Abstract
We have used a virus-binding assay to examine conformational changes that occur when soluble CD4 (sCD4) binds to the surface of intact, native, primary human immunodeficiency virus type 1 virions. The isolates examined belong to seven genetic clades (A to H) and are representative of syncytium-inducing and non-syncytium-inducing phenotypes. Conformational changes in epitopes in the C2, V2, V3, C5, and CD4 binding domain (CD4bd) of gp120 and the cluster I and II regions of gp41 of these viruses were examined using human monoclonal antibodies that are directed at these regions. The studies revealed that sCD4 binding causes a marked increase in exposure of epitopes in the V3 loop, irrespective of the clade or the phenotype of the virus. Sporadic increases in exposure were observed in some epitopes in the V2 region, while no changes were observed in the C2, C5, or CD4bd of gp120 or the cluster I and II regions of gp41.
The human immunodeficiency virus (HIV) envelope glycoprotein is produced as a gp160 precursor that is proteolytically cleaved into a mature noncovalent multimeric complex forming gp120 and gp41 subunits (2, 10). Several antigenic epitopes have been identified within regions of the gp120 and gp41 subunit proteins. One region that has received substantial attention is the V3 loop. The V3 loop of gp120 is an immunodominant region (27), constitutes a target for neutralizing antibodies (18), is implicated in syncytium induction in MT2 cells (4, 9, 21, 25), and contains determinants recognized by the viral coreceptors CXCR4 and CCR5 (1, 5).
It is known that in the course of HIV type 1 (HIV-1) infection, the viral CD4 binding domain (CD4bd) located in the gp120 envelope glycoprotein binds to CD4, which leads to conformational changes in other regions of gp120 and subsequently to coreceptor binding (8, 11, 24, 26). Epitopes that become exposed on gp120 as a result of binding to CD4 could serve as targets for antibodies that might prevent infectivity by the virus. Indeed, studies of cells infected with laboratory-adapted strains of HIV-1 and cells expressing different forms of recombinant gp120 have demonstrated that soluble CD4 (sCD4) binding causes conformational changes leading to exposure of epitopes in the V3 and V2 regions (12, 17, 19, 20, 22, 23). However, studies that have examined the exposure of antigenic epitopes on primary, intact, native virions upon binding to sCD4 are lacking. Identification of epitopes exposed on primary isolates after binding to CD4 could improve our understanding for designing a potent HIV-1 vaccine and new therapeutic strategies.
In a previous study, we examined the antigenic landscape of intact, native HIV-1 isolates using monoclonal antibodies (MAbs) directed at epitopes on the viral envelope (14). We observed that epitopes in the V3 and C5 regions of gp120 and in cluster I of gp41 are well exposed on intact virions compared to epitopes in the V2, C2, and CD4bd of gp120 and in cluster II of gp41. In the present study, we addressed for the first time the issue which epitopes become exposed when primary, intact, native HIV-1 virions bind to sCD4. To address this question, eight HIV-1 group M isolates belonging to seven clades, A (92RW021), B (MNp and JR-FL), C (ZB18), D (MAL), F (CA20), G (VI526), and H (CA13), were preincubated with sCD4 and tested for their abilities to bind to MAbs directed at epitopes in the gp120 and gp41 regions. The viruses were selected to include those that use CXCR4 (MNp, MAL, VI526, and CA13) and CCR5 (92RW021, JR-FL, ZB18, and CA20). These viruses were passaged only in human peripheral blood mononuclear cells (PBMCs), except for MAL, which had been passaged in a transformed cell line (H9 cells) before subsequently being carried in human PBMCs. Virus stocks were produced by infecting 3-day phytohemagglutinin-stimulated HIV-negative donor PBMCs with 1 ml of a p24-positive culture supernatant (15, 16). The p24 concentration in each virus stock was quantitated using a noncommercial p24 enzyme-linked immunosorbent assay (ELISA) (13).
