Abstract
The functional analysis of molecular determinants which control the replication of pestiviruses was considerably facilitated by the finding that subgenomic forms of the positive-strand RNA genome of BVDV (bovine viral diarrhea virus) are capable of autonomous replication in transfected host cells. The prototype replicon, BVDV DI9c, consists of the genomic 5′ and 3′ untranslated regions and a truncated open reading frame (ORF) encoding mainly the nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B. To gain insight into which of these proteins are essential for viral replication and whether they act in cis or in trans, we introduced a large spectrum of in-frame mutations into the DI9c ORF. Tests of the mutant RNAs in terms of their replication capacity and their ability to support translation and cleavage of the nonstructural polyprotein, and whether defects could be rescued in trans, yielded the following results. (i) RNA replication was found to be dependent on the expression of each of the DI9c-encoded mature proteins NS3 to NS5B (and the known associated enzymatic activities). In the same context, a finely balanced molar ratio of the diverse proteolytic processing products was indicated to be crucial for the formation of an active catalytic replication complex. (ii) Synthesis of negative-strand intermediate and progeny positive-strand RNA was observed to be strictly coupled with all functional DI9c ORF derivatives. NS3 to NS5B were hence suggested to play a pivotal role even during early steps of the viral replication pathway. (iii) Mutations in the NS3 and NS4B units which generated nonfunctional or less functional RNAs were determined to be cis dominant. Likewise, lethal alterations in the NS4A and NS5B regions were invariably noncomplementable. (iv) In surprising contrast, replication of functional and nonfunctional NS5A mutants could be clearly enhanced and restored, respectively. In summary, our data provide initial insights into the organization of the pestivirus replication machinery.
Bovine viral diarrhea virus (BVDV) types I and II, border disease virus of sheep, and classical swine fever virus constitute the genus Pestivirus, comprising widely distributed pathogens of ruminants and pigs (reviewed in reference 31). Together with the genera Flavivirus and Hepacivirus (hepatitis C viruses [HCVs]), the pestiviruses are classified in the family Flaviviridae (reviewed in reference 24), all members of which are characterized by an enveloped virion that harbors a single-stranded, linear RNA genome of positive polarity. The genomic RNA, which in the case of pestiviruses has a length of approximately 12.5 kb, consists of a single open reading frame (ORF) and untranslated regions (UTRs) at the 5′ and 3′ ends, respectively. Following infection, it operates initially as a messenger in the cytoplasm. Translation is mediated by an internal ribosomal entry site (IRES) within the 5′ UTR (22) and leads to the synthesis of a polyprotein that is co- and posttranslationally processed into a range of viral proteins. The order of the final maturation products has been determined to be NH2-Npro, core, Erns, E1, E2, p7, NS2-3, NS4A, NS4B, NS5A, NS5B-COOH. Npro, a nonstructural protein of uncertain function, has autoprotease activity and generates its own C terminus (33). Host cell proteases process the constituents of the virion, i.e., the core and the envelope proteins Erns (which exhibits also an RNase activity), E1, and E2 as well as the hydrophobic peptide p7, the latter which is assumed to be also involved in virion assembly (13, 31). Processing of the nonstructural protein NS2-3 (125 kDa) and release of the 80-kDa C-terminal NS3 portion occur at different extents in various pestivirus strains and were demonstrated to be associated with a certain phenotype of virus infection in tissue culture: whereas viruses that replicate without obvious damage to the host cell express predominantly NS2-3, generation of high amounts of NS3 is strictly correlated with cytopathogenicity, i.e., virus-induced lysis of the cell (reviewed in reference 28). Proteolysis of the remainder of the polyprotein that gives rise to the mature nonstructural proteins NS3 to NS5B is catalyzed by a viral proteinase complex consisting of a serine protease domain within the N terminus of NS3 and the essential cofactor NS4A (27, 30, 34, 35).
Replication of the pestiviral genome proceeds in an asymmetric manner similar to that reported for other monocistronic positive-strand RNA viruses. Concomitant with the generation and cleavage of the polyprotein, the nascent viral proteins and hypothetical host components are supposed to associate with the 3′ terminus of the genomic RNA and to form membrane-associated replication complexes. These catalyze the synthesis of a low number of negative-strand (antigenomic) intermediates, which, in a subsequent step, serve as templates for the transcription of an excess of novel positive-strand RNA molecules (4, 10).
Although NS3 to NS5B are expected to represent functional components of the viral replication machinery, how they participate in pestivirus replication is still not understood. Besides the NS3/NS4A protease, two other viral enzymes were demonstrated to be required for the replication process: a nucleoside triphosphatase/RNA helicase activity contained at the C-terminal portion of NS3, and the viral RNA-dependent RNA polymerase (RdRp) which is associated with the NS5B protein (12, 25, 32, 38). Except for its hydrophobic nature, virtually nothing is known about NS4B, and only preliminary data are available on the NS5A protein. Data from infection studies, transient expression experiments, or cell-free in vitro assays revealed that NS5A of BVDV and HCV as well as NS5 of the yellow fever flavivirus are phosphorylated by (probably identical) serine/threonine kinases (23), which suggests that these proteins might share a common function related to their phosphorylation state. NS5A of HCV has attracted attention due to the proposal that it interacts with the protein kinase PKR and thus may play a role in the resistance of certain HCV variants to interferon (9).
Detailed investigations of pestivirus replication have been enabled by the successful composition of stable genomic cDNA copies which are capable of producing infectious RNA transcripts in vitro. Further experiments demonstrated that RNA molecules encompassing mainly the 5′ and 3′ UTRs and the coding region of NS3 to NS5B support both steps of the replication pathway upon transfection into host cells (4). Because of a number of experimental advantages with respect to the full-length viral RNA, the most important of which is the possibility of examining RNA replication independently from the assembly process of the virion, we have been using this BVDV replicon (termed DI9c) to explore the role of individual components of the replication complex. As a general scheme, the viral RNA is mutagenized via the cDNA construct (reverse genetics), and the effects of mutagenesis on translation or replication are monitored by using appropriate in vivo and in vitro assay systems. Thus, RNA motifs formed by the 3′ terminus of the BVDV genome were identified to create a common cis-acting signal, which is suspected to contribute to the negative-strand promoter of the initial replication complex (36). In a similar manner, the genomic 5′ terminus was shown to fold into a stem-loop structure which modulates translation as well as replication of the viral RNA (37). A combination of reverse genetics and biochemical studies indicated the NS3 protein as well as the NS3-associated protease and NTPase/helicase activities to be required at an early stage of the replication pathway and to operate in cis during assembly of the catalytic replication complex (12).
In continuation of this latter work, we compared the previously characterized NS3 mutants with a large set of novel DI9c derivatives bearing mutations in all other genetic units of the ORF with regard to replication, polyprotein processing, and complementation behavior. The results of this study underline the crucial importance of each of the replicon-encoded proteins NS3 to NS5B and permit the proposition of an initial model on cis-and trans-acting factors of the pestivirus replication pathway.
MATERIALS AND METHODS
Cells and viruses.
Cells and culture conditions were described in references 4 and 12. Hygbici cells, i.e., BHK-21 or MDBK cells which contained the noncytopathogenic replicon BVDV Hygbici (see Fig. 4A), were selected in medium supplied with hygromycin B (500 μg/ml). The BVDV isolate has been described previously (7, 26).
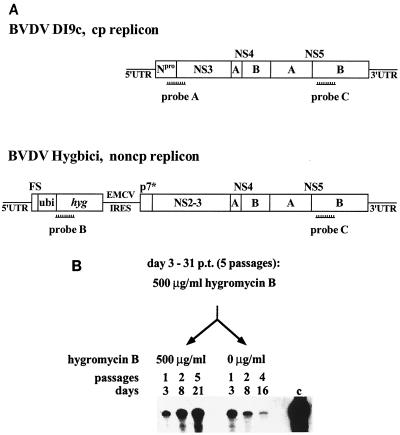
FIG. 4.
