Abstract
Dengue virus infects target cells by attaching to a cell surface receptor through the envelope (E) glycoprotein, located on the surface of the viral membrane. On Vero and BHK cells, heparan sulfate (HS) moieties of proteoglycans are the receptors for dengue virus; however, additional proteins have also been described as putative dengue virus receptors on C6/36, HL60, and BM cells. HS can also act as a receptor for other types of viruses or as an attachment molecule for viruses that require additional host cell molecules to allow viral penetration. In this study we searched for molecules other than HS that could participate in dengue virus infection of Vero cells. Labeled dengue 4 virus bound with high affinity to two molecules of 74 and 44 kDa. Binding of dengue virus to the 74-kDa molecule was susceptible to protease and sodium periodate treatment and resistant to heparinase treatments. Lectins such as concanavalin A and wheat germ agglutinin prevented dengue virus binding to both the 74- and the 44-kDa protein in overlay assays, while phytohemagglutinin P did not affect binding, suggesting that carbohydrate residues (α-mannose or N-acetylglucosamine) are important in virus binding to host cells. Protease susceptibility, biotin labeling, and immunofluorescence with a polyclonal antibody raised against the 74-kDa protein consistently identified the protein on the surfaces of Vero cells. Moreover, the antibody against the 74-kDa protein was able to inhibit dengue virus infection. These data suggest that HS might serve as a primary receptor, probably concentrating virus particles on the surfaces of Vero cells, and then other molecules, such as the 74-kDa protein, might participate as coreceptors in viral penetration. The 74-kDa protein possibly constitutes part of a putative receptor complex for dengue virus infection of Vero cells.
Dengue virus, a mosquito-borne member of the Flaviviridae family, causes a serious febrile illness in humans known as dengue fever and its associated complications: dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (6, 22). Dengue fever affects over 100 million people worldwide, and there are still no vaccines or antiviral agents available (12, 29).
Virus binding to susceptible target cells is the first event required for productive infection. In humans, dengue virus infects monocytes, either through the binding of virus-antibody complexes to the Fc receptor or through the direct interaction of viral proteins with a specific host cell receptor (8, 20). The first mechanism has been studied extensively because DHF and DSS have been associated with an increase in infection due to the virus-antibody complexes that bind Fc-γ receptor-positive cells via the Fc portion of immunoglobulin G (IgG) (11, 20, 25, 26). The second mechanism, which produces the primary infection, has only recently started to be explored in different cell lines (2, 4, 18, 33).
The envelope (E) protein, which is exposed on the surface of the viral membrane, contains structural and functional elements that participate in the virus-host cell receptor interaction (14, 15, 32) and is hence known as the viral attachment protein. By using recombinant E protein, infection of Vero cells by dengue virus serotype 2 (DEN-2) is inhibited, and the binding domain of E protein has been identified between amino acids 281 and 423 (5). However, studies with lectins suggest that carbohydrates such as α-mannose residues present on the E protein also contribute to binding and to penetration into BHK and C6/36 cells (18).
Previous studies designed to identify one or more cellular proteins involved in dengue virus binding and subsequent entry into various susceptible host cells have revealed several candidate molecules. Dengue virus uses an uncharacterized trypsin-susceptible molecule located on the cell surface to bind to monocytic cells and neuroblastoma cells (8, 31), while in Vero and BHK cells, dengue virus binding and entry require the presence of a highly sulfated form of heparan (HS) (4). The four serotypes of dengue virus could bind with different degrees of affinity to the surfaces of HL60 myelomonocytic cells and non-Epstein-Barr virus (EBV)-transformed B cells. Specifically, DEN-2 bound to two molecules of 40 to 45 and 70 to 75 kDa (found on the membranes of HL60 and non-EBV-transformed B cells) in an overlay assay; however the nature, occurrence, and specificity of these molecules have not been sufficiently studied (2). For mosquito cells, putative molecules involved in dengue virus binding to C6/36 cells (from Aedes albopictus larvae) have been described and two glycoproteins of 40 and 45 kDa present on the surfaces of the cells were detected specifically by DEN-4 (33), while an 80-kDa molecule has been shown to be involved in DEN-2 binding to this cell line (28). Although several molecules have been reported to be involved in dengue virus binding and entry into the host cell, at present only three of these have been postulated to play a role in dengue virus infection; HS, which is present on Vero and BHK cells (4, 18), and two glycoproteins of 40 and 45 kDa identified on C6/36 cells (33). Elimination of HS from Vero cells, using glycosaminoglycan (GAG) lyase I or III, considerably reduced dengue virus infection (4), while incubation of C6/36 cells with anti-40- and anti-45-kDa glycoprotein antibodies also inhibited dengue virus infection (33). It is possible that dengue virus uses various cell molecules for binding (receptors) and entry (coreceptors) into different cell lines.
Dengue virus, like other viruses such as herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) (17), varicella-zoster virus (39), pseudorabies virus (PrV) (34), human cytomegalovirus (36), foot-and-mouth disease virus type O (19), human immunodeficiency virus type 1 (HIV-1) (13), and respiratory syncytial virus (RSV) (21), attaches to target cells by interaction of the virion with HS. HS could act as a viral receptor due to its high negative charge, or it could allow access of the virus to additional molecules (coreceptors) needed to penetrate into the host cells, as has already been described for PrV (40), HSV-1 (23), and HIV-1 (13). GAGs, commonly expressed in the extracellular matrix as well as on cell membranes, are characterized by high heterogeneity, which could explain the selective tropism of dengue and other viruses for primate and human cells (4, 16, 17, 31). Vero cells contain large amounts of HS on their surfaces and are highly susceptible to dengue virus infection. However, the participation of coreceptors during dengue virus penetration into these cells has not been investigated.
In this study, we attempted to identify additional molecules on Vero cells that could have affinity for dengue virus and facilitate virus infection. This paper therefore reports the specific binding of dengue virus to two molecules of 74 and 44 kDa on Vero cells. Binding of the virus to the 74-kDa molecule was susceptible to protease and sodium periodate treatments and resistant to treatment by heparinases. Lectins such as concanavalin A (ConA, which binds to α-mannose residues on N-linked high-mannose or hybrid glycans) and wheat germ agglutinin (WGA, which recognizes acetylglucosamine [GlcNAcβ1-4] on N-linked glycans) prevented dengue virus binding to both the 74- and the 44-kDa protein in overlay assays, while phytohemagglutinin-P (PHA-P, which recognizes oligosaccharides) did not affect binding to either of the cellular molecules, suggesting the participation of carbohydrate residues such as α-mannose or N-acetylglucosamine in virus binding to host cells. A polyclonal antibody raised against the 74-kDa protein localized the protein on the surfaces of Vero cells in immunofluorescence assays. The antibodies were also able to inhibit dengue virus infection. These data suggest that during dengue virus infection of Vero cells, HS might serve as a primary receptor, probably concentrating virus particles on the cell surface, and that subsequent viral penetration could require other molecules, such as the 74-kDa protein. It is possible that this protein could be part of a putative receptor complex for dengue virus in Vero cells.
MATERIALS AND METHODS
Cells and virus.
Monolayers of Vero and LLC-MK2 cells (from green and rhesus monkey kidneys, respectively) were grown at 37°C under 5% CO2 in Dulbecco's modified minimal essential medium (DMEM) supplemented with 10% neonatal calf serum, 5,000 U of penicillin, and 5 μg of streptomycin.
