Abstract
A Sindbis virus (SV) variant with a 6K gene partially deleted has been obtained. This SV Del6K virus is defective in the proteolytic processing of virus glycoprotein precursor, transport of glycoproteins to the plasma membrane, and plaque phenotype. A revertant virus (SV Del6K-revQ21L) containing a point mutation in the deleted 6K gene was isolated and characterized. SV Del6K-revQ21L has corrected the defects of proteolytic processing and transport of virus glycoproteins to the plasma membrane, but it still remains attenuated compared to wild-type (wt) SV, exhibiting defects in virus budding. Neither mutant nor revertant viruses are complemented by the coexpression in trans of a wt SV 6K gene.
Alphaviruses are enveloped viruses that contain a single-stranded positive-sense RNA as a genome (23). The alphavirus structural proteins are synthesized from a subgenomic mRNA encoding a polyprotein that is proteolytically processed. The C protein is first synthesized and separated from the rest of the polyprotein by autocatalytic cleavage. Once the C protein has been released to the cytoplasm, further translation of the mRNA continues that is associated with membranes (23). The newly exposed amino terminus contains a signal sequence that interacts with membranes of the endoplasmic reticulum (ER) and directs this portion of the glycoprotein precursor (E3-E2–6K-E1) into the lumen of the ER. The precursor associates with the ER membrane, spanning the lipid bilayer six times. Soon after synthesis, this precursor is cleaved at both ends of the 6K protein by a cellular protease present in the ER, generating the products PE2 (E3 plus E2), 6K, and E1. Subsequently, PE2 and E1 associate to form dimers that migrate together with 6K through the vesicular system to the plasma membrane. PE2 is cleaved by a furin-like protease present in a post-Golgi compartment, giving rise to glycoproteins E3 and E2 (23). Despite the association of the 6K protein with the plasma membrane and its association with E1-E2, very little 6K is incorporated into the released virus particles (6, 18).
The 6K protein is a small hydrophobic polypeptide that is acylated with fatty acids (6, 18). 6K provides the signal sequence for translocation of E1 to the lumen of the ER (15). A Semliki Forest virus (SFV) variant lacking the entire 6K is processed between E2 and E1 (16). E1 is properly translocated to the ER in the 6K-deleted SFV mutant. The major defects of this variant are found in the budding process (16, 17). Similarly, Sindbis virus (SV) variants with single or multiple amino acid substitutions in the 6K gene are defective in virion release, leading to the formation of multinucleated virus particles (5, 7,11, 12). Proper proteolytic processing of the virus glycoproteins is hampered in a SV variant bearing an insertion of 15 amino acids in the 6K protein (22). This SV mutant exhibits a trans-dominant phenotype, but virus particles have a morphology similar to wild-type (wt) virus. On the other hand, the functions of the 6K protein cannot be rescued by the corresponding counterpart from related virus species. Thus, the replacement of the SV 6K gene with the 6K counterpart from Ross River virus produced viruses with a small plaque phenotype and reduced formation of infectious virus (26). This SV variant containing the 6K gene from Ross River virus was able to proteolytically cleave the glycoproteins' precursor and to transport them to the plasma membrane. Although these observations suggest a function for 6K in the release of virions, the precise molecular mechanism by which 6K protein enhances virus particle release remains unknown.
Generation of a 6K deletion variant of SV.
The role of the 6K protein in the SV replication cycle has been analyzed previously by several laboratories using a number of 6K variants. To further assess the role of 6K protein, a variant of SV that lacks a significant portion of the 6K gene was generated (SV Del6K) (Fig. 1A). pT7 SV wt is a full-length cDNA clone of SV. It was constructed by inserting the fragment obtained upon digestion of pT7 TOTO 1102 with SacI/SpeI into pTE3′2J1 (9) digested with SacI/SpeI. This construction changes the Sp6 promoter in pTE3′2J1 to the T7 promoter. Both plasmids were a kind gift of C. M. Rice (Washington University School of Medicine).