Establishment of a cell line containing a persistently replicating BVDV RNA (A) Side-by-side representation of the genome organization of the monocistronic, cytopathogenic (cp) BVDV DI9c replicon and of the bicistronic noncytopathogenic (noncp) BVDV Hygbici replicon. Boxes indicate the protein-coding regions; horizontal lines symbolize the UTR. The BVDV Hygbici replicon mainly corresponds to the bicistronic RNA Bi-NS2ins-, previously reported by Tautz et al. (29). The additional 5′-terminal ORF consists of a short sequence element of the Npro -coding region (FS) which was shown to be important for efficient BVDV IRES function (4, 6, 29) and which comprises the natural translational start codon. Moreover it contains a ubiquitin gene (ubi) and the hygromycin B phosphotransferase gene (hyg) with an artificial translational stop codon. The ubiquitin gene, which was fused in frame to the hyg ORF, enables generation of the authentic N terminus of the phosphotransferase by the activity of cellular ubiquitin C-terminal hydrolases. The second ORF located downstream of the EMCV (encephalomyocarditis virus) IRES encodes the pestiviral NS proteins; p7∗ denotes the 3′-terminal part of the p7-coding unit which comprises the cleavage site for generation of the correct N terminus of NS2. The (antisense) RNA probes used in the differentiating RPAs (see text) are schematically drawn at their positions of hybridization (see text). Probe A was used to specifically detect BVDV DI9c-derived RNAs; probe B was employed for the specific detection of BVDV Hygbici RNA; probe C served to monitor replication of any BVDV CP7-derived RNA. (B) Monitoring replication of BVDV Hygbici RNA in BHK-21 cells. In vitro-synthesized BVDV Hygbici cRNA (5 μg) was transfected into BHK-21 cells. Hygromycin B (500 μg/ml) was added to the culture medium 3 days p.t. From day 3 to 31 p.t., the cells were continuously grown and passaged five times in this medium. At day 31 p.t., the cell population was divided into two aliquots; one portion was cultured in normal medium, and the other was grown under constant selection pressure in antibiotic-containing medium. At the given time points and number of passages after separation, an aliquot of each isolated cell population was analyzed for the presence of the noncytopathogenic replicon using RPA with antisense probe C. Lane c, RNase protection with 300 ng of in vitro-synthesized BVDV Hygbici cRNA (positive control).
Infection of MDBK cells.
Infection of MDBK cells with BVDV isolate NCP7 was performed at a multiplicity of infection of 0.1. The infection was verified by an immunofluorescence (IF) assay utilizing a monoclonal anti-BVDV NS3 antibody (4, 8).
Construction of recombinant plasmids.
Restriction and cloning procedures were done according to standard protocols. Restriction endonucleases and modifying enzymes were obtained from New England Biolabs (Schwalbach, Germany), Promega (Mannheim, Germany), MBI Fermentas (St. Leon-Rot, Germany), Gibco BRL (Eggenstein, Germany), and Roche Diagnostics (Mannheim, Germany). DNA oligonucleotide primers used for mutagenesis, reverse transcription-PCR (RT-PCR), and DNA sequencing (5′ IRD41-labeled primers) were purchased from MWG Biotech GmbH (Ebersbach, Germany).
The full-length BVDV CP7 cDNA clone pA/BVDV/N as well as DI9c cDNA constructs pA/BVDV/D9 and pP/BVDV/D9 are described in references 19 and 37, respectively. Mutagenesis of the DI9c cDNA was performed on both plasmids, which do not differ in the DI9c-coding sequence.
Mutations 1a to 10a have been described previously (4, 12). Mutations 10b and 10c are described in reference 30. The deletion mutations 20, 21, and 26 were made by cutting cDNA cloning intermediates with appropriate restriction endonucleases, removing the excised fragment, blunting, and religation. The insertion mutations were created by a linker-insertion protocol described by Grassmann et al. (12); oligonucleotides used for generation of mutations were BEMLU for mutations 15 and 17, BALU for mutations 11, 13, 18, 24, and 25, NIPLU for mutations 12 and 16, SALU for mutation 14, BESLU plus BESLU-R for mutation 19, and HILU plus HILU-R for mutations 22 and 23 (Table 1). Introduction of each mutation was confirmed by DNA sequencing. DI9c derivative 27 has been described by Yu et al. (37). Mutations 28, 29, 30, and 31 were created by primer-directed PCR mutagenesis techniques applying oligonucleotides BVD CS 11, L2336R, L2683R+/plus L2683R−, and 5ABLSDPs/plus 5ABLSDPas, respectively (Table 1).
TABLE 1.
Oligonucleotides used for linker-insertion mutagenesis, primer-directed mutagenesis, and RT-PCR
| Oligonucleotide | Sequencea (5′→3′) | Nucleotide positions (BVDV CP7) |
|---|---|---|
| BEMLU | acgcgt | |
| BALU | gatcgacgcgtc | |
| NIPLU | cacgcgtgtgca | |
| SALU | tcgagacgcgtc | |
| BESLU | gtgacgacgcgt | |
| BESLU-R | gtcacacgcgtc | |
| HILU | agctacgcgtgg | |
| HILU-R | agctccacgcgt | |
| BVD CS 11 | tttcggcagaagatcttcctaccacttg | 7224–7197 |
| L2336R | cacccactgctcgctcctttagttc | 7413–7389 |
| L2683R+ | gggaagatacgtaaccggtctgggaattatg | 8427–8457 |
| L2683R− | cataattcccagaccggttacgtatcttccc | 8457–8427 |
| 5ABLSDPs | cctataccatgaaggatcctagttggtttcttc | 9916–9948 |
| 5ABLSDPas | gaagaaaccaactaggatccttcatggtatagg | 9948–9916 |
| B38 | cagtccagacaattggc | 8266–8282 |
| B42 | gttgggatcactttagtt | 9270–9287 |
| B49R | agaacaaagggcctttcat | 9469–9451 |
| B53R | cctgcttccgtcattatg | 10372–10355 |
Restriction sites introduced by the mutation are underlined (MluI in BEMLU, BALU, NIPLU, SALU, BESLU, BESLU-R, HILU, and HILU-R; BglII in BVD CS 11; SnaBI in L2683R+ and L2683R−; BamHI in 5ABLSDPs and 5ABLSDPas). Mutated nucleotides are indicated by italics.
For in vitro transcription of the cRNAs with T7 RNA polymerase (Stratagene), all pA/BVDV/D9 and pP/BVDV/D9 derivatives as well as the BVDV CP7 cDNA plasmid were linearized with SmaI.
The bicistronic replicon BVDV Hygbici (schematized in Fig. 4A) corresponds mainly to replicon Bi-NS2ins-, described in detail by Tautz et al. (29); the only difference is that the β-glucuronidase-coding unit of Bi-NS2ins- was replaced by ubiquitin-coding sequence combined in frame with the hyg gene. Thus, the authentic N terminus of the hygromycin B phosphotransferase is expected to be generated by cellular ubiquitin C-terminal hydrolases. To obtain the BVDV Hygbici RNA, the cDNA plasmid was linearized with SmaI and transcribed with SP6 RNA polymerase (NatuTec, Frankfurt am Main, Germany).
The plasmid constructs used to generate radiolabeled probes for the detection of all BVDV-derived RNAs (probe C) and for the specific detection of DI9c-derived RNA molecules (probe A) (see Fig. 4A) by RNase protection assay (RPA) are described in references 4 and 37, respectively. To generate probes used to detect specifically the BVDV Hygbici replicon, the 5′-terminal 353 nucleotide fragment of the hyg gene, originally contained in p3′SS (Stratagene), was inserted into the multiple cloning site of pBluescript II KS(+) (Stratagene). The recombinant plasmid was linearized with SacI and transcribed with T3 RNA polymerase to generate antisense probe (probe B [see Fig. 4A]).
To create Sindbis virus-derived RNA replicons which encode the individual BVDV NS proteins, PCR products containing each NS protein-coding region flanked by artificial translation initiation and termination codons were cloned into the multiple cloning site of pSinRep5 (Invitrogen). Generation of recombinant replicons by in vitro-transcription was carried out as described in reference 12.
DNA sequencing.
Dideoxy sequencing of double-stranded DNA was performed as described previously (36).
In vitro transcription, purification, and transfection of RNA.
Procedures to generate viral cRNAs as well as radiolabeled RNA probes for the RPA are described in references 4 and 36. Transfection of RNA in BHK-21 and MDBK cells was performed according to electroporation protocols described in references 4 and 29, respectively.
RPA.
RPA to measure the replication ability of individual DI9c derivatives was carried out as described in detail by Yu et al. (36) and Grassmann et al. (12). For the specific detection and quantification of de novo-synthesized replication products derived from BVDV DI9c ORF mutants, we used a [32P]UTP-labeled probe which hybridizes to the 3′-terminal Npro/5′-terminal NS3-encoding region of the DI9c replicon (probe A). Specific detection of BVDV Hygbici replication was enabled by a probe hybridizing to the 5′-terminal part of the hyg gene (probe B). To determine the stability of the diverse BVDV DI9c derivatives, RPAs on different regions of the RNA molecules were performed. For this purpose we used probes A and C, which hybridize to distant portions of the DI9c RNA. To standardize different experiments for variations in the transfection efficiency and/or the yield of extracted nucleic acids, a defined amount of plasmid DNA was cotransfected with the viral RNA in each experiment and quantified at the indicated time points posttransfection (p.t.) by using a plasmid-specific probe (12). Quantification of protected RNA fragments was performed with a Fuji bioimaging analyzer.
In vitro translation.
BHK-21 S10 extract and BHK-21 cell translation initiation factors were prepared as described in reference 3. In vitro translation reactions and analysis of the [35S]methionine-labeled reaction products by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were carried out essentially as described by Grassmann et al. (12) and Yu et al. (37). For quantification of translation efficiency, the amounts of the Npro and/or NS3 proteins were determined with a Fuji bioimaging analyzer.