DEN-4 strain H-241, generously donated by Goro Kuno, was propagated in suckling mice and LLC-MK2 cells as previously described (9).
Radiolabeled dengue virus.
Radiolabeling of dengue virus was carried out in infected LLC-MK2 cells. Briefly, subconfluent monolayers of cells (85%) were infected at a multiplicity of infection (MOI) of 0.1 PFU/cell. At 24 h postinfection, the medium was replaced with methionine-free medium (Sigma), containing 5 μC of [35S]-methionine (Dupont)/ml, 10% neonatal calf serum, penicillin, and streptomycin. After 4 to 6 days, the supernatant was harvested and clarified by centrifugation at 10,000 rpm for 10 min in a JA20 Beckman rotor. Viruses were pelleted at 10,000 rpm for 30 min after incubation with 10% (wt/vol) polyethylene glycol 8000 in 1.5 M NaCl at 4°C for 24 h. The pellet was resuspended in 1/10 the original volume with GTNE buffer (50 mM Tris-HCl, 200 mM glycine, 100 mM NaCl, 1 mM EDTA) and clarified by centrifugation at 15,000 rpm for 15 min (JA20 Beckman rotor), and the supernatant was applied to a discontinuous gradient of 60 and 30% (wt/vol) sucrose in GTNE. Sucrose gradients were centrifuged at 27,500 rpm for 2.5 h at 4°C in an SW28 rotor. The visible band that contained the viruses was harvested, diluted (vol/vol) with GTNE, and pelleted at 23,000 rpm for 2 h at 4°C in a 50 Ti rotor. Finally, the viral pellet was resuspended in GTNE containing 1% bovine serum albumin. Radioactive counts per minute were determined in a scintillation counter, and the solution was aliquoted and stored at −20°C.
As a control for the labeled virus, labeled proteins from uninfected LLC-MK2 cells were prepared. Briefly, cells were grown in methionine-free medium (Sigma), containing 5 μCi of [35S]methionine (Dupont)/ml, 10% neonatal calf serum, penicillin, and streptomycin. After 48 h, cells were detached, harvested, and pelleted at 10,000 rpm for 10 min in a JA20 Beckman rotor. Then cells were resuspended in phosphate-buffered saline (PBS) and degraded by 5 to 10 strokes in a Dounce homogenizer. The cell extract was pelleted at 10,000 rpm for 30 min after incubation with 10% (wt/vol) polyethylene glycol 8,000 in 1.5 M NaCl at 4°C for 24 h. The pellet was resuspended in 1/10 the original volume with GTNE buffer and clarified by centrifugation at 15,000 rpm for 15 min (JA20 Beckman rotor), and the supernatant was obtained for analysis. Counts per minute in the supernatant were determined in a scintillation counter, and the supernatant was aliquoted and stored at −20°C.
Cell membrane protein preparation.
Monolayers of Vero cells were detached with PBS–5 mM EDTA (5 ml/75-cm2 flask) for 10 min at room temperature. Afterwards, cells were resuspended in ice-cold buffer M (100 mM NaCl, 20 mM Tris [pH 8], 2 mM MgCl2, 1 mM EDTA, and 1 mM β-mercaptoethanol) and lysed by 5 to 10 strokes in a Dounce homogenizer. Nuclei and debris were removed by centrifugation at 2,500 rpm for 10 min at 4°C in a Sorvall centrifuge. Membrane proteins were pelleted from the supernatant by centrifugation at 18,000 rpm for 30 min at 4°C in a Beckman JA20 rotor and resuspended in buffer M without β-mercaptoethanol. The concentration of protein was quantified by the Bradford method (3).
GAG preparation.
GAGs from Vero cells were prepared as described previously (38).
VOPBA.
To identify Vero cell molecules involved in virus binding, virus overlay protein-binding assays (VOPBAs) were carried out. Briefly, 80 to 100 μg of Vero cell membrane proteins was subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) and transferred to nitrocellulose membranes by using a semidry Bio-Rad blotting apparatus in transfer buffer (50 mM Tris base, 40 mM glycine, and 10% methanol [vol/vol]). The membranes were blocked overnight at 4°C with 5% nonfat milk in PBS and were washed once with PBS, once with 1% nonfat milk in PBS, and finally once with overlay buffer (1% nonfat milk in PBS, 220 mM NaCl). The VOPBA was carried out by incubating the nitrocellulose membrane with 100,000 cpm of radiolabeled DEN-4, with a specific activity of 4,500 to 5,000 cpm/μg of protein, in overlay buffer for 3 h at 37°C. Afterwards, membranes were washed at least six times, for 10 min each time, with PBS at room temperature. Finally, membranes were dried and subjected to autoradiography.
Sodium chlorate treatment.
Subconfluent (90%) Vero cells were maintained for 24 h in low-sulfate DMEM (Gibco); later, sodium chlorate was added to the cells at final concentrations of 10 and 50 mM. After 48 h, the treated cells were detached and washed three times with fresh medium, and cell membrane proteins were prepared as described above.
GAG lyase treatment.
A suspension of Vero cells in buffer G (10 mM phosphate buffer [pH 7.4], 0.14 M NaCl, 3 mM KCl, 0.5 mM MgCl2, 1 mM CaCl2, and 1% neonatal bovine serum), was incubated with 2 U of GAG lyase I/ml or 5 U of GAG lyase III/ml at 37°C for 1 h. Cells were washed twice with fresh medium supplemented with serum, and cell membrane proteins were obtained as described above. The effect of GAG lyase on the cells was demonstrated spectrophotometrically by determining the absorption of α, β-unsaturated uronides (in the supernatant from the treated-cell samples) at 232 nm as previously described (24).
Protease treatment.
Detached Vero cells were resuspended in PBS and then incubated with 10 μg of trypsin, proteinase K, or pronase E/ml at 37°C for 1 h. After incubation, the cells were washed three times with ice-cold fresh DMEM supplemented with 10% neonatal calf serum, and cell viability was monitored by trypan blue exclusion. Finally, membrane proteins were obtained as described above.
In a similar experiment, detached Vero cells in PBS were incubated with 10 μg of proteinase K/ml at 37°C for 1 h. After incubation, the cells were washed three times with ice-cold fresh medium supplemented with 10% newborn calf serum and then reincubated in the same medium at 37°C. Replacement of lost surface protein was determined at different times posttreatment (30, 60, 120, or 180 min), the cells were resuspended in buffer M, and membrane proteins were obtained essentially as described above.
Sodium periodate treatment.
A suspension of Vero cells was treated with 1.5 mM sodium periodate at 37°C for 20 min, and cell membrane proteins were prepared as described above. Additionally, 80 to 100 μg of membrane proteins from Vero cells separated by SDS–10% PAGE and transferred to nitrocellulose membranes was incubated with 10 and 20 mM sodium periodate in 50 mM acetate buffer (pH 4.5) for 1 h at room temperature in the dark, washed three times with PBS, and then incubated with 100,000 cpm of labeled dengue virus.
Lectin incubation.
To analyze the role of carbohydrates in dengue virus binding, membrane proteins from Vero cells, separated by SDS–10% PAGE and transferred to nitrocellulose membranes, were incubated with 100 μg of ConA, WGA, or PHA-P diluted in PBS (pH 7.2) for 1 h at 37°C. The membranes were washed three times with PBS and then incubated with labeled dengue virus as described above.