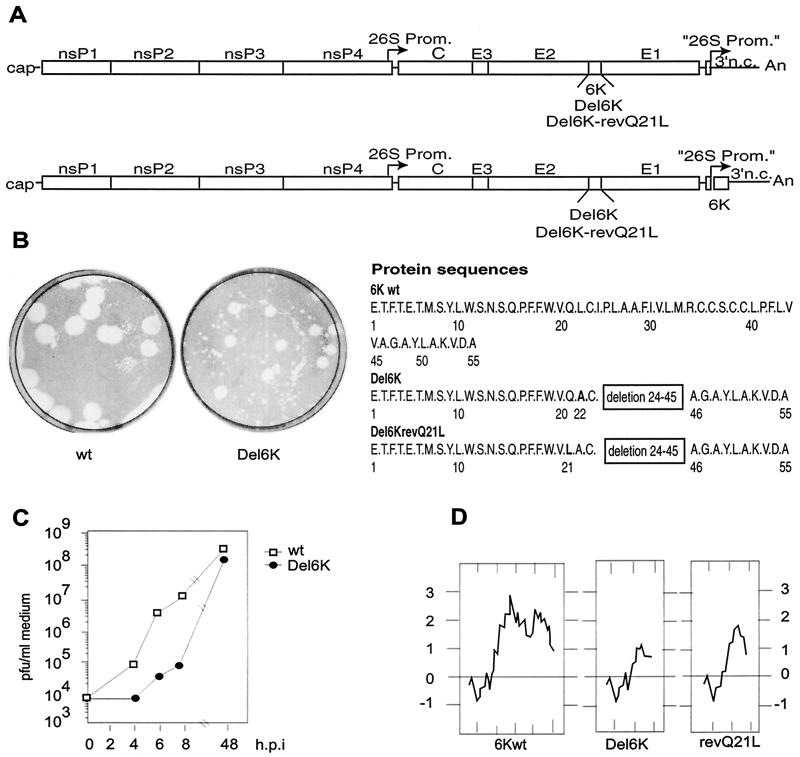
FIG. 1.
(A) Schematic representation of the different RNAs derived from the constructs pT7 SV wt, pT7 SV Del6K, or pT7 SV Del6K-revQ21L (top) and pT7 SV Del6K + 6K or pT7 SV Del6K-revQ21L + 6K (bottom). The sequences of 6K and deleted 6K are shown. (B) Plaques formed by virus wt SV or SV Del6K. The titers of the stocks of wt SV and SV Del6K on BHK cells obtained after RNA electroporation were determined, and the cells were stained at 72 h postinfection. (C) Kinetics of release of wt SV and SV Del6K viruses from infected BHK cells. The titers of viruses released to the medium from BHK cells infected at a concentration of 50 PFU/cell were determined at the times indicated. The viruses used in this experiment correspond to amplifications of initial stocks obtained after RNA electroporation. (D) Hydrophobicity plots of 6K, Del6K and Del6K-revQ21L proteins are according to the Kite-Doolittle hydrophobicity test.
pT7 SV Del6K contains an internal deletion in the sequence encoding the 6K protein. The coding sequence corresponding to amino acids 24 to 45 of the 6K protein has been deleted. In addition, it has the mutation altering TTG nucleotides to GCA, predicting a change from L22 to A.
The PCR product generated using the oligonucleotides 5′ C CCGGGCATATGGGATGGCCACACGAAATAGTACAG 3′ and 5′ GGCGCCGCATGCCTGGACCCAGAAGAACGGCTGACT 3′ was digested with SphI and ligated to the PCR product generated using the oligonucleotides 5′ CCCGGGGCATGCGCCGGCGCCTACCTGGCGAAGGTA 3′ and 5′ GGCGCCGGATCCTTATTAGACGTACGCCTCACTCATCTGGCT 3′ previously digested with SphI. The new product was digested with BssHII and BsiWI and inserted into BssHII/BsiWI sites of pT7 SV wt. The SV Del6K variant obtained retains the two cleavage signals at either end of the 6K protein. Electroporation of BHK cells with the viral RNA synthesized in vitro led to the production of progeny viruses. A plaque assay of the viruses thus obtained indicated that most of the plaques had a minute phenotype, while a significant number exhibited a size about half that of the wt virus (Fig. 1B). Further amplification of SV Del6K obtained after electroporation of BHK cells produced viruses with an intermediate-plaque-size phenotype (results not shown). These viruses already represent revertant SV. The virus yield obtained with these viruses was almost equal to that of wt SV, albeit with a delayed rate of replication (Fig. 1C).
