Abstract
Gene expression in eukaryotes depends upon positioning, mobility and packaging of nucleosomes; thus, we need the detailed information of the human nucleosome core particle (NCP) structure, which could clarify chromatin properties. Here, we report the 2.5 Å crystal structure of a human NCP. The overall structure is similar to those of other NCPs reported previously. However, the DNA path of human NCP is remarkably different from that taken within other NCPs with an identical DNA sequence. A comparison of the structural parameters between human and Xenopus laevis DNA reveals that the DNA path of human NCP consecutively shifts by 1 bp in the regions of superhelix axis location −5.0 to −2.0 and 5.0 to 7.0. This alteration of the human DNA path is caused predominantly by tight DNA–DNA contacts within the crystal. It is also likely that the conformational change in the human H2B tail induces the local alteration of the DNA path. In human NCP, the region with the altered DNA path lacks Mn2+ ions and the B-factors of the DNA phosphate groups are substantially high. Therefore, in contrast to the histone octamer, the nucleosomal DNA is sufficiently flexible and mobile and can undergo drastic conformational changes, depending upon the environment.
INTRODUCTION
DNA in eukaryotic nuclei is organized in a hierarchical scheme of folding and compaction into protein–DNA assemblies, collectively called chromatin (1–3). Genetic processes, such as transcription and replication, depend on chromatin structures, which affect the DNA transaction activity (4–9). Therefore, chromatin structures play a central role in the regulation of the gene expression processes in nuclei. At the lowest level of organization, two copies of each of the four histone proteins, H2A, H2B, H3 and H4, are assembled into an octamer, which is wrapped by two tight superhelical turns of 145–147 bp of DNA to form a nucleosome core particle (NCP) (10). In higher eukaryotes, the addition of the linker histone H1 to the linker DNA forms nucleosomes, corresponding to the basic repeating unit of chromatin.
High-resolution structures of NCPs from different organisms, such as Xenopus laevis (11,12), chicken (13) and yeast (14), are already available. The DNA-dependent divalent cation binding in an NCP of X.laevis (Xla-NCP) was investigated in detail (15). Recently, the structural geometry of the nucleosomal DNA has been evaluated in the Xla-NCP structure at 1.9 Å resolution (16). The overall NCP structures are very similar to each other among the available ones. The histone sequences are highly conserved among eukaryotic organisms (17). However, the crystal structure of a yeast NCP revealed the significant differences in the internucleosome interactions, as compared with those of other NCPs (14). This fact prompts us to investigate the details in the human NCP structure, which may allow us to clarify the differences from other NCPs.
Although the organization of DNA into nucleosomes, which is essentially ubiquitous among genomes, generally results in gene expression control in a repressive manner, its certain modes also contribute to gene-specific transcription. This suggests that nucleosome positioning in gene promoters is crucial for transcriptional regulation in vivo (18–21). The question, therefore, arises as to how the chromatin structure, leading to DNA compaction, permits the site-specific access to regulatory factors and the more extensive exposure to transcription proteins. To answer this question, we need the detailed conformational view of the nucleosomal DNA, which could contribute toward an understanding of basic chromatin properties, such as nucleosome positioning and mobility.
Here, we report the 2.5 Å resolution crystal structure of human NCP, which facilitates a comparison with those of other NCPs determined so far. The overall structures of all of these NCPs are very similar, in agreement with the minor sequence differences between these histones. However, the path of the nucleosomal DNA in human NCP is different from that within Xla-NCP (11), in spite of their identical 146 bp sequences.
MATERIALS AND METHODS
Expression, purification and reconstitution of human NCP
The four core histone genes, forming a cluster (22,23), were amplified by PCR using genomic DNA obtained from HeLa cells. Vectors for the expression of all four recombinant human core histone proteins were constructed by inserting the gene fragments into pET-22b (Novagen). The histone genes in each of the resulting plasmids were sequenced to confirm their integrity. The histones were overexpressed in BL21 (DE3) codonplus RIL (Stratagene) and were purified using the protocols published previously (24,25). The histone proteins were refolded into a histone octamer. The pUC-based vector, a kind gift from Dr K. Luger, contains one-half of a 146 bp palindromic DNA fragment derived from a human α-satellite region (12). The 146 bp palindromic DNA fragment was constructed from this vector and purified as described previously (24,25). The histone octamers were reconstituted into NCPs using the 146 bp palindromic DNA fragment (24,25). Milligram amounts of human NCPs were subjected to heat shifting, followed by subsequent purification using anion exchange chromatography on a Mini Q column (Amersham Biosciences).