RT-PCR analysis.
RT-PCR, carried out as described previously (4, 12), was used to confirm the RPA data, to verify the stability and identity of the transfected DI9c derivatives before as well as after-transfection, and to discriminate RNA replication of different BVDV replicons upon cotransfection (see Fig. 7). Quantitation was performed with two independent sets of RNA standards.
FIG. 7.
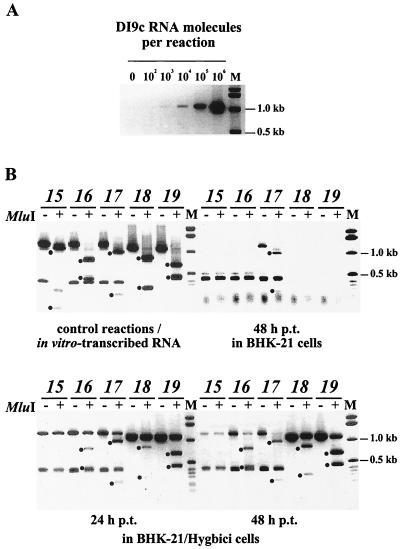
Restoration or enhancement of the replication ability of DI9c NS5A mutants 16 to 19 cannot be explained by RNA recombination. (A) Detection level of quantitative RT-PCR. Applying primers B53 R and B42 (see Materials and Methods), RT-PCR was carried out with different amounts of in vitro-transcribed DI9c RNA. The standard reaction was carried out in parallel with the reactions shown in panel B. The detection limit was thus demonstrated to be 5 fg (corresponding 103 molecules) of DI9c RNA. (B) RT-PCR analysis of the DI9c ORF mutant RNAs after transfection into naive BHK or BHK/Hygbici cells. Mutants 15 to 19 (15 as a negative control) were transfected into normal BHK and BHK/Hygbici cells. The naive cells were harvested at 48 h p.t. (top right); the Hygbici cells were divided into two portions and harvested at 24 and 48 h p.t., respectively (bottom). RT-PCR was carried out on identical amounts of cytoplasmic RNA. Primers B49R (mutants 15 to 17) and B53R (mutants 18 and 19) were used for reverse transcription; primers B49R and B38 (mutants 15–17) and primers B53R and B42 (mutants 18 and 19) were used for the subsequent PCRs (Table 1). As a positive control, the RT-PCRs were performed on the in vitro-transcribed mutant RNAs (top left). The resulting DNA fragments, which were confirmed to comprise the expected portion of the NS5A-coding region by sequencing (data not shown), had sizes of 1.2 kb (mutants 15 to 17) and 1.1 kb (mutants 18 and 19). In addition, RT-PCR with primers B49R and B38 gave rise to an unspecific product of ca. 0.4 kb (see also control reactions). Replication of the mutant DI9c RNAs was confirmed via restriction digest of the RT-PCR products with MluI, because this restriction site was originally introduced into the mutant cDNA constructs (see Materials and Methods and control reactions). Digestion with MluI should yield the following fragments: mutant 15, 1.10 and 0.10 kb; mutant 16, 0.77 and 0.43 kb; mutant 17, 0.96 and 0.24 kb; mutant 18, 0.81 and 0.29 kb; mutant 19, 0.67 and 0.43 kb. Lanes: M, DNA length marker; italic numbers, assays performed on either in vitro-transcribed RNAs (top left) or cytoplasmic RNA fractions of BHK cells (top right) or Hygbici cells (bottom) which had been transfected with DI9c derivatives 15 to 19. Plus signs indicate that the PCR products were digested with MluI. Positions of the 0.5- and 1.0-kb marker fragments are given on the right. The MluI cleavage products are marked by dots.
RESULTS
Mutagenesis of the BVDV DI9c ORF.
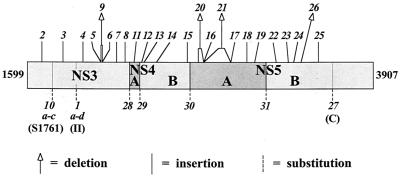
To generate mutations at various defined positions in the BVDV DI9c ORF region, we pursued two strategies. (i) To affect the overall folding of the polyprotein and/or of the mature NS proteins without changing the translational reading frame, short linker insertions or in-frame deletions were introduced into each of the different NS protein-coding units of the RNA (mutants 2 to 9 and 11 and 26). (ii) In a second series, defined amino acid residues were substituted to abolish the known virus-encoded enzymatic activities or to interfere with cleavage at the diverse proteolytic cleavage sites of the NS3-NS4A protease complex within the NS protein precursor. Thus, we modified the catalytic serine of the NS3 protease domain (mutants 10a to 10c), the Walker B motif (motif II) of the NS3 helicase (mutants 1a to 1d), the GDD motif of the NS5B RdRp (mutant 27), and the P1 and/or P1′ positions in the NS3/NS4A (mutant 28), NS4A/NS4B (mutant 29), NS4B/NS5A (mutant 30), and NS5A/NS5B (mutant 31) cleavage sites. All mutations are schematized in Fig. 1; the exact locations and types of mutagenesis used are summarized in Table 2. To compare a comprehensive spectrum of ORF derivatives and to extend our previous data, we included also mutants 1 to 10 and 27 (described in references 4, 12, and 37) in the experiments discussed below.
FIG. 1.
Mutagenesis of the BVDV DI9c nonstructural polyprotein. Mutations were introduced into the cDNA of BVDV DI9c as described in the text. The schematic drawing represents the DI9c NS polyprotein NS3-NS5B, which corresponds to amino acids 1599 to 3907 of the BVDV CP7 polyprotein (19). The individual NS proteins resulting from proteolytic processing of the polyprotein are depicted as different shaded boxes. The positions of insertions, deletions, and substitutions are marked by symbols as indicated at the bottom. The variant RNAs are denoted by italic numbers. The mutagenized conserved amino acid motifs in the protease and ATPase/RNA helicase domains of NS3 and the NS5B RdRp, respectively, are marked by thin vertical lines and denoted according to the nomenclature of Gorbalenya and Koonin (11) and Lai et al. (17). For further details, see Table 2.
TABLE 2.
BVDV DI9c ORF derivatives
| Mutant | Mutation type and positiona | No. of inserted or deleted residues |
|---|---|---|
| NS3 | ||
| Substitutions in the DEYH box [1919–1922] | ||
| 1ab | [1919] D→L | |
| 1bb | [1921] Y→T | |
| 1cb | [1921] Y→F | |
| 1db | [1921] Y→E | |
| Insertions | ||
| 2b | [1693] RV | 2 |
| 3b | [1834] TRQL | 4 |
| 4b | [1966] LTRQ | 4 |
| 5b | [2088] NTRV | 4 |
| 8b | [2099] H | 1 |
| 7b | [2191] DRRV | 4 |
| 8b | [2249] DRRV | 4 |
| Deletion | ||
| 9b | [2090–2099] | 10 |
| Substitutions for S1761 (serine protease) | ||
| 10ac | [1761] S→A | |
| 10b | [1761] S→T | |
| 10c | [1761] S→C | |
| NS4A | ||
| Insertions | ||
| 11 | [2309] DASI | 4 |
| NS4B | ||
| Insertions | ||
| 12 | [2361] HACA | 4 |
| 13 | [2377] TRRS | 4 |
| 14 | [2456] ETRL | 4 |
| 15 | [2668] HA | 2 |
| NS5A | ||
| Insertions | ||
| 16 | [2776] TRVC | 4 |
| 17 | [2955] TR | 2 |
| 18 | [3065] RRVD | 4 |
| 19 | [3114] TRVT | 4 |
| Deletions | ||
| 20 | [2738–2776] | 39 |
| 21 | [2776–2955] | 180 |
| NS5B | ||
| Insertions | ||
| 22 | [3259] TRGA | 4 |
| 23 | [3352] YAWS | 4 |
| 24 | [3379] DASI | 4 |
| 25 | [3545] RRVD | 4 |
| Deletion | ||
| 26 | [3428] | 1 |
| Substitutions in motif C [3635–3637] | ||
| 27d | [3636]D→T + [3637]D→R | |
| Cleavage sites | ||
| Substitutions at P1 and/or P1′ position | ||
| 28 | [2281] L→R(P1) | |
| 29 | [2345] L→R(P1) | |
| 30 | [2692] L→R(P1) | |
| 31 | [3188] L→D(P1) + [3189] S→P(P1′) | |
The numbering scheme refers to that of Fig. 1. Numbers in brackets correspond to the affected positions of the BVDV CP7 nonstructural polyprotein (26). Substitution and deletion mutations are denoted by the affected amino acid(s); insertions mutations are denoted by the preceding amino acid of the BVDV CP7 polyprotein. The number of inserted residues is indicated.
Construction described in reference 12.
Construction described in reference 4.
Construction described in reference 37.
