transform REFs, and appears to disrupt the p16INK4-pRb regulatory pathway (18, 19), we determined whether CtBP binding was involved in the cooperation of EBNA3C with (Ha-)Ras in primary REF immortalization and transformation assays. Multiple experiments (n = 14) performed as described previously (4, 18) are summarized in Table 1. They show a remarkably good correlation between EBNA3C binding to CtBP and the capacity of EBNA3C to cooperate with Ras to produce transformed foci. The two mutants of EBNA3C (ΔPLDLS and ALDAS) that no longer bind CtBP were both severely compromised in their ability to cooperate with (Ha-)Ras. The binding of CtBP to EBNA3C appears to be important for the outgrowth of the primary rodent cells cotransfected with EBNA3C and activated Ras. This again suggests that there is an interaction in vivo and that it has some biological significance. Both mutants of EBNA3C were shown to have a nuclear distribution when expressed transiently in U20S cells (data not shown).
TABLE 1.
Cooperation of EBNA3C with Rasa
| No. of colonies/dish in expt no.:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| Plasmid | ||||||||||||||
| pcDNA3 | 0 | 4 | 3 | 2 | 0 | 2 | 2 | 2 | 0 | 0 | 0 | 1 | 4 | 0 |
| WT3C | 13 | 13 | 20 | 11 | 9 | 14 | 12 | 9 | 13 | 7 | 9 | 18 | 9 | 11 |
| ΔPLDLS | 2 | 5 | 5 | 3 | 5 | 6 | 6 | 4 | 6 | 4 | 0 | 5 | 5 | 2 |
| ALDAS | 0 | 1 | 6 | 5 | 4 | 4 | 3 | 1 | 1 | 2 | 0 | 0 | 3 | 3 |
In each assay, 15 μg of EBNA3C expression plasmid or the pcDNA3 vector was cotransfected with 5 μg of EJ6.6 (Ha-Ras expression vector). Cells were then grown for 21 to 28 days with G418 (400 μg/ml). The highly refractile, morphologically transformed colonies were then counted under a microscope.
Using various in vitro and in vivo assays here, we have convincingly demonstrated that EBNA3C can interact with the cell protein CtBP through a PLDLS motif. The motif is both necessary and sufficient for binding to CtBP; however, the surrounding amino acids affect the affinity of the interaction between EBNA3C and CtBP. Most or perhaps all of the factors that have been shown to interact with CtBP do so through a PLDLS or PLDLS-like motif. Without exception, all the reported CtBP-binding factors have been implicated in transcriptional repression, and unsurprisingly, fusions of CtBP with the Gal4 DNA-binding domain are also capable of repressing transcription when targeted to promoters (15, 28). Since EBNA3C includes a canonical CtBP-binding motif and this mediates a physical interaction between the two proteins, we determined the functional significance of this sequence in two of the phenotypes described for EBNA3C. The roles of PLDLS in the repression of transcription and in the cooperation with (Ha-)Ras were investigated. In an initial series of experiments, a fusion between the C terminus of EBNA3C (aa 580 to 992) and DNA-binding domain of Gal4 was used. Deleting or mutating the PLDLS motif transformed this polypeptide from a modest repressor of transcription (four- to fivefold) into a moderately powerful transactivator (eightfold). This contrived system confirmed, using a functional readout, that EBNA3C and CtBP can interact in vivo through the PLDLS. Furthermore, the results suggest that in addition to recruiting a repression complex to the EBNA3C fusion, the binding of CtBP to the PLDLS motif occludes a domain in EBNA3C that can activate transcription. This cryptic activation sequence has not been characterized here, but it may be the transactivation domain described previously (14).
Despite compelling evidence for a physical and functional interaction between CtBP and the C terminus of EBNA3C, surprisingly, when experiments were performed with the full-length EBNA3C, deletion or mutation of the PLDLS motif made little difference to the activity recorded. This is probably because other factors recruited by Gal4-EBNA3C or unfused EBNA3C are more important. These could include CBF1/RBP-Jκ, an HDAC complex, and an unidentified factor(s) that associates with the region between aa 280 and 525 of EBNA3C (1, 25, 26). Since it is not known whether EBNA3C targets any cellular genes, we cannot rule out the possibility that other EBNA3C-responsive promoters exist and that these could be dependent on the CtBP interaction for repression.
We next determined whether the ability to bind CtBP affected the capacity of EBNA3C to cooperate with (Ha-)Ras and permit the latter to transform primary rodent fibroblasts. Multiple experiments revealed a very good correlation between the ability of EBNA3C to bind CtBP in vitro and its ability to produce transformed foci after cotransfection of REFs with pCDNA3-EBNA3C and the (Ha-)Ras expression vector pEJ6.6.
The results presented in this study suggest at least two mechanisms for how EBNA3C might function. Either EBNA3C binds to CtBP and targets the associated repression complex to a cellular gene(s) and so down regulates a critical function required for the inhibition of cell proliferation, or, because it includes the same CtBP interaction motif as CtIP, the Polycomb proteins, and various other transcription factors involved in the regulation of proliferation and differentiation, EBNA3C could compete effectively with these factors for binding to CtBP. In this way EBNA3C might disrupt or modify repression complexes that are critical for the regulation of genes that drive cell cycle progression and/or differentiation. We cannot rule out either of these possibilities, and we are currently exploring both.
Acknowledgments
We thank G. Chinnadurai (St. Louis) for CtBP plasmids. We are grateful to M. Rowe (Cardiff) for providing the A.10 MAb and A. Otte (Amsterdam) for the anti-CtBP serum. We are also very grateful to Paul Farrell and Roger Watson for helpful comments on the manuscript and to Mark Bain for originally drawing our attention to the PLDLS motif in EBNA3C.