Crystallographic procedures
Crystals of a human NCP were obtained by vapor diffusion, at a protein concentration of 4 mg/ml with 55–58 mM KCl, 70–80 mM MnCl2 and 20 mM potassium cacodylate, pH 6.0 in a drop equilibrated against 35 mM KCl, 40–45 mM MnCl2 and 20 mM potassium cacodylate, pH 6.0. The crystals were transferred into 37 mM MnCl2, 40 mM KCl, 20 mM potassium cacodylate, pH 6.0, 28% (v/v) 2-methyl-2,4-pentanediol and 2% (w/v) trehalose for cryoprotection and improvement of the diffraction resolution. Crystals were flash-frozen in liquid nitrogen directly from the cryoprotectant condition. X-ray diffraction data were collected at the synchrotron radiation source at BL41XU (SPring-8, Hyogo, Japan), at 100 K using a nitrogen stream. Processing of the diffraction images and scaling of the integrated intensities were carried out using the program HKL2000 (26).
Molecular replacement, with the PDB entry 1AOI (12) as the search model, was used to obtain the initial phases via Amore (27). Refinement was performed using CNS (28), and model building into the 2Fo − Fc and Fo − Fc electron density maps was carried out by the use of program O (29). Mn2+ and Cl− ions were initially located in the Fo − Fc electron density maps as solvent peaks (>3σ) with the surrounding molecular environments accommodating one or more potential hydrogen bonds to the peak. These ions were identified as described in a following section. DNA base-pair-step parameters were calculated using the program curves (30). Graphic figures were prepared using the program UCSF chimera (31). The coordinates and structure factors have been deposited in the PDB under ID codes 2CV5.
RESULTS AND DISCUSSION
Overall structure
A human NCP was crystallized under conditions similar to those previously published for the crystal structure of a Xla-NCP (11,12). The human crystal showed the same orthorhombic space group as that of the X.laevis crystal (Table 1). However, the unit cell parameters were substantially different from those of Xla-NCP, with an exchange of the b- and c-axes and a 13% reduction of the unit cell volume, implying a denser packing than that in the Xla-NCP crystal (11,12). The crystal structure was determined by the molecular replacement method, using the previously published Xla-NCP structure (PDB entry 1AOI) (12) as a search model. Data collection and final refinement statistics are summarized in Table 1. Sequence differences between the human and X.laevis histones were discernible in the original 2Fo − Fc electron density map (data not shown).
Table 1.
Data and refinement statistics
| Data collection statistics | |
| Wavelength (Å) | 1.0 |
| Space group | P212121 |
| Unit cell parameters (Å) | a = 99.6, b = 108.4, c = 169.4 |
| No. of unique reflections | 61 700 |
| Resolution (Å)a | 50–2.50 (2.59–2.50) |
| Rmergea,b | 7.8 (49.5) |
| Completeness (%)a | 96.0 (84.5) |
| I/σa | 28.8 (3.3) |
| Refinement statistics | |
| Number of protein residues in the final modelc | 761 |
| Number of base pairs in the DNA | 146 |
| Number of water molecules | 430 |
| Number of Mn ions | 9 |
| Number of Cl ions | 4 |
| Total number of atoms in the final model | 12 475 |
| Rwork/Rfree (%)d | 22.38/27.69 |
| R.m.s.d from ideality | |
| Bond (Å) | 0.0062 |
| Angles (°) | 1.14 |
| Average B-factors (Å2) | |
| Protein | 28.91 |
| DNA | 51.80 |
| Solvent | 33.36 |
aValues in the parentheses are for the final shell.
b, where Ihkl is the intensity measurement for the reflection with indices hkl, and 〈Ihkl〉 is the mean intensity for multiply recorded reflections.
cResidues included in each histone subunit: H3: 38–134, H3′: 37–135, H4: 25–102, H4′: 18–102, H2A: 11–118, H2A′: 15–118, H2B: 30–125, H2B′: 31–124. The remaining portions of the histone tails were too disordered to be included in the final model.
d, where the R-factors were calculated using the working and free reflection sets, respectively. The free reflections comprise a random 10% of the data reserved for unbiased cross-validation throughout the refinement.
Amino acid sequence alignments between the human and X.laevis histones revealed that H2A, H2B and H3 are slightly divergent (92.1, 93.6 and 98.5% identity, respectively), whereas H4 is identical between the two species. This degree of sequence divergence is comparable with that between the primary structures of the human and chicken histones. In agreement with this minor difference, the overall structure of a human NCP (Figure 1) is very similar to those of Xla-NCP (11,12) and chicken (13) NCP. The global structure is also similar to that from yeast (14), although the sequence identity is ∼60–80% between the two species. Therefore, the basic NCP structure is well conserved from yeast to human, despite the considerable differences in the chromatin organization between yeast and higher eukaryotes (32–34).