We next isolated viruses from several intermediate-size plaques of SV Del6K. The region corresponding to 6K was sequenced in 12 of these isolates. Seventy percent of the variant viruses contained a point mutation at nucleotide 9961 (A changed to T) affecting residue 21 (Q was replaced by L) in the deleted 6K gene. This point mutation rendered the truncated 6K protein more hydrophobic (Fig. 1D). To ensure that this was the only significant variation in the revertant Del6K, the region corresponding to the wt 6K gene of SV was replaced with the same region from a variant strain. This virus was designated SV Del6K-revQ21L. Virions obtained by growth of the bulk SV Del6K and the 6K revertant SV Del6K-revQ21L were similar in all respects analyzed, except that the plaques obtained with the 6K revertant were more homogeneous and always exhibited an intermediate-plaque-size phenotype. The virus stock of SV Del6K obtained by several reinfections was also sequenced, showing the point mutation at nucleotide 9961 that leads to the change from Q21 to L.
Glycoprotein synthesis and trafficking in BHK cells electroporated with SV Del6K and SV Del6K-revQ21L RNAs.
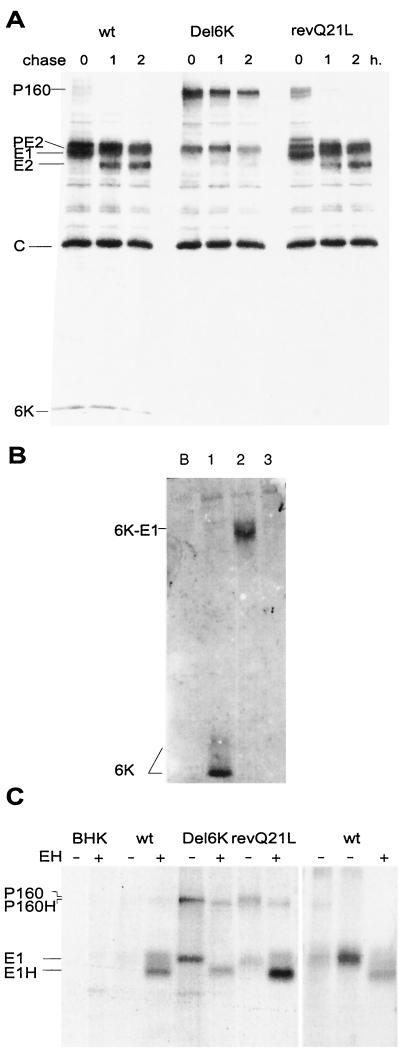
Since electroporation of cells with Del6K RNA followed by virus harvesting leads to the production of revertant viruses with mutations in the partially deleted 6K gene, it was of interest to directly electroporate cells with viral RNAs. Initially, protein synthesis and glycoprotein processing were tested. To this end, BHK cells were electroporated with wt SV, SV Del6K, or SV Del6K-revQ21L RNAs made by in vitro transcription. Electroporated cells were labeled with [35S]methionine-cysteine at 16 h postelectroporation (h.p.e.) for 30 min and chased by incubation in nonradioactive medium for 1 or 2 h (Fig. 2A). In addition to protein C, two major bands corresponding to PE2 and E1 were apparent in BHK cells electroporated with wt SV RNA. The precursor containing E3-E2–6K-E1 was not detected. Mature E2 glycoprotein was detected after a chase. In the case of SV Del6K, however, the precursor E3-E2-Del6K-E1 (P160) was clearly apparent and did not diminish significantly after the 2-h chase period. Part of the precursor was proteolytically processed, but only one band was detected in the region of processed glycoproteins. This band likely corresponds to product PE2 and a fusion product, Del6K-E1. Almost no mature glycoprotein E2 was observed after a chase. Notably, SV Del6K-revQ21L showed a protein pattern similar to wt SV. A thin band was detected above PE2 in the pulse-labeled cells, perhaps corresponding to a fusion product, PE2-Del6K. This product disappeared during a chase, while mature E2 and E1 appeared. According to these data, the SV Del6K variant showed defects in the cleavage of both scissile bonds, E2–6K and 6K-E1, that occur in the ER lumen. Cleavage of 6K-E1 was particularly resistant. This defect is largely corrected in the revertant SV Del6K-revQ21L.
