Abstract
To identify parts of retroviral integrase that interact with cellular DNA, we tested patient-derived human immunodeficiency virus type 1 (HIV-1) integrases for alterations in the choice of nonviral target DNA sites. This strategy took advantage of the genetic diversity of HIV-1, which provided 75 integrase variants that differed by a small number of amino acids. Moreover, our hypothesis that biological pressures on the choice of nonviral sites would be minimal was validated when most of the proteins that catalyzed DNA joining exhibited altered target site preferences. Comparison of the sequences of proteins with the same preferences then guided mutagenesis of a laboratory integrase. The results showed that single amino acid substitutions at one particular residue yielded the same target site patterns as naturally occurring integrases that included these substitutions. Similar results were found with DNA joining reactions conducted with Mn2+ or with Mg2+ and were confirmed with a nonspecific alcoholysis assay. Other amino acid changes at this position also affected target site preferences. Thus, this novel approach has identified a residue in the central domain of HIV-1 integrase that interacts with or influences interactions with cellular DNA. The data also support a model in which integrase has distinct sites for viral and cellular DNA.
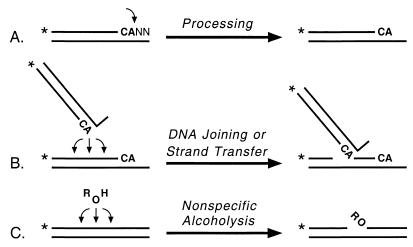
Integration of a DNA copy of the retroviral genome into cellular DNA is essential for retrovirus replication. The retroviral integrase protein catalyzes two endonuclease reactions that are necessary for integration. During processing, integrase prepares the viral DNA for integration by removing two or three nucleotides that follow highly conserved CA bases near the 3′ end of each DNA strand. During DNA joining (or strand transfer), integrase inserts the processed viral DNA ends into cellular DNA in a sequence-independent manner. These in vivo activities can be modeled by using purified integrase proteins and oligonucleotides that mimic either end of viral DNA (4, 16, 18). In particular, human immunodeficiency virus type 1 (HIV-1) integrase processes oligonucleotides derived from HIV-1 DNA by removing two nucleotides that follow the conserved CA (Fig. 1A). Integrase also inserts some of the processed oligonucleotides into various sites on other oligonucleotides, which now act as cellular DNA, to create a set of products that are longer than the substrate (Fig. 1B). As occurs in vivo, almost any site can be used as the target for insertion even though certain sites are preferred (17). Our studies with chimeric proteins between the integrases of HIV-1 and visna virus demonstrated that the central domain of integrase plays a major role in selecting the target sites for insertion of viral DNA ends (22, 23). Moreover, the central region of integrase was solely responsible for the selection of nonviral target sites when the chimeric integrases catalyzed a reaction referred to as nonspecific alcoholysis (20, 22). During this activity, which shares certain characteristics with the DNA joining reaction (23), integrase uses a variety of nucleophiles to nick almost any site in nonviral DNA (Fig. 1C). In fact, the isolated central fragment of HIV-1 integrase can catalyze nonspecific alcoholysis and exhibits the same target site preferences as the full-length protein (21, 23).
FIG. 1.
Integrase assays. The conserved CA bases near the 3′ ends of viral DNA are shown in boldface, the terminal two nucleotides are indicated by NN, and asterisks denote 32P groups. (A) During processing, integrase makes a site-specific nick to form a labeled product two nucleotides shorter than the substrate. (B) During DNA joining or strand transfer, integrase inserts the processed viral DNA ends into any of various sites on either strand of target DNA (the labeled strand is used in the schematic), yielding a set of labeled products longer than the substrate. (C) During nonspecific alcoholysis, integrase uses certain nucleophilic molecules (shown as ROH) to nick nonviral DNA at any of multiple sites.
To identify amino acid residues of integrase that influence target site preferences in nonviral DNA, we screened naturally occurring, patient-derived HIV-1 integrase variants for alterations in the selection of nonviral target sites. We reasoned that a set of natural integrases, because of the genetic diversity of HIV-1, would provide many integrases that differ by a small number of amino acids. We also predicted that many of the proteins would be active for processing and joining because of in vivo selection but hypothesized that biological pressures on noncritical aspects of integrase function, such as the choice of nonviral target sites, would be minimal. Although we realized that some viral sequences recovered from clinical samples might not come from infectious genomes, we had already established that 85% of integrase sequences that we had amplified from HIV-infected patients encoded full-length proteins (33). Moreover, substitutions at any of the seven highly conserved amino acids known to be important for integrase activity were rare. Testing these proteins in functional assays has now validated these hypotheses. In particular, variations in nonviral target site selection were found for many proteins that had wild-type levels of processing and joining activity.
Target site preferences of patient-derived HIV-1 integrases.