Twenty-two MAbs were used to capture viruses that had been preincubated with or without sCD4. These MAbs included seven anti-V3 (447-52D, 908-D, 257-2D, 412-D, 1006-15D, 694/98-D, and 537-D), three anti-V2 (697-D, 830-A, and 1393A), two anti-C2 (1006-30D, and 847-D), three anti-C5 (670-D, 1331A, and 989-D), two anti-CD4bd (448-D and IgG1B12), two anti-gp41 cluster I (246-D and 50-69), and three anti-gp41 cluster II (98-6, 167-D, and 2F5) antibodies. These MAbs have been extensively described elsewhere (7, 14). Human MAb 1418 to parvovirus B19 (6) and 246-D (anti-HIV-1 gp41) were used as negative and positive controls, respectively. Soluble CD4 was purchased from Life Science Products (Boston, Mass.). To determine the effect of sCD4 on epitope exposure, 100 μl of HIV-1 culture supernatant diluted to contain 100 ng of p24/ml was preincubated with or without sCD4 (100 μl at 5 μg/ml) for 1 h at 37°C before the addition and further incubation in microtiter wells coated as previously described with MAbs (100 μl at 10 μg/ml) for 1 h at 37°C (13, 14). Unbound virus was washed away with RPMI 1640, and bound virus was lysed with 250 μl of 1% Triton X-100. p24 was assayed using a noncommercial ELISA as previously described (3). All experiments were performed in duplicate. The cutoff value was determined by calculating the average of the negative control for all eight isolates plus 3 standard deviations. The p24 cutoff value was 23 pg/ml. Thus, only readings of >23 pg/ml were considered positive.
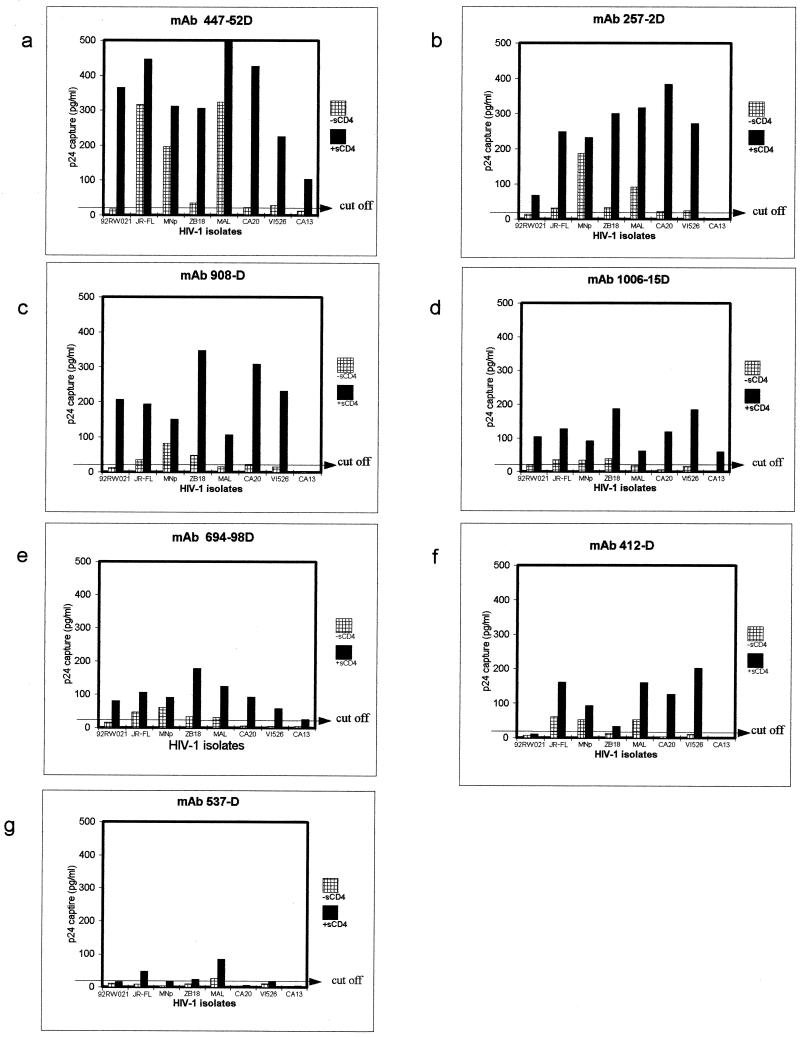
MAbs directed at seven envelope regions (V2, C2, V3, C5, and CD4bd of gp120 and cluster I and II of gp41) were used to identify epitopes that become exposed when gp120 on intact, native virions bind to sCD4. Of the seven regions studied, only epitopes in the V3 loop displayed increased exposure when sCD4 bound to seven of the eight intact, native virions examined (Fig. 1). This was revealed by a 2- to 22-fold increase in binding of anti-V3 MAbs.
FIG. 1.
Effect of sCD4 on exposure of epitopes in the V3 region of primary, intact, native HIV-1 virions. Results are expressed as the value of p24 capture in picograms per milliliter when virus was preincubated with (■) or without ( ) sCD4. Values of >23 pg of p24/ml were considered positive. Values of >500 pg of p24/ml are shown as 500 pg/ml.