Consequences of the various ORF mutations on the replication behavior of BVDV DI9c RNA in vivo.
Functional in vivo studies making use of BVDV replicons were previously determined to be most reproducible with BHK-21 cells (12 and 37). For that reason, we tested the different DI9c ORF derivatives predominantly in these cells. However, to permit infections with helper virus (see below), we evaluated each of the RNAs also in bovine MDBK cells, which, in contrast to BHK cells, are highly susceptible to infection with the parental virus.
The various transcripts were introduced into the cells via electroporation (applying protocols which yield in the average about 80 to 90% of replicon-containing cells), and RNA replication was assayed at different time points p.t. This was done by judging the replication-associated synthesis of NS3 via IF staining (not shown) or by quantifying the amounts of accumulating RNA replication products (negative-strand intermediate and/or progeny positive-strand viral RNA) in the cytoplasmic fraction of the cells by RPA or RT-PCR.
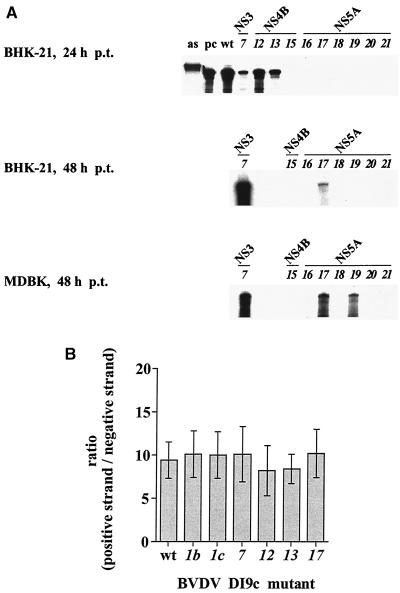
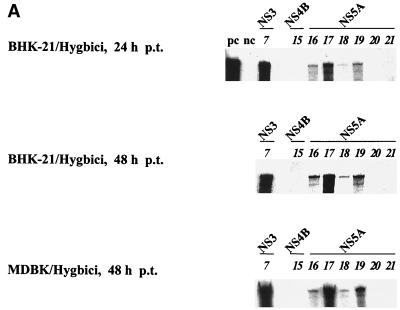
The results of five independent transcription-transfection experiments with all different ORF variants are summarized in Table 3. A representative RPA of transfection experiments with mutants (see below) is shown in Fig. 2A. Considering the protein-coding region as particularly sensitive to mutational changes (see Discussion), the vast majority of interventions were expected to result defective RNAs. In fact, besides three previously characterized DI9c NS3 derivatives (mutants 1b, 1c, and 7), only two NS4B mutants (12 and 13) and two NS5A mutants (17 and 19) were found to support both steps of the replication pathway (Fig. 2 and Table 3). Quantification of the amounts of replication products revealed the replication capacity of the DI9c NS4B RNAs (mutants 12 and 13) to be only moderately compromised and to correspond approximately 60% (mutant 12) or 40% (mutant 13) of that of the wild-type RNA. In contrast, intracellular multiplication of the functional replicons which contained alterations in the NS5A unit (mutants 17 and 19) was extremely low and detectable (by RPA or RT-PCR but not by IF) at only late time points p.t. (e.g., 48 h). Because the wild-type replicon induces a considerable cytopathogenic effect (CPE) at about 30 h p.t. (29), we compared the replication efficiency of mutants 17 and 19 with that of the NS3 mutant 7. Considering that the latter RNA replicates at approximately 10% of the wild-type level (see reference 12 and Fig. 5), the NS5A RNAs were hence estimated to display a replication competence of barely 0.5% (Fig. 2A). As in previous studies, the pattern of data obtained with the different DI9c mutants was found to be largely congruent in BHK and MDBK cells. However, replication of RNAs 17 and 19 was observed to be generally more efficient in MDBK cells, and in a number of transfection experiments with BHK cells, replication of mutant 19 was below the level of detection (Fig. 2; see also Fig. 7).
TABLE 3.
Functional analysis of BVDV DI9c ORF mutantsa
| Mutant | Replicationb | Processingc | Complementabilityd |
|---|---|---|---|
| NS3 | |||
| 1a | − | a | − |
| 1b | + | wt | − |
| 1c | + | wt | − |
| 1d | − | a | − |
| 2 | − | a | − |
| 3 | − | a | − |
| 4 | − | a | − |
| 5 | − | a | − |
| 6 | − | a | − |
| 7 | + | wt | − |
| 8 | − | a | − |
| 9 | − | a | − |
| 10a | − | a | − |
| 10b | − | a | − |
| 10c | − | a | − |
| NS4A | |||
| 11 | − | a | − |
| NS4B | |||
| 12 | + | wt | − |
| 13 | + | wt | − |
| 14 | − | wt | − |
| 15 | − | a | − |
| NS5A | |||
| 16 | − | wt | + |
| 17 | + | wt | + |
| 18 | − | wt | + |
| 19 | ± | wt | + |
| 20 | − | a | − |
| 21 | − | a | − |
| NS5B | |||
| 22 | − | a | − |
| 23 | − | wt | − |
| 24 | − | wt | − |
| 25 | − | wt | − |
| 26 | − | wt | − |
| 27 | − | wt | − |
| Cleavage sites | |||
| 28 | − | a | − |
| 29 | − | a | − |
| 30 | − | a | − |
| 31 | − | a | − |
All RNAs were generated by in vitro transcription and tested as described in the text. Importantly, the diverse DI9c ORF derivatives were confirmed by RPA and RT-PCR to exhibit the same stability as the wild-type replicon under the experimental conditions of the replication and translation assays (data not shown). RT-PCR on cytoplasmic RNA preparations of transfected cells followed by sequencing or restriction analysis of the resulting PCR products was performed to exclude reversion or contamination of mutant RNAs which were recovered as replication competent (for an example, see Fig. 7).
Summary of experimental results obtained during five independent transcription-transfection experiments with each of the diverse DI9c ORF mutants in BHK-21 and MDBK cells. −, replication deficient; +, replication capable. Except for mutant 19 (indicated as +/−), which was found to replicate more efficient in MDBK cells than in BHK cells (see text and Fig. 2), both cell types yielded congruent results.
Summary of five independent in vitro translation-processing experiments performed with each of the diverse DI9c derivatives as templates: “a” indicates that processing of the DI9c mutant-encoded polyprotein was significantly affected with respect to the cleavage profile obtained with the wild-type replicon; “wt” indicates that the mutation exhibited negligible effects on polyprotein cleavage. The overall translation efficiencies of the DI9c ORF mutants did not differ from that of the wild-type RNA (see text and Fig. 3).
Summary of the complementation analysis performed as described in the text. −, not complementable; +, complementable.
FIG. 2.
Replication abilities and ratios of positive-strand RNA to negative-strand RNA of BVDV DI9c ORF mutants. (A) DI9c ORF derivatives (the same set of mutants analyzed in Fig. 3, 5, 6, and 7) were transfected into BHK-21 or MDBK cells. Identical numbers of cells were harvested at the indicated time p.t., and the cytoplasmic fractions were analyzed for newly synthesized positive-strand viral RNA by a quantitative RPA with 32P-labeled antisense probe A (see Materials and Methods and Fig. 4A). The protected fragments were separated on a 5% polyacrylamide gel containing 7 M urea and quantitated by phosphorimaging. (Top) RPA carried out with cytoplasmic RNA of transfected BHK-21 cells at 24 h p.t. Lanes: as, aliquot of input antisense probe; pc, RPA performed on 300 ng of in vitro-transcribed DI9c cRNA (positive control); wt, RPA with cytoplasmic RNA from wild-type DI9c transfected cells; italic numbers, identical experiments carried out with DI9c ORF mutants 7, 12, 13, 15, and 16 to 21 (numbering scheme as in Fig. 1 and Table 2). Mutant 15 was applied as a negative control. The protein-coding regions of DI9c which contain the various mutations are indicated. By quantitative evaluation of five independent transcription-transfection experiments, the relative replication abilities of mutants 12 and 13 were determined to correspond 63% ± 12% and 44% ± 14% of that of the wild-type replicon, respectively. (Middle and bottom) RPA of DI9c derivatives 7 and 15 to 21 at 48 h after transfection into BHK-21 and MDBK cells. Mutant 7 (replicating at 10% ± 4% of the efficiency of the wild-type DI9c) was used as a reference in these experiments because this RNA does not induce a CPE at this time point (for a quantitative analysis, see Fig. 5 and 6). Note that except for two RNAs (1b and 1c), which were analyzed in detail in reference 12, the transfection experiments with all DI9c ORF derivatives listed in Fig. 1 and Table 2 yielded negative results, i.e., blank gels at 24 h as well as at later time points p.t. (these data are summarized in Table 3). The latter mutants were also negative if tested by RT-PCR (data not shown). (B) Using sense and antisense probes during RPA, the ratio of progeny positive-strand RNA to negative-strand RNA intermediate was measured with all replication-competent DI9c ORF derivatives (1b, 1c, 7, 12, 13, and 17) and compared with the wild-type (wt) value. Error bars indicate the mean deviations of five independent transcription-transfection experiments.