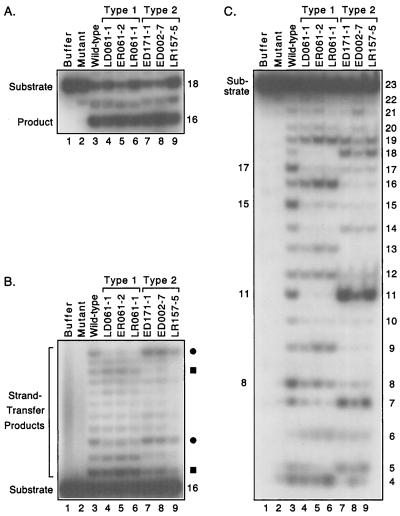
We previously described 102 HIV-1 integrase sequences that were amplified from viral DNA in blood cells or viral RNA in plasma that had been obtained many years ago from 10 HIV-infected hemophiliacs (33). These sequences encode 87 full-length proteins, of which 75 are unique. Each of these 75 proteins has between 4 and 16 amino acid changes compared to the prototypic HIV-1 integrase used in our laboratory. We have now expressed and purified the unique full-length proteins from a bacterial expression system and screened them for enzymatic activity, with a particular focus on the selection of target sites in the biologically relevant DNA joining assay. Purifications were conducted from 10 ml of bacterial cultures, using methods described previously (19, 20) but aided by the use of magnetic Ni2+-nitrilotriacetic acid beads (Qiagen, Chatsworth, Calif). Processing and joining reactions were initially conducted under conditions known to optimize HIV-1 integrase activity, including the use of oligonucleotides derived from the U5 end of HIV-1 DNA and Mn2+ as the divalent cation cofactor (34). Standard 10-μl reaction mixtures contained 0.5 pmol of double-stranded DNA, 25 mM Tris-HCl (pH 8.0), 10 mM dithiothreitol, 1.0 μl of integrase or protein storage buffer, and 10 mM MnCl2; reactions were conducted for 90 min and then analyzed by denaturing polyacrylamide gel electrophoresis and autoradiography. As expected, some of the variant proteins did not purify well and were inactive in these assays. However, 36 of the purified proteins catalyzed DNA joining. Remarkably, most of these proteins made patterns of strand-transfer products that differed from that of the laboratory integrase, reflecting altered target site preferences during DNA joining.
The novel sets of strand-transfer products created by the natural integrases clearly separated into two major patterns. Reactions with representative patient-derived proteins are shown in Fig. 2. These proteins catalyzed processing at levels comparable to that of the laboratory HIV-1 integrase and specifically removed two nucleotides from the 18-mer viral DNA substrate to create a 16-mer product (Fig. 2A). Moreover, some of the processed products from these reactions were inserted into other oligonucleotides to create strand-transfer patterns that differed from that of the laboratory integrase (not shown). To highlight the longer products, we performed joining assays with a preprocessed substrate that was missing the two nucleotides 3′ to the conserved CA. Under these conditions, the laboratory integrase (referred to as wild-type in the figures) creates a distinct pattern of slower-migrating products in which certain bands are more prominent (Fig. 2B, lane 3). However, the patterns created by many of the patient-derived integrases reproducibly had different relative intensities at certain positions. For example, the pattern referred to as type 1 included faint bands at positions that were much darker in reactions with the laboratory integrase (Fig. 2B, lanes 4 to 6, the bands denoted by circles), even though other bands in these lanes were darker than bands in the wild-type pattern. Similarly, the pattern referred to as type 2 differed in intensity at several positions compared with the pattern of the laboratory integrase (Fig. 2B, lanes 7 to 9, e.g., the bands denoted by squares). These patterns were independent of the amount of joining catalyzed by the enzymes and were not affected by the duration of the reactions (data not shown).
FIG. 2.
Target site selection by patient-derived integrases. Autoradiograms of a denaturing 20% polyacrylamide gel are shown. (A) Processing assay. Double-stranded 18-mers derived from one end of HIV-1 DNA were 5′ labeled on the strand that contains the conserved CA and incubated with protein buffer (lane 1), a laboratory HIV-1 integrase that contained an inactivating D116I mutation (lane 2), the wild-type laboratory HIV-1 integrase (lane 3), or six patient-derived HIV-1 integrases that are grouped as type 1 (lanes 4 to 6) or type 2 (lanes 7 to 9) based on the results shown in panel B. The nomenclature for the naturally occurring integrases is explained in Fig. 3. The 18-mer substrate and the 16-mer product are indicated. The amount of processing in lanes 3 to 9 ranged from 32 to 45%. (B) Joining assay. Double-stranded 16-mers representing a preprocessed end of HIV-1 DNA were 5′ labeled on the strand that contains the conserved CA and incubated with the same proteins as in panel A. The bands indicated by circles or squares are reduced in intensity in the type 1 or type 2 pattern, respectively, compared to the wild-type pattern. The 16-mer substrate and strand-transfer products are indicated. The amount of joining in lanes 3 to 9 ranged from 8 to 23%. (C) Nonspecific alcoholysis assay. Double-stranded 23-mers of nonviral sequence were 5′ labeled on one strand and incubated with the same proteins as in panel A in reactions that included 20% glycerol. Sizes in nucleotides of cleavage products are shown on the right; no bands were detected below the position of 4-mers. The 23-mer substrate and prominent products from the laboratory integrase are indicated on the left. The amount of substrate nicked in lanes 3 to 9 ranged from 41 to 59%.