FIG. 2.
(A) Analysis of protein synthesis by pulse-chase labeling. At 16 h.p.e., cells were labeled for 30 min with [35S]methionine-cysteine (lanes 0), and some cultures were chased for 1 (lanes 1) or 2 (lanes 2) h as indicated. (B) Detection of 6K by Western blot analysis using anti-6K antiserum. Samples were collected after 16 h.p.e. Lanes: B, no RNA was added; 1, cells electroporated with RNA from wt SV; 2, RNA from SV Del6K; 3, RNA from SV Del6K-revQ21L. (C) Endo H sensitivity analysis of viral glycoproteins. (Left) Samples of BHK cells labeled with [35S]methionine-cysteine for 30 min at 16 h.p.e. were lysed and treated with (+) or without (-) endo H (EH). (Right) BHK cells electroporated with SV wt RNA were treated as described for the left-hand panel, but after lysis the samples were divided in three aliquots. Lanes: left, mock treated; center, denatured sample; right, denatured and incubated with endo H.
Further insights into the pattern of cleavages between E2–6K and 6K-E1 were obtained by immunodetection. Samples of BHK cells electroporated with the different RNAs were analyzed by Western blotting with previously described anti-6K antibodies (6) (Fig. 2B). Mature 6K was observed only in wt SV electroporated cells, while a fusion band was observed with SV Del6K electroporated cells bound either to PE2 or most probably to glycoprotein E1. This fusion band does not appear with SV Del6K-revQ21L. The cleaved Del6K protein formed by the revQ21L virus was not detected by Western blotting because it was too small to be retained by nitrocellulose membranes. However, synthesis of Del6K protein was demonstrated by immunoprecipitation (data not shown). Moreover, the product P160 present in the pulse-chase experiment with SV Del6K was not recognized by the anti-6K antibody. This result supports the view that the attenuated phenotype of revertant SV Del6K-revQ21L was due to the deficient functioning of the deleted 6K. It also rules out the possibility that this phenotype is a consequence of the permanent fusion of Del6K with glycoprotein E2 or E1.
Additional analysis of the processing of E1 glycoprotein was carried out by examining endo-β-N-acetylglucosaminidase H (endo H) sensitivity. Extracts from electroporated cells were treated with or without endo H glycosidase and immunoprecipitated with anti-E1 antibodies. The anti-E1 antiserum was raised in rabbits immunized with fusion protein MBP-ΔSV E1. Protein MBP-ΔSV E1 contains amino acids 186 to 343 from E1 sequence fused to maltose-binding protein. The immunoprecipitated material was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (Fig. 2C). No bands were detected with wt SV untreated extracts, although a band appeared after endo H treatment. A similar result was seen with SV Del6K-revQ21L extract, but in this case two faint bands were clearly seen in the untreated sample, one corresponding to the P160 precursor and the other to E1. In the case of SV Del6K, these two bands were clearly visible in the untreated sample. Both of these bands migrated faster after endo H treatment, suggesting that they became glycosylated. These results indicate that with the three viral RNAs assayed all forms of E1 become glycosylated. In addition, the experiment revealed differences in the immunoprecipitation behavior of E1 from wt SV or SV Del6K-revQ21L compared to SV Del6K. E1 was not immunoprecipitated from wt SV or SV Del6K-revQ21L untreated samples, but it was immunoprecipitated from SV Del6K cell extracts (Fig. 2C). Perhaps these differences reflect a different conformation of E1. The fact that anti-E1 antibodies immunoreact well with the P160 precursor suggests that the correct association of processed E1 and E2 glycoproteins masks the epitope. In the absence of endo H treatment, E1 was immunoprecipitated from wt SV samples only after denaturation (Fig. 2C).
