Abstract
Rous sarcoma virus RNA contains a negative regulator of splicing (NRS) element that aids in maintenance of unspliced RNA. The NRS binds U1 snRNA at a sequence that deviates from the 5′ splice site consensus by substitution of U's for A's at three positions: −2, +3, and +4. All three of these U's are important for NRS-mediated splicing suppression. Substitution of a single nonconsensus C or G at any of these sites diminished NRS activity, whereas substitution of a single A generated a preferred 5′ splice site within the NRS.
Retroviruses synthesize full-length, viral RNAs that resemble cellular pre-mRNA substrates for splicing. Unlike cellular pre-mRNAs, many of these full-length viral transcripts are transported to the cytoplasm, where they are packaged into viral particles or translated to produce gag and pol gene products. Retroviruses also splice a fraction of their primary transcripts to generate additional mRNA, including env mRNA (reviewed in references 2 and 17).
The regulation of avian retroviral RNA splicing involves the use of suboptimal 3′ splice sites, an exonic splicing enhancer, and a negative regulator of splicing (NRS) (1, 8, 14, 20, 21). The 230-nucleotide (nt) cis-acting NRS element is located within the gag gene, about 300 nt downstream from the 5′ splice site and 4,000 nt from the env 3′ splice site. Insertion of the NRS into a cellular intron inhibits splicing from that site in an orientation- and distance-dependent fashion (15). NRS-mediated inhibition of splicing involves the interaction of U1 snRNP with the 3′ end of the NRS (7, 13). Mutations that interfere with U1 snRNA base-pairing impair NRS activity, and this activity is partially rescued by a compensatory U1 snRNA mutant (7). The U1 snRNP binding site in the NRS partially overlaps a consensus U11 snRNP binding site, and U11 snRNA has also been shown to bind to the NRS (6). However, mutations specifically disrupting the U11 binding sequence did not inhibit NRS activity, suggesting that U11 snRNP binding is not necessary for NRS function (7, 13). The NRS appears to function as a nonproductive 5′ splice site decoy which competes with the authentic viral 5′ splice site upstream for interaction with 3′ splice sites (3, 5).
In the present study, we have carried out a detailed mutational analysis of the 5′ splice site-like sequence in the NRS. We asked whether this sequence or its context was responsible for the failure of splicing from this site. This study revealed that all three nonconsensus U's are important for NRS activity. Point mutations at any of these sites can either neutralize splicing suppression or activate productive splicing at this NRS site.
NRS contains a decoy 5′ splice site.
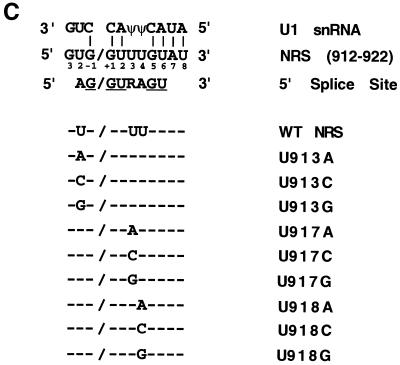
The Rous Sarcoma Virus (RSV) NRS sequence contains a 5′ splice site-like sequence near its 3′ end, and mutations in this region have been shown to impair NRS activity (6, 7, 13, 15). The potential base-pairing between the NRS and the 5′ end of U1 snRNA includes positions −1, +1, +2, +5, and +6 of the 5′ splice site consensus sequence (AG/GURAGU), as shown in the alignment in Fig. 1C. In addition, the NRS sequence at positions +7 and +8 is complementary to the first two encoded bases of U1 snRNA, extending the potential base-pairing. However, nonconsensus bases are observed at NRS positions −2, +3, and +4 of the decoy 5′ splice site. In all three nonconsensus positions, the viral sequence has a U instead of an A (or an R at +3). These U's could result in potential interactions with a U or pseudouridine (Ψ) in U1 snRNA. We investigated the functional significance of these nonconsensus U's by mutating each to the other three nucleotides.
FIG. 1.