The proteins that created the type 1 pattern in Fig. 2B were derived from different clinical samples from one patient, whereas the selected proteins that created the type 2 pattern were from three different patients. Overall, 10 of the 36 natural proteins that catalyzed DNA joining made the type 1 pattern, 10 made the type 2 pattern, and 1 (not discussed further) made a third novel pattern. There was no correlation between the target site patterns and the rate of disease progression. For example, the 15 proteins that exhibited the same preferences as the laboratory integrase were derived from four patients who subsequently died of AIDS and three patients with slow or no progression, and the 10 proteins grouped as type 2 were from two patients who died of AIDS and two slow progressors. In two cases, different integrases from the same individual created a wild-type and a novel pattern (Fig. 3, code numbers 002 and 143). A detailed analysis correlating clinical features with the other activities of these enzymes has been completed (M. Katzman, A. L. Harper, M. Sudol, L. M. Skinner, and M. E. Eyster, submitted for publication).
FIG. 3.
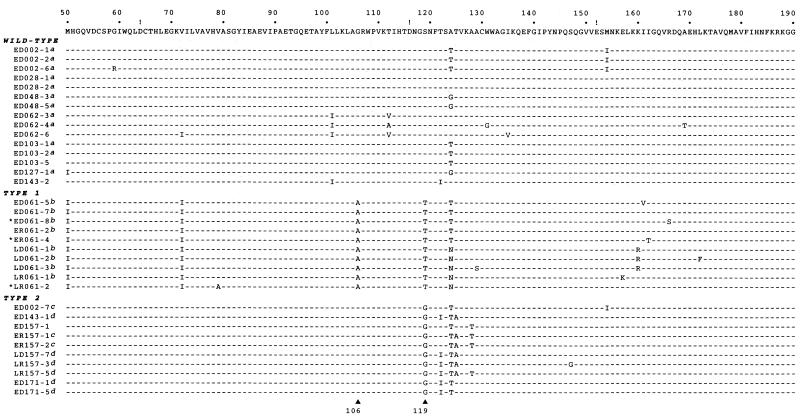
Sequence alignment of central domains of patient-derived integrases. The central region (residues 50 to 190) of the laboratory HIV-1BH10 integrase, which is similar to the HIV-1HXB2 integrase (30, 31), is shown above the patient-derived proteins. The conserved acidic amino acids at positions 64, 116, and 152 are indicated with exclamation points. Identity to the laboratory integrase sequence is indicated by a dash. Proteins are grouped according to the patterns exhibited in DNA joining assays conducted with Mn2+. Confirmation in DNA joining assays with Mg2+ is indicated as follows: a, confirmed as wild type in reactions with Mg2+; b, confirmed as type 1 in reactions with Mg2+; c, created one type of novel pattern in reactions with Mg2+; d, created another type of novel pattern in reactions with Mg2+. Groupings also were confirmed by nonspecific alcoholysis assays for all but the three proteins indicated with asterisks. Residues 106 and 119, which were targeted by site-directed mutagenesis, are indicated by triangles at the bottom. Patient-derived proteins are named with an E or L (indicating whether the samples were obtained early after infection or 5 years later), D or R (indicating whether DNA or RNA was the source for amplification), a three-digit patient code, and a single-digit number identifying the clone (33). The sequences shown match those submitted to GenBank but include three corrections compared to the previously published sequences: S129 in LD061-3, K157 in LR061-1, and G119 in ED002-7.