We are grateful to the Wellcome Trust for providing financial support through programme grant 056822 to M.J.A.
REFERENCES
- 1.Bain M, Watson R J, Farrell P J, Allday M J. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J Virol. 1996;70:2481–2489. doi: 10.1128/jvi.70.4.2481-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd J M, Subramanian T, Schaeper U, La Regina M, Bayley S T, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 Ela protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Criqui-Filipe P, Ducret C, Maira S-V, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and deacetylation. EMBO J. 1999;18:3392–3403. doi: 10.1093/emboj/18.12.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crook T, Parker G A, Rozycka M, Crossland S, Allday M J. A transforming p53 mutant, which binds DNA, transactivates and induces apoptosis, reveals a nuclear:cytoplasmic shuttling defect. Oncogene. 1998;16:1429–1441. doi: 10.1038/sj.onc.1201699. [DOI] [PubMed] [Google Scholar]
- 5.Furusawa T, Moribe H, Kondoh H, Higashi Y. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor dEF1. Mol Cell Biol. 1999;19:8581–8590. doi: 10.1128/mcb.19.12.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fusco C, Reymond A, Zervos A S. Molecular cloning and characterisation of a novel retinoblastoma-binding protein. Genomics. 1998;51:351–358. doi: 10.1006/geno.1998.5368. [DOI] [PubMed] [Google Scholar]
- 7.Harbour J W, Dean D C. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 8.Kempkes B, Pisch D, Zeilder R, Sugden B, Hammerschmidt W. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J Virol. 1995;69:231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 10.Koipally J, Georgopoulos K. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J Biol Chem. 2000;275:19594–19602. doi: 10.1074/jbc.M000254200. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Ting N S Y, Zheng L, Chen P-L, Ziv Y, Shiloh Y, Lee E Y-H P, Lee W-H. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 2000;406:210–215. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Chen P-L, Subramanian T, Chinnadurai G, Tomlinson G, Osborne C K, Sharp Z D, Lee W-H. Binding of CtIP to the BRCT repeats of BRCA1 involved in the transcription regulation of p21 is disrupted upon DNA damage. J Biol Chem. 1999;274:11334–11338. doi: 10.1074/jbc.274.16.11334. [DOI] [PubMed] [Google Scholar]
- 13.Malloy D P, Milner A E, Yakub I K, Chinnadurai G, Gallimore P H, Grand R J A. Structural determinants present in the C-terminal binding protein binding site of adenovirus early region 1A proteins. J Biol Chem. 1998;273:20867–20876. doi: 10.1074/jbc.273.33.20867. [DOI] [PubMed] [Google Scholar]
- 14.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meloni A R, Smith E J, Nevins J R. A mechanism for Rb/p 130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc Natl Aad Sci USA. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 18.Parker G A, Crook T, Bain M, Sara E A, Farrell P J, Allday M J. Epstein-Barr nuclear antigen (EBNA 3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene. 1996;13:2541–2549. [PubMed] [Google Scholar]
- 19.Parker G A, Touitou R, Allday M J. Epstein-Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene. 2000;19:700–709. doi: 10.1038/sj.onc.1203327. [DOI] [PubMed] [Google Scholar]
- 20.Pootinga G, Watanabe M, Parkhurst S M. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Postigo A A, Dean D C. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postigo A A, Ward E, Skeath J B, Dean D C. Zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol Cell Biol. 1999;19:7255–7263. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinlan M P, Whyte P, Grodzicker T. Growth factor induction by the adenovirus type 5 E1A 12S protein is required for immortalization of primary epithelial cells. Mol Cell Biol. 1988;8:3191–3203. doi: 10.1128/mcb.8.8.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan M P, Douglas J L. Immortalization of primary epithelial cells requires first- and second-exon functions of adenovirus type 5 12S. J Virol. 1992;66:2020–2030. doi: 10.1128/jvi.66.4.2020-2030.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radkov S A, Bain M, Farrell P J, West M, Rowe M, Allday M J. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J Virol. 1997;71:8552–8562. doi: 10.1128/jvi.71.11.8552-8562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radkov S A, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday M J. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaeper U, Boyd J M, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterisation of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in a negative modulation of oncogenic transformation. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sewalt R G, Gunster M J, van der Vlag J, Satijn D P, Otte A P. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol. 1999;19:777–787. doi: 10.1128/mcb.19.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian T, La Regina M, Chinnadurai G. Enhanced ras oncogene mediated cell transformation amd tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1A protein. Oncogene. 1989;4:415–520. [PubMed] [Google Scholar]
- 30.Sundquist A, Sollerbrant K, Svensson C. The carboxy-terminal region of adenovirus E1A activates transcription through targeing of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 1998;429:183–188. doi: 10.1016/s0014-5793(98)00588-2. [DOI] [PubMed] [Google Scholar]
- 31.Tomkinson B, Robertson E, Kieff E. Epstein-Barr nuclear proteins EBNA3A and EBNA3C are essential for B lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner J, Crossley M. Cloning and characterisation of mCtBP2, a co-repressor that associates with basic Kruppel-like factors and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ-EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X, Baer R. Nuclear localisation and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J Biol Chem. 2000;275:18541–18549. doi: 10.1074/jbc.M909494199. [DOI] [PubMed] [Google Scholar]
- 35.Yu X, Wu L C, Bowcock A M, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C L, McKinsey T A, Lu J-R, Olson E N. Association of the COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem. 2000;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 37.Zhao B, Sample C E. Epstein-Barr virus nuclear antigen 3C activates the latent membrane promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi1/SpiB binding site. J Virol. 2000;74:5151–5160. doi: 10.1128/jvi.74.11.5151-5160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]