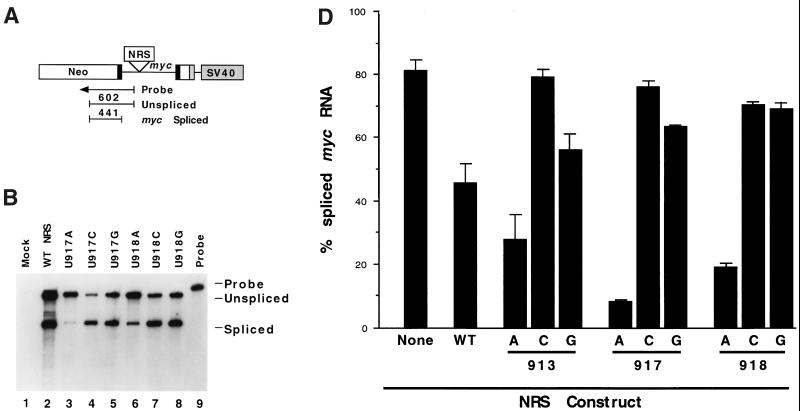
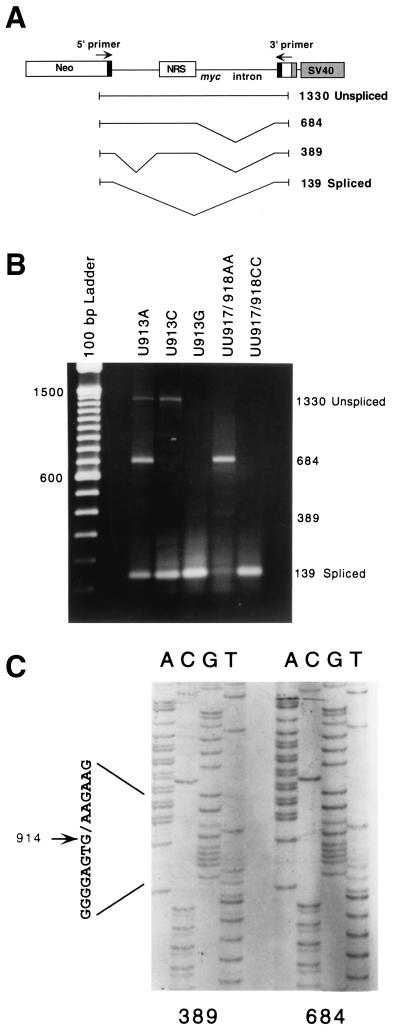
Nonconsensus U's in a 5′ splice site-like sequence are needed for NRS activity. (A) The 230-nt NRS sequence was inserted into the myc intron of pRSVNeo-int at a site 336 nt downstream from the 5′ splice site and 632 nt upstream from the 3 splice site. A riboprobe spanning the myc 5 splice site was used to detect spliced (441 nt) and unspliced (602 nt) RNAs. SV40, simian virus 40. (B) RNase protection assay with the myc 5 splice site probe of transcripts isolated from 293 cells transfected with wild-type (WT) or mutant NRS sequences in pRSVNeo-int. (C) Potential base-pairing of the NRS with U1 snRNA. The 5′ splice site consensus sequence is shown below the NRS sequence, and consensus bases present in the NRS are underlined. NRS mutations are aligned with the wild-type (WT) sequence. (D) Histogram of spliced myc RNA assayed by RNase protection as in Fig. 2A. Error bars represent the standard deviation from the mean for at least three independent transfection experiments.
Nonconsensus uridines in 5′ splice site-like sequence are important for NRS activity.
To assay the effect of NRS mutations on splicing in transfected cells, we inserted NRS mutants into the myc intron of a heterologous splicing construct, pRSVNeo-int (9), as shown in Fig. 1A. The constructs were transiently transfected into the human embryonic kidney 293 cell line. After total cellular RNA was isolated, splicing was assayed by RNase protection with a riboprobe spanning the myc 5′ splice site (Fig. 1A), as previously described (7, 15). This probe allowed us to quantify the level of splicing at this site by comparison of the relative amounts of spliced (exonic) and unspliced protected fragments.
