Abstract
Vaccines to the Plasmodium falciparum reticulocyte binding-like protein homologue 5 (PfRH5) target the blood-stage of the parasite life cycle. PfRH5 has the potential to trigger the production of strain-transcendent antibodies and has proven its efficacy both in pre-clinical and early clinical studies. Vaccine-induced monoclonal antibodies (mAbs) to PfRH5 showed promising outcomes against cultured P. falciparum laboratory strains from distinct geographic areas. Here, we assessed the functional impact of vaccine-induced anti-PfRH5 mAbs on more genetically diverse P. falciparum clinical isolates. We used mAbs previously isolated from single B cells of UK adult PfRH5 vaccinees and used ex-vivo growth inhibition activity (GIA) assays to assess their efficacy against P. falciparum clinical isolates. Next-generation sequencing (NGS) was used to assess the breadth of genetic diversity in P. falciparum clinical isolates and to infer the genotype/phenotype relationship involved in antibody susceptibility. We showed a dose-dependent inhibition of clinical isolates with three main GIA groups: high, medium and low. Except for one isolate, our data show no significant differences in the mAb GIA profile between P. falciparum clinical isolates and the 3D7 reference strain, which harbors the vaccine allele. We also observed an additive relationship for mAb combinations, whereby the combination of GIA-low and GIA-medium antibodies resulted in increased GIA, having important implications for the contribution of specific clones within polyclonal IgG responses. While our NGS analysis showed the occurrence of novel mutations in the pfrh5 gene, these mutations were predicted to have little or no functional impact on the antigen structure or recognition by known mAbs. Our present findings complement earlier reports on the strain transcendent potential of anti-PfRH5 mAbs and constitute, to our knowledge, the first report on the susceptibility of P. falciparum clinical isolates from natural infections to vaccine-induced human mAbs to PfRH5.
Subject terms: Malaria, Vaccines
Introduction
Malaria continues to afflict humankind, with over 200 million cases and more than 600,000 deaths reported in 20221. Combined efforts in both preventive and therapeutic measures have helped reduce the malaria burden to more than a third over the last two decades2,3. This progress has however stalled since 2015 with an observed plateau1. Moreover, recent data showed an increase of 5 million cases globally between 2021 and 20221. With the growing threats associated with the emergence and spread of parasite and vector resistance to antimalarials and insecticides, respectively4, this situation will likely worsen if new strategies are not developed soon. While a very high efficacy malaria vaccine would be a key transformative tool in the malaria elimination toolbox, achieving this goal has been challenged by both the complexity of the parasite life cycle and the extent of genetic diversity, which can limit vaccine efficacy5.
Infection with Plasmodium falciparum, the most lethal species of human malaria parasites, starts with sporozoites inoculated through the bite of an infected female Anopheles mosquito. This stage is targeted by RTS,S/AS01 (Mosquirix™)6 and R21/Matrix-M™7, so far, the only two malaria vaccines recommended by the World Health Organization (WHO) for use in children living in areas with moderate to high malaria transmission1. In phase 3 efficacy trials, the protective efficacy against clinical malaria of R21/Matrix-M, over a 12-month follow-up period, was 67–75% in children (aged 5–36 months) who were fully vaccinated (3 doses, efficacy endpoint measured at 12 months after the third dose)7. On the other hand, phase 3 clinical assessment of RTS,S/AS01 protective efficacy against uncomplicated, over a median 48-months follow-up, was respectively 18.3% and 28.2%in infants (6–12 weeks) and children (5–17 months)6. Moreover, in both age groups a booster dose at 20 months after the third dose yielded efficacy against uncomplicated clinical malaria of 25.9% in infants and 36.3% in children6. A highly effective malaria vaccine that provides durable protection is urgently needed to eliminate malaria. Consequently, a strain-transcendent blood-stage vaccine could complement the pre-erythrocytic efficacy achieved through RTS,S/AS01 and/or R21/Matrix-M™ vaccination. Blood-stage vaccines have been explored in the past, but have often struggled to induce strain-transcendent strongly efficacious immunity. AMA1-based vaccines induce strong antibody responses, however, fail to provide significant protection against clinical malaria and are associated with a limited efficacy in both early clinical trials8–10 and control human malaria infection studies11. AMA1 vaccination has been shown to provide a modest efficacy against clinical malaria (17.4%). This lack of efficacy is attributed to the strain-specific immunity triggered upon AMA1 vaccination, which selects for breakthrough infections from non-matched genotypes circulating in the natural parasite population, as efficacy against clinical malaria with vaccine-type AMA1 was 64.3%12. Likewise, this allele-specific immunity has also been associated to the partial efficacy of RTS,S following phase 3 clinical trial studies13. This highlights the potential impact of current efforts in the development of next-generation blood-stage vaccines, such as the P. falciparum reticulocyte binding protein homologue 5 (PfRH5) that have shown promise in eliciting strain-transcending immune responses14. Early testing of the effect of naturally arising P. falciparum genetic diversity on PfRH5 antibody responses is therefore critical to help achieve this goal.
Vaccines targeting the blood-stage of the parasite life cycle are of particular interest as they target the clinical stage of infection and are aimed at reducing parasite density and severe disease. The PfRH5 protein has emerged as a leading vaccine candidate, with attractive characteristics being both an essential and conserved15,16 promising blood-stage vaccine candidate. PfRH5 is by far the most advanced targeted blood-stage vaccine candidate that has reached clinical development over the last few years17,18. In addition to being strain-transcendent14,19–21, antibodies to PfRH5 have shown protection in non-human primates against heterologous blood-stage challenge22,23. Clinically, PfRH5 immunization triggers higher antibody titers in vaccinated infants and children relative to adults18. Notably, antibodies to PfRH5 are associated with a delayed parasite onset in a controlled human malaria infection (CHMI) in naïve adults, which also correlates with in vitro growth inhibition activity (GIA) of vaccine-induced antibodies24. Moreover, human monoclonal antibodies (mAbs) isolated from PfRH5-vaccinated UK adults have been reported with broadly neutralizing activities against P. falciparum laboratory lines25, however, their potential has not been tested on more diverse clinical parasite isolates circulating in endemic settings.
Here, we assess the inhibitory potential of vaccine-induced anti-PfRH5 human mAbs against genetically diverse P. falciparum clinical isolates from a malaria-endemic setting in Senegal. Parasite isolate susceptibility to antibody inhibition was performed using a flow cytometry-based ex-vivo GIA assay and the breadth of PfRH5 genetic diversity was assessed using targeted deep amplicon sequencing of PfRH5 amplicons. The potential impact of PfRH5-associated genetic diversity was predicted through structural threading of the identified polymorphisms onto the three-dimensional structure of PfRH5 in complex with its binding partner PfCyRPA, its erythrocyte receptor, Basigin, or the human mAbs used in the study.
