Abstract
Senescence and epigenetic alterations stand out as two well-characterized hallmarks of aging. When cells become senescent, they cease proliferation and release inflammatory molecules collectively termed the Senescence-Associated Secretory Phenotype (SASP). Senescence and SASP are implicated in numerous age-related diseases. Senescent cell nuclei undergo epigenetic reprogramming, which intricately regulates SASP expression. This review outlines the current understanding of how senescent cells undergo epigenetic changes and how these alterations govern SASP expression.
Subject terms: Senescence, Cell biology
Introduction
According to the World Health Organization, the global population aged 60 years and above is projected to increase from 1 billion in 2020 to 1.4 billion by 2030 and is expected to double by 2050, reaching 2.1 billion. This indicates a rapid aging of the world population. As aging is correlated with an elevated vulnerability to various common diseases, including diabetes, Alzheimer’s disease (AD), Parkinson’s disease (PD), cardiovascular disease, chronic obstructive pulmonary disease (COPD), osteoporosis (OP), osteoarthritis (OA), and cancer1, it will impose tremendous health and economic burdens on a global scale. Addressing this unprecedented and rapid rise in aging requires an equally unprecedented and swift approach. For this reason, over the past few decades, there has been a rapid surge in aging research aimed at comprehending the aging process at the cellular and molecular biology levels. Biologically, aging arises from the accumulation of a diverse range of molecular and cellular damage over time. There are 12 recognized ‘hallmarks of aging’: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient-sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, altered intercellular communication, disabled macroautophagy, chronic inflammation, and dysbiosis2,3. In this review, we will explore how three of these hallmarks—epigenetic alterations, cellular senescence, and chronic inflammation—are interconnected.
Cellular senescence, discovered in human diploid cells in the 1960s, is characterized by a loss of proliferative capacity4, later linked to telomere shortening. Senescence in adult tissues can be triggered by factors beyond replicative exhaustion, including a persistent DNA damage response (DDR) induced by various internal and external factors such as oncogenic activation, oxidative and genotoxic stress, mitochondrial dysfunction, exposure to ultraviolet or γ-irradiation, and chemotherapy agents5,6. In the study of cellular senescence, various in vitro models are primarily employed using human or murine fibroblast cells. One common model is replicative senescence (RS), which is induced by continuously passaging fibroblasts until they cease proliferation. Besides the RS model, stress-induced premature senescence models are also used. For example, in the oncogene-induced senescence (OIS) model, an activated oncogene (typically, RAS or BRAF) is ectopically expressed in human fibroblasts. Initially, these cells grow rapidly but eventually stop proliferating and become senescent. Additionally, DNA-damaging chemicals and chemotherapeutic drugs, such as Doxorubicin and Etoposide, or ionizing radiation (typically a single dose of 10 Gy X-ray exposure), can trigger therapy-induced senescence (TIS) in normal fibroblast or tumor cells.
The accumulation of senescent cells over an animal’s lifespan contributes to age-related diseases, including renal dysfunction, type-2 diabetes, idiopathic pulmonary fibrosis (IPF), metabolic dysfunction associated fatty liver disease (MAFLD), osteoarthritis, and declining immune function3,5. In addition to growth arrest, senescent cells display distinctive phenotypic changes, encompassing chromatin remodeling, metabolic reprogramming, and the establishment of a multifaceted proinflammatory secretome5,7. This secretome comprises various proinflammatory cytokines, chemokines, growth factors, and proteases, collectively known as the senescence-associated secretory phenotype (SASP)7,8. As a source of chronic inflammation, SASP contributes to the development of ‘inflammaging’, a chronic, sterile, low-grade inflammation during aging that plays a role in the pathogenesis of age-related diseases9.
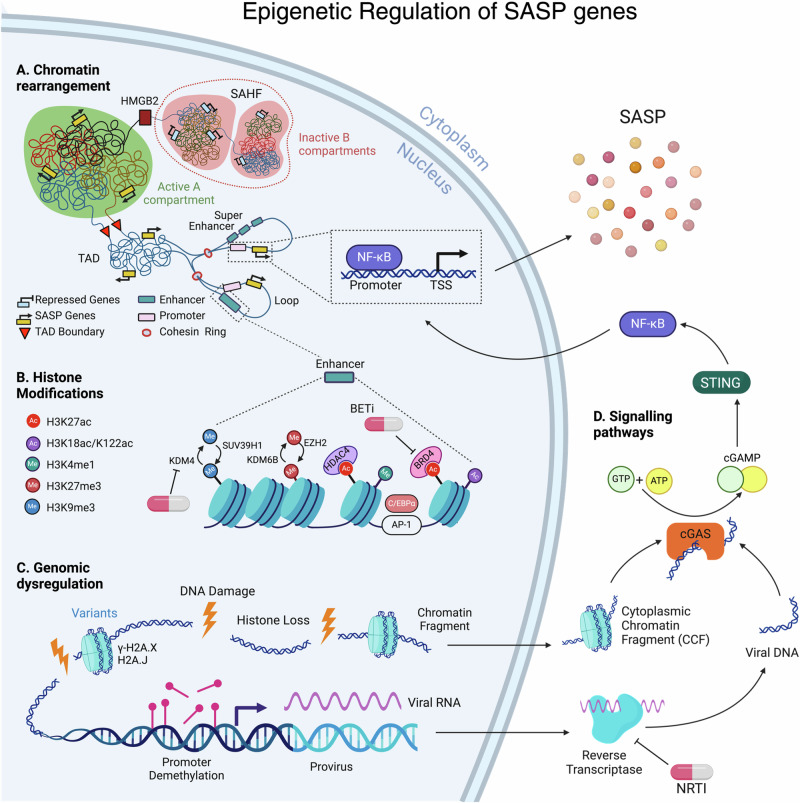
Multiple transcription factors and signaling pathways have been identified as regulators of SASP expression. The transcription factor NF-κB is recognized as a ‘master regulator’ of SASP expression (Fig. 1)10. Senescent cells release their damaged DNA into the cytoplasm, forming cytoplasmic chromatin fragments (CCF)11. Within the cytoplasm, CCF is detected by the cytoplasmic DNA sensor cGAS, which activates the cGAS-STING-NF-κB pathways to induce SASP expression (Fig. 1)12. In addition to the cGAS-STING-NF-κB pathway, several other factors, including DNA damage signaling, C/EBPß, GATA4, and pathways such as p38 MAP-kinase, mTOR, JAK-STAT, and Notch, regulate SASP5,13,14. Multiple lines of evidence suggest that epigenetic alterations in senescent cells also regulate SASP expression.
Fig. 1. Senescent cells exhibit a dynamic epigenetic landscape orchestrating the expression of the Senescence-Associated Secretory Phenotype (SASP).
A Chromatin rearrangement: Within heterochromatic B compartments, long-range interactions often occur, culminating in the formation of Senescence-Associated Heterochromatin Foci (SAHF). HMGB2 plays a pivotal role in preventing the incorporation of SASP gene loci into SAHF structures. Conversely, the euchromatic A compartment fosters SASP enhancer-promoter looping within Topologically Associating Domains (TADs). Here, enhancer and super-enhancer elements interact with promoters, facilitating the binding of transcription factors, notably the master regulator of SASP, NF-κB. B Histone Modifications: Enhancers exhibit enrichment in H3K4me1 and multiple histone acetyl marks, including H3K27ac, H3K18ac, and H3K122ac. The transcription factor Activator Protein 1 (AP-1) acts as a pioneering factor binding to enhancers, while HDAC4 is recruited to H3K27ac loci to regulate chromatin status at AP-1-regulated enhancers. Epigenetic reader BRD4 is recruited to induce NF-κB-regulated SASP genes in senescent cells. Repressive histone marks H3K9me2/3 and H3K27me3 also influence SASP expression levels. C Genomic Dysregulation: During senescence, DNA damage leads to significant histone loss from chromatin and in the formation of cytoplasmic chromatin fragments (CCFs). These damage sites are enriched with H2A.X and H2A.J histone variants. Additionally, DNA demethylation at provirus loci in senescent cells can reactivate endogenous retroviruses (ERVs). The reverse transcriptase (RT) converts viral RNA into DNA in the cytoplasm. D Signaling Pathways: Subsequently, cytoplasmic viral DNA and CCFs can upregulate SASP factors through cGAS/STING-dependent activation of NF-κB. Nucleoside Reverse Transcriptase Inhibitors (NRTI), BET inhibitors and KDM4 inhibitors can be used as senomorphic agents. The SAHF adjacent to the nuclear membrane is for representation purposes only; it does not depict its position within the nucleus.