One isolate, CA13, bound poorly to five of seven anti-V3 MAbs in both the presence and absence of sCD4 but did display increases of 8- and 30-fold in binding with MAbs 447-52D and 1006-15D, respectively, in the presence of sCD4 (Fig. 1a and d). Viruses RW92021, ZB18, CA20, and VI526 bound poorly to the anti-V3 MAbs in the absence of sCD4, but in the presence of sCD4 79% of these virus-anti-V3 MAb combinations showed increased binding. While viruses JR-FL, MNp, and MAL bound to most of the anti-V3 MAbs in the absence of sCD4, their binding was significantly increased in 86% of the virus-MAb combinations tested in the presence of sCD4. In sum, 44 of 56 virus-MAb combinations showed substantially increased binding to anti-V3 MAbs in the presence of sCD4.
Conversely, while one MAb, 537-D, showed only small increases in binding to virions after preincubation with sCD4, all other anti-V3 MAbs displayed increased binding to the majority of sCD4-treated virions.
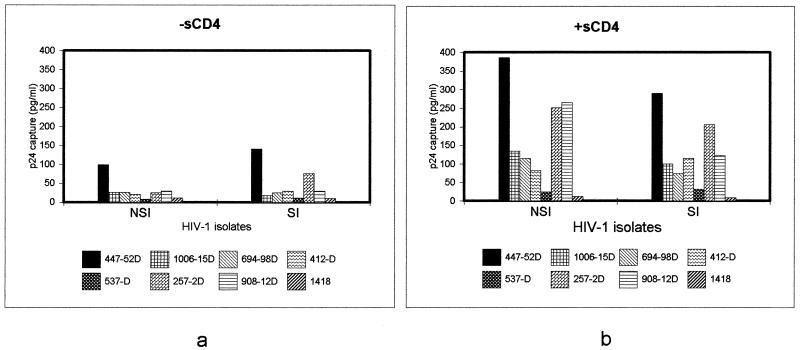
Because the V3 loop has been implicated in the induction of syncytium formation and coreceptor usage, we analyzed whether there are any differences in the exposure of V3 epitopes between non-syncytium-inducing (NSI) and syncytium-inducing (SI) viruses, which are CCR5-tropic and CCR4-tropic, respectively. The results (Fig. 2) clearly demonstrate that a similar increase in exposure of V3 loop epitopes was observed for both categories of viruses when preincubated with sCD4.
FIG. 2.
Exposure of epitopes in the V3 loops of NSI (CCR5-trophic) and SI (CXCR4-trophic) viruses in the absence (a) and presence (b) of sCD4. Four NSI viruses (92RW021, JR-FL, ZB18, and CA20) and four SI viruses (MNp, MAL, VI526, and CA13) were tested. The p24 values shown (in picograms per milliliter) represent the average binding of each MAb with four NSI and SI viruses in the absence and presence of sCD4. The data on which the averages depicted in this figure are based are shown in Fig. 1.
Overall, no increase in exposure was observed with epitopes in the remaining regions of the viral envelope examined (data not shown). However, sporadic increases in binding were observed using anti-V2 MAbs 697-D and 830-A. For example, MAb 697-D showed a 7-fold and 5-fold increase in binding with JR-FL and VI526, respectively, when virus was preincubated with sCD4. MAb 830-A also showed a 5-fold increase in binding with VI526.
These results clearly demonstrate that, upon preincubation of virus with sCD4, conformational changes occur mainly in the V3 loop. Though some epitopes in the gp41 cluster I and C5 region are well exposed on the intact, native virions prior to incubation with sCD4 (13, 14), these epitopes are not better exposed upon preincubation with sCD4 (data not shown). The fact that conformational changes occur mainly in the V3 loop region leading to enhanced exposure of epitopes when virus binds to sCD4 supports the critical biological role of the V3 region in the infectious process of the virus.
The results of this study suggest that conformational changes in regions of the envelope other than the V3 may not be of biologic relevance at the initial step after the virus binds to sCD4. However, conformational changes in other envelope regions may occur when the virus binds to its coreceptors. Further studies are needed to determine other epitopes that may become exposed on the viral envelope after binding to coreceptors. Such epitopes could serve as potential targets for antiviral agents that prevent entry of the virus into host cells, thereby preventing infectivity.
Acknowledgments
This study was supported by grants from the National Institutes of Health (AI 44302, AI 47053, and TW 01254).