Results
P. falciparum clinical isolates show broad susceptibility to PfRH5 vaccine-induced mAbs
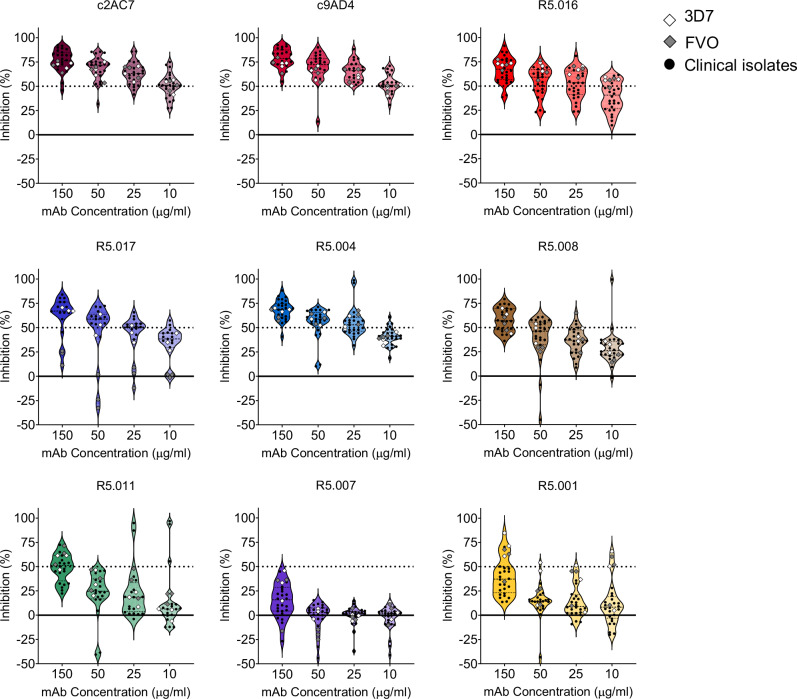
We used a panel of previously characterized human mAbs isolated from single-cell-sorted plasmablasts of UK adult volunteers immunized with a viral-vectored PfRH5-based vaccine25. Additionally, we included two well-characterized anti-PfRH5 mouse mAbs chimeric with human Fc (c2AC7 and c9AD4)22,26. To assess the susceptibility of P. falciparum clinical isolates to PfRH5-vaccine-induced antibodies, we tested four different concentrations (150, 50, 25, and 10 µg/ml) of the mAbs on freshly collected P. falciparum clinical isolates from malaria symptomatic patients using a flow cytometry-based ex-vivo GIA assay27. These concentrations were chosen to incorporate the EC50 values of the most potent mAbs that were previously tested against laboratory lines25. P. falciparum clinical isolates were co-incubated with mAbs during the first ex-vivo cycle and seeded at parasitemia of 0.5-1%. Cultures were harvested upon complete reinvasion of the parasites and the data acquired by flow cytometry. A total of 23 isolates were tested in ex-vivo GIAs, of which, 22 had successful reinvasion rates, while no relationship was observed between the parasites’ multiplication rate (PMR) and their initial parasitemia and or the GIA starting parasitemia (Supplementary Fig. 1). At these tested concentrations, the clinical isolates showed a dose-dependent susceptibility to the individual mAbs, with the chimeric mAbs, c2AC7 and c9AD4, showing the highest inhibition rates across isolates resulting in mean percent GIAs of 80.9% and 77.6% and 53.9% and 50.5%, respectively at the highest and lowest concentrations. The human vaccine-induced mAbs, R5.004, R5.016 and R5.017 showed the highest inhibition across all concentrations, with the least inhibitory mAbs being R5.001 and R5.007, while R5.008 and R5.011 showed intermediate phenotypes (Fig. 1); this is in line with previously reported data using the 3D7 reference laboratory line25. Based on these results, we stratified the mAbs into three groups referred to as GIA-high (GIA ≥ 75% at 150 µg/ml), GIA-medium (75% > GIA ≥ 50% at 150 µg/ml) or GIA-low (GIA < 50% at 150 µg/ml). Colors shown in Fig. 1 were chosen to specifically match that of the previously reported epitope bins of the respective mAbs25. Notably, the trend of parasite susceptibility to individual mAbs was conserved across all test concentrations, with a relatively similar inhibition profile observed across all isolates. The only exception to this trend was isolate 400113, which showed a reduced percent GIA across all mAbs and test concentrations (Fig. 2).
Fig. 1. Ex-vivo assessment of P. falciparum clinical isolates’ susceptibility to anti-PfRH5 mAbs.
Growth inhibitory activities (GIA) of the panel of both vaccine-induced human and chimeric mouse mAbs were tested at different concentrations against ex-vivo cultured P. falciparum clinical isolates (black dots) or in-vitro cultured laboratory lines (diamond shapes). Shown within the violin plots are the mean percent GIA (dashed lines) and the 25th and 75th quartiles (dotted lines) of the clinical isolates. Colors were chosen to specifically match that of the previously reported epitope bins of the respective mAbs25.
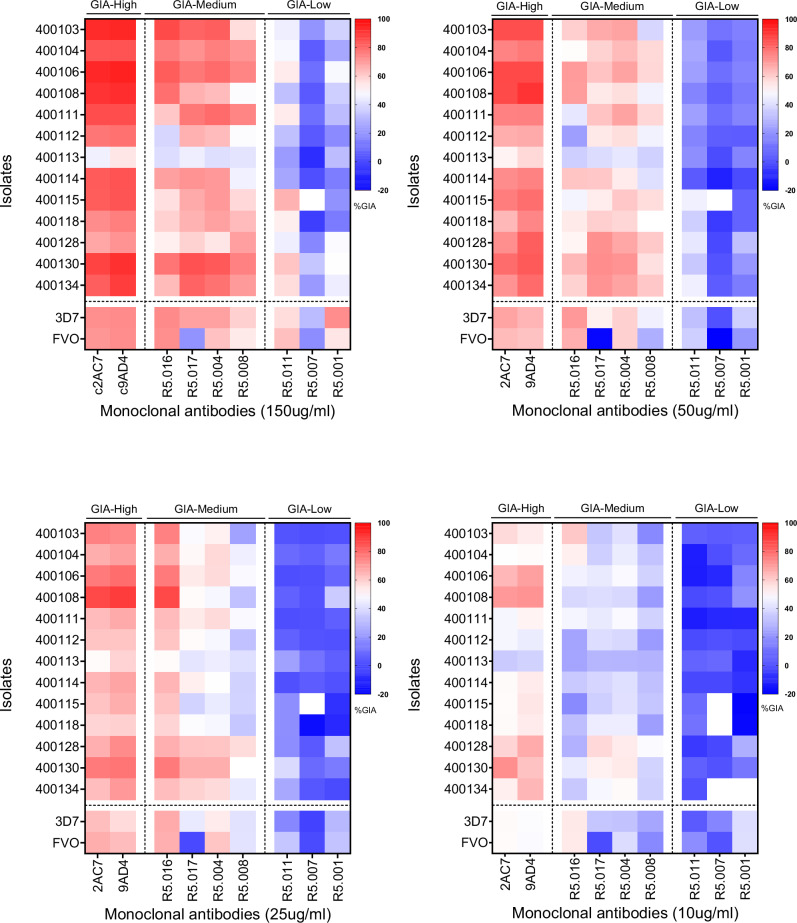
Fig. 2. Classification of the GIA profiles of PfRH5-vaccine-induced mAbs against P. falciparum clinical isolates.
Heat maps presenting the percent GIA data of mAbs to PfRH5 tested at different concentrations (150; 50; 25 and 10 µg/ml). The GIA data from Fig. 1 were used to classify the mAbs in this panel into three main groups, GIA-high (GIA ≥ 75% at 150 µg/ml), GIA-medium (75% > GIA ≥ 50% at 150 µg/ml) or GIA-low (GIA < 50% at 150 µg/ml) separated by vertical dotted lines. Horizontal dotted lines depict the separation between P. falciparum clinical isolates and laboratory lines, presented in the y-axis of the graphs.
To further ascertain the clinical isolates’ susceptibilities to the panel of mAbs, we compared their percent GIA data with that of the laboratory lines 3D7 (reference strain) and FVO, for which well-characterized phenotypic data have been published28. GIA data for both 3D7 and FVO were acquired using the same batches and concentrations of mAbs and, for each strain, three independent assays were performed in duplicates on different days. We used two-way ANOVA analysis to assess the differences in percent GIA across isolates and within each test concentration and found no significant differences in mean percent GIA among the population of clinical isolates relative to 3D7. The two-way ANOVA was associated to the Dunnett’s multiple comparison test to assess the pairwise differences in %GIA between 3D7 and the individual clinicals isolates. This analysis revealed only two isolates (400113 and 400130) showing significant differences in %GIA of clinical isolates relative to 3D7 at the highest concentration (150 ug/ml), while these differences were more pronounced at the lower concentrations with 13, 11 and 12 isolates showing significant %GIA relative to 3D7, respectively at 50, 25 and 10 ug/ml concentrations. Of these isolates, only 400103 showed significant difference across all test concentrations. Encouragingly, none of the antibodies showed significant differences across all test concentrations, (Supplementary Table 1). When comparing FVO to the reference 3D7, significant differences in percent GIA were observed across all concentrations in the presence of the R5.017 mAb, thereby confirming an earlier report in which this difference was attributed to the immune escape phenotype associated with the PfRH5 S197Y mutation in the FVO strain25. Otherwise, significant differences in percent GIA between FVO and 3D7 were only observed when comparing the results for mAb R5.001 at 150 µg/ml (p = 0.046) and the R5.008 at 50 µg/ml (p = 0.025 (Supplementary Table 1).