Figure 1.
The overall structure of a human NCP. The upper ribbon model is the crystal structure of a human NCP. The arrow above NCP indicates a pseudo 2-fold axis passing through the center of the structure. The bottom figure displays the axial view of the particle. Histone chains are colored blue for H3, green for H4, yellow for H2A and red for H2B. The DNA is shown in ribbon traces for the 146 bp DNA phosphodiester backbones (brown for the I chain and forest green for the J chain). The Mn2+ and Cl− ions are depicted by cyan and silver balls, respectively.
The histone octameric structures of human NCP and other NCPs [2.0 Å resolution structure of Xla-NCP (PDB code 1KX3) (11), yeast NCP (PDB code 1ID3) (14) and chicken NCP (PDB code 1EQZ) (13)] were compared by superimposing residues (H3: 38–134, H3′: 38–135, H4: 25–102, H4′: 18–102, H2A: 14–118, H2A′: 15–118, H2B: 32–125, H2B′: 31–124) in each human histone subunit onto their equivalents from other species; thus, an overall root-mean-square deviation (r.m.s.d.) was estimated to be as small as 1.3 Å (1.30, 1.29 and 1.41 Å for the above NCPs, respectively). However, the corresponding DNA duplexes, wrapping the histone octamers, exhibit substantial discrepancies with an r.m.s.d. of ∼3.0 Å (3.04, 2.92 and 2.98 Å, respectively), in spite of their completely identical DNA sequences (11,13,14). The detailed structures of the histone–DNA interactions are also conserved from yeast to human, including the participation of identical amino acids. Notably, the path of the nucleosomal DNA, coiling around the histone octamer, is dramatically altered in human NCP as compared with those of other organisms.
Alteration of the nucleosomal DNA path
The two 73 bp DNA halves (−72 to 0 and 1 to 73, see Figure 2A) of the superhelix in the crystal structure of a human NCP adopted remarkably different structures, with an r.m.s.d. value of 4.19 Å (all atoms). This value is substantially larger than those of other NCP structures, such as 2.55 Å for Xla-NCP (PDB code 1KX3) (11), 2.44 for yeast NCP (PDB code 1ID3) (14) and 2.67 for chicken NCP (PDB code 1EQZ) (13), demonstrating that the human nucleosomal DNA takes a different path. The superposition of 18 or 19 bp DNA segments between the human and X.laevis structures allowed us to identify the local conformational change in human NCP. As shown in Figure 2A, two segments (−53 to −18 and 37 to 73, see the legend) present strikingly higher r.m.s.d. values than the others, indicating that the human nucleosomal DNA exhibits a local change in comparison with that of Xla-NCP.
Figure 2.
Comparison of the DNA parameters between human and X.laevis NCPs. (A) B-factors of phosphates, metal binding sites, r.m.s.d. values and twist base-pair-step parameter of the 146 bp DNA compared between human and X.laevis NCPs. The base pair numbers (−72 to 73) and SHL (−7 to 7) labels are the same as those in the earlier structures (11,12). The primary bound-phosphate groups are indicated above the base sequence by the pointers showing the strand direction (dark, 3′→5′; light, 5′→3′) and the interacting histone motif (L1, loop1; L2, loop 2; A1, α1). B-factors (Å2) are plotted for the 5′ phosphate group of each base (brown for the I chain and forest green for the J chain). Mn2+ binding sites are indicated above the base sequence by the yellow and light blue arrows (solid, binding the DNA major groove; broken, binding the DNA minor groove) in the structures of human and X.laevis NCPs (PDB code 1KX3) (11), respectively. The r.m.s.d. values below the base sequence were calculated by the superposition of 18 or 19 bp between two NCPs. The twist base-pair-step parameters were plotted as open circles and closed squares for the human (yellow line) and X.laevis (light blue line) nucleosomal DNA, respectively. The pink, orange and green ellipsoids of the twist parameters represent the corresponding regions illustrated by those in Figures 3A and B and Figure 4A. (B–D) The simplified stereo model of the DNA duplexes in the regions surrounded by the ellipsoids in (A). The differences in helical twist are shown by the superposition between human (yellow) and X.laevis (light blue; PDB code 1KX3) (11) NCPs, based on the histone octamers alone. Numbers presented for each base pair are the same as those in Figure 2A.