We first tested the effect of single point mutations in the NRS at positions −2, +3, and +4 (nt 913, 917, and 918; Fig. 1C). The substitution of A for U at any of these positions resulted in a dramatic reduction in splicing from the myc 5′ splice site (Fig. 1B, lanes 3 and 6, and Fig. 1D). Quantitation of the results revealed that U913A was 28% spliced, U917A was 8% spliced, and U918A was 19% spliced at this site (Fig. 1D). In comparison, the construct bearing the wild-type NRS was 45% spliced, and the construct lacking an NRS spliced 81% of its RNA at the myc 5′ splice site.
The substitution of either C or G for U had the opposite effect, increasing splicing from the myc 5′ splice site. The results were as follows: U913C, 79% spliced; U917C, 75% spliced; U913G, 56% spliced; U917G, 63% spliced; U918C, 71% spliced; and U918G, 69% spliced (Fig. 1D). Thus, single base changes at −2, +3, or +4 gave similar results: mutations to an A decreased myc splicing, while changes to a C or G increased splicing at this site.
Mutations in U1 binding site sequence in NRS can activate splicing within NRS.
We next investigated the possibility that some of these NRS mutations had activated a cryptic 5′ splice site sequence within the NRS. RNA spliced from a 5′ splice site within the NRS or in other regions of the intron would be indistinguishable from unspliced RNA with the myc 5′ splice site probe. Therefore, a second probe which spanned the NRS sequence and extended 70 nt downstream into the myc intron was used in RNase protection assays to determine if cryptic splicing was occurring within this region (Fig. 2A).
FIG. 2.
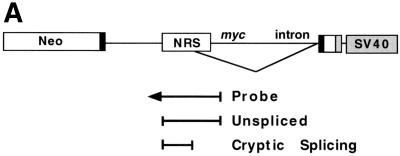
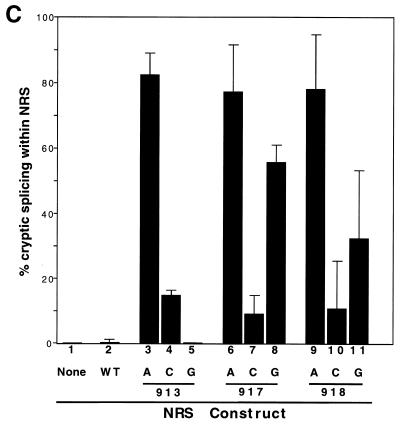
Point mutations that improve the 5′ splice site consensus sequence activate splicing within the NRS. (A) Probe designed to detect splice sites within the NRS. This probe encompasses the entire NRS and extends 70 nt downstream into the myc intron. (B) Wild-type (WT) and mutant NRS sequences were analyzed by RNase protection analysis with the NRS-specific probe to determine relative amounts of cryptically spliced and unspliced RNA. (C) Summary of the transfection data using these same constructs. Error bars represent the standard deviation from the mean for at least three independent transfection experiments.
Assay of the wild-type NRS construct with the NRS-specific probe showed only unspliced RNA (Fig. 2B, lane 2). (RNA spliced at the myc splice sites would not hybridize with this probe.) In contrast, the mutations that changed the sequence from U to A at position −2, +3, or +4 all showed very high levels of splicing within the NRS. This was represented by a new doublet band in the RNase protection analyses (Fig. 2B, lanes 3 and 6). This spliced RNA represented 77 and 78% of the RNA generated by constructs U917A and U918A, respectively, and detected with the NRS probe (Fig. 2C). Similarly, U913A appeared to be 83% spliced (Fig. 2C), and the double point mutant UU917/918AA (7) yielded 88% spliced RNA with this NRS-specific probe (data not shown).
The base changes from U to G at positions 917 and 918 also resulted in splicing within the NRS at levels of 56 and 32%, respectively (Fig. 2B, lanes 5 and 8; Fig. 2D). The U917Gmutant is closer to the 5′ splice site consensus than the wild-type sequence. In contrast, no splicing within the NRS was observed with U913G (Fig. 2C). The substitutions from U to C at positions −2, +3, and +4 resulted in very little splicing within the NRS (15, 10, and 12%, respectively; Fig. 2B, lanes 4 and 7; Fig. 2C).
Identification of splice sites within NRS.