The altered target site preferences of the patient-derived proteins were confirmed with the nonspecific alcoholysis assay. Although integrase can use glycerol or certain other alcohols as the nucleophile to nick nonviral DNA at any internal position, certain sites are reproducibly preferred by different integrases (21). For example, the laboratory HIV-1 integrase prefers to nick at positions located 8, 11, 15, and 17 nucleotides from the 5′ end of the 23-mer substrate used in this assay (Fig. 2C, lane 3). Significantly, the novel alcoholysis patterns made by the natural integrases shown in Fig. 2 resulted in the same grouping of the proteins as occurred with the DNA joining assay. In particular, the three natural proteins that were grouped as type 1 for DNA joining made an alcoholysis pattern that included a very prominent band at position 16 (Fig. 2C, lanes 4 to 6). In contrast, the three proteins that were grouped as type 2 for DNA joining made a novel alcoholysis pattern that included a strong band at position 11, as did the laboratory enzyme, but very weak bands at positions 8, 15, and 17 (Fig. 2C, lanes 7 to 9). The patterns in Fig. 2C resulted from the unique alcoholysis activity of integrase, and not from contaminating nucleases, because attachment of glycerol to the appropriate sites was confirmed in assays that used a 3′-labeled substrate (data not shown) (21). Overall, the nonspecific alcoholysis assay confirmed the categorization of 32 of the 35 proteins listed in Fig. 3, whereas three proteins that were type 1 by the joining assay were inactive or unclassifiable in the alcoholysis assay (Fig. 3, asterisks).
Because of the importance of the central domain of integrase in the selection of target sites in nonviral DNA (13, 20–23, 32), we compared the sequences of this region between the natural proteins (Fig. 3). This alignment suggested amino acids that might be contributing to the observed patterns. For example, every protein grouped as type 1 has alanine at residue 106 and threonine at residue 119, whereas proteins with the wild-type or type 2 pattern do not share these amino acids. Similarly, every protein grouped as type 2 has glycine at residue 119, whereas no other protein has this amino acid. In contrast, isoleucine at residues 50 and 72 is common to the type 1 proteins, but two proteins grouped as wild type also have Ile at one of these residues (i.e., ED127-1 and ED062-6). Similarly, although all proteins grouped as type 2 have threonine at position 124, some integrases with a wild-type or type 1 pattern also have Thr at this position (e.g., ED002-1 and ED061-5). The sequences of four other proteins that would have been classified as wild type (1 protein), type 1 (1 protein), or type 2 (2 proteins) based solely on their patterns in the alcoholysis assay are not shown in Fig. 3 but are consistent with these observations. Thus, this analysis suggested that residues 106 and 119 contribute to the selection of target sites in nonviral DNA.
Single amino acid substitutions in the laboratory HIV-1 integrase.
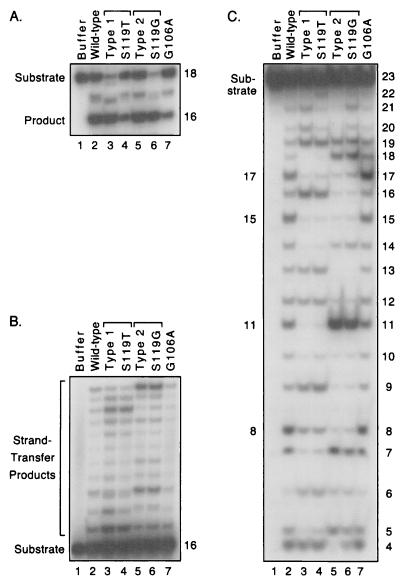
To examine the role of particular protein residues in the selection of target sites, we replaced single amino acids in the laboratory integrase by using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, Calif.). The entire integrase-coding region of all new proteins was confirmed by sequencing. Based on the above discussion, we made single amino acid changes of Gly-106 to Ala or of Ser-119 to Thr or Gly. We first established that each of the new proteins specifically processed the HIV-1 viral DNA 18-mer substrate to a 16-mer product (Fig. 4A). In addition, each of these enzymes catalyzed DNA joining (Fig. 4B). Moreover, the S119T protein created a pattern that closely matched the type 1 pattern made by a patient-derived integrase that included this amino acid among 13 differences from the laboratory enzyme (Fig. 4B, lanes 3 and 4). Similarly, the S119G protein made a pattern that was identical to the type 2 pattern made by a natural integrase that included this change among eight differences (Fig. 4B, lanes 5 and 6). Thus, these substitutions at residue 119 account for the novel patterns made by the naturally occurring integrases. In contrast, substitution at residue 106 did not explain the type 1 pattern because the G106A protein made a strand-transfer pattern indistinguishable from that of the laboratory integrase (Fig. 4B, lanes 2 and 7).
FIG. 4.
Site-directed mutations explain the novel target site patterns. Details are as in the legend to Fig. 2. For each panel, the substrates were incubated with protein buffer (lane 1), the laboratory integrase (lane 2), a patient-derived integrase that makes the type 1 pattern (ED061-7, lane 3), the laboratory integrase with an S119T substitution (lane 4), a patient-derived integrase that makes the type 2 pattern (ED171-1, lane 5), the laboratory integrase with an S119G substitution (lane 6), or the laboratory integrase with a G106A substitution (lane 7). The amount of products formed in lanes 2 to 7 ranged from 25 to 52% in panel A, 8 to 27% in panel B, and 40 to 52% in panel C.