Production of a polyclonal antibody against the 74-kDa protein.
Cell membrane proteins were run on an SDS–10% PAGE gel. The 74-kDa band was excised from the gel and used to immunize BALB/c mice six times intraperitoneally at 15-day intervals. Mouse sera were collected 10 days after the last immunization. Immunoglobulins were purified in protein G columns (Gibco BRL), dialyzed against PBS, and lyophilized. Sera were tested by Western blotting for the presence of antibody against the 74-kDa protein
Western blot assay.
Vero cell proteins were subjected to SDS-PAGE and transferred to nitrocellulose membranes as described above. Membranes were blocked in PBS containing 5% (wt/vol) nonfat milk overnight at 4°C and washed three times in 0.5% (wt/vol) Tween 20 in PBS. The anti-74-kDa serum diluted 1:1,000 in PBS was added to the membrane and incubated overnight at 4°C. After incubation and washings, the secondary antibody, anti-mouse IgG conjugated to alkaline phosphatase (diluted 1:4,000 in PBS) was added and incubated at room temperature for 2 h. Color was developed by the addition of 5-bromo-4-chloro-3-indolylphosphate toluidinium (BCIP) and nitroblue tetrazolium chloride (NBT). The reaction was stopped with water.
Inhibition of infection by polyclonal anti-74-kDa antibodies and by sodium chlorate, heparinase I, and heparinase III treatments.
Inhibition of dengue virus infection by sodium chlorate, heparinases, and the anti-74-kDa antibody was evaluated in subconfluent monolayers of Vero cells.
For evaluation of sodium chlorate inhibition, Vero cells washed with PBS (pH 7.2) were grown either in standard medium (untreated) or with low-sulfate DMEM in the presence of 50 mM sodium chlorate for 48 h. Later, cells were washed twice with serum-free DMEM and incubated for 1 h at 37°C with DEN-4 (at an MOI of 0.5) in fresh medium with 10% newborn calf serum. After two washes with fresh medium without serum, cells were maintained with DMEM supplemented with 10% newborn calf serum, penicillin, and streptomycin and incubated for 48 h at 37°C. Indirect immunofluorescence was performed by using the anti-dengue virus monoclonal antibody 4-E (1:25) (Instituto Pierre Kouri, Havana, Cuba) as the primary antibody and a fluorescein isothiocyanate (FITC)-coupled goat anti-mouse IgG as the secondary antibody (1:175) (Zymed). Fluorescent focus units in untreated and treated cells were determined. The number of fluorescent focus units in untreated cells was taken as 100%.
For evaluation of inhibition by heparinases, Vero cells washed with PBS (pH 7.2) were either left untreated or treated with heparinase I or heparinase III for 1 h at 37°C. Then cells were washed, infected, incubated, and assayed as described above for the sodium chlorate assay.
For evaluation of inhibition by polyclonal anti-74-kDa antibodies, Vero cells washed with PBS (pH 7.2) were preincubated in the absence or presence of preimmune serum or a polyclonal antibody against the 74-kDa protein for 1 h at 37°C. Cells were then washed, infected, incubated, and assayed as described above for the sodium chlorate assay.
The effect of the anti-74-kDa antibody on poliovirus infection was evaluated in subconfluent monolayers of Vero cells as follows. Vero cells washed with PBS (pH 7.2) were preincubated in the absence or presence of preimmune serum or the anti-74-kDa-protein antibody for 1 h at 37°C. Subsequently, cells were washed twice with serum-free DMEM and incubated for 1 h at 37°C with poliovirus type 3 in fresh medium with 1% newborn calf serum. After two washes with fresh medium without serum, cells were maintained with DMEM supplemented with 10% newborn calf serum, penicillin, and streptomycin and incubated for 48 h at 37°C. Results of a plaque assay with the supernatant obtained from Vero cells incubated in the absence of the anti-74-kDa-protein antibody were taken as 100% infectivity.
Indirect immunofluorescence assay.
Subconfluent (85%) Vero cells were plated in 16-microwell tissue culture chambers (Lab-Tek), and indirect immunofluorescence assays were performed as described previously (33) with the following modifications: the anti-74-kDa antibody was diluted 1:50 in PBS with 10% normal goat serum, and an FITC-coupled goat anti-mouse antibody was used at 1:175 (Zymed).
Fluorescent focus units.
The fluorescent focus units (comprising approximately 45,000 cells, with 12% of the infected cells stained at 48 h postinfection) were estimated in duplicate in all the fields showing the fluorescence signal. Each individual cell showing a signal represents a focus unit. The stained cells were examined using a UV Olympus microscope, model BX 60. The number of fluorescent focus units was determined manually in at least two separate experiments using the Image-Pro Plus by Media Cybernetics, L.P.
Biotinylated membrane proteins.
Detached Vero cells were washed with PBS and centrifuged at 1,000 rpm for 10 min at 4°C (JA20 Beckman rotor). Cells were resuspended at a concentration of 2 × 107/ml in biotinylation buffer (50 mM Na2CO3–NaHCO3 [pH 8.5] and 150 mM NaCl) and chilled on ice for 15 min. Then a final concentration of 50 g of biotin/ml (10 mg/ml in dimethyl sulfoxide) was added, and cells were incubated on ice for another 30 min. The reaction was stopped with 10 mM NH4Cl. Cells were washed three times with PBS (pH 7.2), and biotinylated membrane proteins were obtained as described above.
RESULTS
Identification of dengue virus-binding molecules on Vero cells.
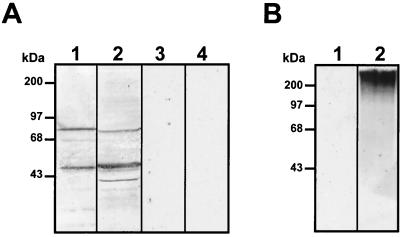
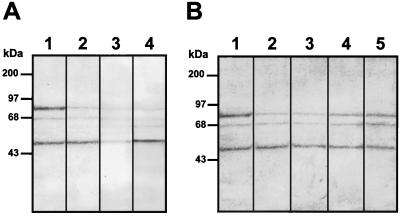
To analyze if dengue virus uses additional molecules, other than HS, to infect Vero cells, we first determined whether some molecules on Vero cells could bind with high affinity to DEN-4 using an overlay assay. Membrane proteins from mosquito cells (C6/36 cells), with high affinity for DEN-4 (33) as well as GAGs isolated from Vero cells, were used as a positive control. Nitrocellulose membranes to which cellular proteins or GAGs had been transferred were incubated with [35S]methionine-labeled DEN-4 (Fig. 1A, lanes 1 and 2) or with [35S]methionine-labeled proteins from uninfected cells (Fig. 1A, lanes 3 and 4). Under hypertonic conditions (220 mM NaCl), we were able to detect 40-, 45-, and 72-kDa proteins in C6/36 extracts (Fig. 1A, lane 2), confirming previous reports (33). In contrast, DEN-4 bound to three molecules of 44, 68, and 74 kDa from Vero cell membrane extracts (Fig. 1A, lane 1) and to GAG molecules from Vero cells with molecular sizes greater than 150 kDa (Fig 1B, lane 2). The bands of 44- and 74-kDa molecules were the most intense, suggesting that a greater amount of labeled dengue virus bound to these two molecules. Labeled proteins from uninfected cells were unable to bind to cellular molecules from mosquito cells (Fig. 1A, lane 4), Vero cell extracts (Fig. 1A, lane 3), or GAGs (Fig. 1B, lane 1). The presence of the 44- and 74-kDa molecules in the VOPBA performed with membrane cell extracts and their absence in the GAG preparation from Vero cells suggest that 44- and 74-kDa molecules might not contain GAGs.