The NRS probe used in Fig. 2 allowed identification of cryptic splice sites within the NRS or 70 nt downstream of it. To extend this analysis to include the entire 1,200-nt intron containing the NRS, we performed reverse transcription (RT)-PCR using primers in both myc exons. Unspliced RNA would be expected to generate a 1,330-bp RT-PCR product, and normally spliced myc RNA would yield a 139-bp product (Fig. 3A). These were the only products generated from the wild-type NRS construct, confirming that no cryptic splicing occurred (data not shown). In contrast, mutants UU917/918AA, U917A, U918A, and U913A generated a novel product of 684 bp and a reduced amount of the 139-bp spliced product (Fig. 3B and data not shown). The UU917/918AA, U917A, and U918A mutants also generated small amounts of a 389-bp product. Small amounts of the 684-bp product were also observed with U917G and U918G (data not shown). In contrast, mutants U913C, U913G, and UU917/918CC did not generate any of these novel RT-PCR products, yielding mainly the 139-bp product, generated by splicing at the normal myc splice sites (Fig. 3B).
FIG. 3.
RT-PCR analysis of splicing of NRS mutants. (A) Diagram of the splicing reporter construct and the normally and cryptically spliced products observed. The numbers on the right represent transcript lengths (in base pairs). (B) RT-PCR products were generated from RNA isolated from transfected cells using primers in the exons as shown in panel A. (C) Sequencing of cryptic products revealed that both used the NRS U1 binding site as a 5′ splice site. The 389-bp product also had an upstream intron of 313 nt, extending from the myc 5′ splice site, as shown in panel A.
To identify these novel splice sites, the 389-bp and 684-bp RT-PCR products were cloned and sequenced. The predominant 684-bp product was found to be generated by splicing from an activated cryptic 5′ splice site within the NRS at nt 915 to the normal myc 3′ splice site (Fig. 3C). The minor 389-bp product was doubly spliced, utilizing the myc 5′ splice site, together with a cryptic 3′ acceptor site in the myc intron (315 nt from the myc 5′ splice site, just upstream of the NRS insertion site), as well as the NRS 5′ splice site and the myc 3′ splice site (Fig. 3A). This double splicing event created a new 220-nt internal exon and may be a result of exon bridging between the activated 5′ splice site at nt 915 and the cryptic 3′ splice site upstream. Consistent with our results, McNally and McNally (13) observed activation of a 5′ splice site at nt 915 with a triple mutant having all three nonconsensus U's mutated to A's to form a consensus splice site. However, they did not report activation of a cryptic 3′ splice site with this mutant.
Competition between two splice sites.
The data presented above suggested that alternative splicing was occurring with some of the NRS mutants, with competition between the upstream myc 5′ splice site and an activated 5′ splice site within the NRS. The experiments shown in Fig. 1 used the myc donor probe and generated data on splicing at this site only; the apparent “unspliced” RNA in this assay includes any RNA spliced at a downstream site. In contrast, the NRS probe data presented in Fig. 2 give the relative amount of RNA spliced within the NRS or directly downstream of it; any RNA spliced at the myc 5′ splice site would not hybridize to this probe. To give an overall picture of the distribution of the RNA species between the alternatively spliced products and unspliced RNA, we analyzed the data as shown in Fig. 4.
FIG. 4.
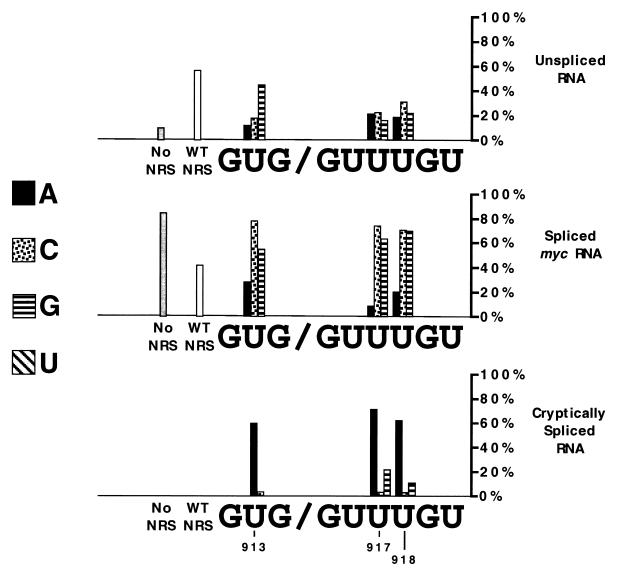
Summary of effects of NRS point mutations on RNA splicing. To give an overall view of the distribution of RNA, calculations were based on data using both riboprobes. The amount of splicing at the myc donor site was first determined directly from the data in Fig. 1. We then calculated the distribution of the remaining RNA between cryptically spliced RNA (splicing within the NRS) and unspliced RNA based on data with the NRS probe (Fig. 2).