Epigenetic alterations, which involve modifications to the genome that don’t alter the DNA sequence, play a significant role in modulating the aging process. These modifications encompass abnormal chromatin remodeling, changes in DNA methylation patterns, histone modification, reduction in overall levels of core histones, accumulation of histone variants, and the dysregulated functioning of non-coding RNAs believed to exert their influence primarily at the level of transcription in gene expression15–18. This review will explore how epigenetic changes are orchestrated in senescent cells and how they regulate SASP. Additionally, it will discuss how epigenetic-modulating drugs can inhibit SASP expression, acting as senomorphics.
However, the regulation of SASP is not universally applicable across all senescence models. This diversity introduces layers of complexity, as different senescence-inducing agents and cellular contexts activate distinct pathways, including epigenetic mechanisms, to control SASP expression. Moreover, SASP is comprised of numerous molecules that vary across different senescence models19 and in vivo scenarios, reflecting the complexity of its regulation. Understanding the interplay between epigenetic regulation and the diverse array of molecules comprising SASP enriches our comprehension of the multifaceted nature of cellular senescence.
Reorganization of heterochromatin is essential for the SASP
The intricate packaging of DNA into chromatin results in spatial separation within the nucleus, forming distinct compartments known as compartments A and B20. This compartmentalization is essential for regulating gene expression and maintaining genomic stability20. Euchromatin, primarily located in compartment A, is associated with active gene transcription and is less condensed, allowing for easier access by transcription factors and regulatory proteins20. In contrast, heterochromatin, mainly situated in compartment B, is characterized by densely packed chromatin and typically contains silenced or repressed genes20 (Fig. 1). Compartment switching can impact gene expression by transitioning from A to B or B to A21. In both OIS and RS nuclei, B compartments frequently merge with adjacent A compartments, forming BA-switching regions22,23. The binding of the structural maintenance of chromosomes (SMC) complex condensin is notably increased at these B-to-A-switching regions, coinciding with the upregulation of SASP genes22. These regions exhibit significant enrichment of gene groups related to SASP. For instance, the gene encoding the master regulator of SASP, NF-κB, is predominantly located in A compartments23, remaining active, along with other senescence genes, correlating with their heightened expression22,23. Senescent cells show changes in chromatin interactions22–25. In RS cells, these changes involve less long-range and more short-range interactions23. In contrast, OIS is associated with a decrease in short-range interactions at heterochromatic regions and an increase in long-range contacts with other repressive regions. This increase in long-range interactions within heterochromatic B compartments often leads to chromatin compaction22,24,25 and the formation of structures known as Senescence-Associated Heterochromatin Foci (SAHF) (Fig. 1)26.
Dissection of the formation of SAHF revealed that each chromosome condenses into a single SAHF focus27. SAHFs originate from late-replicating genome regions marked with heterochromatin, including H3K9me2, H3K9me3, and H3K27me3, along with crucial markers like H4K20me3, H4K16ac, and DNA methylation, indicating a complex organization of repressive chromatin within SAHFs28,29. Their structure reflects broader chromatin organization principles30, with concentric layers: H3K9me3 and H4K20me3 delineate the interior, surrounded by H3K27me3, while active marks such as H3K4me3 and H3K36me3 occupy the perichromatin space, distinct from the interior31. Several studies have shown that HIRA, a known chromatin regulator, together with other members of the complex, ASF1a and UBN1, are essential for SAHF formation27,32–34. Preventing the translocation of HIRA to PML nuclear bodies, structures ranging from 0.2–1.0 μM in diameter containing the PML protein and various other proteins, inhibits the formation of SAHF32. Similarly, disrupting PML bodies themselves also hinders SAHF formation34. Although SAHF are not essential for cell cycle arrest35,36, their disruption correlates with a decrease in SASP expression. This is evident when SAHF formation is disrupted either through HIRA depletion37 or a reduction in nuclear pore density35. However, the correlation or the necessity of SAHF for SASP expression is yet to be understood. The function of SAHFs becomes further perplexing, particularly as their formation is not readily observed in vivo, as noted for HGPS patient cells or preneoplastic lesions expressing other senescence markers38. Furthermore, even in vitro, mouse embryo fibroblasts (MEFs) and mouse skin fibroblasts fail to form robust punctate SAHFs in response to OIS or RS models39.These observations further raise questions about the requirement of SAHFs in SASP expression.
The high mobility group box proteins HMGBs, abundant nuclear proteins characterized by their distinctive HMG-box DNA-binding domains, have the capability to distort DNA through processes like unwinding, bending, or looping40. HMGBs experience depletion in senescent cells and aging tissues40,41. This depletion of HMGB2 triggers a shift between constitutive and facultative heterochromatin, coinciding with an increase in SASP genes in replicative senescence40. Although in replicative senescence, HMGB2 binding remodeling does not impact SASP loci40, during oncogene-induced senescence (OIS), chromatin-bound HMGB2 effectively binds to SASP gene loci41. This binding protects against the spread of repressive heterochromatin into SASP gene loci, thereby preventing the incorporation of SASP gene loci within SAHF structures and subsequently silencing SASP gene expression (Fig. 1)41. However, depletion of HMGB2 does not affect SAHF formation in either the RS or OIS models40,41.
In contrast to the facultative heterochromatin formation of SAHF, the constitutive peri/centromeric satellite heterochromatin undergoes striking decompaction in various senescent cell models independently of SAHF formation, a phenomenon coined as senescence-associated distension of satellites (SADS)42. This decompaction in the pericentrosome of certain chromosomes leads to new DNA-DNA interactions in specific genomic regions, interacting with inflammatory SASP gene loci, altering the chromatin state of these regions, and influencing the regulation of SASP genes43. Therefore, the compartmental switching of euchromatin and heterochromatin (BA switching), the formation of facultative heterochromatin (SAHF), and the decompaction of constitutive heterochromatin (SADS) together play intricate roles in proliferation arrest and SASP regulation in senescent cells. In conclusion, the intricate DNA packaging into chromatin and its dynamics in senescent cells highlight the complexity of SASP regulation.