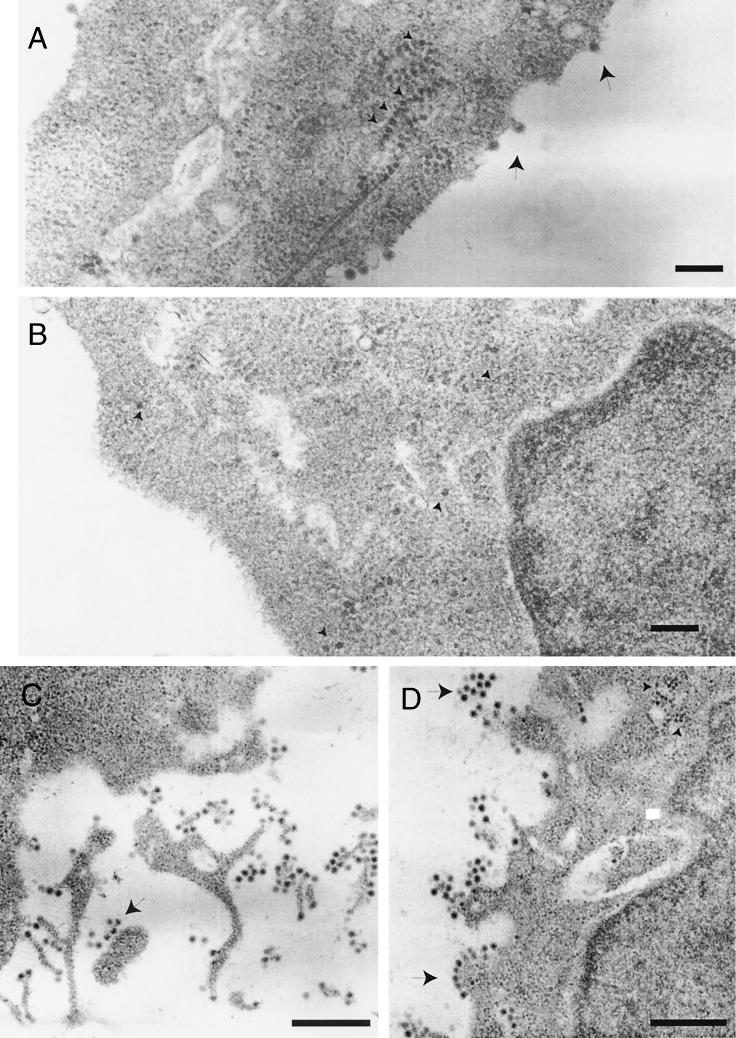
After precursor processing, alphavirus glycoproteins are delivered to the plasma membrane ready for virus budding. Subcellular localization of E1 or its precursor products was analyzed in BHK cells electroporated with wt SV, SV Del6K, and SV Del6K-revQ21L RNAs. Immunofluorescence analysis with anti-E1 antibodies was carried out with permeabilized or unpermeabilized cells to determine the proportion of E1 delivered to the plasma membrane. E1 immunofluorescence was clearly visible at the plasma membrane of unpermeabilized wt SV or SV Del6K-revQ21L cells, but not when SV Del6K was tested (results not shown). The presence of E1 (or its precursors) was seen in all three cases when cells were permeabilized to the anti-E1 antibodies (results not shown). In addition, we examined electroporated cells by electron microscopy. At 16 h.p.e., the cells were fixed, dehydrated, embedded, and examined. Representative micrographs are shown in Fig. 3. The main feature of cells electroporated with SV Del6K RNA is the virtual absence of virus budding from the plasma membrane. Only a few cells showed virus budding, while the nucleocapsids were scattered in the cytoplasm. No cytopathic vacuoles in BHK cells electroporated with SV Del6K RNA were observed. In contrast, cells electroporated with SV Del6K-revQ21L RNA showed a picture very similar to wt SV. Virus budding occurred in these cells, and aggregates of nucleocapsids and cytopathic vacuoles were observed in the cytoplasm. However, the revertant virus particles are associated with membranes to a greater extent than wt SV particles after budding. The last event of virus budding, i.e., pinching off, is inefficient in SV Del6K-revQ21L. Chains and groups of viruses associated with membranes are commonly observed.