The bending behavior of the nucleosomal DNA path in Xla-NCP was successfully evaluated by comparing its base-pair-step parameters (roll, tilt, shift, slide and twist) with the ideal superhelical DNA (16). According to the same computing procedure, we compared the bending behaviors of the nucleosomal DNA path between human NCP and Xla-NCP, and thereby revealed their structural discrepancies at the base pair level. The roll parameter is almost equivalent between two NCPs, although slight differences exist in two segments (−33 to −21 and 52 to 61), which belong to each region of the altered DNA path (data not shown). The tilt parameter is remarkably different in entire DNA regions (data not shown). The shift, slide and twist parameters of human NCP indicate that two segments (−53 to −18 and 37 to 73) are quite different from those of Xla-NCP (Figure 2A). Particularly, the twist parameters provide critical information about the alteration of the supercoiling DNA paths. In human NCP, superhelix axis location (SHL) −5 (−57 to −46; Figures 2A and 3A, pink ellipsoid and circle) and SHL 5 (46 to 57; Figures 2A and 3B, green ellipsoid and circle) exhibit twists of 9.8 and 9.5 bp per turn, respectively. In contrast, the corresponding values in Xla-NCP are 10.7 and 10.6 bp per turn, respectively. This result indicates that the human DNA path has a much tighter twist than the X.laevis DNA path, and thus the human base pairs are shifted by 1 bp to the 3′-side of the I chain (Figure 2B and D). On the other hand, the twist of SHL −2 (−27 to −16; Figures 2A and 3A, orange ellipsoid and circle) is 10.8 bp per turn, while Xla-NCP shows 9.7 bp per turn. Therefore, in this region, the human DNA path is looser, by 1 bp, in the twist than the X.laevis DNA path (Figure 2C). This indicates that 1 bp shifts, caused by the tighter twist around SHL −5 and SHL 5, were compensated by the looser twist around SHL −2 and 1 bp extension around SHL 7 at the terminus, respectively. These consecutive base pair shifts can essentially account for the high r.m.s.d. values and the parameter differences between human and X.laevis NCPs. Similar differences in the DNA pathway were also observed in comparison with the NCP structures from chicken (13) and yeast (14) (data not shown).
Figure 3.
Comparison of the DNA structures between human and X.laevis NCPs. The best superposition between human (yellow) and X.laevis (light blue; PDB code 1KX3) (11) NCPs, based on the histone octamers alone, reveals the discrepancy in the DNA structures. (A and B) Partial NCP structure, viewed as the upper display in Figure 1, but showing only 77 bp and associated proteins. The black arrows indicate the directions and ranges of the consecutive 1 bp shifts to the 3′-side, in comparison with the DNA in Xla-NCP. The pink, orange and green circles correspond to those in Figures 2A and 4A. Each SHL label of −1 to −7 or 1 to 7 represents one further DNA double helix turn from SHL0, which is located at the central base pair numbering as zero in Figure 2A. The close-up inset indicates a H2B tail, whose positional change may induce the local alteration of the DNA path. In the close-up stereo view, all of the atoms of a human NCP are shown as normal sticks, where the phosphorus, oxygen and nitrogen atoms are colored cyan, red and blue, respectively, and all of the Xla-NCP atoms are shown as light blue sticks. (C) The side stereo view, corresponding to the lower display in Figure 1, but representing the superhelical DNA pathway alone. The broken ellipsoids, shadowed yellow and light blue, represent the space between two DNA duplexes within human and X.laevis NCPs, respectively. The black arrows indicate the expanding distance between the same sugar–phosphate backbones of two NCPs.
The regions around SHL −5, −2 and 5 do not contact the histones, and their conformations appear to be relatively susceptible to the environment. Thus, it is not surprising that the alteration of the human nucleosomal DNA path is concentrated within these regions. However, the B-factors of the DNA backbones at SHL −5 and 5 are lower in human NCP than those of NCPs from other species, including X.laevis (Figure 2A). This suggests that some factors constrain the local alterations in these regions of the human nucleosomal DNA. Conversely, the B-factors at SHL −2 have higher values than the other regions.