The above results were confirmed when the proteins were tested in the nonspecific alcoholysis assay. In particular, the S119T protein made a pattern that closely matched the type 1 alcoholysis pattern made by a patient-derived integrase that included this amino acid (Fig. 4C, lanes 3 and 4). Similarly, the S119G protein made a pattern that was very similar to the type 2 pattern made by a natural integrase that included this amino acid (Fig. 4C, lanes 5 and 6). These results were confirmed by using a 3′-labeled substrate and by using a different nonviral DNA substrate that was 45 bp long (data not shown). In contrast, residue 106 did not contribute to the type 1 pattern because the G106A protein made an alcoholysis pattern identical to that of the laboratory integrase (Fig. 4C, lanes 2 and 7). Although the relevance of nonspecific alcoholysis to the retrovirus life cycle is unknown, our conclusions are strengthened by finding the same results with two assays that use very different nucleophilic donor molecules. Thus, we conclude that residue 119 either interacts with or influences interactions with nonviral DNA during catalysis.
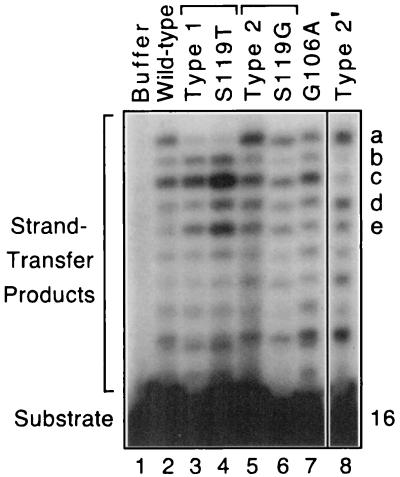
The above assays used Mn2+ as the divalent cation cofactor; however, Mg2+ may be more relevant within cells. Although HIV-1 integrase is usually inactive with Mg2+ in standard assays, our preparations of the laboratory HIV-1 integrase catalyze Mg2+-dependent DNA joining in the presence of dimethyl sulfoxide (28). Interestingly, the strand-transfer pattern created under these conditions (Fig. 5, lane 2) differs slightly from that seen with Mn2+ (Fig. 4B, lane 2). Thus, all of the patient-derived proteins that were active with Mn2+ were tested in joining reactions using Mg2+. The results, as indicated in the legend to Fig. 3, confirmed the groupings obtained previously. In particular, 12 of the 15 proteins that were classified as wild type in joining reactions conducted with Mn2+ created the same pattern as the laboratory integrase in reactions with Mg2+ (the other three proteins were inactive with Mg2+). Similarly, 8 of the 10 proteins classified as type 1 in joining reactions with Mn2+ made a novel but similar pattern with Mg2+ (e.g., Fig. 5, lane 3; the other two proteins were inactive with Mg2+). Finally, 9 of the 10 proteins classified as type 2 in joining reactions with Mn2+ were active with Mg2+, and each made a new pattern distinct from the wild-type or type 1 pattern. Interestingly, these nine proteins made two different new patterns (Fig. 5, lanes 5 and 8), suggesting that other amino acids worked in conjunction with residue 119 to influence the selection of target sites (see below). More importantly, even under Mg2+-dependent conditions, the S119T and S119G proteins made strand-transfer patterns that matched those of natural integrases that included these changes (Fig. 5, lanes 3 and 4 and lanes 5 and 6, respectively), whereas the G106A protein made a pattern similar to that of the laboratory enzyme (Fig. 5, lanes 2 and 7). Thus, the findings with Mg2+ paralleled those with Mn2+, even though the sets of preferred target sites differed as a function of the divalent metal.
FIG. 5.
DNA joining reactions conducted with Mg2+. Reactions were performed as described in the legend to Fig. 2B except that Mn2+ was replaced by 3 to 10 mM Mg2+ plus 30% dimethyl sulfoxide. Lanes 1 to 7 tested the same enzymes as in the legend to Fig. 4 except that the patient-derived protein for the type 2 pattern in lane 5 was ED002-7. Lane 8 shows the other novel pattern created by some of the type 2 proteins in reactions with Mg2+ (type 2′; the reaction shown used the LR157-5 protein); only one intervening gel lane was removed between lanes 7 and 8. Bands a to e distinguish the patterns as follows: the most intense bands in the upper portions of the lanes for the wild-type, type 1, type 2, and type 2′ patterns are a/c, c/e, a/c/d/e, and a/d, respectively. Unequal amounts of counts per minute were loaded into the various lanes to display the patterns at comparable intensities. The amount of joining catalyzed by the proteins in lanes 2 to 8 ranged from 0.5 to 8%.