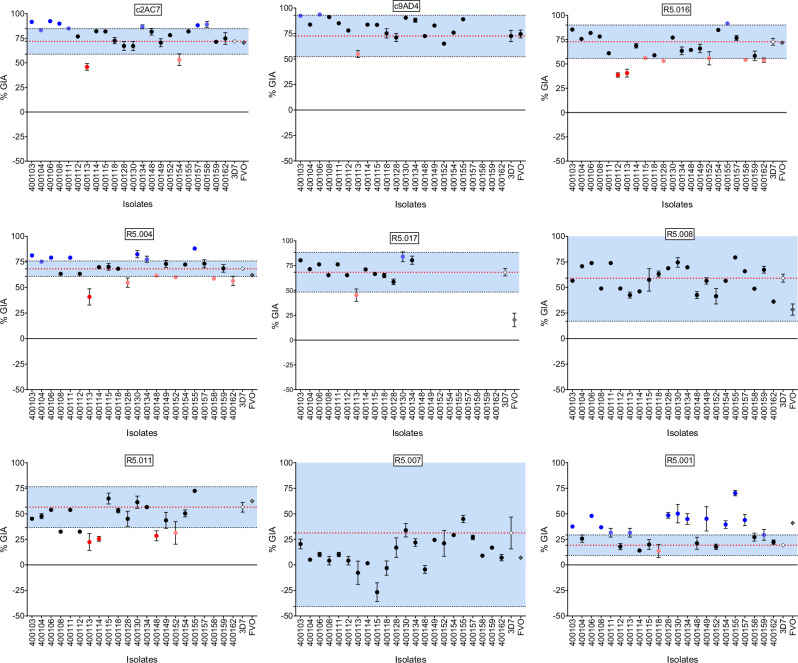
Taking into consideration the earlier observed immune evasive phenotype of FVO relative to 3D7 in the presence of the R5.017 mAb, we defined a conservative range of values for the variability inherent in the assay, using our replicate data from 3D7. We defined a GIA difference as biologically relevant (i.e. loss of GIA susceptibility) if an isolate demonstrated a percent GIA different from that of the mean percent GIA of 3D7 ± 3 SD (standard deviations) from the 3 experimental replicates for any mAb test concentration. Such an approach was previously devised to determine the error of the assay for GIA in the presence of anti-PfRH5 antibodies29. Of note, here we did not include many factors that could increase variability, such as blood donors, etc. and as such our cut-offs may be very conservative. Encouragingly, while we observed some variation in the clinical isolates’ susceptibility to anti-PfRH5 mAbs, none of the isolates showed a complete loss of GIA susceptibility across all mAbs tested here (Fig. 3). Interestingly, R5.016 showed the greatest variability in GIA and multiple isolates showed lower GIA activity across all test concentrations (Fig. 3 and Supplementary Fig. 3A-C). Moreover, at the highest concentration (150 µg/ml), both R5.016 and R5.004 showed multiple isolates with reduced GIA susceptibilities, while the percent GIA of the bulk of these isolates (2 of 7 for R5.016 and 5 of 6 for R5.004) was at the borderline of the set threshold (Fig. 3).
Fig. 3. Variation of percent GIA ranges and immune susceptibility profiles of P. falciparum clinical isolates to PfRH5-vaccine-induced mAbs at 150 µg/ml.
Dot plots of %GIA to anti-PfRH5 mAbs (Y-axis) of P. falciparum clinical isolates (X-axis). The %GIA susceptibility ranges in blue are defined as the mean 3D7%GIA (red dotted line) ± 3 SD 3D7%GIA (black dotted lines). Black dots represent isolates laying within the defined susceptibility ranges. Red dots represent isolates reflecting a reduced GIA susceptibility phenotype (bright red) or laying at the borderline (dim red) of the lower limit of the defined susceptibility threshold. Blue dots represent isolates showing an increased GIA susceptibility (bright blue) or laying at the borderline (dim blue) of the upper limit of the defined susceptibility threshold.
Antibody combinations enhance GIA activity against P. falciparum clinical isolates
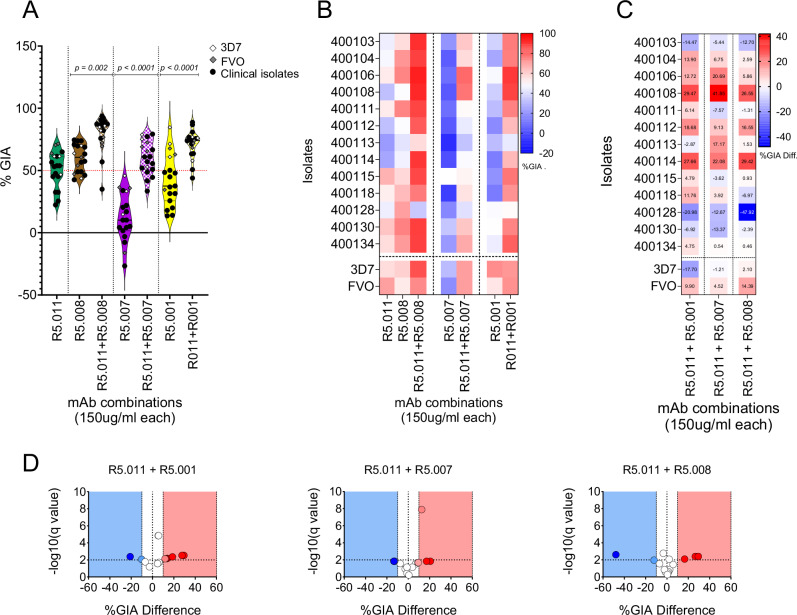
Having shown the broad neutralizing activities of PfRH5-vaccine-induced antibodies on P. falciparum clinical isolates, we sought to investigate the earlier reported synergistic property of R5.011 relative to mAbs with GIA-medium and GIA-low phenotypes. In these assays, R5.011 was combined in equal concentrations (150 µg/ml each) with either R5.001, R5.007 or R5.008 in ex-vivo GIA assays or in vitro GIA assays of P. falciparum clinical isolates or laboratory lines, respectively. Notably, there was a significant increase in the mean percent GIA of the antibody combinations relative to that of R5.001, R5.007 or R5.008 alone (p < 0.0001; Wilcoxon matched-pairs signed rank test). The same was true when comparing the mean percent GIA of the antibody combinations to that of R5.011 alone. Similar to the single mAb data, there were no significant differences between the mean percent GIA of clinical isolates to that of 3D7, however, there was a greater range of GIA values for the clinical isolates (Fig. 4A). Across all isolates, the combination R5.011-R5.007 showed more significant differences in GIA profiles relative to 3D7, while only two isolates, 400113 and 400128 showed significantly lower GIA profiles relative to 3D7 across all combinations (One-way ANOVA and Dunnett’s multiple comparison test) (Fig. 4B). Having shown that R5.011 was able to potentiate the activities of the least potent mAbs in inhibiting growth of P. falciparum clinical isolates, we next sought to investigate whether these combinations were synergistic and, given the limited number of test concentration combinations, we opted for the Bliss additivity model as described previously30. The predicted Bliss additivity percent GIA was calculated from the percent GIAs of the individual mAbs in a given combination and compared to the percent GIA of the corresponding test combination. Differences between predicted and actual percent GIA data were used to define the combination GIA profile. We used the earlier reported thresholds31 to categorize combinations as synergistic (Difference %GIA ≥ 10), additive (-10 ≥ Difference %GIA ≥ 10) or antagonistic (Difference %GIA ≥ -10). Notably, these combinations mostly resulted in either synergistic of additive interactions between the mAbs against most of the isolates tested here, except for 400128 where all combinations were antagonistic (% GIA Difference -20.98 (p = 0.002; t-test), -12.67 (p = 0.006; t-test) and -47.91 (p = 0.002; t-test), respectively for R5.011-R5.001, R5.011-R5.007 and R5.011-R5.008). Across all isolates, the combination of R5.001-R5.011 resulted in more synergistic or additive interactions, while only two isolates (400108 and 400114) showed synergy in all combinations (Fig. 4C-D).