The 1 bp shift in the two DNA regions (SHL −5 to −2, SHL 5 to 7; black arrows in Figure 3A and B) is clearly illustrated by the best superposition of the histone octameric structures alone between human and X.laevis NCPs. In the altered region (SHL −5 to −2) of human NCP, the phosphate groups, bound to the histones, are also shifted by 1 bp to the 3′-side in comparison with that of Xla-NCP (Figures 2A and 3A). However, the interactions of the DNA backbones with the histones are invariant, as revealed from the involvement of the DNA-binding motifs of α1 in H2B, α1 in H2A, loop1 in H2A, loop2 in H2B, loop2 in H4 and loop1 in H3. Similarly, the phosphate groups in the SHL 5 to 7 region move to the 3′-side, while maintaining the same binding modes by loop1 in H2B, loop2 in H2A and αN and tail in H3 (Figures 2A and 3B).
Crystal packing
A notable difference between the human and X.laevis NCP crystal structures is the molecular arrangement (Figure 4), which is associated with the tighter packing within the human crystal. As reported previously (35), the packing of nucleosomes within a crystal does not necessarily reflect the molecular arrangements in the condensed chromatin. However, it is still interesting to examine the packing differences in terms of the internucleosome positioning. For instance, the Xla-NCP structure revealed that the histone H4 tail contacts H2A/H2B of a neighboring particle (11,12). A recent study, using disulfide cross-linking between these histones, found that the chromatin fiber comprises two stacks of nucleosomes, in accord with the two-start model (36).
Figure 4.
Crystal packing of human NCP. (A) The upper stereo model shows the human NCP crystal packing, viewed approximately perpendicular to the crystallographic c-axis. The bottom figure displays the side view of the same molecular arrangements. Short arrows show the approximate locations of the three crystallographic axes. Only the DNA backbone is shown for clarity. The pink, orange and green circles are equivalent with the regions illustrated by those in Figures 2A and Figures 3A and B. The black arrows indicate the directions and ranges of the consecutive 1 bp shifts to the 3′-side. (B–D) The close-up stereo views of the boxed area in (A) show the discrepancy in the DNA structures by the superposition between human (yellow) and X.laevis (light blue; PDB code 1KX3) (11) NCPs, based on the histone octamers alone. The gray arrows represent the modification of the DNA pathway by the inter-particle collides.
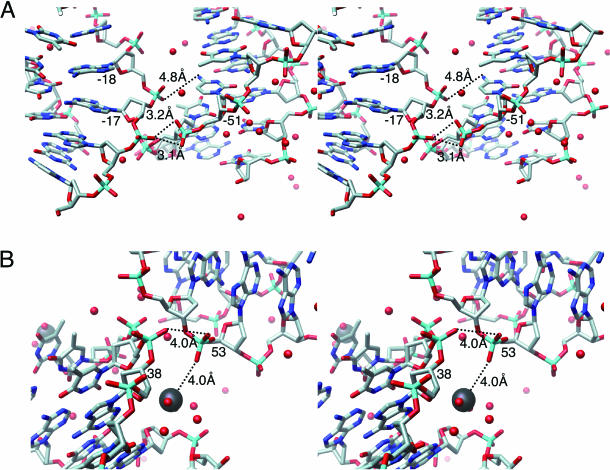
It is thus interesting to investigate human NCP arrangements in the crystal lattice in comparison with other crystals. Remarkable differences were observed in the DNA–DNA contacts between human NCPs. The DNA duplex at SHL −2 (Figure 4A, orange circles) collides with the DNA at SHL −5 (Figure 4A, pink circles) of the adjacent particle. This collision causes the tighter and looser twist in each DNA path (Figure 4B and C, respectively). In detail, it is likely that this inter-particle contacts occur through water-mediate interactions between the cytosine base at the location −51 and the phosphate group at the location −18 and between the two phosphate groups at the location −17 and −51 (Figure 5A). The similar groove–backbone interaction was observed in the crystal packing of DNA duplex dodecamers (37,38). In this packing, the duplexes were adjusted to lie close together by the reciprocal fit of the sugar–phosphate backbone into the major groove. Therefore, it is also likely that the self-fitting of DNA molecules by these interactions modifies the DNA structure within the tighter packing of human NCP.
Figure 5.
Details of the DNA–DNA contact within the human NCP crystal. (A) Detailed stereo view of the DNA–DNA contact shown in Figure 4B and C. (B) Detailed stereo view of the DNA–cation–DNA contact shown in Figure 4D. All of the atoms of the DNA duplexes are shown as normal sticks, where the phosphorus, oxygen and nitrogen atoms are colored cyan, red and blue, respectively. The Mn2+ ions and the water molecules are depicted by black and red balls, respectively. Broken lines and distances between two atoms show possible interactions through water molecules. Numbers presented for each base pair are the same as those in Figure 2A.