Alterations in large-scale 3D chromatin landscapes regulate the SASP
Besides the global levels, such as interchromosome contacts and A/B compartments, chromatin’s higher-order structure is organized at an intermediate scale ranging from tens of kilobases to a few megabases. Within this scale, chromatin is arranged into spatial domains known as topologically associated domains (TADs) and loops44. TADs represent regions of the genome where DNA sequences interact more frequently with each other compared to sequences outside the domain. These domains are believed to regulate gene expression by enabling enhancer-promoter looping within the same TAD, where enhancer elements interact with promoters to influence gene transcription by facilitating the binding of transcription factors (TFs)44. The regulation of SASP involves rewiring the enhancer-promoter (EP) network, where hyper-connected enhancers to the promoters of SASP correspond to increased SASP transcription (Fig. 1)45,46. Human methyltransferase-like 14 (METTL14), together with methyltransferase-like 3 (METTL3), forms a crucial heterodimer complex (METTL3–METTL14) that catalyzes N6-methyladenosine (m6A) methylation in mammalian nuclear RNAs47. Despite their primary role in RNA methylation, the METTL3-METTL14 complex also facilitates senescence-associated EP contact and SASP expression in OIS cells48. Their genome-wide redistribution in senescent cells promotes SASP expression through chromatin looping in an enzymatic activity-independent manner48. Upon senescence entry, enhancers identified by the H3K27ac marker of active enhancers undergo global remodeling at the TAD scale40,49,50. This occurs in replicative (RS)40, oncogene-induced (OIS), and DNA-damaging drug (Etoposide)-induced senescence (TIS) in human fibroblasts50, as well as in RS in murine fibroblasts49. The high mobility group box protein, HMGB1 and HMGB2, delineate the boundaries of TAD/loop domains40,51. Consequently, when HMGB1 is lost from the nucleus upon senescence entry, it influences chromatin topology, thereby contributing to changes in gene expression, including SASP40,51. Chromatin accessibility increased significantly in senescent cells52,53. In both RS and OIS cells, accessibility surged in gene-poor regions such as enhancers, intergenic, intronic, and repeat regions while diminishing in gene-rich areas52,53. This increased accessibility in enhancers correlated with the upregulated genes, including SASP factors like IL1A53. In OIS, the chromatin-binding protein HMGA1 (High Mobility Group AT-hook 1) plays a pivotal role by binding to AT-rich DNA regions, thereby influencing chromatin structure and promoting a more open and accessible chromatin state53. Super-enhancers (SEs) consist of large clusters of neighboring enhancers with high-fold enhancer activity, which collectively drive cell-type-specific gene expression54. Activation of super-enhancers (SEs) could potentially explain the robust expression of SASP genes in OIS and TIS cells50. However, it remains to be determined how super-enhancer (SE) regions remain outside of SAHF and if HMGB2, similar to SASP loci41, protects the SE regions from sequestering in SAHF. Similar to senescent cells, hyper-connected age-specific enhancer communities drive transcriptional changes enriched in essential pathways in the livers of aged mice46.
In summary, alterations in large-scale 3D chromatin landscapes, such as TADs and loops, play a crucial role in regulating the SASP. This regulatory mechanism involves enhancer-promoter looping within TADs, with factors like METTL3-METTL14 complex and HMGB1/2 influencing chromatin topology and accessibility to drive SASP expression during senescence.
Histone H3 lysine modifications and pioneer transcription factors play a pivotal role in regulating SASP
Histone modifications are a central component of chromatin, crucial for gene regulation. They play essential roles in various biological processes, particularly in their involvement in enhancer-promoter interactions and facilitating transcription. Alterations in histone modifications are closely linked to aging and senescence. Active enhancers uphold an open chromatin state to maintain their functionality, preventing repressive chromatin recruitment. They exhibit characteristics including enrichment in histone modifications such as H3K4me1 and H3K27ac, along with the presence of enhancer RNAs (eRNAs), a class of noncoding RNA transcribed at active enhancers55. Conversely, enhancers can be silenced through various chromatin-based mechanisms, including DNA methylation, H3K27 and H3K9 trimethylation (H3K27me3, H3K9me3)55,56. Promoters of actively transcribed genes exhibit enriched H3K4me3 and acetylation on histone H3 and H456. Additionally, actively transcribed genes often demonstrate elevated levels of H3K36me3 and H3K79me3 within the gene body56. In senescent cell genomes, the most notable chromatin state transitions occurred at enhancers, with promoters being minimally affected50,57. The majority of enhancer activation occurred de novo from previously unmarked chromatin regions57. Senescence-activated super-enhancers (SEs) exhibit enrichment in H3K4me1 and multiple histone acetyl marks, such as H3K27ac, H3K18ac, H3K122ac, and H4K5ac (Fig. 1)58. They actively transcribe enhancer RNAs (eRNAs), gain binding of the histone acetyltransferase p300, and directly drive senescence-specific gene expression58.
Pioneer factors are transcription factors that possess the distinct capability to initiate the opening of closed chromatin59. The transcription factor (TF) activator protein 1 (AP-1) acts as a pioneering factor that binds to enhancers associated with SASP and other senescence-related genes, initiating the opening of closed chromatin and subsequent gene activation in RS and OIS models57. Remarkably, depletion of the AP-1 superfamily member c-Jun alone is adequate to partially reverse the cell cycle arrest57. HDAC4, a class IIa histone deacetylase that is catalytically inactive but functions as an epigenetic reader, is swiftly recruited to H3K27ac loci to monitor the acetylation status of H3K27 (Fig. 1)60. HDAC4 oversees the chromatin status at AP-1 regulated SEs through interaction with class I HDAC360. Upon entry into senescence, the buffering activity of HDAC4 is diminished due to its degradation, allowing the AP-1 mediated senescence transcriptional program to be unleashed60. Consequently, while depletion of AP-1 affects the expression of NF-κB-regulated SASP genes57, depletion of HDAC4 enhances their expression60. Additionally, the BET protein BRD4, an epigenetic reader recognizing H3K27ac, and the transcription factor C/EBPα bind to a subset of these senescence-activated enhancers flanking key SASP genes, thereby facilitating their expression49,50. Therefore, H3K27ac is one of the crucial histone modifications, where the pioneering factor AP-1 and epigenetic reader BRD4 are recruited to ultimately induce the NF-κB-regulated SASP genes in senescent cells (Fig. 1).
A redistribution of H3K27me3 and H3K4me3 consequently has a significant impact on gene expression. H3K4me3 is activating, whereas H3K27me3 is a repressive histone modification. H3K27me3 and H3K4me3 tend to co-localize in regions termed “mesas,” while H3K27me3 is depleted from regions referred to as “canyons” in replicative senescent cells61. EZH2 and MLL1 (also known as KMT2A) are histone lysine methyltransferases responsible for H3K27me3 and H3K4me3, respectively. Depleting or inhibiting EZH2 leads to the removal of H3K27me3 from SASP loci, enabling the derepression of SASP genes in multiple models, which is also proposed as a potent approach for cancer immunotherapy61–64. Similarly, the overexpression of the histone H3K27 demethylase enzyme JMJD3 (or KDM6B) specifically reduces H3K27me3 levels at the IL-6 and IL-8 gene loci, elevating the expression of these genes along with other SASP factors65. Inhibiting or depleting MLL1 decreases SASP expression. However, the overall reduction in H3K4me3 at SASP loci was moderate and did not closely align with the significant decreases in SASP transcription. Instead, MLL1 inhibition or depletion reduces SASP expression by preventing activation of the DNA-damage response (DDR) pathway66. H3K9me2/3, along with H3K27me3, are repressive histone marks. Loss of H3K9me2 around the major SASP gene IL-6 and IL-8 promoters correlates with the induction of the expression of these SASP factors67. JMJD2 (or KDM4), the histone demethylase enzyme, targets H3K9, while SUV39H1 is a histone methyltransferase that specifically trimethylates H3K9. The expression of KDM4 protein is increased, and SUV39H1 is decreased in senescent cells, which are associated with a diminished methylation status of H3K9 and enhanced SASP expression (Fig. 1)68,69. In OIS cells, there is a widespread rise in transcription-activating histone marks H3K79me2/370. DOT1L, the sole methyltransferase for H3K79, is upregulated in senescent cells, responsible for this enrichment of H3K79me2/370. This elevation of H3K79me2/3 at the IL1A gene locus, but not other SASP factors, corresponds to an increase in IL1A transcription and protein expression at the cell surface70. By inducing IL1A expression, DOT1L sustains a positive feedback loop that promotes the transcription of multiple SASP factors71. The delicate balance among various histone H3 modifications and the expression of their corresponding enzymes are pivotal in regulating SASP expression across different senescent models.