Fig. 4. Assessment of GIA in combinatorial assays of anti-PfRH5 mAbs tested against P. falciparum clinical isolates (N = 13).
A Violin plots showing percent GIA from combinations of R5.011 (green) with either R5.001 (yellow), R5.007 (purple) or R5.008 (brown). Depicted in the plots are the mean percent GIA (plain black line) and the 25th and 75th quartiles (dotted black lines). Here, mAbs were combined in equal concentrations and incubated with equal volumes of either P. falciparum clinical isolates (black dots) or laboratory lines (diamond shapes). The resulting GIA was compared to the single antibody at the same concentration. All assays were performed in duplicates and the data are presented as means from the two replicates. Statistical differences between antibody combinations and single antibody treatments were computed in GraphPad Prism using the Wilcoxon matched-pairs signed rank test. B Heat map showing the GIA profiles of antibody combinations (x-axis) against P. falciparum parasites (y-axis). The dotted vertical lines separate the different antibody combinations, while P. falciparum clinical isolates are separated from the laboratory lines by the horizontal dotted line. C heat maps and D Volcano plots depicting the difference between the Bliss additivity predicted percent GIA and the actual percent GIA from mAb combinations. Bliss analysis was calculated as previously described35 and the multiple unpaired t-test was used to assess the statistical difference. Volcano plots show −log10 of the q-values.
Next-generation sequencing shows a high level of PfRH5 genetic diversity in P. falciparum clinical isolates from Kédougou
Given the extreme diversity of malaria parasites and the potential to impact on vaccine efficacy and immune evasion, we further assessed the breadth of PfRH5-associated polymorphisms, in addition to the multiplicity of infection (MOI) per isolate in our parasite population. PfRH5 genomic sequences were amplified from individual samples and subjected to next-generation sequencing using the Illumina NovaSeq 6000 platform, while the msp1 and msp2 genes were used to assess the number of clones per isolate (referred to as MOI). Pfrh5 sequences were successfully obtained from 50 P. falciparum clinical isolates, of which 22 have complete GIA phenotypic data (Supplementary Fig. 5). PfRH5-associated single nucleotide polymorphisms (SNPs) were called relative to the 3D7 reference using the Geneious prime software. The median MOI of our study population was 2.59, with individual patient infections harboring 1 to 5 distinct P. falciparum genotypes (Fig. 5A). Interestingly, our data showed the reference allele to be the minor allele, representing only 26% (13/50) of our study population, while 74% (27/50) of the parasites presented at least one SNP in the PfRH5 locus (Supplementary Fig. 5). Overall, a total of fifty-five SNPs were identified within our parasite population, with C203Y being the most prevalent identified in 33 isolates (66.4%), while the majority of the other identified SNPs were rare and identified in single isolates (2%). Moreover, a total of twelve SNPs were present within the sampled parasites with phenotypic data. Of these SNPs, five were novel (K51R, I60V, D243N, F505Y and Y254C) and all but one (K51R) occurred in a single isolate (Fig. 5B). We next sought to assess the allele frequencies within complex samples. Given the difficulty to accurately measure intra-sample allele frequency within polyclonal infections, we opted to use the Novaseq 6000 sequencing platform, which offers an in-depth sequencing of the targeted amplicons to enable the discovery of rare variants within polyclonal infections. The relationship between the clinical isolates’ initial parasitemia and the number of sequencing reads mapped to the PfRH5 reference sequence was not correlated (Supplementary Fig. 4). Consequently, the variant read frequencies herein reported were calculated as the frequency of the number of reads with any given SNP over the total number of reads mapped to the reference sequence at the SNP position. As seen with the SNP prevalence, the highest variant read frequencies were associated with the C203Y mutation, with frequencies ranging from 4.1 to 100%. Interestingly, of the novel SNPs, D243N presented the highest variant read frequency (99.7%) and was present in a monogenomic infection (MOI = 1) representing the least susceptible isolate 400113 (Fig. 5C). However, except for C203Y, none of the SNPs reported here occurred at a high population prevalence and high variant frequency at the same time (Fig. 5B-C).
Fig. 5. PfRh5-associated genetic diversity in P. falciparum clinical isolates from Kédougou (N = 22).
A Summary table showing the multiplicity of infections (MOI) per isolate determined through msp1-2 typing as well as the number of SNPs at each position (bold characters) and their respective read frequencies (shown in brackets). B The prevalence of PfRH5-associated SNPs was calculated as the percentage of SNPs detected within the total phenotyped sample population (N = 22). PfRH5 sequencing was performed from pfrh5 amplicons using the Illumina NovaSeq 6000 sequencing platform and variant analysis was performed using the Geneious Prime software version 23.1.1. SNPs displayed to the right of the dotted line represent SNPs present in a single sample. C SNP variant read frequencies in genomically complex samples were determined from the sequencing data outputs and calculated as the percentage of the variant read coverage relative to the coverage at the variant position. The red dotted line depicts the 2% threshold for variant allele frequency. The graphs were plotted using the GraphPad Prism version 1.0.2 software.
Structure‑based insights on potential SNP function
Taking into consideration the existing relationship between genetic diversity and immune evasion, as seen in the case of the PfRH5 S197Y mutation associated with the FVO escape to the R5.017 mAb, we assessed the functional impact of the mutations herein reported on PfRH5 complexing with either binding partner PfCyRPA, receptor basigin (BSG) or mAbs. Using a combination of in-silico prediction tools, we threaded these polymorphisms onto the structure of PfRH5 in complex with either PfCyRPA, BSG and some of the mAbs used in this study for which crystal structures have already been solved and reported25,32,33. Individual FASTA files with PfRH5 and individual SNP amino acid sequences were threaded through the crystal structure and the impact of the mutant versions of the protein was evaluated for predicted binding affinity of PfRH5 to its binding partner PfCyRPA, its erythrocyte receptor, BSG, or the test human mAbs. The structure of PfRH5 used here comprises a truncated version of the protein in which the flexible N-terminus (residues E26 - Y139) and internal disordered loop (residues N248 - M296) regions were removed to enable crystallization28. Consequently, only seven out of the twelve SNPs reported here could be accurately mapped onto the PfRH5 structure by our model (Fig. 6A). Notably, these SNPs were evenly distributed between the N- and C-termini of the protein with C203Y, the most prevalent SNP reported here, occurring at the tip of the second helix around the BSG binding site, and F505Y present around the PfRH5-PfCyRPA interaction site (Fig. 6A). The difference in binding energy between the mutant and the reference allele of PfRH5 was predicted by FoldX version 5.0 and was used to estimate the functional impact of each SNP on protein folding, or binding dynamics with PfCyRPA, BSG, or mAbs (Supplementary Table 2). Except for D249G, for which no structural information was available, all previously reported SNPs were predicted to have a mild impact on PfRH5 by either altering its conformation and stability (K221R, E306G, V371I and I407V) or enhancing its binding to BSG (Fig. 6B-H). In contrast, while no functional prediction was possible for majority of the novel SNPs (K51R, I60V, Y254C and F505Y), the impact of D243N, although near the interface of PfRH5-PfCyRPA, was not predicted to impact the structure. Moreover, none of the SNPs were predicted to bind within or near the known binding epitopes of the profiled mAbs.
Fig. 6. Structural threading of PfRH5-associated polymorphisms onto the BSG-PfRH5-PfCyRPA-mAbs complex.
A Distribution of identified SNPs, red spherical shapes, onto the PfRH5 structure. The model presented here was built by superposing the structure of PfRH5 in complex with BSG (4U0Q), PfCyRPA (6MPV), 9AD4 (4U0R), R5.004 and R5.016 (6RCU) and R5.011 and R5.016 (6RCV). BSG, PfRH5 and PfCyRPA ribbons are shown by light aqua, grey and light orange, respectively. Ribbons of the 9AD4, R5.004, R5.011 and R5.016 mAbs are depicted in dark red, blue, green and light red, respectively. B–H Sub panels highlighting the predicted effect of selected SNPs, the black dotted lines represent hydrogen bonds or salt bridges. From top-left to bottom-right: SNPs that potentially impact binding of PfRH5 and BSG (B), to enhance binding of PfRH5 to PfCyRPA (C), with no predicted function (D) or to alter the structure of PfRH5 E-G.