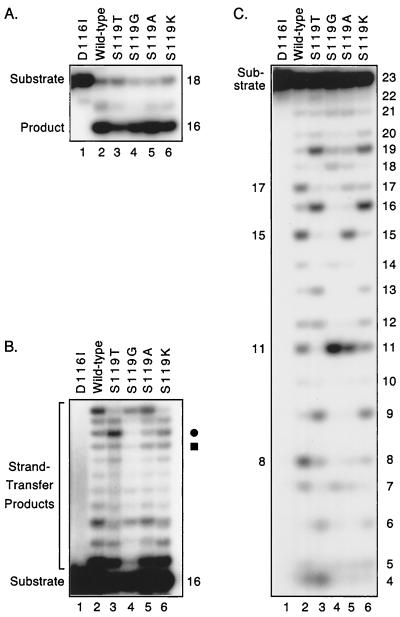
Given the different strand-transfer and alcoholysis patterns that resulted from the S119T and S119G substitutions, we made other changes at residue 119 in the laboratory integrase. Substitution of alanine or lysine at this position resulted in proteins that efficiently catalyzed Mn2+-dependent processing, joining, and nonspecific alcoholysis (lanes 5 and 6 in Fig. 6A, B, and C, respectively). In addition, the new proteins exhibited target site preferences in the latter two assays that differed from those of the wild-type or other point-substituted proteins (lanes 2 to 4 in Fig. 6B and C). In some cases, the patterns differed only slightly in the joining assay. For example, the relative intensities of the bands indicated by the circle and square in Fig. 6B are the only differences between the laboratory and S119A proteins (lanes 2 and 5) or between the S119T and S119K proteins (lanes 3 and 6), but these differences were reproducibly observed in multiple experiments. Similarities also were evident between the laboratory and S119A proteins and between the S119T and S119K proteins in joining reactions conducted with Mg2+ (not shown). However, the distinct target site preferences of these proteins are readily apparent in the alcoholysis assay. In particular, the S119A protein differed from the laboratory integrase by nicking inefficiently at positions 8 and 17 despite similar preferences for positions 11 and 15 (Fig. 6C, lanes 2 and 5). Similarly, the S119T and S119K proteins differed by their relative usage of positions 4 and 11 (Fig. 6C, lanes 3 and 6). These differences were confirmed in alcoholysis assays that used a 3′-labeled substrate (not shown). Thus, multiple substitutions at residue 119 influenced the interactions between HIV-1 integrase and nonviral DNA. These data, which were obtained with point-substituted proteins, are consistent with our previous studies that used protein fragments or chimeric integrases to map nonviral target site selection to the central domain of integrase (20–23). Together, the results suggest that the central domain of integrase interacts with cellular DNA during integration.
FIG. 6.
Other substitutions at position 119 in HIV-1 integrase. Details are as in the legend to Fig. 2. For each panel, substrates were incubated with the laboratory integrase that contained an inactivating D116I mutation (lane 1), the wild-type laboratory integrase (lane 2), or the laboratory integrase with the indicated amino acid substitutions (lanes 3 to 6). The circle and square in panel B identify bands that are discussed in the text. The amount of products formed in lanes 2 to 6 ranged from 49 to 67% in panel A, 4 to 15% in panel B, and 38 to 46% in panel C.
Implications.
Although other regions in HIV-1 integrase may influence interactions with cellular DNA (13, 20, 22, 23, 32), our data prove that changes at one particular residue in the central region were responsible for the novel patterns observed in this study. Residue 119 is a serine in the consensus sequence for North American subtype B of HIV-1 and in the consensus sequence or isolates of other HIV-1 subtypes (24). However, many of the natural integrases recovered from the hemophiliacs in our study had Thr or Gly at position 119 (33). Other amino acids found at this position in some HIV-1 integrases include Pro and Arg (24). Our data show that HIV-1 integrase tolerated several amino acids at this position, including Ser, Thr, Gly, Ala, and Lys, without a significant effect on the efficiency of its catalytic activity. However, as we had hypothesized, certain changes affected noncritical aspects of integrase function. The finding that changes at residue 119 do not affect specificity for viral DNA ends during processing but do affect selectivity for nonviral DNA sites during joining suggests that integrase has two sites for binding DNA (i.e., one for viral DNA and one for cellular DNA). We previously made a similar suggestion based on the finding that preferred target sites during DNA joining differ when Mg2+ substitutes for Mn2+, even though specific processing of viral DNA is supported by both metals (28). Competition studies also are consistent with two DNA sites on the enzyme (7, 9, 29).