This inter-particle contact may simultaneously generate the sequential 1 bp shift, which ranges from SHL −5 to −2 (Figure 4A, black arrows). This alteration of the human DNA path expands by ∼2 Å between two DNA duplexes, wrapping around the histone octamer, as compared with that of Xla-NCP (Figure 3C). The resultant space accommodates a short segment of a DNA strand in the adjacent NCP, because of the tighter crystal packing (Figure 4A, green circle). This partial insertion of DNA also generates the tighter twist in the DNA path at SHL 5 of the neighboring NCP (Figure 4D) and another consecutive 1 bp shift from SHL 5 to 7 at the terminus (Figure 4A, black arrow). In detail, it is likely that the intermolecular contacts occur through water-mediate interactions between the Mn2+ ion bound to the minor groove and the phosphate group at the location 53 and between the two phosphate groups at the location 38 and 53 (Figure 5B). The DNA–cation–DNA contact may modify the DNA structure by the reciprocal fit of the sugar–phosphate backbone into the minor groove. Although experimental data are unavailable at the moment, a similar change in the DNA path may take place in vivo, coupled with the contacts between NCPs in the compacted state of human chromatin.
Effects of histone sequence differences
The amino acids of human histones that differ from those of X.laevis homologs are located within the common octameric core structure. Notably, only Ala116 in H2A lies on the molecular surface, and it is too far away to interact with the neighboring NCP. Ser38 and Val39 in H2B are close to the histone–DNA contact region, but do not appear to participate in the contact. Then, what causes the alteration of the DNA path and the uniquely compact packing in human NCPs? It is likely that Lys30, Arg31 and Ser32 in the H2B tail (triplet residues: Arg30, Lys31 and Thr32 in Xla-H2B) perturb the DNA path. In fact, the superposition between human and X.laevis NCPs revealed that human Arg 31 structurally clashes with the DNA backbone in Xla-NCP (SHL −5; Figure 3A, stereo view). In the symmetric part of the human H2B tail (SHL 5; Figure 3B, stereo view), the DNA path is also altered by the steric hindrance between the DNA and the H2B tail, which has a different conformation from that of the other H2B tail.
Notably, the corresponding triplets in the H2B tail (Lys30, Arg31 and Ser32 in human; Arg30, Lys31 and Thr32 in X.laevis; Ser32, Lys33 and Val34 in yeast; and Lys30, Lys31 and Ser32 in chicken) occupy identical positions in the 3D structures, although the equivalent H2B segment does not affect the nucleosomal DNA path in either chicken (13) or yeast (14). However, the triplet residues are highly conserved among the human H2B variants (17). In addition, it has been shown that the deletion of the yeast H2B tail segment, containing the triplet residues, significantly decreases plasmid superhelical density, suggesting a release from the DNA constraints within the nucleosome (39). It is thus possible that the conformational change in the histone tail locally perturbs the DNA path, although it is unlikely to generate the entire alteration around SHL −5 to −2 and 5 to 7 (Figure 3).
Ion-binding sites
Specific ion-binding sites in the protein and DNA structures were identified from the residual peaks (>3σ) in the Fo − Fc difference maps, with information on the composition and the metal ion coordination geometry. This approach allowed us to identify 13 bound ions in a human NCP. Five are bound to the protein alone and eight are associated with the DNA. One peak corresponds to a Mn2+ ion bound between particles in the Xla-NCP crystal structure (11). Therefore, this peak was identified as a Mn2+ ion, which is coordinated by the carboxylic oxygen of H3-D77 from one particle, the peptide carbonyl oxygen of H2B-V45 from the other and two water molecules. Four chloride ions were identified, as two peaks of the H3-α3 helices near the center of the DNA and two peaks of the H2B-α3 helix and H2A-α2 helix, in agreement with the Xla-NCP structure at 1.9 Å resolution (PDB code 1KX5) (11).
Eight peaks observed in the DNA region could be identified as Mn2+ ions, because no other DNA-bound metal ions were present in the crystallization solution, except for potassium ions. In addition, the distances of the metal ions to the ligands ranged from 2.24 to 2.89 Å, which are much shorter than the minimal K+ to ligand distance of 2.9 Å observed in oligonucleotide DNA (40). The seven Mn2+ ions bound in the NCP-DNA major groove interacted exclusively with the N7 and O6 atoms of the guanines at GG, GC and GA steps, either directly or via the water molecules in their coordination shell. Of these sites, six at the base pair locations of −60, −47, 3, 35, 48 and 61 were the same binding sites (Figure 2A) as those in the 2.0 Å structure of Xla-NCP (PDB code 1KX3) (11). The single remaining Mn2+ binding site, at the base pair location of −5, was observed in the 1.9 Å structure of Xla-NCP (PDB code 1KX5) (11,15). However, in comparison with Xla-NCP, the positions of the two Mn2+ ions (at −47 and 61) were shifted, presumably because of coupling with the alteration of the DNA path.