FIG. 5.
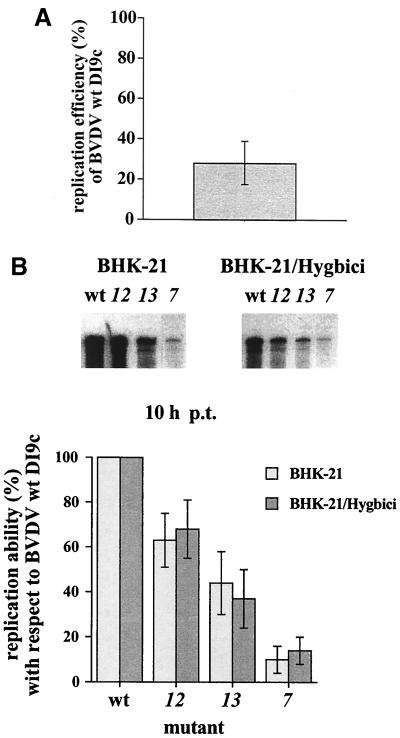
Complementation experiments with BHK-21/Hygbici cells to enhance the replication of the functional DI9c NS3 and NS4B mutants. (A) General replication competence of wild-type (wt) DI9c in BHK-21/Hygbici cells. Identical amounts of DI9c cRNA were transfected into BHK-21 cells and BHK-21/Hygbici cells. Equal numbers of cells were harvested at 10 h p.t., and identical amounts of the cytoplasmic fractions were analyzed by RPA using the DI9c-specific probe A (Fig. 4A and panel B). The major protected bands were quantified by analysis with a phosphorimager. The replication efficiency of DI9c RNA in Hygbici cells was calculated with respect to its replication competence in normal BHK cells (set as 100%). The average value of three independent transfection experiments is depicted as a column diagram; the mean deviation is indicated by the error bar. The same experiments performed at different time points (prior to the onset of a DI9c-induced CPE) and with MDBK cells yielded congruent results (data not shown). (B) Comparison of replication abilities of the functional mutants 7, 12, and 13 in BHK-21/Hygbici and normal cells. The wild-type DI9c replicon (wt) and the DI9c ORF mutants 7, 12, and 13 were transfected into Hygbici and normal cells. Equal numbers of cells were harvested at 10 h p.t., and identical amounts of the cytoplasmic fractions were analyzed by DI9c-specific RPA (see above). A representative RPA is shown at the top (mock controls yielded a blank gel and are not shown). The same experiments analyzed at different time points yielded congruent results. As indicated in panel A, replication of the DI9c derivatives was found to be generally lower in Hygbici cells. The signals obtained for the different replicons were quantified, and their relative replication abilities in each cell line were calculated with respect to the wild-type replicon (estimated as 100% replication competent). The average values of three independent transfection experiments are presented in the column diagram below. Mean deviations are indicated by error bars. Experiments with other functional DI9c NS3 derivatives (1b and 1c) yielded analogous results; i.e., a significant enhancement of the replication ability could not be observed in BHK-21/Hygbici cells (data not shown).
Interestingly, when we measured the amounts of replication products of the replication-competent DI9c ORF derivatives at different time points p.t., the ratio of positive-strand RNA to negative-strand RNA was found to be unchanged with regard to the wild-type value (Fig. 2B). Accordingly, synthesis of both replication products appeared to be simultaneously affected by the alterations in the NS3, NS4B, and NS5A units (see Discussion).
Effects of mutagenesis of the DI9c ORF on synthesis and cleavage of the replicon-encoded polyprotein.
Mutations affecting the DI9c ORF were generally envisaged to keep the IRES domain intact and thus expected to have no effect on translation of the viral RNA. However, for a correct interpretation of the complementation data described below, we needed to eliminate this possibility for each of the variant RNAs. Moreover, it was found important to assess the impact of the different mutations on the NS3/NS4A-mediated processing of the nonstructural polyprotein.
To address these issues, we used a suitable in vitro translation assay made up of cytoplasmic and initiation factor fractions of BHK-21 cells. This system was proven to promote synthesis as well as processing of the DI9c-encoded polyprotein in a manner which reflects the in vivo situation rather accurately (12, 37). As exemplified in Fig. 3, translation, if programmed with wild-type RNA, gives rise to a reproducible pattern of protein bands on SDS-PAGE. Side-by-side electrophoresis of individually translated cleavage products of the DI9c polyprotein (not shown) and mutagenesis of the different cleavage sites revealed that most of these bands could be identified as the mature NS proteins or certain intermediates of polyprotein proteolysis (Fig. 3; see also below).
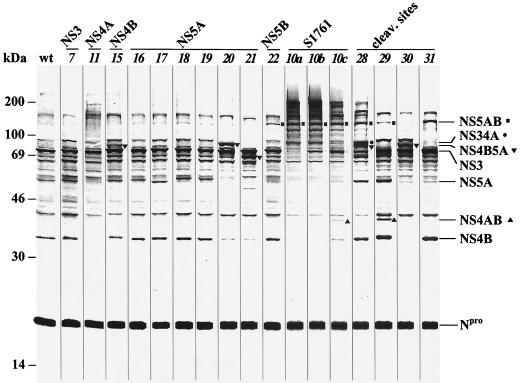
FIG. 3.
In vitro translation analysis of BVDV DI9c ORF mutants. In vitro translation reactions (see text) were programmed with in vitro-transcribed wild-type DI9c (wt) and a selection of mutant ORF RNAs (numbering refers to that in Fig. 1 and Table 2); the representative SDS-polyacrylamide gel shows the translation/processing data of those mutant RNAs which were further analyzed in Fig. 5 to 7. Also shown are data for RNAs which, besides the previously described NS3 mutants (see reference 12 and Table 3), were anomalous with respect to polyprotein processing (see text). As in Fig. 2, the mutagenized regions of the applied DI9c derivatives are indicated. Molecular masses of marker proteins (M) are given on the left; positions of some of the mature BVDV-encoded proteins and uncleaved precursor proteins are marked on the right (for a better orientation, NS5AB is additionally marked with a square, NS34A is marked with a dot, NS4B5A is marked with an inverse triangle, and NS4AB is marked with a triangle). The proteins were identified as described in the text and/or by calculation of molecular weights. As a consequence of deletions or insertions, the migration behavior of some of the mature proteins was found to be modified (for example, mutants 16 and 17). For quantification of translation efficiencies, the amounts of Npro protein were measured with a phosphorimager. In five independent translation assays, the mean deviation in the level of translation was determined to be approximately 10% for each RNA. Significant variations in the rate of protein synthesis were not observed with each of the different RNAs, irrespective of whether translation was performed for shorter or longer intervals (data not shown). Note that translation or proteolysis was found to be unaffected for a number of mutants which were not analyzed on this gel. These data are summarized in Table 3.
Five translation experiments based on independent extract and initiation factor isolates were performed to compare the synthesis rate and proteolysis of the polyprotein of the wild-type DI9c with those of the different ORF derivatives. By measuring the amount of the self-releasing Npro protein and/or NS3 as the most prominent processing products, the level of translation was established to be the same for the wild-type and all mutant ORF RNAs; slight variations in the quantity of the in vitro-translated viral proteins were confirmed to be causally related to experimental deviations rather than differences in the efficiency of protein synthesis from the various RNAs (a representative SDS-PAGE is shown in Fig. 3; all data are summarized in Table 3).
The translation experiments further revealed that the polyprotein cleavage pattern of several DI9c ORF mutants clearly differed from the profile obtained with the wild-type RNA. As expected, these included RNAs which expressed an inactive or less active NS3 protease (see mutants 10a to 10c in Fig. 3) as well as variants which encoded a modified protease cofactor NS4A (mutant 11) or polyproteins with altered cleavage sites (mutants 28 to 31). However, also insertions in the immediate neighborhood of cleavage sites gave rise to a dectable enrichment of precursor proteins (mutant 15, NS4B5A; mutant 22, NS5AB), and some deletion mutations had a similar effect; for example, mutations 20 and 21, where 39 and 180 amino acids, respectively, had been deleted in the NS5A protein, led to a significant accumulation of NS4B5A precursor (Fig. 3). The degree of cleavage inhibition was found to vary dramatically between different mutants. While proteolysis was nearly absent with mutant 10a, where the catalytic serine (S1761) of the NS3 protease had been replaced by alanine, slight extents of cis and trans cleavages were detectable if S1761 was replaced by threonine (10b) and cysteine (10c), respectively (Fig. 3 and reference 30). Similar tendencies were observed with other NS3 mutations (for details, see reference 12) and with the diverse cleavage site alterations. Mutants 28 (P1, NS3/NS4A) and 29 (P1, NS4A/4B) still enabled a certain degree of proteolysis at the mutated cleavage sites, as indicated by the release of mature NS3 and NS4B, respectively. In contrast, mutants 30 (P1, NS4B/NS5A) and 31 (P1/P1′, NS5A/NS5B) generated only NS4B5A and NS5AB but none of the mature proteins (Fig. 3). Remarkably, the entire spectrum of RNAs which exhibited a modified polyprotein cleavage profile were unable to replicate (Table 3). Proteolysis of the nonstructural precursor hence appears as a fine-tuned process which is highly susceptible to mutational interventions (see Discussion).