Discussion
PfRH5 is currently the lead blood-stage malaria vaccine candidate, with recent promising clinical trial outcomes17,18,24,34. Malaria vaccine development, however, has long been hampered by the extensive genetic diversity of naturally circulating parasite populations. Early testing of the functional impact of natural genetic diversity is critical to the development of a strain-transcendent PfRH5-based malaria vaccine. While vaccine-induced mAbs to PfRH5 are associated with varying levels of neutralizing activity against P. falciparum laboratory lines from different geographical locations25, these mAbs had not been tested on P. falciparum clinical isolates, the ultimate target of any given malaria vaccine. Importantly, we and others have recently reported an emergence of rare PfRH5 variants from naturally circulating P. falciparum clinical isolates across Africa35–39. It is critical to determine if such variants, while rare, could constitute vaccine-resistant parasites that could expand under vaccine-selective pressure.
Having earlier reported the near-exclusive invasion inhibition in clinical isolates using low concentrations of antibodies to the PfRH5 receptor, Basigin37, here, we aimed to assess the effectiveness of a previously characterized panel of vaccine-induced anti-PfRH5 mAbs on P. falciparum clinical isolates. The mAbs used here had been classified into different groups based on their GIA profiles25, Our present findings complement these reports and constitute, to our knowledge, the first report on the susceptibility of P. falciparum clinical isolates ex vivo from natural infections to vaccine-induced anti-PfRH5 mAbs. Using a flow cytometry-based ex-vivo GIA assay, we showed these mAbs have broadly neutralizing activity resulting in a dose-dependent inhibition of P. falciparum clinical isolates; and further, in addition to the earlier reported dose-dependent susceptibility of P. falciparum parasites to anti-PfRH5 mAbs22,25, our findings accurately reproduced the categorization of these mAbs into three main groups with distinct GIA profiles25. Despite the observed dose-dependent inhibition of P. falciparum clinical isolates by vaccine-induced mAbs to PfRH5, it is worth highlighting the observed variations in the isolates’ susceptibilities to individual mAbs. However, the breadth of these variations was somewhat expected to increase as the concentration decreases, but also with regards to the mAbs with low or no GIA activities. Encouragingly, GIA appears to be enhanced in polyclonal responses as the combined activities of clones in a polyclonal pool can result in a GIA EC50 at a concentration as low as 8.2 µg/ml17, while the comparison of polyclonal responses from the different PfRH5 vaccines resulted in very similar GIA EC50s of 10 µg/ml40. The quantitative levels of vaccine-induced polyclonal antibodies induced by PfRH5 vaccine candidates have also been shown to greatly exceed that of the naturally acquired PfRH5-specific IgGs from naturally exposed Ghanaian and Kenya adults, while vaccinees who received the regimen ChAd63 RH5 (5 ×1010 vp) and MVA RH5 (2 ×108 pfu) at an 8-week interval, showed serum antibody levels above pre-boost threshold up to day 140 post vaccination17. A more recent study has also reported that vaccine-induced antibodies to the protein-based RH5.1 vaccine could not only be maintained for up to 643 days postvaccination, but also could be enhanced through a delayed-fractional dose vaccination regimen24. Interestingly, we also found a comparable level of susceptibility of the clinical isolates relative to the reference clone 3D7, which harbors the vaccine allele used for immunization, therefore consolidating earlier studies reporting the clear evidence of PfRH5-associated strain-transcending functional activity14,20,23,25. This outcome is particularly promising as, if confirmed, will help rule out the earlier observed and most feared strain-specific efficacy associated with previous vaccine candidates41.
Given the extremely rapid and efficient process of merozoite invasion of erythrocytes, any malaria vaccine targeting the blood-stage of the parasite’s life cycle needs to trigger higher titers of antibodies or a combination of antibodies that can synergize together to achieve protection even at low concentrations. While the former is difficult to achieve23, there is recent evidence of synergistic interactions between anti-PfRH5 antibodies leading to enhanced potency25,31. Consequently, we assessed the earlier reported synergistic profile of R5.01125 and found that this mAb was able to potentiate the neutralizing activities of the least potent mAbs. Interestingly, while the majority of these interactions were synergistic or additive, we also noted some antagonistic interactions between R5.011 with either R5.001, R5.007 or R5.008. These contrasting results, however, involved only two clinical isolates, which showed antagonistic relationships across all combinations and warrant further investigation to better ascertain the contributing factors to the observed phenotypes. Encouragingly, while we observed some variation in the clinical isolates’ susceptibility to anti-PfRH5 mAbs, relative to our set threshold, none of these showed a complete loss of GIA susceptibility across all mAbs. The observed differences in percent GIA between clinical isolates and the 3D7 reference clone, although not significant, could possibly be attributed to donor-to-donor erythrocyte variance, a major contributor in the GIA error of the assay (EoA)29. Notably, with respect to our set range of starting parasitemia (0.5-1%), parasites were diluted with fresh erythrocytes from a single donor in 13 out of the 22 assays performed, while the remaining assays were directly performed with homologous patient-derived erythrocytes.
Given parasite genotype can impact susceptibility to vaccine-induced antibodies, we sought to investigate the breadth of PfRH5 genetic diversity in our study population. As in our previous study, we implemented a deep amplicon sequencing of the PfRH5 gene using a sensitive discovery threshold of 2% variant allele frequency36. This analysis resulted in the discovery of 12 SNPs, the majority of which were present at low population prevalence, and detected in single isolates. Interestingly, we observed a deficit of the reference allele (the vaccine allele), represented in only one-third of the isolates, while the most common allele was represented by the C203Y mutation, therefore confirming previous findings36,37,42,43. Moreover, 5 out of these 12 SNPs were novel and, to our knowledge, have not been previously reported. Furthermore, as our genotyping experiment revealed an average of 3 parasite genotypes per isolate; we sought to assess the frequency of the identified SNPs within the complex mixture of parasite genomes per isolate using our previously described approach36. This analysis revealed that the majority of the SNPs reported here were present at low variant read frequencies (<5%) in polygenomic infections, except for C203Y, which most of the time occurred at high variant read frequencies (Mean = 75.11%, ranges 4.1 – 100%). Interestingly, out of the 70 SNPs identified in our previous study as novel36, only four (K51E, K221R, D249G and E306G) were found in this study. Notably, most of the novel SNPs identified in our studies occurred in regions of the protein reported to be internally disordered and not included in the crystal structure of PfRH528. These regions include the N-terminal domain of the protein (residues E26 - Y139), which is cleaved through plasmepsin X activity44, and the internal disordered loop (residues N248 - M296), both of which appear to routinely induce non-neutralizing antibodies22,25,26,45. These observations warrant further investigation on the spatiotemporal mapping of PfRH5-associated polymorphisms and genetic relatedness of circulating parasite populations to ascertain the contribution of this diversity in the susceptibility of the parasites to vaccine-induced antibodies.
As recently reported by Farrell et al.46, PfRH5 does not change its conformation upon binding to PfCyRPA or BSG and, given the previous reports that neutralizing antibodies to PfRH5 primarily recognize conformational epitopes25,31, we sought to assess the impact of the identified SNPs on PfRH5 structure or its binding with PfCyRPA or BSG, as well as their functional impact on PfRH5 recognition by neutralizing antibodies. To this end, the identified SNPs were mapped onto the PfRH5 crystal structure in complex with binding partner PfCyRPA, receptor BSG or neutralizing mAbs for which crystal structures are available. Expectedly, no information was observed for K51E, K51R, I60V, D249G and Y254H, as the sequences mapping to these regions were truncated in the PfRH5 sequence used to generate the protein’s crystal structure. Nonetheless, our prediction data revealed that the remaining SNPs could affect PfRH5 in different ways, either by altering its structure, or enhancing its binding to CyRPA or BSG. Interestingly, no SNP was found to lie within known antibody-binding epitopes. We also noted the binding site of E306G to occur on the opposite face of PfRH5 to the R5.011 binding epitope, while the structural prediction showed no impact of this SNP on antibody binding. This observation, combined with that of isolate 400128 (in which this mutation is present), which along with 400113 showed moderately antagonistic interactions in combinatorial experiments, warrants further investigation. However, the functional impact of D243N mutation (present in 400113) was predicted as unknown by our model, hence the need for further functional assays (e.g. use of transgenic lines) to ascertain the contribution of this mutation to the reduced GIA susceptibility with 400413.