FIG. 1.
VOPBA with [35S]methionine-labeled dengue virus. (A) Total protein from C6/36 cells (lanes 2 and 4) and membrane protein from Vero cells (lanes 1 and 3) were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated with 100,000 cpm of labeled dengue virus (lanes 1 and 2) or 100,000 cpm of labeled proteins from uninfected cells (lanes 3 and 4) in the presence of 220 mM, NaCl. (B) Vero cell GAGs were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated with 100,000 cpm of labeled dengue virus (lane 2) or 100,000 cpm of labeled proteins from uninfected cells (lane 1) in the presence of 220 mM NaCl.
The specificity of the interaction between dengue virus and the 44- and 74-kDa molecules from Vero cells was demonstrated by competition assays with unlabeled dengue virus and blue eyes syndrome virus (an enveloped member of the family Paramyxoviridae). Proteins transferred to nitrocellulose membranes were incubated with a 5- or 10-fold excess of cold dengue virus before incubation with labeled dengue virus (Fig. 2A, lanes 2 and 3). Binding of labeled DEN-4 to the 44- and 74-kDa molecules was considerably reduced in the presence of a 5-fold excess of unlabeled dengue virus and practically absent in the presence of a 10-fold excess, suggesting that binding of virus to the 44- and 74-kDa molecules was specific. When a 5- or 10-fold excess of unlabeled blue eyes syndrome virus was used as a heterologous competitor, binding of the labeled DEN-4 to 44- and 74-kDa molecules was not altered (Fig. 2B, lanes 2 and 3).
FIG. 2.
Competition experiment with unlabeled dengue (A) and blue eyes syndrome (B) viruses. Membrane proteins obtained from Vero cells were subjected to SDS–10% PAGE, transferred to a nitrocellulose membrane, and preincubated in the presence of labeled virus (lanes 1) or in the presence of 5- and 10-fold excesses (lanes 2 and 3, respectively) of unlabeled dengue virus or unlabeled blue eyes syndrome virus prior to incubation with 100,000 cpm of labeled dengue type 4 virus. Molecular size markers are indicated on the left.
Dengue virus-binding proteins do not contain HS.
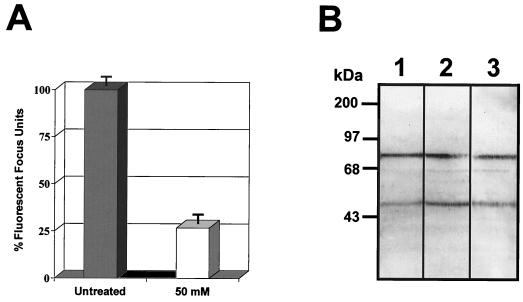
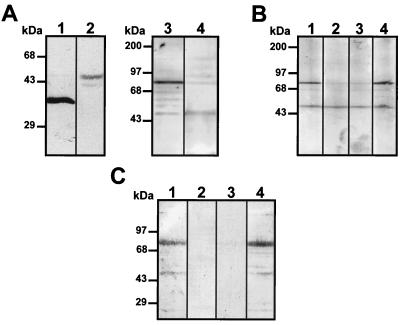
To provide evidence that the dengue virus binding molecules do not contain HS, two types of experiments were performed. First, we reduced the level of sulfate present in GAG of Vero cells, and second, we eliminated HS by heparinase I and III treatments. To reduce the GAG sulfation level, Vero cells were grown in the presence of sodium chlorate (4). Under this condition cells became round and reduced their contact with each other, and infection with dengue virus was inhibited by as much as 73.2% (Fig. 3A) as previously described (4). However, this reaction was reversible, and optimal DEN-4 binding was restored when sulfate was added to the treated cells. Membrane proteins from Vero cells grown in the presence of sodium chlorate for 48 h were used in an overlay assay. The reduced sulfation of GAG did not affect dengue virus binding to the 74- and 44-kDa proteins (Fig. 3B, lanes 2 and 3), indicating that if these molecules contain sulfated GAGs, the sulfated GAGs are not involved in DEN-4 binding.
FIG. 3.
Effect of sodium chlorate on dengue virus-binding molecules. (A) Effect of low-sulfate medium and 50 mM sodium chlorate on dengue virus infection of Vero cells. Florescent focus units in untreated and treated cells were determined. The number of fluorescent focus units obtained with untreated cells was taken as 100%. For the procedure, see Materials and Methods. (B) Membrane proteins extracted from Vero cells grown in standard medium (lane 1) or in low-sulfate DMEM in the presence of 10 and 50 mM sodium chlorate (lanes 2 and 3, respectively) were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated with 100,000 cpm of [35S]methionine-labeled dengue virus. Molecular size markers are indicated on the left.
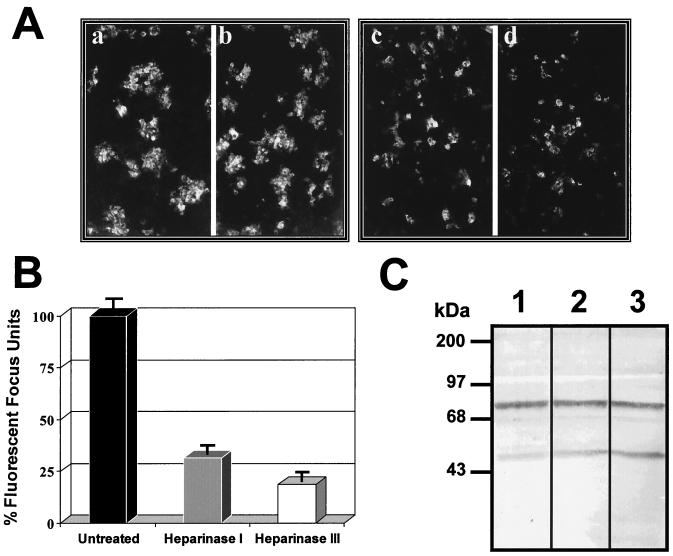
To confirm the absence of HS in the 74- and 44-kDa molecules, Vero cells were treated with heparinase I or III and then infected with DEN-4. Both heparinases I and III efficiently eliminate HS and caused a significant reduction in dengue virus binding and infection of Vero cells (4). After treatment with the heparinases, cells were infected with DEN-4 for 48 h and incubated with a monoclonal antibody against DEN-4 and a second anti-mouse antibody coupled to FITC. The number of immunofluorescent foci counted after treatment with heparinase I (Fig. 4Ac) or heparinase III (Fig. 4Ad) was lower (Fig. 4B) than that noted with untreated cells (Fig. 4Aa and b). In addition, treatment with either heparinase reduced the amount of fluorescent foci by as much as 80% (Fig. 4B), indicating that HS is necessary for dengue virus infection, consistent with the reports of Chen et al. (4) and Hung et al. (18). However, neither migration of the 74- and 44-kDa molecules nor binding of labeled dengue virus was altered after heparinase treatment (Fig. 4C, lanes 2 and 3), suggesting that HS is not a component of either of the two molecules.