On the other hand, numerous RNAs with substitutions or insertions in the NS3 (1b, 1c, and 7), NS4B (12 to 14), NS5A (16 to 19), or NS5B (23 to 27) unit yielded polyprotein cleavage patterns which were virtually unaltered compared with that of the wild-type replicon (Fig. 3 and Table 3). We attribute the replication debility of these RNAs to the inactivation of either mature viral proteins or cleavage intermediates which are essentially involved in the formation of the active replication complex (see Discussion). Coordinated expression and processing of each of the DI9c-encoded nonstructural proteins NS3 to NS5B was thus indicated to be absolutely essential for catalysis of the pestivirus replication pathway.
Preparation of BVDV replicon-carrying cell lines.
Given that all mutations in the NS3 protein were suggested to be strictly cis dominant (12), it was interesting to test if other nonstructural proteins would behave similarly or if they could possibly be complemented in trans. The latter case implies that coexpression of intact viral proteins should rescue the replication ability of defective DI9c ORF derivatives and/or enhance the function of less effective RNAs, respectively.
As the conditions of coexpression were assumed to be of critical importance for the efficient replacement of defective components of the viral replication complex, we used a number of different experimental approaches in an effort to complement the diverse DI9c mutants. Remarkably, numerous initial attempts were not successful, though the diverse applied helper systems were confirmed by IF to provide significant amounts of the desired protein(s). Thus, complementation was never detectable during experiments where the mutant RNAs had been cotransfected with either wild-type DI9c, other ORF mutants, full-length BVDV genome, or Sindbis virus replicons expressing individual NS proteins. Negative results were also obtained if MDBK cells were infected with noncytopathogenic virus and subsequently transfected with the DI9c ORF variants (data not shown). As a promising alternative, we decided to set up cell lines which should persistently express BVDV RNA, because a similar system had been used successfully during complementation studies with the flavivirus Kunjin virus (14, 15). For this purpose, we used a bicistronic replicon construct which, in addition to the viral proteins NS2-3 to NS5B, encoded hygromycin B phosphotransferase, an enzyme that inactivates the translation inhibitor hygromycin B (Fig. 4A). In accordance with the fact that this RNA, hereafter referred to as the Hygbici replicon, expresses uncleaved NS2-3 protein, it was shown to be noncytopathogenic and to replicate in an episomal-like manner in transfected host cells (see reference (29) and the introduction). To ensure a maximum of potential complementation efficiency, a population of cells which homogeneously carry the Hygbici replicon should be raised. Accordingly, BHK and MDBK cells were transfected with the in vitro-transcribed Hygbici replicon RNA and continuously passaged in the presence of 500 μg of hygromycin B per ml of culture medium. As judged by IF and RPA analysis, a few splittings were sufficient to obtain cultures where virtually 100% of the cells contained the viral RNA (Fig. 4B; IF data not shown). Conversely, removal of the antibiotic led to a significant loss of the replicon after a couple of weeks (Fig. 4B). We concluded that the presence of the BVDV RNA is generally disadvantageous for the host cell but can be maintained under the constant pressure of an efficient selection marker. The bicistronic pestiviral noncytopathogenic replicon was thus demonstrated to represent a suitable tool for persistent expression of the viral nonstructural proteins as well as of a foreign protein (see Discussion).
Defective NS5A can be complemented in trans.
Next, we wanted to evaluate the utility of the established Hygbici cell lines to recover the function of cotransfected mutant DI9c ORF RNAs. To search for positive complementation, we devised an RPA method which was based on a riboprobe hybridizing with the transition region of the Npro and NS3 units of DI9c RNA. This probe was confirmed to permit the exclusive detection of DI9c RNA (and of all DI9c ORF derivatives) without cross-reacting with the Hygbici RNA. Conversely, employing a probe complementary to the hygromycin B-coding region, an appropriate screening assay was set up for the bicistronic RNA as well (see Materials and Methods, Fig. 4A, and below).
As a pilot experiment, we introduced wild-type DI9c RNA into BHK cells which contained the Hygbici replicon (BHK/Hygbici cells). Application of the differentiating RNase protection procedures revealed that replication of the monocistronic replicon and of the bicistronic, persisting replicon proceeded simultaneously in the transfected cells. This finding was confirmed by specific RT-PCR tests (see below). However, with respect to control experiments where we transfected an identical quantity of transcript into naive BHK cells, the overall amount of accumulating DI9c RNA was determined to be considerably lower in Hygbici cells (roughly 30% of that of the control [Fig. 5]). Associated with this observation, DI9c was found to reach its maximum level of replication at a rather early stage (ca. 12 h p.t.) and to induce CPE more rapidly than in normal cells (Fig. 6 and data not shown). We reasoned that these observations mirrored interference of the replication of both viral RNAs, a phenomenon which can be commonly observed during cotransfection or coinfection of related viral RNAs and which is suspected to be caused by competition of the catalytic complexes for a limited host factor and/or a certain cell compartment (12).
FIG. 6.
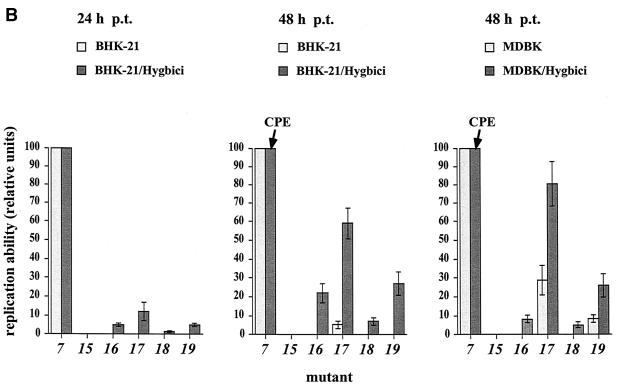
Complementation experiments with BHK-21/Hygbici and MDBK/Hygbici cells. (A) Replication abilities of a selected set of DI9c ORF derivatives (the same mutants as analyzed in Fig. 2) in BHK-21/Hygbici and MDBK/Hygbici cells. As in the previous experiments, mutant 7 was used as an internal, positive control (see text) and mutant 15 was applied as a negative control. The representative RPA experiments were performed at 24 h and/or 48 h after transfection of the indicated cell type. Identical numbers of cells were harvested, and aliquots of cytoplasmic RNA preparations were monitored with the DI9c-specific riboprobe (Fig. 4A) for de novo-synthesized positive-strand RNA. Lanes: pc, RPA with 200 ng of in vitro-synthesized DI9c cRNA (positive control); nc, RPA of the cytoplasmic fraction of mock-transfected Hygbici cells (negative control); italic numbers, identical experiments carried out with cells which had been transfected with the diverse ORF mutants (numbering scheme as described above). (B) Comparison of the replication abilities of mutant RNAs 7 and 16 to 19 in Hygbici and normal cells. RNAs 16 to 19, 7, and 15 (negative control) were transfected into naive BHK-21 and MDBK cells (Fig. 2) or BHK-21/Hygbici and MDBK/Hygbici cells (A). Identical amounts of cells and cytoplasmic extracts were analyzed by RPA at the indicated time points p.t.; the major protected bands were quantified by phosphorimager analysis. The relative replication abilities of mutants 16 to 19 were determined with respect to mutant 7, which served as a reference replicon (and set to 100 relative units of replication ability). The average values of three independent transcription-transfection experiments are depicted as a column diagram; error bars indicate mean deviations. Note that mutant 7 induced a slight CPE in both types of Hygbici cells at 48 h p.t. (as indicated by the arrow). Therefore, the replication capacities of mutants 16 to 19 could be quantified definitively at 24 h p.t. However, the 48-h values clearly show restoration and enhancement of the replication abilities of mutants 16 to 19 in BHK-21/Hygbici and MDBK/Hygbici cells, repectively.
Keeping these data in mind, we next tested the replication efficiency of DI9c ORF derivatives which were previously defined to be fairly replication competent in naive BHK cells in BHK/Hygbici cells, i.e., NS3 mutants 7 (exhibiting ca. 10% of wild-type activity) 1b (ca. 80%) and 1c (ca. 60%) and NS4B mutants 12 (ca. 60%) and 13 (ca. 40%) (Fig. 2). Due to the early onset of the DI9c-induced CPE in Hygbici cells, we quantified the amount of newly synthesized positive-strand RNA at time points between 8 and 12 h p.t. Within the limits of accuracy given by the detection system, the replication behavior of the DI9c NS3 and NS4B RNAs was thus determined to be not noticeably changed (i.e., enhanced) in Hygbici cells (Fig. 5; 1b and 1c data not shown). Identical results were obtained when we assayed the replication of the mutant RNAs at earlier or later time points p.t., though in the latter case it was, for the aforementioned reasons, necessary to standardize with mutant 7 RNA (data not shown). We deduced from these data that replication complexes which assembled a defective NS3 or NS4B component could not be functionally recovered.