The transcriptional changes observed during senescence are underpinned by the balance between active and repressive chromatin marks. These dynamic chromatin rearrangements are orchestrated by factors such as pioneer transcription factors, epigenetic readers, and chromatin remodeling enzymes. Their interplay shapes the SASP profile, suggesting potential implications for aging and age-related diseases. The intricate involvement of histone modifications in SASP expression highlights the potential for repurposing various histone-modifying enzyme inhibitors used for other diseases as senomorphic agents (Table 1).
Table 1.
Epigenetic modulators under clinical trials for different disease conditions
| Target | Drug | Used in diseases | Safety | Status (clinical trial) | References (clinicaltrials.gov) |
|---|---|---|---|---|---|
| BRD4 | BMS-986158, BMS-986378 (CC-90010) | Pediatric Cancer | Not reported | Phase 1, On going | NCT03936465 |
| BET | Molibresib (GSK525762) | Solid tumors | Tolerated with some toxicity issues | Phase I and II, Completed | NCT01587703 |
| BET | Apabetalone (RVX-208) | Cardiovascular disease | Well tolerated | Phase II, Completed | |
| KDM4 | TACH101 | Advanced or Metastatic Cancer | Not reported | Phase 1, On going | NCT05076552 |
| DOT1L | EPZ-5676 | Leukemia | Well tolerated | Phase 1, Completed | |
| NRTI | Emtricitabine | Alzheimer’s disease (AD) | A prescription medicine used in Human Immunodeficiency Virus-1 (HIV-1) infection | Phase 1, On going | NCT04500847 |
SASP expression is correlated with loss of canonical histones
The abundance of histones regulates chromatin movement and decompaction. In senescence, loss of canonical histones and heterochromatin occur72–74, which is linked to decreased biosynthesis and increased histone mRNA and protein degradation11,72,73,75,76. This decrease in histone HMGB1 not only plays a crucial role in maintaining nucleosome assembly, as mentioned earlier, but its nuclear level is also closely correlated with total histone levels77. Hence, the loss of nuclear HMGB1 in senescent cells is another contributing factor to histone loss and SASP expression51,78. Additionally, DNA damage induces substantial histone loss (20–40%) from chromatin79. Therefore, damaged chromatin experiences increased mobility, decompaction, and flexibility due to histone loss79. H4 and H3 enrichment decreased at the promoters of SASP-related genes and cell cycle inhibitor genes in a peroxide-induced senescence model in human fibroblasts, suggesting that the loss of histones in senescence-associated gene regions further promotes nucleosome decomposition and upregulates senescence-associated gene transcription76. In therapy-induced senescent cells, persistent DNA damage foci, referred to as ‘DNA segments with chromatin alterations reinforcing senescence’ (DNA-SCARS)80 might potentially play a role in evicting histones from nucleosomes, thereby making the chromatin more flexible for transcriptional reprogramming (Fig. 1). Additionally, DNA-SCARS could be precursors to the formation of CCF (Fig. 1)11. However, the connection between histone loss and CCF remains unknown. While DNA-SCARS and CCF are known to play pivotal roles in SASP expression12,80, and the loss of histones is correlated with SASP expression, the specific role of histone loss in SASP expression remains undefined. Studies on histone loss in different aged tissues are limited. Notably, naive CD4 + T cells in older adults have shown reduced histone expression and increased SASP expression. Surprisingly, knocking down the transcription factor NPAT, which controls core histone gene expression in young activated CD4+ T cells also led to increased expression of multiple SASP components further suggesting a link between histone loss and SASP expression81.
In conclusion, the intricate interplay between histone abundance, chromatin dynamics, and SASP expression underscores the multifaceted role of histone loss in cellular senescence, paving the way for further exploration into its mechanistic underpinnings.
Histone variants
In addition to the loss of canonical histones, aged and senescent cells are also characterized by a unique landscape of histone variants. These variants are isoforms of histones with specialized roles in the regulation of transcription, DNA repair, and chromatin maintenance82,83. They also facilitate several aspects of the senescence phenotype, such as proliferation arrest, formation of SAHF, and SASP expression84–87. Canonical histones, comprising H2A, H2B, H3, and H4, are synthesized during the S phase, whereas histone variants are typically deposited into the DNA in a replication-independent manner82,88,89. Consequently, several of these variants tend to accumulate in post-mitotic and senescent cells, while the abundance of canonical histones decreases15,72,85,90.
A variant of histone H3 has been implicated in senescence and aging. H3.3 differs from its canonical H3.1 and H3.2 counterparts by only five and four amino acids89. In post-mitotic cells, H3.3 accumulates with age and progressively replaces H3.1 and H3.290–92. In senescent cells, there is also an increase in the ratio of H3.3 to total H387, and its histone chaperone HIRA plays a role in SAHF formation32, while also maintaining chromatin dynamics87. H3.3 can undergo N-terminal tail cleavage by the lysosomal protease Cathepsin L1 to remove posttranslational modifications controlling cell cycle regulators93. Both H3.3 and its cleaved product H3.3cs1 are increased in various senescent cell types11,87,93,94 and can trigger senescence when ectopically expressed93. Interestingly, BRD4 plays a protective role against histone H3.3 clipping, especially at genomic loci associated with senescence-related genes during hematopoiesis95. Conversely, the absence of BRD4 triggers H3.3 clipping induction, leading to its association with senescence-specific gene loci and the emergence of H3K27ac, H3K122ac, and H3K4me3 modifications95. Consequently, this enhances chromatin accessibility and facilitates the transcription of senescence-specific genes, including SASP95. Intriguingly, BRD4 binding at enhancers is necessary for SASP induction in OIS, while its depletion in normal hematopoiesis induces senescence and SASP expression in hematopoietic cells. Another H3 variant called CENP-A is downregulated or displaced during senescence42,96,97. CENP-A is normally present in centromeres, and its loss is thought to contribute to cell-cycle arrest due to the decompaction of centromeric DNA42,96, likely correlated with SASP expression, as observed with the decompaction of pericentromeric heterochromatin in senescent cells43.
The H2A family has the greatest number of histone variants, of which four play a defined role in senescence. H2A.X and H2A.J associate with sites of DNA damage and contribute to the activation of SASP (Fig. 1)80,85,98. The level of H2A.J rises in human senescent fibroblasts85,99, in aged human skin100, and in different mouse tissues, when exposed to persistent DNA damage85,101. H2A.J is enriched in SASP promoters85, facilitating changes in chromatin accessibility that induce SASP expression101. Another variant H2A.X is 95% identical to H2A but has a longer C-terminal which can be phosphorylated on the Ser-139 residue (γ-H2A.X)102. The latter serves as a beacon to recruit DNA repair proteins at the sites of double-stranded DNA breaks, induces p53 expression, and subsequently activates the senescence signaling pathways80,102–105. γ-H2A.X increases during aging in various mouse and human tissues91,106–109, likely as a result of persistent DNA damage (or DNA-SCARS) and deprotected telomeres80,103,110. CCF is enriched in γ-H2A.X11, and the depletion of H2A.X impairs senescence and SASP in human fibroblasts following irradiation80. Consequently, DNA damage and the presence of H2A.X are well-recognized hallmarks of senescence111. Lastly, the variant macroH2A was shown to accumulate in senescent cells and in aged mouse and primate tissues86,112,113. During senescence, this variant is thought to repress proliferation and stabilize SAHF32,112, as well as contribute to the proper timing of SASP expression86,114. The increased expression of macroH2A in senescent cells activates a specific subset of senescent genes, leading to growth arrest and induction of key SASP genes32,86,114,115. However, the removal of macroH2A from SASP loci during OIS suggests that the induction of SASP genes may not directly result from macroH2A binding but rather from other underlying causes associated with growth arrest86,115.