The four Mn2+ binding sites at −34, −26, 27 and 65, which were fully occupied by the metal in Xla-NCP (11), were vacant in the human NCP structure. Three of them (at −34, −26 and 65) are located in the altered DNA path (Figure 2A). Metal ions bound to DNA generally contribute to the stabilization of the preexisting conformation (41,42). Consistently, in the region of −37 to −18, which lacks Mn2+ ions in human NCP, the DNA phosphates have substantially high B-factors (Figure 2A). This instability may be connected with the alteration of the DNA path.
A unique Mn2+ ion binding site was found in the minor groove at only one location 40 in the entire human NCP structure (Figure 2A). A similar Mn2+ binding mode in the minor groove was observed at different base pair position 13 in the Xla-NCP structure (11,15). This Mn2+ ion in an Xla-NCP is bound between the minor groove and the H2B-αC helix belonging to the neighboring NCP (15). As described above, the Mn2+ ion in a human NCP is very close to the phosphate group at the location 53 between adjacent NCPs (Figure 5B). Internucleosomal histone–cation–DNA or DNA–cation–DNA interactions, as observed previously (15,43), may mediate chromatin compaction. This DNA–cation–DNA contact probably contributes to the tight crystal packing of a human NCP.
CONCLUSION
We have determined the crystal structure of a human NCP at 2.5 Å resolution. Our findings confirm that the overall NCP structures are conserved between human and other eukaryotes, in spite of the considerable differences in the chromatin organization between the eukaryotes (32–34). However, the path of the nucleosomal DNA exhibits a remarkable alteration in the human NCP crystal structure. This alteration appears to be caused by a combination of several factors, such as the conformation of a H2B tail, the conversion of metal binding sites and the different crystal packing, suggesting that the nucleosomal DNA conformation is more flexible and sensitive to the environment than we expected.
The extensive alteration of the human DNA path induces the space expansion between two DNA duplexes within a single NCP structure. The post-translational modification of the histone tail may additionally contribute to the further expansion. It is intriguing to envisage that the resultant space is linked to interactions with some activators and chromatin remodeling factors. The demonstration of this hypothesis awaits further studies, such as the crystal structure determinations of NCPs complexed with some factors. However, it was previously proposed that nucleosome repositioning by 2 or 3 bp contributes to a subtle conformational change, but highly effective mechanism for gene regulation (44). Furthermore, although the nucleosomes are still packaged in the first event of the transcriptional activation, the chromatin is permissive for the association of the activator HNF-1α, TBP and TFIIB (21,45). Thus, the DNA path alteration, accompanied by 1 bp shift, may be important in the transcriptional mechanism recruiting some factors, such as the activators, transcription factors and chromatin remodeling factors, onto NCPs.
Acknowledgments
The authors thank Drs Takuji Oyama and Daisuke Tsuchiya for their support in X-ray crystallography and helpful discussions, and Dr Masahide Kawamoto for data collection at the SPring-8. The authors acknowledge Dr Toshiyuki Shimizu for helpful discussions about the DNA base-pair-step parameters and Dr Noriyuki Suka for helpful discussions. The authors are grateful to Dr Karolin Luger for the kind gift of the pUC-based vector, which generated the 146 bp human α-satellite DNA. This work was partly supported by a research grant endorsed by the Japan New Energy and Industrial Technology Development Organization (NEDO). Funding to pay the Open Access publication charges for this article was provided by Biomolecular Engineering Research Institute.
Conflict of interest statement: None declared.