This supposition was plainly strengthened when we tested also the remaining spectrum of mutants in BHK/Hygbici cells. As summarized in Table 3, DI9c variants with lethal alterations in the NS3, NS4A, NS4B, or NS5B region as well as RNAs which encoded modified polyprotein cleavage sites remained replication deficient in Hygbici cells (for example, mutant 15 in Fig. 6).
In contrast, replication of NS5A mutants 16, 18, and 19, which are inactive in parental BHK cells, was clearly restored if these RNAs were transfected into Hygbici cells. This becomes evident in the experiments shown in Fig. 6 and 7, where de novo synthesis of progeny positive-strand RNA molecules could be unambiguously verified and quantitated by either RPA or RT-PCR. Consistently, if we measured the replication ability of NS5A mutant 17 in Hygbici and normal cells, this RNA was found to amplify at least 10-fold more efficiently in the former cells (Fig. 6). Congruent results were obtained if the entire set of DI9c ORF variants was tested in MDBK/Hygbici cells. While all other RNAs were also defective in these cells (data not shown), replication of mutants 16 and 18 could be partially rescued and replication of mutants 17 and 19 was found to be significantly improved (Fig. 6).
To eliminate the possibility that the positive signals monitored with RNAs 16 to 19 (Fig. 6 and 7) were caused by reversion of the originally introduced alterations, the DNA fragments obtained during RT-PCR from cytoplasmic RNA preparations of transfected Hygbici cells were either sequenced (data not shown) or digested with a certain restriction enzyme. The latter control was enabled by the fact that engineering of each mutation was accompanied by introduction of an additional restriction site into the DI9c cDNA (see Materials and Methods and Table 1). As shown in Fig. 7, each mutation in the NS5A coding unit was thus ascertained to be retained in the course of the transfection experiments with Hygbici cells. This is consistent with the view that the monocistronic and bicistronic RNAs replicate concurrently in the transfected cells. Retention of the original mutations as well as the fact that transfection of Hygbici cells with replication-deficient DI9c variants never led to the appearance of novel replicating RNA species (for example, mutant 15 in Fig. 6 and 7) supported the conclusion that the above results could not be explained by RNA recombination events. We interpreted the finding that the deleterious effect of mutations 16 to 19 could be (at least partially) compensated for by a helper replicon in such a way that the putative function(s) of NS5A during the replication process can be provided in trans.
DISCUSSION
The purpose of this study was to gain initial information on the involvement of the different viral NS proteins in replication of the pestivirus genome. The discovery that RNA molecules encompassing essentially the UTRs and the coding units of NS3, NS4A, NS4B, NS5A, and NS5B operate as autonomous RNA replicons evidently demonstrated that this region of the ORF encodes all protein factors sufficient to catalyze the entire replication pathway of the viral genome (4). This observation raised several novel questions, the most obvious of which concerned whether all five units are also necessary for RNA replication and if the diverse proteins act in cis and/or in trans during assembly of the viral replication complex. To approach these issues, we pursued a strategy which in previous works proved successful for the identification and functional analysis of molecular determinants of the multiplication process of BVDV: a variety of mutations were introduced into the ORF region of the BVDV-derived DI9c replicon, and the effects of mutagenesis were evaluated by assay procedures that allowed us to measure the synthesis of positive- and negative-strand RNAs on the one hand and to monitor translation and processing of the NS polyprotein on the other hand (12, 37).
As an initial and generally expected result, all components that ought to be involved in the maturation process of the NS proteins were shown to be indispensable for both replication steps (see also below): besides an altered proteinase activity, also a mutated NS4A protease cofactor and modified cleavage sites were clearly lethal for the RNA replication process while leading to a more or less pronounced accumulation of precursor proteins (Fig. 3 and Table 3; see also reference 12). The fact that marginal changes of the molar ratio of the proteolytic cleavage products correlated with replication-deficient RNA phenotypes suggests that even slight differences of the kinetics of proteolysis interfere severely with the activity of the catalytic replication complex. Proteolytic maturation of the viral polyprotein was thus affirmed as a finely balanced process that is closely connected with the RNA replication pathway.
For the majority of the DI9c ORF variants, reduction or absense of RNA replication could not be attributed to an altered polyprotein cleavage profile (Fig. 3 and Table 3). Accordingly, the debility of these RNAs was explained either by the (intended) inactivation of the individual mature NS proteins and/or functional cleavage intermediates or by the (unintended) destruction of hypothetical cis-acting RNA elements which may be localized in the respective coding regions. The following indications clearly support the former notion, though it is impossible to exclude for each case that RNA signals were modified as well. (i) All interventions were demonstrated to have consequences for neither the translation rate nor the stability of the viral RNA molecules. (ii) Inhibition of RNA replication was observed with mutations that mapped at different parts of the protein-coding units (Table 3) and which were accordingly expected to affect different functional areas of the respective corresponding proteins. Along on this line, the replication deficiency of the NS3 and NS5B mutant 1b, 1c, or 27 could be directly traced back to a diminished helicase, ATPase, or RdRp activity, respectively, while, for example, mutant 7 was shown to affect an NS3 domain of yet unknown function (see reference 12; RdRp data not shown). (iii) The strongest evidence that proteins rather than RNA signals were inactivated derived from the complementation data showing that insertion mutations 16 to 19, which are located at different parts of the NS5A unit, could be all rescued in trans (Fig. 6 and see below). Taken together, these data favor the conclusion that catalysis of the RNA replication process requires the proper expression and structural integrity of all five mature NS proteins and of the associated enzymatic activities, respectively. Thus, NS4B and NS5A, the functional roles of which remain to be determined, are for the first time reported to be essentially involved in the pestiviral replication process.
A further interesting aspect of this work concerns the finding that with all replication-competent DI9c ORF mutants, the ratio of positive-strand to negative-strand RNAs was found to be the same as with the wild-type replicon (Fig. 2). Independently of the time point p.t. (at the earliest 5 h p.t.), synthesis of negative-strand intermediate could be measured only with the simultaneous synthesis of progeny positive-strand RNA by RNase protection or RT-PCR (the latter not shown). Considering also previous data (references 4, 12, 36, 37), we conclude that all functional DI9c mutants hitherto characterized behave identically in this respect; i.e., irrespective of whether mutations affected the protein-coding region or the UTRs, synthesis of negative- and positive-strand RNA was found to be symmetrically reduced. Two scenarios are conceivable to interpret this result. First, the initial amount of negative-strand RNA may be below the detection limit of RT-PCR (Fig. 7). Second and more likely, these data may indicate a fundamental difference between the multiplication strategy of pestiviruses (and possibly other related members of the Flaviviridae family) and that of picorna- and togaviruses, where both replication steps can be uncoupled (2, 18). The latter scenario would suggest that the majority of pestivirus-encoded components are essentially recruited at a very early stage of the replication pathway. This would be consistent with the cis- dominance of most ORF mutations and would substantiate the idea that the pestivirus replication complex represents a finely adjusted as well as a rather closed-up functional entity.
The most important results of this report derived from numerous experiments aimed at defining whether the BVDV DI9c-encoded proteins act in cis or in trans during RNA replication. From several tested helper systems, only cell lines which carried a noncytopathogenic replicon and continuously produced functional BVDV replication complexes were found to deliver viral proteins such that the function of cotransfected defective DI9c NS5A variants could be restored (Fig. 6). Importantly, this was observed with different cell types, and RNA recombination between the modified regions of the DI9c derivatives and homologous counterparts of the persistent Hygbici replicon could be largely excluded (Fig. 6 and 7). To this extent, our observations are compatible with data published for the Kunjin flavivirus (14, 15). However, in contrast with Kunjin virus, all mutations which modified the NTPase/helicase- or RdRp-coding regions of the BVDV RNA were found to be noncomplementable. Likewise, complementation was detected neither with the NS4B mutants nor with the entire set of RNAs exhibiting an altered polyprotein processing profile (Fig. 5 and Table 3). Thus, a broad spectrum of mutations (in the majority small insertions or single nucleotide exchanges) which affected different, discrete areas of the NS3, NS4A, NS4B, and NS5B proteins were determined to be cis dominant. Since all of these interventions kept the translational reading frame intact (Fig. 3), this phenomenon cannot be explained by a “cis-translation required region” as has been postulated for a portion of the poliovirus ORF (21). It is hence tempting to assume that these proteins and/or their respective precursors are functioning preferentially in cis. Though complementation assays performed differently might yield other results (for a detailed discussion of this issue, see reference 1), a constellation in which the majority of the BVDV nonstructural proteins including the key enzymes are operating in cis appears reasonable. For example, the inability to complement NS4B may be explained in such a way that the presumed hydrophobic interaction of this protein with membranes can occur only when it is nascent or newly synthesized. This could ensure a correct topology of the assembling replication complex. On the same line, cis activity of NS3, NS4A, and NS5B, which implies a restricted ability of the correctly folded proteins to associate with other RNA molecules than the original translation template, could ensure a privileged replication of functional viral genomes and partly compensate for the absence of a polymerase proofreading activity. Taken together, these data support the idea according to which translation, maturation, and activity of the BVDV nonstructural proteins are tightly coupled (see above) as a mode to coordinate the assembly of the functional viral replication machinery and to ensure its specificity and fidelity.