FIG. 4.
Effects of heparinases on dengue virus-binding molecules. (A) Vero cells washed with PBS (pH 7.2) were either left untreated (a and b) or treated with heparinase I (c) or III (d) for 1 h at 37°C. For the procedure, see Materials and Methods. (B) Fluorescent focus units with untreated and heparinese I- or III-treated cells. The number of focus units obtained with untreated cells was taken as 100%. (C) Membrane proteins from Vero cells that were either left untreated (lane 1) or treated with heparinase I (lane 2) or heparinase III (lane 3) were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated with 100,000 cpm of labeled dengue virus. Molecular size markers are indicated on the left.
Protease susceptibility of dengue virus-binding molecules.
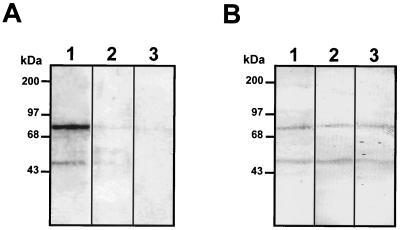
To initially characterize the 74- and 44-kDa molecules, an overlay assay was performed with membrane proteins obtained after treatment of Vero cells with different proteases. Interaction between labeled dengue virus and the 74-kDa molecule was reduced after treatment with proteinase K (Fig. 5A, lane 2), trypsin (Fig. 5A, lane 3) or pronase E (Fig. 5A, lane 4), while binding to the 44-kDa molecule was reduced only in the presence of trypsin (Fig. 5A, lane 3). These data suggest that the 74-kDa molecule is a protein. The high susceptibility of the 74-kDa molecule to protease treatment suggests that it may be located on the surfaces of Vero cells.
FIG. 5.
Protease treatment of Vero cells and binding to dengue virus. (A) Membrane proteins obtained from Vero cells that were either left untreated (lane 1) or treated with 10 μg of proteinase K (lane 2), trypsin (lane 3), or pronase E (lane 4)/ml were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated with 100,000 cpm of labeled dengue virus. (B) Binding inhibition induced by proteinase K treatment. Vero cells that were either left untreated (lane 1) or treated with proteinase K (10 μg/ml) for 1 h at 37°C were washed and incubated at 37°C in complete medium for 30 (lane 2), 60 (lane 3), 120 (lane 4), and 180 (lane 5) min. Membrane proteins isolated from treated cells were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated with 100,000 cpm of labeled dengue virus. Molecular size markers are indicated on the left.
When cells are exposed to protease treatment, the proteins located on the surfaces are more susceptible to disruption than the unexposed proteins. Lost protein molecules are usually replaced on the cell membrane. To analyze if replacement of the 74-kDa molecule occurred, Vero cells were treated with 10 μg of proteinase K/ml for 1 h at 37°C, washed three times, resuspended in fresh medium, and incubated for different times before membrane proteins were obtained. Binding of labeled dengue virus to the 74-kDa molecule increased after 120 and 180 min (Fig. 5B, lanes 4 and 5) up to approximately 50% of the untreated cells. This result further suggests that the 74-kDa molecule is exposed on the surfaces of Vero cells and that the time to replacement of the lost molecule on the cell surface usually exceeds 120 mim.
Carbohydrate characterization of dengue virus-binding molecules.
To continue the characterization of the dengue virus-binding molecules, the role of carbohydrate in dengue virus binding was analyzed using Vero and C6/36 cells treated with sodium periodate in an overlay assay. Sodium periodate is a useful chemical agent which destroys the carbohydrate moiety without altering protein or lipid epitopes. Sodium periodate causes an oxidation of vicinal hydroxyl groups on sugars to dialdehydes at acidic pHs (37). Labeled dengue virus bound to the 40- and 45-kDa proteins in untreated C6/36 cells (Fig. 6A, lane 2), and to a single 38-kDa molecule in extracts of sodium periodate-treated cells (Fig. 6A, lane 1) as described previously (33). However, binding of dengue virus to the 74-kDa protein from Vero cells was strongly reduced after treatment of cells with sodium periodate (Fig. 6A, lane 4). These data suggest that the 40- and 45-kDa proteins from C6/36 are glycoproteins and that the oxidation of vicinal hydroxyl groups on sugars induced by the sodium periodate treatment might have reduced the size of the molecules but not their abilities to bind dengue virus. On the other hand, the ability of dengue virus to bind to the 74-kDa molecule of Vero cells suggests that this molecule is a glycoprotein, since the carbohydrate moiety is important in this interaction. However, since oxidation of vicinal hydroxyl groups on sugars induced by the sodium periodate treatment could modify the ability of dengue virus to bind to the molecule, it is not clear if this treatment modified the molecular size of the 74-kDa molecule. In addition, in C6/36 cells, the carbohydrate moiety may not be relevant to the interaction with dengue virus, because the 38-kDa protein bound efficiently with dengue virus even after periodate treatment, as previously demonstrated (33). Nevertheless, the carbohydrate residue may be important in viral attachment to the 74-kDa protein of Vero cells, because dengue virus binding was considerably inhibited after sodium periodate treatment. To further analyze the susceptibility of virus binding to treatment with sodium periodate, untreated Vero membrane proteins transferred to nitrocellulose membranes were treated with 0, 10, and 20 mM sodium periodate (Fig. 6B, lanes 1 to 3). Binding of labeled dengue virus to the 74-kDa protein was reduced after treatment with sodium periodate, confirming that a significant component of the carbohydrate moiety is important for viral attachment.
FIG. 6.
Carbohydrate characterization of dengue virus-binding molecules. (A) Membrane proteins obtained from C6/36 cells (lanes 1 and 2) or Vero cells (lanes 3 and 4) that were either left untreated (lanes 2 and 3) or treated with sodium periodate (lanes 1 and 4) were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated with 100,000 cpm of labeled dengue virus. (B) Membrane proteins from Vero cells were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated in the presence of 0 (lane 1), 10 (lane 2), and 20 (lane 3) mM sodium periodate in acetate buffer or in PBS (lane 4) before incubation with 100,000 cpm of labeled dengue virus. (C) Binding inhibition induced by lectins. Membrane proteins from Vero cells transferred to a nitrocellulose membranes were incubated either without lectin (lane 1) or with 100 μg of ConA (lane 2), WGA (lane 3), or PHA-P (lane 4) for 1 h at 37°C before incubation with 100,000 cpm of labeled dengue virus. Molecular size markers are indicated on the left.
To characterize the nature of the carbohydrate present in the 74-kDa molecule, membrane proteins from Vero cells were incubated with different lectins. Binding of the labeled dengue virus to the 74-kDa protein was inhibited by ConA (Fig. 6C, lane 2) and WGA (Fig. 6C, lane 3); however, PHA-P did not alter attachment to the 74-kDa molecule (Fig. 6C, lane 4). These results suggest that carbohydrate residues such as α-mannose or N-acetylglucosamine are present in the 74-kDa molecule and that they could be involved in dengue virus binding.