Although this study provides interesting insights on the efficacy of vaccine-induced mAbs to PfRH5 on P. falciparum clinical isolates, it also has some limitations that are important to note. First, the data reported here are gathered from a relatively small sample size from sites with similar transmission patterns and therefore might only cover some of the diversity with regards to areas with different endemicity. Moreover, we used a flow cytometry-based assay to collect phenotypic data using ex-vivo cultured isolates, which represent the closest parasite populations to that from natural infections. Although this strategy provides more reliable data with regards to the parasite in vivo phenotype relative to data obtained from culture-adapted parasites or laboratory lines, its limitation remains the fact that it is practically impossible to obtain data from distinct technical replicates of the assay, therefore, leaving room for other extrinsic sources of variation to affect assay outcome. One way to address this is to repeat the assays with culture adapted isolates, which still has limitations as it is known that only dominant clones of a given isolate will grow out and adapt47, therefore this does not guarantee data reproducibility. To mitigate such possible outcomes, and based on our previous data on potential sources of variation48,49, all assays were conducted using erythrocytes from a single unexposed donor for parasite dilution where needed, while all antibody dilutions and culture media were prepared fresh and used within 24 hours. Furthermore, as the goal was to test as many isolates as possible with a reasonable range of concentrations spanning previously defined EC50s for laboratory isolates, we were unable to test a wider range of mAb concentrations, and therefore unable to fully compare these data to those obtained with laboratory lines using the same mAb panel. On the other hand, as a result of the limited amounts of antibodies available, we were unable to test an even number of isolates for all individual mAbs or mAb combinations. Notwithstanding, the data described here revealed a limited impact of these challenges on the assay outcomes, as the bulk of the clinical isolates tested here presented relatively similar susceptibility profiles relative to the 3D7 reference line. Another limitation of this study worth highlighting is related to our sequencing strategy. Indeed, here we opted for deep targeted sequencing, which is a powerful tool to accurately identify rare variants occurring at very low frequencies within the parasite populations, however, due to its resulting short reads, we were unable to accurately assemble full-length PfRH5 gene haplotypes within a given polygenomic infection to compare their genetic relatedness to earlier reported parasite haplotypes35–39.
In summary, this study provides, to our knowledge, the first compelling evidence of the susceptibility of P. falciparum clinical isolates to vaccine-induced mAbs targeting the leading blood-stage malaria vaccine candidate antigen PfRH5, as well as the additive interaction between antibodies with different GIA profiles. Moreover, although our data seem to highlight the lack of functional impact of the identified diversity in the study population, the results from our in-silico prediction of the functional impact of these polymorphisms on the parasites’ susceptibility warrant further biochemical and functional genetic studies to ascertain their contribution on the overall parasite phenotype.
Methods
Sample collection and patient socio-demographic characteristics
This study was conducted in Kédougou, the Southeastern region of Senegal, where malaria transmission is highly endemic with a high transmission intensity associated with the rainy season from June to December. The study protocol was approved by National Ethics Committee of Senegal (CNERS) (SEN19/36), the regulatory board of the Senegalese Ministry of Health and the Institutional Review Board of the Yale School of Public Health (2000025417); and informed consent was obtained from all participants and/or their legal guardians. A total of twenty-two patients, aged 10 to 63 years, presenting with a positive P. falciparum RDT and microscope results were recruited from two healthcare centers, Bandafassi (N = 7) and Dalaba (N = 15), between July – August 2022. The participant eligibility to the study was assessed by the local healthcare practitioner and informed consent was obtained from patients who tested positive for P. falciparum single infection on a Pf/Pan (HRP/pLDH) Ag combo rapid diagnostic test (RDT) (Accessbio). A subsequent confirmation of P. falciparum mono-infection was performed using a Pf/VOM (HRP/pLDH) Ag combo rapid diagnostic test (RDT) (Accessbio). A venous blood draw (5 ml) was performed in EDTA vacutainers (BD Bioscience) from all enrolled patients and transported to the laboratory for processing and diagnostic confirmation by microscopy. Infected erythrocyte samples with confirmed P. falciparum infection were separated from other blood components and used for parasite culturing, ex-vivo GIA assays and parasite DNA isolation. The parasitaemia of the collected samples was estimated using thin blood smears and expressed as the percentage of the number of infected erythrocytes over the total number of counted erythrocytes.
Monoclonal antibody production
The generation of the mAbs used here has previously been described25. Briefly, the variable heavy and light chains of the antibody-producing genes were amplified from single-cell-sorted plasmablasts isolated from peripheral blood mononuclear cells (PBMCs) of vaccinees from the first-in-human viral-vectored PfRH5 vaccine trial (NCT02181088)17. Cognate, paired heavy and light chains were cloned into separate human IgG vectors and transformed into E. coli Mix & go competent cells. The panel of mAbs used in this study included previously well-characterized human-vaccine-induced antibodies (R5.001, R5.004, R5.007, R5.008, R5.011, R5.016 and R5.017) as well as two chimeric mAbs (c2AC7 and c9AD4). These mAbs were expressed by transfection of Expi293F cells with paired plasmids encoding matched heavy and light chains using the ExpiFectamine 293 (Invitrogen) reagents and purified from culture supernatants using a protein G column. Purified antibodies were subsequently buffer-exchanged and resuspended at 1 mg/ml in incomplete parasite medium (RPMI 1640 containing 25 mM HEPES, 0.1 mg/ml Hypoxanthine and 50 μg/mL Gentamicin) and stored at −20 °C until use.
Parasite culturing and growth inhibition activity assays
P. falciparum clinical isolates were cultured at 4% hematocrit in complete parasite medium (RPMI 1640 containing 25 mM HEPES, 0.1 mg/ml Hypoxanthine, 0.5% Albumax II, 2 mg/mL sodium bicarbonate and 50 μg/mL Gentamicin and 5% normal human serum). Human erythrocytes of blood group O+ from a single donor were used for all cultures and GIA assays. For ex-vivo GIA assays, parasites were seeded at 0.5-1% parasitemia at 2% hematocrit and co-incubated at ring stage with different concentrations of human mAbs against PfRH5 (150, 50, 25 and 10 μg/ml), while a mouse IgG was used as an isotype control in all assays. Assays were set up in duplicates in a total volume of 40 µL in flat bottom 96 well half-well area plates with additional smear wells to monitor parasite growth; and incubated at 37 °C in an atmosphere of 5% O2, 5% CO2 and balanced with N2. Cultures were harvested after a reinvasion rate of at least 95% was reached in the control wells. Parasite cultures were transferred into U-bottom 96 well plates and washed with 1X PBS-3% BSA (100 μl/well) at 1500 rpm for 5 minutes. The red blood cell pellets were subsequently stained with 1/2000 Sybr Green in PBS for 20 minutes at room temperature in a shaking plate and washed twice with 1X PBS-3% BSA at 1500 rpm for 5 minutes. The pellets were finally resuspended into 200 µl 1X PBS and data were acquired using a Cytoflex cytometer (Beckman Coulter), where 100,000 events were recorded per well. A similar gating strategy was applied to all data sets to discriminate the population of infected erythrocytes from the total number of cells (Supplementary Fig. 2). Erythrocyte singlets were gated as “Erythrocytes”. Histograms of erythrocyte counts (y axis) and the fluorescence intensity of SYBR Green, FL1 (FITC) positive infected erythrocytes (x axis). Parasitemia was quantified as percentage of infected erythrocytes relative to the total erythrocytes. Infected erythrocytes are gated as M1-M5, with M1 representing singly infected, M2 representing doubly, M3 representing triply, M4 representing quadruple infected erythrocytes, and M5 representing total parasitemia. Flow cytometry data outputs were analyzed to determine parasitemia in each well using the Flow Jo_v10.8.1 software. Finally, the FlowJo outputs were exported as Excel files and the percent GIA for each well was calculated from the invasion rate of each well relative to that of the IgG isotype control (Mouse IgG1 kappa mAb, abcam ab81032) at any given concentration. For each concentration, parasitemia from the duplicate test wells were averaged and invasion inhibition, relative to isotype control of the matched concentration, was calculated as follows: %GIA = 100- [(Average Percent invasion in anti-PfRH5 mAb wells)/ (Average parasitemia in IgG1 isotype control wells) * 100]. The graphs were plotted using the GraphPad Prism version 10.0.2 software.