In contrast with the cis dominance of the above-described mutations, complementation was readily detectable with insertions in the NS5A coding region (16 to 19), which had a severe replication phenotype but no impact on the proteolysis of the polyprotein (Fig. 2 and 3). As illustrated in Fig. 6 and 7, complementation implied that the replication ability of nonfunctional mutants such as RNAs 16 and 18, which generated no signal in the RPA and RT-PCR (detection limit of the latter assay, 103 RNA molecules) at early as well as at late time points p.t., could be partially rescued and that the function of poorly replicating RNAs such as mutants 17 and 19 was clearly enhanced. Thus, besides its role as an obligate determinant of the pestivirus replication complex, the NS5A protein was demonstrated to be capable of operating in trans. Note that for the mentioned reasons it is currently impossible to distinguish whether this trans activity involves the mature NS5A and/or polyprotein intermediates or higher-order protein complexes containing the NS5A portion. The striking restrictedness of complementable mutations on the NS5A unit may argue for the mature protein as a trans-acting factor. Conversely, studies on the hyperphosphorylation of NS5A of the pestivirus-related HCV indicate the formation of an NS3-NS5A multisubunit complex, which presumably functions in trans (16, 20).
The ability of NS5A to act in trans suggests a particular role of this protein during RNA replication and/or other stages of the viral life cycle. A rather attractive hypothesis concerns the possibility that the protein exerts its function(s) through interaction with cellular factors. Thus, it may be involved in the recruitment of essential host components of the replication complex. Alternatively, NS5A might represent a regulatory element which, at a certain stage of the translation/maturation process, comes into contact with the cellular translation machinery to coordinate the switch from translation to replication. The fact that NS5A (or NS5) of different Flaviviridae members was found to be phosphorylated implies an interaction of the protein with cellular kinases (23). Though phosphorylation of the HCV NS5A is apparently not essential for RNA replication (5), this interesting biochemical trait, which could be directly related to the trans activity of the protein, may have implications for host range and/or pathogenic effects of viral infection (5, 9, 23). The establishment of a quick and reliable complementation system provides an important starting point as well as a powerful tool that will help to elucidate the biochemical functions of NS5A and to dissect as it participates in replication and/or viral pathogenesis.
ACKNOWLEDGMENTS
C.W.G. and O.I. were funded by the SFB 535 “Invasionsmechanismen und Replikationsstrategien von Krankheitserregern” from the Deutsche Forschungsgemeinschaft at Justus-Liebig-Universität Giessen.
We thank H.-J. Thiel for support.
REFERENCES
- 1.Agol V I, Paul A V, Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. doi: 10.1016/s0168-1702(99)00037-4. [DOI] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:269–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 3.Barton D J, Flanegan J B. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J Virol. 1993;67:822–831. doi: 10.1128/jvi.67.2.822-831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens S-E, Grassmann C W, Thiel H-J, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA-replicon. J Virol. 1998;72:2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight K J, Kolykhalov A A, Rice C M. Efficient initiation of HCV RNA replication in cell culture. Science. 2001;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 6.Chon S K, Perez D R, Donis R O. Genetic analysis of the internal ribosome entry segment of bovine viral diarrhea virus. Virology. 1998;251:370–382. doi: 10.1006/viro.1998.9425. [DOI] [PubMed] [Google Scholar]
- 7.Corapi W V, Donis R O, Dubovi E J. Monoclonal antibody analyses of cytopathic and noncytopathic viruses from fatal bovine viral diarrhea virus infections. J Virol. 1988;62:2823–2827. doi: 10.1128/jvi.62.8.2823-2827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corapi W V, Donis R O, Dubovi E J. Characterization of a panel of monoclonal antibodies and their use in the study of the antigenic diversity of bovine viral diarrhea virus. Am J Vet Res. 1990;51:1388–1394. [PubMed] [Google Scholar]
- 9.Gale M, Jr, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 10.Gong Y, Trowbridge R, Macnaughton T B, Westaway E G, Shannon A D, Gowans E J. Characterization of RNA synthesis during a one-step growth curve and of the replication mechanism of bovine viral diarrhoea virus. J Gen Virol. 1996;77:2729–2736. doi: 10.1099/0022-1317-77-11-2729. [DOI] [PubMed] [Google Scholar]
- 11.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 12.Grassmann C W, Isken O, Behrens S-E. Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J Virol. 1999;73:9196–9205. doi: 10.1128/jvi.73.11.9196-9205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada T, Tautz N, Thiel H-J. E2–p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J Virol. 2000;74:9498–9506. doi: 10.1128/jvi.74.20.9498-9506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh A A, Kenney M T, Westaway E G. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol. 1998;72:7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khromykh A A, Sedlak P L, Westaway E G. cis- and trans-acting elements in flavivirus RNA replication. J Virol. 2000;74:3253–3263. doi: 10.1128/jvi.74.7.3253-3263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch J O, Bartenschlager R. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J Virol. 1999;73:7138–7146. doi: 10.1128/jvi.73.9.7138-7146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai V C, Kao C C, Ferrari E, Park J, Uss A S, Wright-Minogue J, Hong Z, Lau J Y. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J Virol. 1999;73:10129–10136. doi: 10.1128/jvi.73.12.10129-10136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemm J A, Rümenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B M. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neddermann P, Clementi A, De Francesco R. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J Virol. 1999;73:9984–9991. doi: 10.1128/jvi.73.12.9984-9991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA-virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 22.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 23.Reed K E, Gorbalenya A E, Rice C M. The NS5A/NS5 proteins of viruses from three genera of the family flaviviridae are phosphorylated by associated serine/threonine kinases. J Virol. 1998;72:6199–6206. doi: 10.1128/jvi.72.7.6199-6206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 931–959. [Google Scholar]
- 25.Steffens S, Thiel H-J, Behrens S-E. The RNA-dependent RNA polymerases of different members of the Flaviviridae family exhibit similar properties under in vitro conditions. J Gen Virol. 1999;80:2583–2590. doi: 10.1099/0022-1317-80-10-2583. [DOI] [PubMed] [Google Scholar]
- 26.Tautz N, Meyers G, Stark R, Dubovi E J, Thiel H-J. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J Virol. 1996;70:7851–7858. doi: 10.1128/jvi.70.11.7851-7858.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tautz N, Meyers G, Thiel H-J. Pathogenesis of mucosal disease, a deadly disease of cattle caused by a pestivirus. Clin Diagn Virol. 1998;10:121–127. doi: 10.1016/s0928-0197(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 29.Tautz N, Harada T, Kaiser A, Rinck G, Behrens S-E, Thiel H-J. Establishment and characterization of cytopathogenic and noncytopathogenic pestivirus replicons. J Virol. 1999;73:9422–9432. doi: 10.1128/jvi.73.11.9422-9432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tautz N, Kaiser A, Thiel H-J. NS3 serine protease of bovine viral diarrhea virus: characterization of active site residues, NS4A cofactor domain, and protease-cofactor interactions. Virology. 2000;273:351–363. doi: 10.1006/viro.2000.0425. [DOI] [PubMed] [Google Scholar]
- 31.Thiel H-J, Plagemann P G W, Moenning V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 1059–1074. [Google Scholar]
- 32.Warrener P, Collett M S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiskerchen M, Belzer S K, Collett M S. Pestivirus gene expression: the first protein product of the bovine viral diarrhea virus large open reading frame, p20, possesses proteolytic activity. J Virol. 1991;65:4508–4514. doi: 10.1128/jvi.65.8.4508-4514.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiskerchen M, Collett M S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991;184:341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine protease: polyprotein cleavage sites, cofactor requirements and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H, Grassmann C W, Behrens S-E. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J Virol. 1999;73:3638–3648. doi: 10.1128/jvi.73.5.3638-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Isken O, Grassmann C W, Behrens S-E. A stem-loop motif formed by the immediate 5′ terminus of the bovine viral diarrhea virus genome modulates translation as well as replication of the viral RNA. J Virol. 2000;74:5825–5835. doi: 10.1128/jvi.74.13.5825-5835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong W, Gutshall L L, Del Veccio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]