REFERENCES
- 1.Horn P.J., Peterson C.L. Molecular biology. Chromatin higher order folding—wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 2.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 4.Becker P.B. Nucleosome sliding: facts and fiction. EMBO J. 2002;21:4749–4753. doi: 10.1093/emboj/cdf486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker P.B., Horz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 6.Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Urnov F.D. Methylation and the genome: the power of a small amendment. J. Nutr. 2002;132:2450S–2456S. doi: 10.1093/jn/132.8.2450S. [DOI] [PubMed] [Google Scholar]
- 9.Cosgrove M.S., Boeke J.D., Wolberger C. Regulated nucleosome mobility and the histone code. Nature Struct. Mol. Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 10.Luger K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 11.Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 12.Luger K., Maeder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 13.Harp J.M., Hanson B.L., Timm D.E., Bunick G.L. Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta. Crystallogr. D. Biol. Crystallogr. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 14.White C.L., Suto R.K., Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davey C.A., Richmond T.J. DNA-dependent divalent cation binding in the nucleosome core particle. Proc. Natl Acad. Sci. USA. 2002;99:11169–11174. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richmond T.J., Davey C.A. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 17.Baxevanis A.D., Landsman D. Histone Sequence Database: new histone fold family members. Nucleic Acids Res. 1998;26:372–375. doi: 10.1093/nar/26.1.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson R.T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog. Nucleic Acid Res. Mol. Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe A.P., Kurumizaka H. The nucleosome: a powerful regulator of transcription. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:379–422. doi: 10.1016/s0079-6603(08)60832-6. [DOI] [PubMed] [Google Scholar]
- 20.Wyrick J.J., Holstege F.C., Jennings E.G., Causton H.C., Shore D., Grunstein M., Lander E.S., Young R.A. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 21.Cosma M.P. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 22.Albig W., Kardalinou E., Drabent B., Zimmer A., Doenecke D. Isolation and characterization of two human H1 histone genes within clusters of core histone genes. Genomics. 1991;10:940–948. doi: 10.1016/0888-7543(91)90183-f. [DOI] [PubMed] [Google Scholar]
- 23.Zhong R., Roeder R.G., Heintz N. The primary structure and expression of four cloned human histone genes. Nucleic Acids Res. 1983;11:7409–7425. doi: 10.1093/nar/11.21.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luger K., Rechsteiner T.J., Richmond T.J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 25.Dyer P.N., Edayathumangalam R.S., White C.L., Bao Y., Chakravarthy S., Muthurajan U.M., Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Navaza J. AmoRe: an automated package for molecular replacement. Acta. Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 28.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta. Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 31.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Horz W., Zachau H.G. Deoxyribonuclease II as a probe for chromatin structure. I. Location of cleavage sites. J. Mol. Biol. 1980;144:305–327. doi: 10.1016/0022-2836(80)90093-5. [DOI] [PubMed] [Google Scholar]
- 33.Horz W., Miller F., Klobeck G., Zachau H.G. Deoxyribonuclease II as a probe for chromatin structure. II. Mode of cleavage. J. Mol. Biol. 1980;144:329–351. doi: 10.1016/0022-2836(80)90094-7. [DOI] [PubMed] [Google Scholar]
- 34.Pineiro M., Puerta C., Palacian E. Yeast nucleosomal particles: structural and transcriptional properties. Biochemistry. 1991;30:5805–5810. doi: 10.1021/bi00237a025. [DOI] [PubMed] [Google Scholar]
- 35.Finch J.T., Brown R.S., Richmond T., Rushton B., Lutter L.C., Klug A. X-ray diffraction study of a new crystal form of the nucleosome core showing higher resolution. J. Mol. Biol. 1981;145:757–769. doi: 10.1016/0022-2836(81)90313-2. [DOI] [PubMed] [Google Scholar]
- 36.Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R.R., Richmond T.J. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 37.Timsit Y., Moras D. DNA self-fitting: the double helix directs the geometry of its supramolecular assembly. EMBO J. 1994;13:2737–2746. doi: 10.1002/j.1460-2075.1994.tb06567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timsit Y., Moras D. Self-fitting and self-modifying properties of the B-DNA molecule. J. Mol. Biol. 1995;251:629–647. doi: 10.1006/jmbi.1995.0461. [DOI] [PubMed] [Google Scholar]
- 39.Lenfant F., Mann R.K., Thomsen B., Ling X., Grunstein M. All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J. 1996;15:3974–3985. [PMC free article] [PubMed] [Google Scholar]
- 40.Tereshko V., Wilds C.J., Minasov G., Prakash T.P., Maier M.A., Howard A., Wawrzak Z., Manoharan M., Egli M. Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments. Nucleic Acids Res. 2001;29:1208–1215. doi: 10.1093/nar/29.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm R.H., Kennepohl P., Solomon E.I. Structural and Functional Aspects of Metal Sites in Biology. Chem. Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 42.Glusker J.P. Structural aspects of metal liganding to functional groups in proteins. Adv. Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 43.Schwarz P.M., Felthauser A., Fletcher T.M., Hansen J.C. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Campa C., Politis P., Moreau J.L., Kent N., Goodall J., Mellor J., Goding C.R. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol. Cell. 2004;15:69–81. doi: 10.1016/j.molcel.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Soutoglou E., Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–1904. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]