PCR amplification and next-generation analysis
DNA was extracted from ring-infected red blood cell pellets using the QIAmp DNA Blood Mini Kit (Qiagen) following the manufacturer’s instructions. The PfRH5 gene was amplified as previously described37 using the high-fidelity platinum taq (Invitrogen) and the amplicon size was resolved on a 1% agarose gel electrophoresis. Successfully amplified products were subsequently subjected to Next-generation Sequencing using the Illumina NovaSeq 6000 platform using the following procedure. Briefly, PCR amplicons were bead-purified and quantified by Qubit and Bioanalyzer before library preparation using unique dual indexes (UDIs), which associate each sample with a unique dual index for easy identification after sequencing. Indexed samples were subsequently bead-purified to select for that fragmented DNA of 200-300 bp sizes, which were then quantified with qPCR using Illumina’s KAPA Library Quantification Kit and normalized to a concentration of 4 nM. These normalized samples were pooled into 8 sets of 12 samples each and bead-purified once more to further select DNA with the desired range of sizes, which were run on another KAPA qPCR to assess concentration. The concentration of these 8 sub-pools was normalized to 4 nM and combined in equal parts to form one final pool, which was submitted to the Yale Center for Genome Analysis (YCGA) for High Throughput Next Generation Sequencing with a NovaSeq 6000 with targeted coverage of 500,000 reads per sample.
Analysis of natural genetic diversity and structural threading
For each sample demultiplexed forward and reverse sequencing reads were obtained from the sequencing platform and imported into the Geneious prime software, where matched sequencing reads were paired using the Illumina, paired end-setting and subsequently trimmed using the BBDuk plug-in, with a minimum quality score of (Q) 30 and a minimum length of 75 base pairs set, as reads around 100 base pairs were expected. Trimmed sequences were mapped to the 3D7 reference PfRH5 sequence (PF3D7 0424100). Sequence mapping was set for two iterations and coverage criteria were set at 1000 reads. The criteria for single nucleotide polymorphism (SNP) calling was set to a minimum variant frequency of 0.02 (2%) and 1000 read coverage. At least 3 individuals performed sequence data and SNP analysis for each sample using defined guidelines to ensure data quality. The structure of PfRH5 in complex with BSG (4U0Q), CyRPA (6MPV), 9AD4 (4U0R), R5.004 and R5.016 (6RCU) and R5.011 and R5.016 (6RCV) have been solved and are publicly available in Protein Data Bank (PDB). Identified SNPs were threaded onto the structure of PfRH5 in complex with either Basigin, CyRPA or monoclonal antibodies. The BSG-PfRH5-PfCyRPA-mAbs complex was built by superposing the PfRH5 in the BSG-PfRH5, PfCyRPA-PfRH5 and PfRH5-mAb complexes. PyMOL version 2.3.2 was used to predict the effect and to plot the structural location of each SNP50. Missing residues in antibodies, Basigin, and CyRPA were modeled in Maestro. FoldX v5 was used to predict the effects of SNPs on the stability of PfRH5 and binding affinity of antibodies, Basigin, and CyRPA.
Statistical analysis
All data presented here were analyzed using GraphPad Prism version 10.0.2 (GraphPad Software Inc., California, US). The Fisher Exact test was used to compute differences between categorical variables. Mean GIA differences between isolates and across concentrations were assessed using Two-way ANOVA and, where significant, the Dunnett’s multiple comparison test was used for pairwise comparisons. Differences in GIA activities of antibody combinations were tested using the Wilcoxon matched-pairs signed rank test, while the Spearman rank correlation was used to assess relationships between variables. All tests were performed two-tailed and only a p-value of at least 0.05 was considered significant.
Supplementary information
Acknowledgements
We would like to thank Adama Diallo and Gerald Keita from Bandafassi, Souleymane Ngom from Dalaba, and all the healthcare workers at these sites for their partnership with Institut Pasteur Dakar. We would also like to thank the people of Kédougou for their participation and invaluable contributions to this work. We would like to thank the Yale Center for Genome Analysis (YCGA) and acknowledge the support of the NIH HPC equipment grant (1S10OD030363-01A1). We would like to acknowledge the generosity and continued support of the FlowJo Africa program in their commitment to supporting African researchers. This work was supported by G4 group funding (G45267, Malaria Experimental Genetic Approaches & Vaccines) from the Institut Pasteur de Paris and Agence Universitaire de la Francophonie (AUF) and has been produced with the financial assistance of the European Union (Grant no. DCI-PANAF/2020/420-028), through the African Research Initiative for Scientific Excellence (ARISE), pilot program. ARISE is implemented by the African Academy of Sciences with support from the European Commission and the African Union Commission. The contents of this document are the sole responsibility of the authors and can under no circumstances be regarded as reflecting the position of the European Union, the African Academy of Sciences, and the African Union Commission. AKB is supported by the Fogarty International Center of the NIH (K01 TW010496), National Institute of Allergy and Infectious Diseases of the NIH (R01 AI168238), LGT is supported by an ARISE grant from the African Academy of Sciences (ARISE-PP-FA-056). Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 1S10OD030363-01A1 to YCGA. This study was additionally supported by National Institute of Allergy and Infectious Diseases (NIAID) grant R61 AI176583-01 to ZS.
Author contributions
Project conception, management and coordination: L.G.T., A.K.B. Data collection and experimentation: L.G.T., A.B., R.L., M.N.P., A.C., F.D., S.D.S., A.T., B.D.S., A.M., A.K.B. Data analysis: L.G.T., A.B., M.N.P., Y.G., Z.S., A.B., A.K.B. Contribution on reagents, materials, and analysis tools: S.J.D., K.M.c, D.P., S.P.D., A.M., I.V.W., L.S. Paper writing: L.G.T. and A.K.B. All authors read and approved the manuscript.
Data availability
Sequencing Reads associated with this study have been deposited in the NCBI SRA with the BioProject Accession: PRJNA1101747. GIA data and associated genotype data have been deposited in the Dryad database and are publicly available at: 10.5061/dryad.8931zcrzv.
Competing interests
KMc and SJD are inventors on patent applications relating to RH5 malaria vaccines and/or antibodies. All other authors have declared that no conflict of interest exists.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-024-00986-x.
References
- 1.Organization, W. H. World malaria report 2023. (2023).
- 2.WHO, U. and. Reversing the Incidence of Malaria 2000–2015. WHO Glob. Malar. Program (2015).