In conclusion, the replacement of canonical histones with their variants is a common characteristic of aging and senescence. These changes facilitate several aspects of the senescence phenotype, such as SAHF formation or SASP expression. In addition, the half-life of some variants can extend to several months116,117, making them vulnerable to deleterious chemical modifications over time118–120. These changes negatively impact histone function and may contribute to epigenetic dysregulation during aging121,122.
DNA methylation
DNA methylation is a critical biological process capable of regulating gene expression by either recruiting proteins involved in gene repression or inhibiting the binding of transcription factors to DNA123. Hannum et al. and other researchers developed a widely used biomarker of aging by leveraging age-associated alterations in the mammalian DNA methylome124,125. This approach yields an estimated age for various tissues and organs, commonly referred to as the epigenetic methylation clock124,125. Older studies indicated that aging and senescence result in global CpG methylation losses and focal gains at promoter CpG islands126,127, while recent advancements in next-generation sequencing and bisulfite methods for measuring DNA 5mC content consistently fail to observe significant age-related changes in global DNA methylation127. Enzymes involved in DNA methylation, such as DNMT and TET proteins, also demonstrate no obvious overall effect of age on expression127. Comparison of different types of senescence showed that only RS is accompanied by epigenetic methylation aging of the cell, while OIS and DNA damage-induced senescence is not128,129. The overall hypomethylation in RS cells can be explained by the decline of methyltransferase DNMT1 activity130. In contrast to the global hypermethylation at promoter CpG islands, it was also shown that many inflammation-associated SASP genes were hypomethylated131. Therefore, DNA methylation seems to play an important role in regulating the senescent transcriptome and SASP. Although DNA methylation is typically associated with less accessible chromatin at enhancers55, whether senescence-induced DNA hypomethylation plays a role in enhancer activation for SASP genes in RS cells remains to be elucidated. Nevertheless, the DNA demethylation and decondensation of heterochromatin associated with aging and senescence can trigger the transcription of Endogenous retroviruses (ERVs), thereby activating SASP factors132.
Retrotransposable elements and endogenous retroviruses (ERVs) are responsible for ‘inflammaging’
Retrotransposable elements, also known as retrotransposons, are mobile genetic sequences capable of relocating from one genomic site to another via an RNA intermediate. Two main types are notable: Long INterspersed Elements (LINEs), which possess their own proteins crucial for retrotransposition, and Short INterspersed Elements (SINEs), which are brief, non-coding RNAs exploiting the LINE protein machinery. Although within the human genome, LINE-1 (L1) constitutes approximately 17%, totaling around 500,000 copies133,134, only a small fraction, approximately 80–100 copies, are retrotransposition-competent134. Increased expression of these retrotransposition-competent L1s are also regarded as a characteristic feature of aging135. L1 activity leads to elevated cytoplasmic L1 cDNA levels via L1 reverse transcriptase during aging and senescence135,136. This process triggers a type I interferon response through cGAS cytosolic DNA sensing, consequently promoting ‘inflammaging’ in various tissues similar to SASP135,136. The Retinoblastoma (Rb) family of proteins plays a crucial role in regulating cellular senescence. RB1, a member of this family, is notably abundant in the 5′UTR of L1 loci in proliferating cells, facilitating the heterochromatinization of these loci136. The enzyme Sirtuin 6 (SIRT6), known for its protein deacetylase and mono-ADP ribosyltransferase activities, is also enriched in the 5′UTR of L1 loci137. SIRT6 mono-ADP ribosylates the nuclear corepressor protein KAP1, promoting its interaction with the heterochromatin factor HP1α137. The binding of RB1 and SIRT6 at the 5′UTR of L1 aids in the packaging of L1 elements into transcriptionally repressive heterochromatin136,137. However, during senescence, the level of RB1 declines, and in aging and in response to DNA damage, SIRT6 becomes depleted from L1 loci, derepressing the heterochromatin and leading to the activation of these retroelements, previously silenced136,137. The depletion of RB1 and SIRT6 coincides with a decrease in H3K9me3 and H3K27me3 repressive marks in these regions136,137 Given that retrotransposition can be induced by DNA damage138, DNA-SCARS could potentially serve as one of the upstream factors triggering L1 activation during senescence.
Human Endogenous retroviruses (HERVs) are ancient infection remnants in primates that have integrated into the genome, constituting approximately 8% of the human genetic material132. HERVs exist as proviruses within the host genome, containing sequences necessary for the awakening process and retrovirus-like particle (RVLP) formation132. However, these proviruses lack the elements for replication, rendering them unable to multiply independently132. In senescent cells, age-related DNA demethylation126,127 and heterochromatin decompaction can activate HERV provirus transcription (Fig. 1), leading to RVLP protein synthesis139. HERV expression is heightened in aged human tissues and the serum of elderly individuals. Similar to L1, HERV RNA undergoes reverse transcription by viral reverse transcriptase in the cytoplasm of senescent cells, triggering cGAS/STING-dependent upregulation of SASP factors132,139. Interestingly, HERV viral particles released from senescent cells or found in the serum of older individuals can infect healthy cells via their envelope protein139. This infection can stimulate innate immune responses in infected cells, contributing to the development of a senescence phenotype in a paracrine manner, thus causing increased SASP expression139. Inhibition of L1 or HERV reverse transcriptase by nucleoside reverse transcriptase inhibitor (NRTI) alleviates the interferon-mediated inflammation in multiple tissues in mice and SASP in cells, respectively135,136,139. In conclusion, epigenetic processes in senescent cells, such as heterochromatin decompaction and hypomethylation, regulate retrotransposable elements, thereby influencing their activity and impact on cellular function during aging by promoting inflammation.
Therapeutic approaches: epigenetic modulators as potential senomorphics
Senotherapy holds promise in potentially slowing down the aging process overall, thereby positively impacting a myriad of age-related illnesses140. There are two primary approaches to achieving this. First, senolytics involve eliminating senescent cells through targeted drug interventions or immunotherapy. Numerous senolytic agents are currently available, with some undergoing clinical trials. The second approach, senomorphics, involves disrupting the expression of SASP factors, which play a role in age-related diseases. However, one potential concern is that disrupting SASP expression may increase the risk of cancer by impairing the immune surveillance of senescent cells against cancer cells140. Surprisingly, both the absence and excessive presence of senescent cells can be linked to a worse prognosis, while moderate senescence is associated with the best prognosis141. Additionally, several clinical studies have demonstrated that some senolytics can induce toxicity and kill not only senescent cells but also other healthy cells142. Therefore, rather than eliminating senescent cells, attenuating SASP expression through the inhibition of epigenetic modifications may be a preferable method of intervention. Interestingly, disrupting the epigenetics of SASP may not affect the growth arrest of senescent cells. Knocking down epigenetic regulators such as MLL166, KDM468, DOT1L70, BRD450, and HMGB241 appears to prevent the emergence of SASP while leaving cell cycle arrest unaffected. Inhibitors and degraders targeting KDM4, DOT1L, and BRD4 are currently undergoing clinical trials for cancer treatment (Table 1). Upon assessing safety and tolerance levels, these drugs could potentially be repurposed as senomorphics. Additionally, repurposing HIV drugs may offer potential benefits for enhancing health in old age by targeting L1 and HERV-associated inflammation135,136,139.