Whether residue 119 affects target site selection by directly binding to nonviral DNA or by indirectly affecting protein conformation is unknown. However, other data suggest that amino acids near residue 119 also interact with cellular DNA. For example, the proteins classified as type 2 in the joining assay with Mn2+ created two novel strand-transfer patterns with Mg2+. Although all of these proteins have glycine at residue 119, the two Mg2+-dependent patterns segregated with the presence of isoleucine at residue 122 (Fig. 3). Substitutions at residues 117 and 120 in the related HIV-2 integrase also were shown to affect target site preferences during DNA joining reactions conducted with Mn2+ (36). Moreover, contact between residue 117 of HIV-1 integrase and cellular DNA has been suggested by cross-linking and structural data (14). In contrast, our data show that certain amino acid substitutions at 22 other residues in the central domain of integrase did not affect target site selection during DNA joining (i.e., residues 50, 59, 72, 79, 101, 106, 112, 124, 125, 128, 129, 131, 135, 147, 154, 157, 160, 161, 162, 166, 169, and 172, as deduced from Fig. 3 and 4). Thus, merely substituting any amino acid within the catalytic domain of integrase is not sufficient to alter the preferences of the enzyme for certain target sites in nonviral DNA.
Residue 119 is the second of four amino acids in α-helix 2 near the middle of the central domain of HIV-1 integrase. It is close to Asp-64, Asp-116, and Glu-152, the highly conserved acidic residues that form the D,D-35-E motif of the active site. These four residues (i.e., 64, 116, 119, and 152) are on the surface of the protein, which is involved in many critical functions. For example, the first two conserved residues of the D,D-35-E motif coordinate the required Mn2+ or Mg2+ cofactor in crystals of HIV-1 and Rous sarcoma virus integrase (1, 2, 12, 26), and some data suggest that all three conserved active-site residues may coordinate two metal ions (1, 25). In addition, data from mutational, chemical, or genetic studies suggest that residues 136, 143, 148, 152, and 159 interact with the ends of viral DNA (5, 10, 11, 15, 27). These sites are within or near a disordered, flexible surface loop that extends from residue 141 approximately to residue 150 (6). Recently, computerized docking of an 18-bp viral DNA end to two-domain crystals of HIV-1 integrase placed the viral DNA end near residue 159 and more-internal viral DNA positions near residues 186 to 219 (3). Other data implicate residues 114, 117, 120, 121, 143, 146, 147, and 148 in interactions with the attacking nucleophile (8, 35, 37). Thus, residue 119 may be appropriately situated to interact with cellular DNA during insertion of viral DNA ends. Although it would not be surprising if some residues of integrase participate in more than one function, especially within the context of the multimeric enzyme complex, current data (including those presented in this report) support a model that includes the following: the metal cofactor is coordinated by residues 64 and 116, the attacking nucleophilic group interacts with residues near 116 and 152, the viral DNA end is bound near residue 159, and cellular DNA is bound near residue 119. Thus, future studies that explore how amino acids near residue 119 affect the selection of nonviral target DNA sites have the potential to define the binding site for cellular DNA and would identify a new target for antiviral drug design. The effects of changes at these positions on the preintegration complexes that mediate integration in vivo also await further study.
Acknowledgments
This work was supported by Public Health Service grant R21 AI47216 from the National Institute of Allergy and Infectious Diseases, by W. W. Smith Charitable Trust Research grant A9804, by the Julius H. Caplan Foundation (in honor of Helen Caplan), and by a Dean's Feasibility Grant from the Pennsylvania State University College of Medicine.
We thank M. Elaine Eyster for providing the clinical samples from which the HIV-1 integrases were derived.