- 3.Cibulskis, R. E. et al. Malaria: Global progress 2000 - 2015 and future challenges. Infectious Diseases of Poverty10.1186/s40249-016-0151-8 (2016). [DOI] [PMC free article] [PubMed]
- 4.Balikagala, B. et al. Evidence of Artemisinin-Resistant Malaria in Africa. N. Engl. J. Med. 10.1056/nejmoa2101746 (2021). [DOI] [PubMed]
- 5.Duffy, P. E. & Patrick Gorres, J. Malaria vaccines since 2000: progress, priorities, products. npj Vaccines5, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rts, S. C. T. P. Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet386, 31–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datoo, M. S. et al. Safety and efficacy of malaria vaccine candidate R21/Matrix-M in African children: a multicentre, double-blind, randomised, phase 3 trial. Lancet 533–544. 10.1016/S0140-6736(23)02511-4 (2024) [DOI] [PubMed]
- 8.Thera, M. A. et al. Safety and immunogenicity of an AMA1 malaria vaccine in Malian children: Results of a phase 1 randomized controlled trial. PLoS One5, (2010). [DOI] [PMC free article] [PubMed]
- 9.Thera, M. A. et al. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: Results of a phase 1 randomized controlled trial. PLoS One3, (2008). [DOI] [PMC free article] [PubMed]
- 10.Polhemus, M. E. et al. Phase I dose escalation safety and immunogenicity trial of Plasmodium falciparum apical membrane protein (AMA-1) FMP2.1, adjuvanted with AS02A, in malaria-naïve adults at the Walter Reed Army Institute of Research. Vaccine25, 4203–4212 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Payne, R. O. et al. Demonstration of the blood-stage plasmodium falciparum controlled human malaria infection model to assess efficacy of the p. falciparum apical membrane antigen 1 Vaccine, FMP2.1/AS01. J. Infect. Dis. 10.1093/infdis/jiw039 (2016). [DOI] [PMC free article] [PubMed]
- 12.Thera, M. A. et al. A Field Trial to Assess a Blood-Stage Malaria Vaccine. N. Engl. J. Med.365, 1004–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neafsey, D. E. et al. Genetic Diversity and Protective Efficacy of the RTS,S/AS01 Malaria Vaccine. N. Engl. J. Med.373, 2025–2037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas, A. D. et al. The blood-stage malaria antigen PfRH5 is susceptible to vaccine-inducible cross-strain neutralizing antibody. Nat. Commun. 2, (2011). [DOI] [PMC free article] [PubMed]
- 15.Baum, J. et al. Reticulocyte-binding protein homologue 5 - An essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum. Int. J. Parasitol.39, 371–380 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez, M., Lustigman, S., Montero, E., Oksov, Y. & Lobo, C. A. PfRH5: A novel reticulocyte-binding family homolog of Plasmodium falciparum that binds to the erythrocyte, and an investigation of its receptor. PLoS One3, 1–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne, R. O. et al. Human vaccination against Plasmodium vivax Duffy-binding protein induces strain-transcending antibodies. JCI insight10.1172/jci.insight.93683 (2017) [DOI] [PMC free article] [PubMed]
- 18.Silk, S. E. et al. Superior antibody immunogenicity of a viral-vectored RH5 blood-stage malaria vaccine in Tanzanian infants as compared to adults. Med4, 668–686.e7 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Williams, A. R. et al. Enhancing Blockade of Plasmodium falciparum Erythrocyte Invasion: Assessing Combinations of Antibodies against PfRH5 and Other Merozoite Antigens. PLoS Pathog.8, e1002991 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustamante, L. Y. et al. A full-length recombinant Plasmodium falciparum PfRH5 protein induces inhibitory antibodies that are effective across common PfRH5 genetic variants. Vaccine31, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy, K. S. et al. Multiprotein complex between the GPI-anchored CyRPA with PfRH5 and PfRipr is crucial for Plasmodium falciparum erythrocyte invasion. Proc. Natl Acad. Sci. Usa.112, 1179–1184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douglas, A. D. et al. A defined mechanistic correlate of protection against Plasmodium falciparum malaria in non-human primates. Nat. Commun.10, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas, A. D. et al. A PfRH5-based vaccine is efficacious against heterologous strain blood-stage plasmodium falciparum infection in Aotus monkeys. Cell Host Microbe17, 130–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minassian, A. M. et al. Reduced blood-stage malaria growth and immune correlates in humans following RH5 vaccination. Med2, 701–719.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alanine, D. G. W. et al. Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies. Cell178, 216–228.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douglas, A. D. et al. Neutralization of Plasmodium falciparum Merozoites by Antibodies against PfRH5. J. Immunol.192, 245–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bei, A. K. et al. Research Article A flow cytometry-based assay for measuring invasion of red blood cells by Plasmodium falciparum. 234–237 10.1002/ajh.21642 (2010). [DOI] [PMC free article] [PubMed]
- 28.Wright, K. E. et al. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature515, 427–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura, K. et al. Assessment of precision in growth inhibition assay (GIA) using human anti-PfRH5 antibodies. Malar. J.22, 1–14 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azasi, Y. et al. Bliss’ and Loewe’s additive and synergistic effects in Plasmodium falciparum growth inhibition by AMA1-RON2L, RH5, RIPR and CyRPA antibody combinations. Sci. Rep.10, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett, J. R. et al. Analysis of the Diverse Antigenic Landscape of the Malaria Invasion Protein RH5 Identifies a Potent Vaccine-Induced Human Public Antibody Clonotype. bioRxiv 2023.10.04.560576 (2023). [DOI] [PMC free article] [PubMed]
- 32.Wong, W. et al. Structure of Plasmodium falciparum Rh5–CyRPA–Ripr invasion complex. Nature10.1038/s41586-018-0779-6 (2019). [DOI] [PubMed]
- 33.Wright, G. J. & Rayner, J. C. Plasmodium falciparum Erythrocyte Invasion: Combining Function with Immune Evasion. PLoS Pathog.10, 1–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen, C. M. et al. Protein/AS01B vaccination elicits stronger, more Th2-skewed antigen-specific human T follicular helper cell responses than heterologous viral vectors. Cell Rep. Med2, 100207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ajibaye, O. et al. Genetic polymorphisms in malaria vaccine candidate Plasmodium falciparum reticulocyte-binding protein homologue-5 among populations in Lagos, Nigeria. Malar. J.19, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangou, K. et al. Structure-guided insights into potential function of novel genetic variants in the malaria vaccine candidate PfRh5. Sci. Rep.12, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, A. J. et al. Assessing the functional impact of PfRh5 genetic diversity on ex vivo erythrocyte invasion inhibition. Sci. Rep.11, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ndwiga, L. et al. The Plasmodium falciparum Rh5 invasion protein complex reveals an excess of rare variant mutations. Malar. J.20, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waweru, H. et al. Limited genetic variations of the Rh5-CyRPA-Ripr invasion complex in Plasmodium falciparum parasite population in selected malaria-endemic regions, Kenya. Front. Trop. Dis.4, 1–9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silk, S. E. et al. Blood-stage malaria vaccine candidate RH5.1/Matrix-M in healthy Tanzanian adults and children; an open-label, non-randomised, first-in-human, single-centre, phase 1b trial. Lancet Infect. Dis. 10.1016/S1473-3099(24)00312-8 (2024). [DOI] [PubMed]
- 41.Neafsey, D. E., Taylor, A. R. & MacInnis, B. L. Advances and opportunities in malaria population genomics. Nat. Rev. Genet.22, 502–517 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manske, M. et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature10.1038/nature11174 (2012). [DOI] [PMC free article] [PubMed]
- 43.Patel, S. D. et al. Plasmodium falciparum merozoite surface antigen, PfRH5, elicits detectable levels of invasion-inhibiting antibodies in humans. J. Infect. Dis.208, 1679–1687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Triglia, T. et al. Plasmepsin X activates the PCRCR complex of Plasmodium falciparum by processing PfRh5 for erythrocyte invasion. Nat. Commun. 10.1038/s41467-023-37890-2 (2023). [DOI] [PMC free article] [PubMed]
- 45.King, L. D. W. et al. Preclinical Development of a Stabilized RH5 Virus-Like Particle Vaccine that Induces Improved Anti-Malarial Antibodies (2024). [DOI] [PMC free article] [PubMed]
- 46.Farrell, B. et al. Structure of the PfRCR complex which bridges the malaria parasite and erythrocyte during invasion. bioRxiv (Springer US, 10.1038/s41586-023-06856-1 2023).
- 47.Thiam, L. G., Ansah, F., Niang, M., Awandare, G. A. & Aniweh, Y. Short-term cryopreservation and thawing have minimal effects on Plasmodium falciparum ex vivo invasion profile. Front. Cell. Infect. Microbiol. 10.3389/fcimb.2022.997418 (2022). [DOI] [PMC free article] [PubMed]
- 48.Thiam, L. G. Investigating Factors Influencing Variation in P. Falciparum Invasion Phenotyping Assays. (University if Ghana, Legon, 2019).
- 49.Thiam, L. G. et al. Blood donor variability is a modulatory factor for P. falciparum invasion phenotyping assays. Sci. Rep.11, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrodinger, L. The PyMOL Molecular Graphics System (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing Reads associated with this study have been deposited in the NCBI SRA with the BioProject Accession: PRJNA1101747. GIA data and associated genotype data have been deposited in the Dryad database and are publicly available at: 10.5061/dryad.8931zcrzv.