Perspectives
In recent years, advancements in chromosome conformation capture techniques, such as Hi-C, have expanded our understanding of the 3D genome organization of senescent cells. The compartmentalization of the genome, mapping of SEs, and the formation of enhancer-promoter loops have all contributed to regulating SASP. However, the majority of contacts obtained through Hi‐C are pairwise, and interactions among three or more chromatin sites (multi‐way) can only be deduced from the two‐way data. Capturing multi‐way chromosomal interactions using long-read sequencing techniques, which directly sequences DNA multivalent fragments joined by proximity‐based ligation, can provide deeper insights into the 3D genome, TAD formation, and multi-way enhancer-promoter interactions in senescent cells, ultimately aiding in understanding the regulation of SASP. These new insights provide a better understanding of senescence and SASP. SAHF formation and SASP expression are correlated. However, it remains unclear whether SAHF is necessary for SASP expression and how SASP gene loci and enhancer regions maintain their position within the euchromatin compartment rather than becoming heterochromatinized and sequestered within SAHF. CCF, a crucial upstream regulator of SASP expression, involves the ejection of damaged DNA, most likely DNA-SCARS, into the cytoplasm. CCF is characterized by enrichment with heterochromatin marks and the absence of euchromatin marks11. This suggests that CCF may emerge from the SAHF region or from the damaged region that is undergoing heterochromatinization. Losing a significant portion of DNA also alters the genome integrity of senescent cells. However, the question remains regarding how the 3D genome organization maintains genome stability in senescent cells and dictates the formation of CCF from heterochromatin regions. DNA methylation in senescence remains enigmatic and highly influenced by context. HMGA Proteins play a crucial role in SAHF formation143, and DNMT1 is essential for this process by regulating HMGA2 expression24. Consequently, DNMT1 is associated with 3D genome rewiring in oncogene-induced senescent cells24. However, the direct involvement of CpG methylation in SASP expression remains to be understood. Nonetheless, a recent study has illuminated that DNA demethylation triggers the activation of HERV transcription139. This finding underscores the pivotal role of DNA methylation in senescence and the modulation of SASP. Moreover, it raises intriguing questions about whether manipulating DNA methylation during senescence could potentially revert 3D genome organization and gene expression.
The primary aim of all senescence-associated research is to translate findings into potential therapeutic interventions that promote healthy aging or other therapies for disease. However, a significant hurdle lies in discovering senomorphic agents without causing unintended side effects. This challenge arises because the SASP is not entirely detrimental in physiological contexts. The SASP plays a pivotal role as a signaling mechanism for various immune cells. Its involvement in immune-mediated clearance of senescent cells contributes to suppressing tumor initiation, facilitating tumor regression, and supporting essential processes during development144. Additionally, the SASP provides numerous benefits associated with acute senescence, such as aiding in wound healing and restoring tissue homeostasis, particularly in conditions like liver fibrosis144. In essence, while inhibiting SASP appears beneficial in the aging process and age-associated diseases, its positive effects in acute senescence, as well as its context-dependent nature and multifaceted effects, present challenges for the use of senomorphics. Another additional disadvantage of senomorphics is that the necessity for continuous administration to regulate SASP expression could potentially result in more side effects than those observed with intermittently dosed senolytics145.
In conclusion, the discovery of SASP has shifted the paradigm of senescence research beyond its conventional focus on permanent proliferation arrest and SAHF formation properties. SASP can elucidate many aspects of senescence, whether beneficial or detrimental, bridging the gap between senescence’s tumor-suppressive role in the short term and its tumor-promoting role in the long term. Deciphering its role and regulation holds tremendous potential for understanding aging and age-associated disease physiology. SASP is regulated in multiple ways, depending on the context and the senescence-inducing agent, with epigenetic regulation being no exception. Not all pathways of epigenetic regulation discussed here are universal for all senescence models, adding to the complexity and intrigue surrounding SASP regulation. The use of epigenetic modulators as senomorphics faces similar challenges due to this variability. Identifying an epigenetic modulator effective across all types of senescence models while ensuring safety and efficacy is crucial. Utilizing drug repurposing data and conducting preclinical and clinical trials will aid in determining the trade-off of using senomorphic agents, balancing their beneficial and detrimental effects associated with SASP. The Cellular Senescence Network (SenNet) Program, which aims to comprehensively identify and characterize variations in senescent cells (https://sennetconsortium.org), has the potential to address these variability issues and pinpoint safe and effective therapeutic pathways for senescence and aging.
Acknowledgements
This study was funded by NIA grants 5P01AG031862, 5R01AG071861 (To P.D.A.), the California Institute for Regenerative Medicine grant EDUC4-12813 (To R.A.). The funders played no role in the writing of this manuscript. We utilized ChatGPT 3.5 solely for editing the English language in this review; it was not employed to generate or alter any scientific content. The Graphic is Created with BioRender.com.
Author contributions
N.D. and P.D.A. conceptualized the review. N.D., R.A., A.E., A.G., and P.D.A. wrote the review. N.D. and R.A. conceptualized the figure. N.D., R.A., A.E., and A.G. created the figure. N.D. and A.G. created the table.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nirmalya Dasgupta, Email: ndasgupta@lji.org.
Peter D. Adams, Email: padams@sbpdiscovery.org
References
- 1.Guo, J. et al. Aging and aging-related diseases: from molecular mechanisms to interventions and treatments. Signal Transduct Target Ther.7, 391 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell186, 243–278 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Hayflick, L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res.37, 614–636 (1965). [DOI] [PubMed] [Google Scholar]
- 5.McHugh, D. & Gil, J. Senescence and aging: causes, consequences, and therapeutic avenues. J. Cell Biol.217, 65–77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herranz, N. & Gil, J. Mechanisms and functions of cellular senescence. J. Clin. Invest.128, 1238–1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol.75, 685–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coppe, J. P., Desprez, P. Y., Krtolica, A. & Campisi, J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol.5, 99–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franceschi, C., Garagnani, P., Parini, P., Giuliani, C. & Santoro, A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol.14, 576–590 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Chien, Y. et al. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev.25, 2125–2136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov, A. et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol.202, 129–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature550, 402–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao, X., Wang, C. & Zhang, R. Chromatin basis of the senescence-associated secretory phenotype. Trends Cell Biol.32, 513–526 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi, A. et al. Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat. Commun.9, 1249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunet, A. & Berger, S. L. Epigenetics of aging and aging-related disease. J. Gerontol. A: Biol. Sci. Med. Sci.69, S17–S20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feser, J. & Tyler, J. Chromatin structure as a mediator of aging. FEBS Lett.585, 2041–2048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Sullivan, R. J. & Karlseder, J. The great unravelling: chromatin as a modulator of the aging process. Trends Biochem. Sci.37, 466–476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibney, E. R. & Nolan, C. M. Epigenetics and gene expression. Heredity (Edinb.)105, 4–13 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Basisty, N. et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol.18, e3000599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasaki, O. et al. Involvement of condensin in cellular senescence through gene regulation and compartmental reorganization. Nat. Commun.10, 5688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Criscione, S. W. et al. Reorganization of chromosome architecture in replicative cellular senescence. Sci. Adv.2, e1500882 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sati, S. et al. 4D genome rewiring during oncogene-induced and replicative senescence. Mol. Cell78, 522–538 e529 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandra, T. et al. Global reorganization of the nuclear landscape in senescent cells. Cell Rep.10, 471–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narita, M. et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell113, 703–716 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Zhang, R., Chen, W. & Adams, P. D. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell Biol.27, 2343–2358 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandra, T. et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol. Cell47, 203–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, D. M. et al. Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol.17, 158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olan, I., Handa, T. & Narita, M. Beyond SAHF: An integrative view of chromatin compartmentalization during senescence. Curr. Opin. Cell Biol.83, 102206 (2023). [DOI] [PubMed] [Google Scholar]
- 31.Miron, E. et al. Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci. Adv.6, eaba8811 (2020). [DOI] [PMC free article] [PubMed]
- 32.Zhang, R. et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell8, 19–30 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Banumathy, G. et al. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol. Cell Biol.29, 758–770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye, X. et al. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell Biol.27, 2452–2465 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boumendil, C., Hari, P., Olsen, K. C. F., Acosta, J. C. & Bickmore, W. A. Nuclear pore density controls heterochromatin reorganization during senescence. Genes Dev.33, 144–149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosar, M. et al. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a). Cell Cycle10, 457–468 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Dasgupta, N. et al. Histone chaperone HIRA, promyelocytic leukemia (PML) protein, and p62/SQSTM1 coordinate to regulate inflammation during cell senescence. Mol. Cell. 84, 3271–3287.E8 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, A. S. & Dreesen, O. Biomarkers of Cellular Senescence and Skin Aging. Front Genet9, 247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy, A. L. et al. Senescent mouse cells fail to overtly regulate the HIRA histone chaperone and do not form robust senescence associated heterochromatin foci. Cell Div.5, 16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zirkel, A. et al. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell70, 730–744.e736 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Aird, K. M. et al. HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J. Cell Biol.215, 325–334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanson, E. C., Manning, B., Zhang, H. & Lawrence, J. B. Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence. J. Cell Biol.203, 929–942 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyata, K. et al. Chromatin conformational changes at human satellite II contribute to the senescence phenotype in the tumor microenvironment. Proc. Natl Acad. Sci. USA120, e2305046120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen, A. S., Cattoglio, C., Darzacq, X. & Tjian, R. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus9, 20–32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olan, I. et al. Transcription-dependent cohesin repositioning rewires chromatin loops in cellular senescence. Nat. Commun.11, 6049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, L. et al. Dynamic enhancer interactome promotes senescence and aging. bioRxiv10.1101/2023.05.22.541769 (2023).