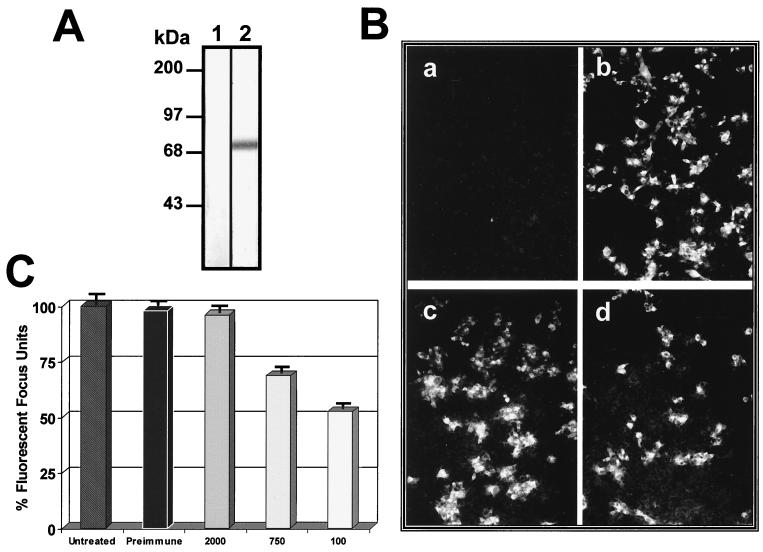
To analyze the role of the 74-kDa molecule in dengue virus infection, we produced a polyclonal antibody against the protein molecule. In the Western blot assays, the antibody detected a single band of 74 kDa on membrane extracts of Vero cells (Fig. 7A, lane 2). In another experiment, the polyclonal antibody was incubated with Vero cells before infection with DEN-4. Viral infection was then monitored by immunofluorescence using a monoclonal antibody against DEN-4, 48 h after infection. Incubation with immune IgG (Fig. 7Bd) reduced infection of cells by over 50% as determined by the fluorescent focus units (Fig. 7C), while the presence of preimmune IgG (Fig. 7Bb) did not alter virus infection of cells, as observed in cells preincubated with complete fresh medium (Fig. 7Bc and C).
FIG. 7.
Infection inhibition induced by a polyclonal antibody against the 74-kDa protein. (A) Western blot assay of membrane extracts from Vero cells. Nitrocellulose membranes with membrane proteins from Vero cells were incubated with preimmune serum (lane 1) or with an anti-74-kDa protein polyclonal antibody (lane 2) at a dilution of 1:1,000. A secondary antibody, goat anti-mouse IgG coupled to alkaline phosphatase, was used at a dilution of 1:4,000. Molecular size markers are indicated on the left. (B) Vero cells washed with PBS (pH 7.2) were preincubated in the absence (a and c) or presence of preimmune serum (b) or the anti-74-kDa-protein antibody (d) at a dilution of 1:100 for 1 h at 37°C. Subsequently, cells were washed twice with serum-free DMEM and incubated for 1 h at 37°C with DEN-4 (at an MOI of 0.5) (b to d) in fresh medium with 10% newborn calf serum. For the procedure, see Materials and Methods. (C) Fluorescent focus units without antibodies, with preimmune serum, or with the anti-74-kDa protein antibody (at the final dilutions given along the X axis). The number of focus units obtained with cells incubated in the absence of antibody was taken as 100%.
To demonstrate the specificity of inhibition of dengue virus infection by the anti-74-kDa antibody, the antibody was preincubated with Vero cells before infection with poliovirus. None of the concentrations of the anti-74-kDa antibody that inhibited dengue virus infection was able to inhibit poliovirus infection (Table 1).
TABLE 1.
Pattern of inhibition of infectivity of dengue virus and polioviruses by different concentrations of the anti-74-kDa antibodya
| Virus | % Inhibition of infectivity by anti-74-kDa antibody at the following concnb:
|
|||
|---|---|---|---|---|
| 0 | 100 | 750 | 2,000 | |
| Dengue virus | 0 | 54.3 ± 4.5 | 34.1 ± 4.0 | 0 |
| Poliovirus | 0 | 0 | 0 | 0 |
For procedures, see Materials and Methods.
Expressed as the reciprocal of the antibody titer.
Surface localization of the 74-kDa molecule on Vero cells.
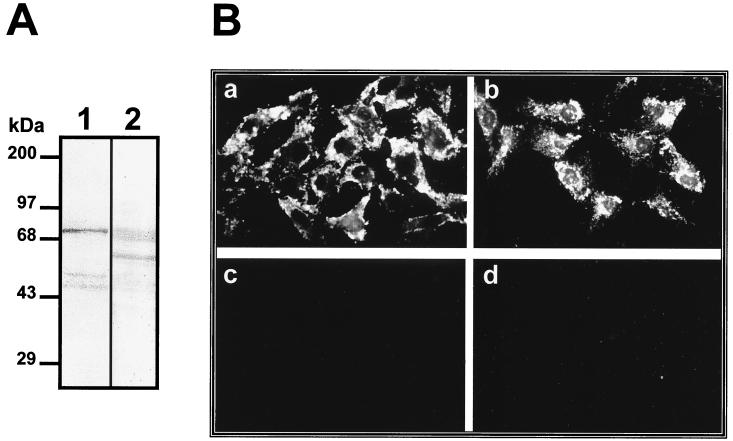
Because our results have consistently indicated that the 74-kDa molecule is located on the surface of the cell, a biotinylated membrane extract of Vero cells was prepared to localize the protein molecule on the cell surface. Proteins transferred to nitrocellulose membranes were incubated with avidinperoxidase to identify cell surface proteins and were also incubated with labeled dengue virus in an overlay assay. The 74-kDa molecule detected by labeled dengue virus (Fig. 8A, lane 1) comigrated with one of the biotinylated bands (Fig. 8A, lane 2), suggesting that the 74-kDa glycoprotein is indeed present on the surface of the cell.
FIG. 8.
Localization of the 74-kDa molecule on Vero cells. (A) Biotinylated membrane proteins from Vero cells were subjected to SDS–10% PAGE, transferred to nitrocellulose membranes, and incubated with 100,000 cpm of labeled dengue virus (lane 1) or with avidin-peroxidase at a dilution of 1:1,000 (lane 2). Molecular size markers are indicated on the left. (B) Non-ethanol-treated (a and c) and ethanol-treated (b and d) Vero cells were incubated with immune serum (a and b) or preimmune serum (c and d) against the 74-kDa protein. As a secondary antibody an FITC-conjugated goat anti-mouse IgG was used.
The localization of the 74-kDa protein on Vero cells was confirmed by immunofluorescence. The polyclonal antibody against the 74-kDa protein was able to react with the surfaces of non-ethanol-treated Vero cells (Fig. 8Ba) and ethanol-treated Vero cells (Fig. 8Bb). The ability of the anti-74-kDa-protein antibody to react with non-ethanol-treated cells indicates that this protein is located on the surfaces of Vero cells.
DISCUSSION
Some viruses such as herpes simplex virus gain entry into target cells through a multistep process that includes an initial attachment to the surface mediated by HS, followed by other stages that require the cooperation of different cell surface molecules that act as coreceptors.