FIG. 3.
Electron microscopy of BHK cells electroporated with wt SV (A), SV Del6K (B), and SV Del6K-revQ21L (C and D) RNAs. At 16 h.p.e., the cells were processed for electron microscopy analysis. Thin sections were cut and stained with uranyl acetate and lead citrate. Small arrows, nucleocapsids; large arrows, virus particles. Large bars, 500 nm; small bars, 200 nm.
Lack of complementation of SV 6K in trans.
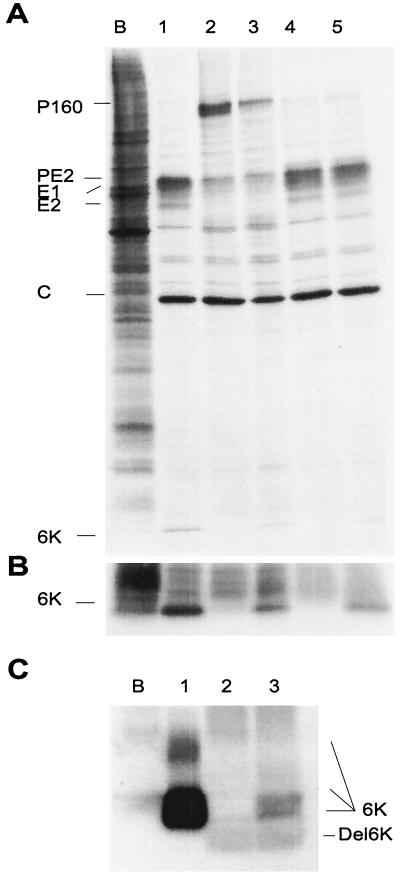
The revertant virus SV Del6K-revQ21L partially recovered the capacity to process the P160 precursor proteolytically. In addition, the viral glycoproteins associate with the cell membrane, in contrast to what happens with SV Del6K. However, SV Del6K-revQ21L still shows defects, with an intermediate-plaque-size phenotype. This defect may be due to the impaired activity of the deleted 6K. Since SV Del6K-revQ21L is able to cleave P160, it was of interest to test whether wt 6K provided in trans complemented the plaque size of the revertant virus. To this end, the SV 6K gene was placed under a second subgenomic promoter (see Fig. 1A) and cloned into either pT7 SV Del6K or pT7 SV Del6K-revQ21L to generate the constructs pT7 SV Del6K + 6K and pT7 SV Del6K-revQ21L + 6K, respectively. Both of them contain the 6K sequence downstream of the duplicated 26S promoter. They were constructed by subcloning the 6K sequence generated by PCR using the oligonucleotides 5′ GCCCGGATCCTTATTAGGCGTCTACCTTCGCCAG 3′ and 5′ CCCGGGCCATGGAAACGTTCACCGAGACC 3′ via subcloning into the NcoI/BamHI sites present in pH3′2J1 (9) in ApaI/XhoI sites of pT7 SV Del6K or pT7 SV Del6K-RevQ21L. The plaques produced by cells electroporated with RNA derived from these constructs were similar in size to those of the parental SV Del6K and SV Del6K-revQ21L. Next, the synthesis of viral proteins and, particularly, of 6K was examined. Thus, cells were electroporated with viral RNAs and labeled with [35S]methionine-cysteine at 16 h.p.e. The pattern of proteins synthesized by all viruses bearing the additional 6K gene was unaltered (Fig. 4A). 6K is synthesized at lower levels than in wt SV, but the 6K protein can be seen by labeling (Fig. 4B). In addition, the amount of 6K was examined by immunoprecipitation with anti-6K antibodies. In this case, the experiment was made with BHK cells infected with viruses obtained by RNA electroporation. The immunoprecipitated proteins were analyzed by SDS-polyacrylamide gel electrophoresis (Fig. 4C). With this method, it was possible to detect the processed Del6K protein. The protein 6K analyzed by high-resolution gels (20% acrylamide) migrates as two bands and a smear (Fig. 4C). It is possible that the 6K forms aggregates that are resistant to denaturing agents such as dithiothreitol and SDS.
FIG. 4.