- 47.Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol.10, 93–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, P. et al. m(6)A-independent genome-wide METTL3 and METTL14 redistribution drives the senescence-associated secretory phenotype. Nat. Cell Biol.23, 355–365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan, Y. et al. Senescence-activated enhancer landscape orchestrates the senescence-associated secretory phenotype in murine fibroblasts. Nucleic Acids Res.48, 10909–10923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tasdemir, N. et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov.6, 612–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sofiadis, K. et al. HMGB1 coordinates SASP-related chromatin folding and RNA homeostasis on the path to senescence. Mol. Syst. Biol.17, e9760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Cecco, M. et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell12, 247–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parry, A. J. et al. NOTCH-mediated non-cell autonomous regulation of chromatin structure during senescence. Nat. Commun.9, 1840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barral, A. & Dejardin, J. The chromatin signatures of enhancers and their dynamic regulation. Nucleus14, 2160551 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gates, L. A., Foulds, C. E. & O’Malley, B. W. Histone marks in the ‘driver’s seat’: functional roles in steering the transcription cycle. Trends Biochem. Sci.42, 977–989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez-Zamudio, R. I. et al. AP-1 imprints a reversible transcriptional programme of senescent cells. Nat. Cell Biol.22, 842–855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sen, P. et al. Histone acetyltransferase p300 induces de novo super-enhancers to drive cellular senescence. Mol. Cell73, 684–698 e688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balsalobre, A. & Drouin, J. Pioneer factors as master regulators of the epigenome and cell fate. Nat. Rev. Mol. Cell Biol.23, 449–464 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Di Giorgio, E. et al. HDAC4 degradation during senescence unleashes an epigenetic program driven by AP-1/p300 at selected enhancers and super-enhancers. Genome Biol.22, 129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah, P. P. et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev.27, 1787–1799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yasuda, T. et al. Inflammation-driven senescence-associated secretory phenotype in cancer-associated fibroblasts enhances peritoneal dissemination. Cell Rep.34, 108779 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Chibaya, L. et al. EZH2 inhibition remodels the inflammatory senescence-associated secretory phenotype to potentiate pancreatic cancer immune surveillance. Nat. Cancer4, 872–892 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu, L. et al. Induction of senescence-associated secretory phenotype underlies the therapeutic efficacy of PRC2 inhibition in cancer. Cell Death Dis.13, 155 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perrigue, P. M. et al. The histone demethylase jumonji coordinates cellular senescence including secretion of neural stem cell-attracting cytokines. Mol. Cancer Res.13, 636–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Capell, B. C. et al. MLL1 is essential for the senescence-associated secretory phenotype. Genes Dev.30, 321–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi, A. et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol. Cell45, 123–131 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Zhang, B. et al. KDM4 orchestrates epigenomic remodeling of senescent cells and potentiates the senescence-associated secretory phenotype. Nat. Aging1, 454–472 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li, R. et al. Aging-related decrease of histone methyltransferase SUV39H1 in adipose-derived stem cells enhanced SASP. Mech. Ageing Dev.215, 111868 (2023). [DOI] [PubMed] [Google Scholar]
- 70.Leon, K. E. et al. DOT1L modulates the senescence-associated secretory phenotype through epigenetic regulation of IL1A. J. Cell Biol.220, e202008101 (2021). [DOI] [PMC free article] [PubMed]
- 71.Laberge, R. M. et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol.17, 1049–1061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O’Sullivan, R. J., Kubicek, S., Schreiber, S. L. & Karlseder, J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol.17, 1218–1225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Funayama, R., Saito, M., Tanobe, H. & Ishikawa, F. Loss of linker histone H1 in cellular senescence. J. Cell Biol.175, 869–880 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Debès, C. et al. Ageing-associated changes in transcriptional elongation influence longevity. Nature616, 814–821 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaygun, H. & Marzluff, W. F. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol.25, 6879–6888 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin, C. et al. Arginine hypomethylation-mediated proteasomal degradation of histone H4-an early biomarker of cellular senescence. Cell Death Differ.27, 2697–2709 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Celona, B. et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol.9, e1001086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davalos, A. R. et al. p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. J. Cell Biol.201, 613–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hauer, M. H. et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol.24, 99–107 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Rodier, F. et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci.124, 68–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim, C. et al. Histone deficiency and accelerated replication stress in T cell aging. J Clin. Invest131, e143632 (2021). [DOI] [PMC free article] [PubMed]
- 82.Henikoff, S. & Smith, M. M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol.7, a019364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Talbert, P. B. & Henikoff, S. Histone variants at a glance. Journal of Cell Science134, jcs244749 (2021). [DOI] [PMC free article] [PubMed]
- 84.Crouch, J., Shvedova, M., Thanapaul, R. J. R. S., Botchkarev, V. & Roh, D. Epigenetic regulation of cellular senescence. Cells11, 672 (2022). [DOI] [PMC free article] [PubMed]
- 85.Contrepois, K. et al. Histone variant H2A.J accumulates in senescent cells and promotes inflammatory gene expression. Nat. Commun.8, 14995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen, H. & Ruiz, P. D. a. MacroH2A1 and ATM play opposing roles in paracrine senescence and the senescence-associated secretory phenotype. Mol. Cell59, 719–731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rai, T. S. et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev.28, 2712–2725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kamakaka, R. T. & Biggins, S. Histone variants: deviants? Genes Dev.19, 295–310 (2005). [DOI] [PubMed] [Google Scholar]
- 89.Szenker, E., Ray-Gallet, D. & Almouzni, G. The double face of the histone variant H3.3. Cell Res.21, 421–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tvardovskiy, A., Schwämmle, V., Kempf, S. J., Rogowska-Wrzesinska, A. & Jensen, O. N. Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res.45, 9272–9289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Piña, B. & Suau, P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol.123, 51–58 (1987). [DOI] [PubMed] [Google Scholar]
- 92.Maze, I. et al. Critical role of histone turnover in neuronal transcription and plasticity. Neuron87, 77–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duarte, L. F. et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat. Commun.5, 5210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rogakou, E. P. & Sekeri-Pataryas, K. E. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp. Gerontol.34, 741–754 (1999). [DOI] [PubMed] [Google Scholar]