Molecules involved in virus entry share some properties such as (i) binding with high affinity to the virus, (ii) surface location on the target cells, and (iii) inhibition of viral infection when incubated with the specific antibody. HS has been described as a dengue virus receptor in Vero cells (4). In this study, we identified a 74-kDa protein located on the surfaces of Vero cells that may also be required for dengue virus infection. High-affinity binding of dengue virus to the protein molecule was demonstrated by overlay assays under hypertonic conditions (220 mM KCl), and the specificity of the interaction was proven in competition experiments with unlabeled dengue virus and blue eyes syndrome virus.
To investigate whether the dengue virus-binding molecules contained HS, overlay assays were performed. Binding of dengue virus to the 44- and 74-kDa molecules was not altered when sulfation of HS was reduced with sodium chlorate or when Vero cells were treated with heparinases, suggesting that these molecules may not contain HS.
The susceptibility of the 74-kDa binding molecule to protease and sodium periodate treatments possibly suggests its glycoprotein nature and surface localization on the cell. Dengue virus also recognized a 44-kDa molecule which was not susceptible to protease treatment with pronase E and proteinase K or to sodium periodate, suggesting that the molecule may not be a glycoprotein or that it may not be present on the surfaces of Vero cells. The susceptibility of the 74-kDa molecule in the membrane fraction after protease treatment has provided strong evidence to suggest that the 74-kDa protein is exposed on the Vero cell surface, whereas the 44-kDa molecule may not be. To further support the surface localization of the 74-kDa protein, an overlay assay with biotinylated proteins was performed. The 74-kDa protein comigrated with a protein detected by avidin-peroxidase. Finally, a polyclonal antibody raised against the 74-kDa protein identified the molecule on the surfaces of non-ethanol-treated Vero cells, further suggesting that it is present on the cell membrane.
Binding of dengue virus to surface proteins has already been reported in other cell lines including HepG2, BHK, HL60, BM, human monocytes, and C6/36 cells (2, 8, 18, 27, 33). Interestingly, the molecular weights of the proteins detected in this study with Vero cells resemble those of the two molecules of 40 to 45 and 70 to 75 kDa which bind DEN-2 in HL60 and BM cells (2). However, the characteristics of these protein molecules and their roles in virus infections have not been sufficiently studied.
In U937 cells (a human monocytic cell line) dengue virus binding was reduced after protease and sodium periodate treatments (our unpublished data). These data reveal that one of the molecules involved in virus attachment to the monocytic cell line could be a glycoprotein. In contrast, binding of dengue virus to the 40- and 45-kDa glycoproteins of C6/36 cells (from larvae of A. albopictus) was resistant to sodium periodate treatment (33), although the molecular weight of the protein obtained after treatment was lower, indicating that the nature of the receptor could be different in different susceptible host cells, as has been demonstrated with other viruses such as HIV (1, 7).
Identification of the carbohydrate residues participating in the interaction between dengue virus and the 74-kDa molecule was carried out in overlay assays in the presence of different lectins. The inhibition of dengue virus binding to the 44- and 74-kDa molecules after incubation with ConA and WGA suggests that at least α-mannose or N-acetylglucosamine carbohydrate residues participate in dengue virus attachment. These results are supported by the work of Hung et al. (18), who reported a reduction in plaque formation of BHK cells when ConA and WGA were preincubated with either dengue virus or cells before the virus-cell interaction. The reduction of dengue virus infection in BHK and Vero cells due to ConA and WGA suggests that these two cell lines may possess similar glycoproteins, containing α-mannose or N-acetylglucosamine residues, that bind dengue virus and might be involved in viral entry, such as the 74-kDa protein detected in Vero cells.
Conclusive evidence for the participation of the 74-kDa protein in dengue virus infection was obtained through the inhibition of viral infection by the polyclonal antibody raised against the 74-kDa protein. Viral infection was reduced by more than 50%, indicating that the 74-kDa protein is an important molecule for dengue virus infection, while infection by an unrelated poliovirus was not altered. This is probably the third piece of evidence to indicate that this protein participates in dengue virus infection.
From the results of this study, it is tempting to conclude that HS may be interacting with dengue virus through a motif present in the E protein which has affinity for GAGs, thereby permitting viral attachment, as was previously postulated (4). Subsequently, entry of dengue virus into cells might require the interaction of the virus with a glycoprotein bearing α-mannose or N-acetylglucosamine residues, such as the 74-kDa protein characterized in this study. Further studies will be necessary to isolate the putative coreceptors involved in dengue virus infection of Vero cells. This will facilitate the design of new antiviral agents and vaccines that could interfere with viral entry into target cells. Attempts are being made in our laboratory to completely characterize the 74-kDa protein and its probable relationship with the 44-kDa protein in order to determine whether they constitute parts of a putative receptor complex for dengue virus in Vero cells. Experiments to introduce the cDNA of the 74-kDa protein into a nonpermissive cell and to successfully infect this with DEN-4 are also being contemplated.
ACKNOWLEDGMENTS
We thank Fernando Medina Salvador Chavarria and Angel Marrufo Olivo for technical assistance. We also thank Martha Espinosa, Leopoldo Flores, Lorena Gutiérrez, Saka S. Baba, Miguel Vargas, and Carlos Argüello for critical comments on the manuscript.
This work was supported by a grant from the Consejo Nacional de Ciencia y Tecnología. J. J. Martínez-Barragán had a scholarship from the Consejo Nacional de Ciencia y Tecnología.