The wild-type NRS generated about 60% unspliced RNA and 40% spliced RNA, with no apparent cryptic splicing at sites within the NRS. Some of the mutations, including U913C, U917C, U918C, and 918G, abolished splicing suppression, yielding more than 70% splicing from the myc 5′ splice site. An interesting group of mutations, including U913A, U917A, and U918A, yielded mainly RNA spliced from an activated 5′ splice site within the NRS. These mutants all exhibited reduced amounts of normally spliced and unspliced RNAs. Lastly, mutant U917G showed about 60% splicing from the myc site and 20% from the NRS site (Fig. 4).
Thus, it appears that the inhibition of splicing at the myc 5′ splice site observed with all three of the U-to-A mutations (Fig. 1) was primarily due to the generation of new spliced products rather than to maintenance of unspliced RNA. These data suggest a correlation between mutations that improve the 5′ splice site consensus sequence within the NRS and activation of splicing at this site. Surprisingly, all of the U-to-C mutations, which would maintain a suboptimal splice site sequence, inactivated the NRS. U-to-G mutations also diminished NRS activity, with U917G also splicing within the NRS.
We previously demonstrated that the Rous sarcoma virus NRS intronic sequence binds U1 snRNP at a 5′ splice site-like sequence near the 3′ end of the element. The importance of U1 snRNP binding for NRS activity was demonstrated by genetic suppression experiments (7). However, it was not known why splicing did not occur at this site, which has a five of eight match to the consensus 5′ splice site sequence, deviating with a U instead of an A at −2, +3, and +4. Analysis of a 5′ splice site sequence data base (18; http://fruitfly.org/seq_tools/splice.html) did not reveal any examples of this NRS sequence (UG/GUUUGU) in a catalog of more than 2,000 nonredundant human 5′ splice sites, suggesting that this sequence may not be functional as a 5′ splice site. Alternatively, there may be contextual problems that prevent splicing at this site.
To determine sequence requirements for NRS splicing suppression, we have mutated each of the nonconsensus U's to the three other bases. Surprisingly, mutation of any one of the U's to a C led to a nearly normal splicing phenotype, abolishing the NRS splicing suppression. When any of the U's was mutated to an A, the NRS site became the preferred 5′ splice site. Mutation of the +3 or +4 site to a G led to a lower level of activation of this cryptic 5′ splice site within the NRS. Therefore, making any one of these three sites fit the consensus led to splicing at this site in preference to the upstream 5′ splice site. It may be important that there is an ASF/SF2 binding site within the NRS (12), located between the two alternative 5′ splice sites (myc site and cryptic NRS site). This has been shown to facilitate U1 snRNP binding to the NRS (13) and may promote use of the downstream splice site (4).
It appears that U's specifically, rather than any nonconsensus base, are important for NRS splicing inhibition. Each of the NRS nonconsensus U's would oppose a U (−2) or a Ψ (+3 and +4) in the U1 snRNA sequence. U-U interactions have been observed in tRNA and found to be more stable than most non-Watson-Crick base-pairing interactions, including C-U (19). It is interesting that the yeast 5′ splice site consensus sequence also has a U at +4 (10), which could interact with the Ψ in yeast U1 snRNA (11). The U at +4 has also been found to be important for stable binding of U1 snRNP in randomization-selection experiments (D. Libri, personal communication). Alternatively, these U's may be important for binding of another factor to the NRS. There is the potential for base-pairing of 12 of 13 nt of U6 snRNA (nt 33 to 45) to this region of the NRS, and the U's at +3 and +4 would stabilize this interaction.