- 95.Yang, H. et al. Loss of BRD4 induces cell senescence in HSC/HPCs by deregulating histone H3 clipping. EMBO Rep.24, e57032 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maehara, K., Takahashi, K. & Saitoh, S. CENP -A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol. Cell. Biol.30, 2090–2104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hédouin, S., Grillo, G., Ivkovic, I., Velasco, G. & Francastel, C. C. E. N. P.-A. chromatin disassembly in stressed and senescent murine cells. Sci. Rep.7, 42520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Isermann, A., Mann, C. & Rube, C. E. Histone variant H2A.J marks persistent DNA damage and triggers the secretory phenotype in radiation-induced senescence. Int. J. Mol. Sci.21, 9130 (2020). [DOI] [PMC free article] [PubMed]
- 99.Lowe, D. J. et al. Chronic irradiation of human cells reduces histone levels and deregulates gene expression. Sci. Rep.10, 2200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rübe, C. E. et al. Human skin aging is associated with increased expression of the histone variant H2A.J in the epidermis. npj Aging Mech. Dis.7, 7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abd Al-Razaq, M. A. et al. Role of histone variant H2A.J in fine-tuning chromatin organization for the establishment of ionizing radiation-induced senescence. Cells12, 916 (2023). [DOI] [PMC free article] [PubMed]
- 102.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem.273, 5858–5868 (1998). [DOI] [PubMed] [Google Scholar]
- 103.Bernadotte, A., Mikhelson, V. M. & Spivak, I. M. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging (Albany NY)8, 3–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol.146, 905–916 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Georgoulis, A., Vorgias, C. E., Chrousos, G. P. & Rogakou, E. P. Genome Instability and \gammaH2AX. Int. J. Mol. Sci.18, 1979 (2017). [DOI] [PMC free article] [PubMed]
- 106.Sedelnikova, O. A. et al. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol.6, 168–170 (2004). [DOI] [PubMed] [Google Scholar]
- 107.Siddiqui, M. S., François, M., Fenech, M. F. & Leifert, W. R. Persistent \gammaH2AX: a promising molecular marker of DNA damage and aging. Mutat. Res. Rev. Mutat. Res.766, 1–19 (2015). [DOI] [PubMed]
- 108.Wang, C. et al. DNA damage response and cellular senescence in tissues of aging mice. Aging Cell8, 311–323 (2009). [DOI] [PubMed] [Google Scholar]
- 109.White, R. R. & Vijg, J. Do DNA double-strand breaks drive aging? Mol. Cell63, 729–738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hovest, M. G., Brüggenolte, N., Hosseini, K. S., Krieg, T. & Herrmann, G. Senescence of human fibroblasts after psoralen photoactivation is mediated by ATR kinase and persistent DNA damage foci at telomeres. Mol. Biol. Cell17, 1758–1767 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hernandez-Segura, A., Nehme, J. & Demaria, M. Hallmarks of cellular senescence. Trends Cell Biol.28, 436–453 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Kreiling, J. A. et al. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell10, 292–304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Borghesan, M. et al. DNA hypomethylation and histone variant macroH2A1 synergistically attenuate chemotherapy-induced senescence to promote hepatocellular carcinoma progression. Cancer Res.76, 594–606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kozlowski, M. & Ladurner, A. G. ATM, macroh2a.1, and SASP: the checks and balances of cellular senescence. Mol. Cell59, 713–715 (2015). [DOI] [PubMed] [Google Scholar]
- 115.Sun, D. et al. MacroH2A impedes metastatic growth by enforcing a discrete dormancy program in disseminated cancer cells. Sci. Adv.8, eabo0876 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Commerford, S. L., Carsten, A. L. & Cronkite, E. P. Histone turnover within nonproliferating cells. Proc. Natl Acad. Sci. USA79, 1163–1165 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Toyama, B. H. et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell154, 971–982 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baldensperger, T. et al. Comprehensive analysis of posttranslational protein modifications in aging of subcellular compartments. Sci. Rep.10, 7596 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hauck, A. K. et al. Histone carbonylation is a redox-regulated epigenomic mark that accumulates with obesity and aging. Antioxidants (Basel, Switzerland)9, 1210 (2020). [DOI] [PMC free article] [PubMed]
- 120.Lindner, H., Sarg, B., Hoertnagl, B. & Helliger, W. The microheterogeneity of the mammalian H1(0) histone. Evidence for an age-dependent deamidation. J. Biol. Chem.273, 13324–13330 (1998). [DOI] [PubMed] [Google Scholar]
- 121.Zheng, Q. et al. Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun.10, 1289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fedintsev, A. & Moskalev, A. Stochastic non-enzymatic modification of long-lived macromolecules—a missing hallmark of aging. Ageing Res. Rev.62, 101097 (2020). [DOI] [PubMed] [Google Scholar]
- 123.Jin, B., Li, Y. & Robertson, K. D. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer2, 607–617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell49, 359–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol.14, R115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Day, K. et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol.14, R102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Unnikrishnan, A. et al. Revisiting the genomic hypomethylation hypothesis of aging. Ann N Y Acad Sci.1418, 69–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lowe, D., Horvath, S. & Raj, K. Epigenetic clock analyses of cellular senescence and ageing. Oncotarget7, 8524–8531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.MacKenzie, D. J. et al. DNMT3B oncogenic activity in human intestinal cancer is not linked to CIMP or BRAFV600E mutation. iScience23, 100838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lopatina, N. et al. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J. Cell Biochem.84, 324–334 (2002). [DOI] [PubMed] [Google Scholar]
- 131.Sakaki, M. et al. Potential roles of DNA methylation in the initiation and establishment of replicative senescence revealed by array-based methylome and transcriptome analyses. PLoS ONE12, e0171431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dasgupta, N. & Adams, P. D. Is aging a “Retro“spective event? Cell186, 233–235 (2023). [DOI] [PubMed] [Google Scholar]
- 133.Gorbunova, V. et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature596, 43–53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brouha, B. et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA100, 5280–5285 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Simon, M. et al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab.29, 871–885.e875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature566, 73–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Van Meter, M. et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun.5, 5011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hagan, C. R., Sheffield, R. F. & Rudin, C. M. Human Alu element retrotransposition induced by genotoxic stress. Nat. Genet.35, 219–220 (2003). [DOI] [PubMed] [Google Scholar]
- 139.Liu, X. et al. Resurrection of endogenous retroviruses during aging reinforces senescence. Cell186, 287–304.e226 (2023). [DOI] [PubMed] [Google Scholar]
- 140.Childs, B. G. et al. Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug Discov.16, 718–735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Domen, A. et al. Cellular senescence in cancer: clinical detection and prognostic implications. J. Exp. Clin. Cancer Res.41, 360 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang, L., Lankhorst, L. & Bernards, R. Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer22, 340–355 (2022). [DOI] [PubMed] [Google Scholar]
- 143.Narita, M. et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell126, 503–514 (2006). [DOI] [PubMed] [Google Scholar]
- 144.Birch, J. & Gil, J. Senescence and the SASP: many therapeutic avenues. Genes Dev.34, 1565–1576 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chaib, S., Tchkonia, T. & Kirkland, J. L. Cellular senescence and senolytics: the path to the clinic. Nat. Med.28, 1556–1568 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]