REFERENCES
- 1.Bujacz G, Alexandratos J, Wlodawer A, Merkel G, Andrake M, Katz R A, Skalka A M. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J Biol Chem. 1997;272:18161–18168. doi: 10.1074/jbc.272.29.18161. [DOI] [PubMed] [Google Scholar]
- 2.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure. 1996;4:89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen J C H, Krucinski J, Miercke L J W, Finer-Moore J S, Tang A H, Leavitt A D, Stroud R M. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding: Proc. Natl Acad Sci USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 5.Du Z, Ilyinskii P O, Lally K, Desrosiers R C, Engelman A. A mutation in integrase can compensate for mutations in the simian immunodeficiency virus att site. J Virol. 1997;71:8124–8132. doi: 10.1128/jvi.71.11.8124-8132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 7.Ellison V, Brown P O. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc Natl Acad Sci USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espeseth A S, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed J Y, Young S, Hamill T, Cole J L, Hazuda D J. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci USA. 2000;97:11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerton J L, Ohgi S, Olsen M, Derisi J, Brown P O. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J Virol. 1998;72:5046–5055. doi: 10.1128/jvi.72.6.5046-5055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldgur Y, Dyda F, Hickman A B, Jenkins T M, Craigie R, Davies D R. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc Natl Acad Sci USA. 1998;95:9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulaouic H, Chow S A. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuer T S, Brown P O. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins T M, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 17.Katzman M, Katz R A. Substrate recognition by retroviral integrases. Adv Virus Res. 1999;52:371–395. doi: 10.1016/s0065-3527(08)60307-3. [DOI] [PubMed] [Google Scholar]
- 18.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzman M, Sudol M. In vitro activities of purified visna virus integrase. J Virol. 1994;68:3558–3569. doi: 10.1128/jvi.68.6.3558-3569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzman M, Sudol M. Nonspecific alcoholysis, a novel endonuclease activity of human immunodeficiency virus type 1 and other retroviral integrases. J Virol. 1996;70:2598–2604. doi: 10.1128/jvi.70.4.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzman M, Sudol M. Mapping viral DNA specificity to the central region of integrase by using functional human immunodeficiency virus type 1/visna virus chimeric proteins. J Virol. 1998;72:1744–1753. doi: 10.1128/jvi.72.3.1744-1753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katzman M, Sudol M, Pufnock J S, Zeto S, Skinner L M. Mapping target site selection for the non-specific nuclease activities of retroviral integrase. Virus Res. 2000;66:87–100. doi: 10.1016/s0168-1702(99)00126-4. [DOI] [PubMed] [Google Scholar]
- 24.Kuiken C, McCutchan F, Foley B, Mellors J W, Hahn B, Mullins J, Marx P, Wolinsky S, Korber B. Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1999. [Google Scholar]
- 25.Lins R D, Adesokan A, Soares T A, Briggs J M. Investigations on human immunodeficiency virus type 1 integrase/DNA binding interactions via molecular dynamics and electrostatics calculations. Pharmacol Ther. 2000;85:123–131. doi: 10.1016/s0163-7258(99)00059-5. [DOI] [PubMed] [Google Scholar]
- 26.Maignan S, Guilloteau J-P, Zhou-Liu Q, Clément-Mella C, Mikol V. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J Mol Biol. 1998;282:359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- 27.Mazumder A, Neamati N, Pilon A A, Sunder S, Pommier Y. Chemical trapping of ternary complexes of human immunodeficiency virus type 1 integrase, divalent metal, and DNA substrates containing an abasic site: implications for the role of lysine 136 in DNA binding. J Biol Chem. 1996;271:27330–27338. doi: 10.1074/jbc.271.44.27330. [DOI] [PubMed] [Google Scholar]
- 28.Morgan A L, Katzman M. Subterminal viral DNA nucleotides as specific recognition signals for human immunodeficiency virus type 1 and visna virus integrases under magnesium-dependent conditions. J Gen Virol. 2000;81:839–849. doi: 10.1099/0022-1317-81-3-839. [DOI] [PubMed] [Google Scholar]
- 29.Pemberton I K, Buc H, Buckle M. Displacement of viral DNA termini from stable HIV-1 integrase nucleoprotein complexes induced by secondary DNA-binding interactions. Biochemistry. 1998;37:2682–2690. doi: 10.1021/bi971893j. [DOI] [PubMed] [Google Scholar]
- 30.Ratner L, Fisher A, Jagodzinski L L, Mitsuya H, Liou R-S, Gallo R C, Wong-Staal F. Complete nucleotide sequence of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 31.Ratner L, Haseltine W, Patarea R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–283. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 32.Shibagaki Y, Chow S A. Central core domain of retroviral integrase is responsible for target site selection. J Biol Chem. 1997;272:8361–8369. doi: 10.1074/jbc.272.13.8361. [DOI] [PubMed] [Google Scholar]
- 33.Skinner L M, Lamers S L, Sanders J C, Eyster M E, Goodenow M M, Katzman M. Analysis of a large collection of natural HIV-1 integrase sequences, including those from long-term nonprogressors. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:99–110. doi: 10.1097/00042560-199810010-00001. [DOI] [PubMed] [Google Scholar]
- 34.Skinner L M, Sudol M, Harper A L, Katzman M. Nucleophile selection for the endonuclease activities of human, ovine, and avian retroviral integrases. J Biol Chem. 2001;276:114–124. doi: 10.1074/jbc.M007032200. [DOI] [PubMed] [Google Scholar]
- 35.van den Ent F M I, Vos A, Plasterk R H A. Mutational scan of the human immunodeficiency virus type 2 integrase protein. J Virol. 1998;72:3916–3924. doi: 10.1128/jvi.72.5.3916-3924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Gent D C, Oude Groeneger A A M, Plasterk R H A. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Gent D C, Oude Groeneger A A M, Plasterk R H A. Identification of amino acids in HIV-2 integrase involved in site-specific hydrolysis and alcoholysis of viral DNA termini. Nucleic Acids Res. 1993;21:3373–3377. doi: 10.1093/nar/21.15.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]