Expression of SV 6K gene in trans. (A) [35S]methionine-cysteine protein labeling from 16 to17 h.p.e in BHK cells electroporated with different RNAs. Lanes: B, no RNA was added; 1, wt SV; 2, SV Del6K; 3, SV Del6K + 6K; 4, SV Del6K-revQ21L; 5, SV Del6K-revQ21L + 6K. (B) Overexposure of the lower part of the field shown in panel A. (C) Immunoprecipitation analysis with anti-6K antibodies of lysates obtained from cells infected with mock-infected BHK cells (lane B), wt SV virus (lane 1), SV Del6K-revQ21L virus (lane 2), and SV Del6K-revQ21L + 6K virus (lane 3).
Conclusions.
The main defects associated with the SV Del6K variant described in this work appeared to be a consequence of inefficient proteolytic processing of the viral glycoprotein precursor. The revertant SV Del6K-revQ21L virus has resolved the defects of cleavage by the signalase and the transport of viral glycoproteins to the plasma membrane. This reversion can be ascribed to the substitution Q21L that renders the Del6K protein more hydrophobic. This reversion to a higher hydrophobicity of the Del6K protein may permit stronger association of Del6K with membranes, which may favor the signalase activity. Nevertheless, the revertant SV Del6K-revQ21L still has an attenuated phenotype, with defects in the release of virions from cells. These findings support another function for the 6K protein besides providing the signal sequences to signalase.
It was proposed that the 6K protein promotes membrane deformation and bending during the budding process (5, 17). Further evidence for the functioning of 6K comes from membrane permeabilization upon expression of the SFV 6K in Escherichia coli cells (21). The 6K protein of SV behaves similarly in this respect (unpublished results). This permeabilizing activity promotes local changes in membrane potential that would generate forces favoring the budding process. In this regard, there is evidence that budding of alphavirus particles is influenced by ionic gradients (14, 25). However, little is known about the molecular basis underlying this phenomenon. Another possible role for the 6K protein during budding is that it promotes lateral interactions between glycoprotein heterodimers or between spikes by 6K-6K interactions. In this way, 6K could favor glycoprotein concentration in the cytoplasmic membrane before its dissociation from viral glycoproteins. The coexpression of a genuine 6K protein did not revert the SV Del6K or SV Del6K-revQ21L phenotypes. The absence of complementation by 6K expressed in trans could be ascribed to one of the following factors. (i) Association with the PE2-E1 heterodimers is impaired. (ii) The 6K generated in trans contains an additional methionine in its N-terminal end. (iii) The 6K does not fold correctly in the cellular membranes, since it is expressed out of the context of the viral polyprotein.
Other animal viruses encode proteins with physical or biological similarities to 6K. These physical characteristics include a low molecular weight in very hydrophobic proteins that oligomerize and interact with membranes. The main biological function of these proteins is to participate in budding of virus particles, although they are excluded from mature virions. Proteins with these features are collectively termed viroporins (3). Typical viroporins are Vpu from human immunodeficiency virus (8, 13), M2 from influenza virus (10, 27), SH from respiratory syncitial virus (19), 2B from picornaviruses (1, 2, 24), or protein E from coronavirus (4, 20). Both M2 and Vpu form ion channels in biological membranes (3). Comparative studies of the biological functioning of these proteins will indicate to what extent they share analogous functions during the replicative cycle of animal viruses.
Acknowledgments
DGICYT project number PB94-0148 and the institutional grant to the CBM of Fundación Ramón Areces are acknowledged for financial support.