We thank John Sullivan for the primary HIV-1 isolate (MNp) and Guido van der Groen for the primary isolates CA20, CA13, and VI526. The MAbs were provided through the support of the New York University Center for AIDS Research Immunology Core (AI 27742), with the exception of IgG1b2 and 2F5, which were obtained from Denis Burton and the Medical Research Council Reagent Program, respectively.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Allan J S, Coligan J E, Barin F, McLane M F, Sodroski J G, Rosen C A, Haseltine W A, Lee T H, Essex M. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228:1092–1094. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 3.Bastiani L, Laal S, Zolla-Pazner S, Kim M. Host cell-dependent alterations in envelope components of human immunodeficiency virus type 1 virions. J Virol. 1997;71:3444–3450. doi: 10.1128/jvi.71.5.3444-3450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng-Mayer C, Shioda T, Levy J A. Host range, replicative, and cytopathic properties of human immunodeficiency virus type 1 are determined by very few amino acid changes in tatand gp120. J Virol. 1991;65:6931–6941. doi: 10.1128/jvi.65.12.6931-6941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 6.Gigler A, Dorsch S, Hemauer A, Williams C, Kim S, Young N S, Zolla-Pazner S, Wolf H, Gorny M K, Modrow S. Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. J Virol. 1999;73:1974–1979. doi: 10.1128/jvi.73.3.1974-1979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorny M K, Gianakakos V, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to HIV. Proc Natl Acad Sci USA. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 10.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 11.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 12.McKeating J A, Cordell J, Dean C J, Balfe P. Synergistic interaction between ligands binding to the CD4 binding site and V3 domain of human immunodeficiency virus type 1 gp120. Virology. 1992;191:732–742. doi: 10.1016/0042-6822(92)90249-o. [DOI] [PubMed] [Google Scholar]
- 13.Nyambi P N, Gorny M K, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J Virol. 1998;72:9384–9391. doi: 10.1128/jvi.72.11.9384-9391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nyambi P N, Mbah H A, Burda S, Williams C, Gorny M K, Nadas A, Zolla-Pazner S. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J Virol. 2000;74:7096–7107. doi: 10.1128/jvi.74.15.7096-7107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyambi P N, Nkengasong J, Peeters M, Simon F, Eberle J, Janssens W, Fransen K, Willems B, Vereecken K, Heyndrickx L, Piot P, van der Groen G. Reduced capacity of antibodies from patients infected with human immunodeficiency virus type-1 (HIV-1) group O to neutralize primary isolates of HIV-1 group M viruses. J Infect Dis. 1995;172:1228–1237. doi: 10.1093/infdis/172.5.1228. [DOI] [PubMed] [Google Scholar]
- 16.Nyambi P N, Willems B, Janssens W, Fransen K, Nkengasong J, Peeters M, Vereecken K, Heyndrickx L, Piot P, van der Groen G. The neutralization relationship of HIV type 1, HIV type 2, and SIVcpz is reflected in the genetic diversity that distinguishes them. AIDS Res Hum Retrovir. 1996;13:7–17. doi: 10.1089/aid.1997.13.7. [DOI] [PubMed] [Google Scholar]
- 17.Pinter A, Honnen W J, Tilley S A. Conformational changes affecting the V3 and CD4-binding domains of human immunodeficiency virus type 1 gp120 associated with envprocessing and with binding of ligands to these sites. J Virol. 1993;67:5692–5697. doi: 10.1128/jvi.67.9.5692-5697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rusche J R, Javaherian K, McDanal C, Petro J, Lynn D L, Grimaila R, Langlois A, Gallo R C, Arthur L O, Fischinger P J, Bolognesi D P, Putney S D, Matthews T J. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc Natl Acad Sci USA. 1988;85:3198–3202. doi: 10.1073/pnas.85.9.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–416. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 22.Stamatatos L, Cheng-Mayer C. Structural modulations of the envelope gp120 glycoprotein of human immunodeficiency virus type 1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J Virol. 1995;69:6191–6198. doi: 10.1128/jvi.69.10.6191-6198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved HIV-type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 25.Westervelt P, Trowbridge D B, Epstein L G, Blunberg B M, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsett A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD-4 induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 27.Zwart G, Langeduk H, Van Der Hoek L, DeJong J, Wolfs T F W, Ramautarsing C, Bakker M, DeRonde A, Goudsmit J. Immunodominance and antigenic variation of the principal neutralization domain of HIV-1. Virology. 1991;181:481–489. doi: 10.1016/0042-6822(91)90880-k. [DOI] [PubMed] [Google Scholar]