In summary, the NRS is capable of interacting with several snRNPs. Single point mutations in the NRS which improve the 5′ splice site consensus sequence make the NRS the preferred 5′ splice site. We propose that the NRS functions as a decoy 5′ splice site and may prevent cryptic as well as productive splicing in the virus (C. T. O'Sullivan, unpublished data). The NRS requires three nonconsensus U's in the 5′ splice site-like sequence to effectively maintain unspliced RNA for export to the cytoplasm (16).
Acknowledgments
R. E. Paca and C. S. Hibbert contributed equally to this work.
This work was supported by Public Health Service research grant RO1 CA48746 from the National Cancer Institute. R.E.P. and C.O.S. were supported in part by NIH predoctoral training grant 52T32G07231.
We thank Raymond Fernalld for technical assistance and members of the Beemon laboratory for review of the manuscript.
REFERENCES
- 1.Arrigo S, Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffin J M. Retroviridae: the viruses and their replication, P. 1767–1847. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. [Google Scholar]
- 3.Cook C R, McNally M T. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J Virol. 1999;73:2394–2400. doi: 10.1128/jvi.73.3.2394-2400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eperon I C, Makarova O V, Mayeda A, Munroe S H, Caceres J F, Hayward D G, Krainer A R. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gontarek R R. A Rous sarcoma virus intronic element negatively regulates splicing by binding snRNPs. Ph.D. thesis. Baltimore, Md: Johns Hopkins University; 1994. [Google Scholar]
- 6.Gontarek R R, McNally M T, Beemon K. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 1993;7:1926–1936. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- 7.Hibbert C S, Gontarek R R, Beemon K L. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA. 1999;5:333–343. doi: 10.1017/s1355838299981347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katz R A, Skalka A M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990;10:696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linial M. Creation of a processed pseudogene by retroviral infection. Cell. 1987;49:93–102. doi: 10.1016/0092-8674(87)90759-8. [DOI] [PubMed] [Google Scholar]
- 10.Long M, de Souza S J, Rosenberg C, Gilbert W. Relationship between “proto-splice sites” and intron phases: evidence from dicodon analysis. Proc Natl Acad Sci USA. 1998;95:219–223. doi: 10.1073/pnas.95.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massenet S, Motorin Y, Lafontaine D L, Hurt E C, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol. 1999;19:2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNally L M, McNally M T. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J Virol. 1996;70:1163–1172. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNally L M, McNally M T. U1 small nuclear ribonucleoprotein and splicing inhibition by the Rous sarcoma virus negative regulator of splicing element. J Virol. 1999;73:2385–2393. doi: 10.1128/jvi.73.3.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNally M T, Beemon K. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J Virol. 1992;66:6–11. doi: 10.1128/jvi.66.1.6-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNally M T, Gontarek R R, Beemon K. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology. 1991;185:99–108. doi: 10.1016/0042-6822(91)90758-4. [DOI] [PubMed] [Google Scholar]
- 16.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabson A B, Graves B J. Synthesis and processing of viral RNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 205–261. [PubMed] [Google Scholar]
- 18.Reese M G, Eeckman F H, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 19.SantaLucia J, Jr, Kierzek R, Turner D H. Stabilities of consecutive A∗C, C∗C, G∗G, U∗C, and U∗U mismatches in RNA internal loops: evidence for stable hydrogen-bonded U∗U and C∗C+ pairs. Biochemistry. 1991;30:8242–8251. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- 20.Stoltzfus C M, Fogarty S J. Multiple regions in the Rous sarcoma virus src gene intron act in cis to affect the accumulation of unspliced RNA. J Virol. 1989;63:1669–1676. doi: 10.1128/jvi.63.4.1669-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Stoltzfus C M. A suboptimal src 3′ splice site is necessary for efficient replication of Rous sarcoma virus. Virology. 1995;206:1099–1107. doi: 10.1006/viro.1995.1033. [DOI] [PubMed] [Google Scholar]