REFERENCES
- 1.Bhat S, Spitalnik S L, Gonzalez-Scarano F, Silberberg D H. Galactosyl ceramide or a derivative is an essential component of the neural receptor for human immunodeficiency virus type 1 envelope glycoprotein gp120. Proc Natl Acad Sci USA. 1991;88:7131–7138. doi: 10.1073/pnas.88.16.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielefeldt-Ohmann H. Analysis of antibody-independent binding of dengue viruses and dengue virus envelope protein to human myelomonocytic cells and B lymphocytes. Virus Res. 1998;57:63–79. doi: 10.1016/s0168-1702(98)00087-2. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Maguire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Maguire T, Marks R M. Demonstration of binding of dengue virus envelope protein to target cells. J Virol. 1996;70:8765–8772. doi: 10.1128/jvi.70.12.8765-8772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chungue E, Deubel V, Cassar O, Laille M, Martin P M V. Molecular epidemiology of dengue 3 viruses and genetic relatedness among dengue 3 strains isolated from patients with mild or severe form of dengue fever in French Polynesia. J Gen Virol. 1993;74:2765–2770. doi: 10.1099/0022-1317-74-12-2765. [DOI] [PubMed] [Google Scholar]
- 7.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature (London) 1984;312:763–768. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 8.Daughaday C C, Brandt W E, McCown J M, Russell P K. Evidence for two mechanisms of dengue virus infection of adherent human monocytes: trypsin-sensitive virus receptor and trypsin-resistant immune complex receptors. Infect Immun. 1981;32:469–473. doi: 10.1128/iai.32.2.469-473.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould E A, Clegg J C. Growth, titration and purification of alphaviruses and flaviviruses. In: Mahy B W J, editor. Virology: a practical approach. Oxford, United Kingdom: IRL Press; 1991. pp. 43–78. [Google Scholar]
- 10.Gutierrez A L, Denova-Ocampo M, Racaniello V R, del Angel R M. Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J Virol. 1997;71:3826–3833. doi: 10.1128/jvi.71.5.3826-3833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halstead S B, Larsen K, Kliks S, Peiris J S M, Cardosa J, Porterfield J S. Comparison of P388D1 mouse macrophage cell line and human monocytes for assay of dengue-2 infection-enhancing antibodies. Am J Trop Med Hyg. 1983;32:157–163. doi: 10.4269/ajtmh.1983.32.157. [DOI] [PubMed] [Google Scholar]
- 12.Halstead S B. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 13.Harrop H A, Rider C C. Heparin and its derivatives bind to HIV-1 recombinant envelope glycoproteins, rather than to recombinant HIV-1 receptor, CD4. Glycobiology. 1998;8:131–137. doi: 10.1093/glycob/8.2.131. [DOI] [PubMed] [Google Scholar]
- 14.Heinz F X, Auer G, Stiasny K, Holzmann H, Mandl C, Guirakhoo F, Kunz C. The interactions of the flavivirus envelope proteins: implications for virus entry and release. Arch Virol Suppl. 1994;9:339–348. doi: 10.1007/978-3-7091-9326-6_34. [DOI] [PubMed] [Google Scholar]
- 15.Helenius A. Alphavirus and flavivirus glycoproteins: structures and functions. Cell. 1995;81:651–653. doi: 10.1016/0092-8674(95)90523-5. [DOI] [PubMed] [Google Scholar]
- 16.Henchal E A, Putnak J R. The dengue viruses. Clin Microbiol Rev. 1990;3:376–396. doi: 10.1128/cmr.3.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold B C, Gerber S I, Belval B J, Siston A M, Shulman N. Differences in the susceptibility of herpes simplex virus types 1 and 2 to modified heparin compounds suggest serotype differences in viral entry. J Virol. 1996;70:3461–3469. doi: 10.1128/jvi.70.6.3461-3469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung S-L, Lee P-L, Chen H-W, Chen L-K, Kao C-L, King C-C. Analysis of the steps involved in dengue virus entry into host cells. Virology. 1999;257:156–167. doi: 10.1006/viro.1999.9633. [DOI] [PubMed] [Google Scholar]
- 19.Jackson T, Ellard F M, Ghazaleh R A, Brookes S M, Blakemore W E, Corteyn A H, Stuart D I, Newman J W, King A M. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70:5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kliks S. Antibody-enhanced infection of monocytes as the pathogenic mechanism for severe dengue illness. AIDS Res Hum Retrovir. 1990;6:993–998. doi: 10.1089/aid.1990.6.993. [DOI] [PubMed] [Google Scholar]
- 21.Krusat T, Streckert H J. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142:1247–1254. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- 22.Kurane I, Rothman A L, Livingstone P G, Green S, Gagnon S J, Janus J, Innis B L, Nimmannitya S, Nisalak A, Ennis F A. Immunopathologic mechanisms of dengue hemorrhagic fever and dengue shock syndrome. Arch Virol. 1994;9:54–64. doi: 10.1007/978-3-7091-9326-6_7. [DOI] [PubMed] [Google Scholar]
- 23.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linker A, Hovingh P. Heparinase and heparitinase from Flavobacteria. Methods Enzymol. 1972;28:902–911. [Google Scholar]
- 25.Littaua R, Kurane I, Ennis F A. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J Immunol. 1990;144:3183–3186. [PubMed] [Google Scholar]
- 26.Mady B J, Erbe D V, Kurane I, Fanger M W, Ennis F A. Antibody-dependent enhancement of dengue virus infection mediated by bispecific antibodies against cell surface molecules other than Fcγ receptors. J Immunol. 1991;147:3139–3144. [PubMed] [Google Scholar]
- 27.Marianneau P, Mégret F, Olivier R, Morens D M, Deubel V. Dengue 1 virus binding to human hepatoma HepG2 and simian Vero cell surfaces differs. J Gen Virol. 1996;77:2547–2554. doi: 10.1099/0022-1317-77-10-2547. [DOI] [PubMed] [Google Scholar]
- 28.Muñoz M L, Cisneros A, Cruz J, Das P, Tovar R, Ortega A. Putative dengue virus receptors from mosquito cells. FEMS Microbiol Lett. 1998;168:251–258. doi: 10.1111/j.1574-6968.1998.tb13281.x. [DOI] [PubMed] [Google Scholar]
- 29.Putnak J R. Progress in the development of recombinant vaccines against dengue and other arthropod-borne flaviviruses. In: Kurstak E, editor. Modern vaccinology. New York, N.Y: Plenum Press; 1994. pp. 231–252. [Google Scholar]
- 30.Putnak J R, Kanesa-Thasan N, Innis B L. A putative cellular receptor for dengue viruses. Nat Med. 1997;3:828–829. doi: 10.1038/nm0897-828. [DOI] [PubMed] [Google Scholar]
- 31.Ramos Castañeda J, Imbert J L, Barrón B L, Ramos C. A 65-kDa trypsin-sensible membrane cell protein as a possible receptor for dengue virus in cultured neuroblastoma cells. Neurovirology. 1997;3:435–440. doi: 10.3109/13550289709031189. [DOI] [PubMed] [Google Scholar]
- 32.Rey F A, Heinz F X, Mandl C, Kunz C, Harrison S C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature (London) 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 33.Salas-Benito J S, del Angel R M. Identification of two surface proteins from C6/36 cells that bind dengue type 4 virus. J Virol. 1997;71:7246–7252. doi: 10.1128/jvi.71.10.7246-7252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trybala E, Bergstrom T, Spillmann D, Svennerholm B, Flynn S J, Ryan P. Interaction between pseudorabies virus and heparin/heparan sulfate. Pseudorabies virus mutants differ in their interaction with heparin/heparan sulfate when altered for specific glycoprotein C heparin-binding domain. J Biol Chem. 1998;273:5047–5052. doi: 10.1074/jbc.273.9.5047. [DOI] [PubMed] [Google Scholar]
- 35.Walther W, Stein U, Uckert W. Rapid method of total RNA mini-preparation from eucaryotic cells. Nucleic Acids Res. 1993;21:1682. doi: 10.1093/nar/21.7.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe S. The receptor and pathways for human cytomegalovirus entry. Nippon Rinsho. 1998;56:44–49. [PubMed] [Google Scholar]
- 37.Woodward M P, Young W W, Jr, Bloodgood R A. Detection of monoclonal antibodies specific for carbohydrate epitopes using periodate oxidation. J Immunol Methods. 1985;78:143–153. doi: 10.1016/0022-1759(85)90337-0. [DOI] [PubMed] [Google Scholar]
- 38.Yanagishita M, Midura R J, Hascall V C. Proteoglycans: isolation and purification from tissue cultures. Methods Enzymol. 1987;138:279–289. doi: 10.1016/0076-6879(87)38023-1. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Z M, Gershon M, Ambron R, Gabel C, Gershon A. Infection of cells by varicella zoster virus: inhibition of viral entry by mannose 6-phosphate and heparin. Proc Natl Acad Sci USA. 1995;92:3546–3550. doi: 10.1073/pnas.92.8.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zsak L, Sugg N, Ben-Porat T, Robbins A K, Whealy M E, Enquist L W. The gIII glycoprotein of pseudorabies virus is involved in two distinct steps of virus attachment. J Virol. 1991;65:4317–4324. doi: 10.1128/jvi.65.8.4317-4324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]