REFERENCES
- 1.Barco A, Carrasco L. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3349–3364. doi: 10.1002/j.1460-2075.1995.tb07341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barco A, Carrasco L. Identification of regions of poliovirus 2BC protein that are involved in cytotoxicity. J Virol. 1998;72:3560–3570. doi: 10.1128/jvi.72.5.3560-3570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrasco L. Modification of membrane permeability by animal viruses. Adv Virus Res. 1995;45:61–112. doi: 10.1016/S0065-3527(08)60058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corse E, Machamer C E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaedigk-Nitschko K, Ding M X, Levy M A, Schlesinger M J. Site-directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology. 1990;175:282–291. doi: 10.1016/0042-6822(90)90210-i. [DOI] [PubMed] [Google Scholar]
- 6.Gaedigk-Nitschko K, Schlesinger M J. The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology. 1990;175:274–281. doi: 10.1016/0042-6822(90)90209-a. [DOI] [PubMed] [Google Scholar]
- 7.Gaedigk-Nitschko K, Schlesinger M J. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183:206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 8.González M E, Carrasco L. The human immunodeficiency virus type 1 Vpu protein enhances membrane permeability. Biochemistry. 1998;37:13710–13719. doi: 10.1021/bi981527f. [DOI] [PubMed] [Google Scholar]
- 9.Hahn C S, Hahn Y S, Braciale T J, Rice C M. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci USA. 1992;89:2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughey P G, Compans R W, Zebedee S L, Lamb R A. Expression of the influenza A virus M2 protein is restricted to apical surfaces of polarized epithelial cells. J Virol. 1992;66:5542–5552. doi: 10.1128/jvi.66.9.5542-5552.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanova L, Le L, Schlesinger M J. Characterization of revertants of a Sindbis virus 6K gene mutant that affects proteolytic processing and virus assembly. Virus Res. 1995;39:165–179. doi: 10.1016/0168-1702(95)00083-6. [DOI] [PubMed] [Google Scholar]
- 12.Ivanova L, Lustig S, Schlesinger M J. A pseudo-revertant of a Sindbis virus 6K protein mutant, which corrects for aberrant particle formation, contains two new mutations that map to the ectodomain of the E2 glycoprotein. Virology. 1995;206:1027–1034. doi: 10.1006/viro.1995.1025. [DOI] [PubMed] [Google Scholar]
- 13.Klimkait T, Strebel K, Hoggan M D, Martin M A, Orenstein J M. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M L, Stollar V. A mutant of Sindbis virus which is released efficiently from cells maintained in low strength medium. Virology. 1995;210:237–243. doi: 10.1006/viro.1995.1340. [DOI] [PubMed] [Google Scholar]
- 15.Liljeström P, Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J Virol. 1991;65:147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loewy A, Smyth J, Von Bonsdorff C-H, Liljeström P, Schlesinger M J. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J Virol. 1995;69:469–475. doi: 10.1128/jvi.69.1.469-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lusa S, Garoff H, Liljeström P. Fate of the 6K membrane protein of Semliki Forest virus during virus assembly. Virology. 1991;185:843–846. doi: 10.1016/0042-6822(91)90556-q. [DOI] [PubMed] [Google Scholar]
- 19.Pérez M, García-Barreno B, Melero J A, Carrasco L, Guinea R. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology. 1997;235:342–351. doi: 10.1006/viro.1997.8696. [DOI] [PubMed] [Google Scholar]
- 20.Raamsman M J, Locker J K, de Hooge A, de Vries A A, Griffiths G, Vennema H, Rottier P J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanz M A, Pérez L, Carrasco L. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J Biol Chem. 1994;269:12106–12110. [PubMed] [Google Scholar]
- 22.Schlesinger M J, London S D, Ryan C. An in-frame insertion into the Sindbis virus 6K gene leads to defective proteolytic processing of the virus glycoproteins, a trans-dominant negative inhibition of normal virus formation, and interference in virus shut off of host-cell protein synthesis. Virology. 1993;193:424–432. doi: 10.1006/viro.1993.1139. [DOI] [PubMed] [Google Scholar]
- 23.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Kuppeveld F J M, Hoenderop J G J, Smeets R L L, Willems P H G M, Kijkman H B P M, Galama J M D, Melchers J G. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997;16:3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waite M R, Pfefferkorn E R. Inhibition of Sindbis virus production by media of low ionic strength: intracellular events and requirements for reversal. J Virol. 1970;5:60–71. doi: 10.1128/jvi.5.1.60-71.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao J S, Strauss E G, Strauss J H. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J Virol. 1996;70:7910–7920. doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zebedee S L, Lamb R A. Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proc Natl Acad Sci USA. 1989;86:1061–1065. doi: 10.1073/pnas.86.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]