Abstract
The question of whether translation initiation factor eIF4E and the complete eIF4G polypeptide are required for initiation dependent on the IRES (internal ribosome entry site) of hepatitis A virus (HAV) has been examined using in vitro translation in standard and eIF4G-depleted rabbit reticulocyte lysates. In agreement with previous publications, the HAV IRES is unique among all picornavirus IRESs in that it was inhibited if translation initiation factor eIF4G was cleaved by foot-and-mouth disease L-proteases. In addition, the HAV IRES was inhibited by addition of eIF4E-binding protein 1, which binds tightly to eIF4E and sequesters it, thus preventing its association with eIF4G. The HAV IRES was also inhibited by addition of m7GpppG cap analogue, irrespective of whether the RNA tested was capped or not. Thus, initiation on the HAV IRES requires that eIF4E be associated with eIF4G and that the cap-binding pocket of eIF4E be empty and unoccupied. This suggests two alternative models: (i) initiation requires a direct interaction between an internal site in the IRES and eIF4E/4G, an interaction which involves the cap-binding pocket of eIF4E in addition to any direct eIF4G-RNA interactions; or (ii) it requires eIF4G in a particular conformation which can be attained only if eIF4E is bound to it, with the cap-binding pocket of the eIF4E unoccupied.
It is now generally accepted that picornavirus RNAs are translated by a mechanism of internal initiation, in which the ribosome enters directly at an internal site within the RNA rather than scanning from the physical 5′ end (reviewed in reference 2). The 5′ untranslated region (UTR) of the viral RNA has an IRES (internal ribosome entry site) about 450 nucleotides (nt) in length which is necessary and sufficient to promote internal ribosome entry and internal initiation. On the basis of primary and secondary structure conservation, the picornavirus IRESs can be divided into one minor and two major groups: (i) hepatitis A virus (HAV); (ii) entero- and rhinoviruses; and (iii) cardio-, aphtho-, and parechoviruses. Internal initiation of translation on the IRESs of the two major groups is thought to require all of the canonical initiation factors that are involved in the scanning mechanism except that eIF4E is completely redundant and the requirement for eIF4G can be fulfilled by just the C-terminal two-thirds fragment of this protein (26, 27). Notably, the activity of these IRESs is not inhibited (and may even be actually stimulated in certain circumstances) when eIF4G is cleaved by entero- or rhinovirus 2A protease or foot-and-mouth disease virus (FMDV) L-protease (5, 6, 7, 28). These viral proteases cleave eIF4G to give (i) an N-terminal one-third fragment which has the site for interaction of eIF4G with eIF4E, the only translation initiation factor that binds directly to 5′ caps, and also a site for binding poly(A)-binding protein; and (ii) a C-terminal two-thirds fragment which has two distinct sites for interaction with eIF4A, the RNA helicase initiation factor, and a site for binding eIF3 (14, 17, 18).
In sharp contrast to the two major types of picornavirus IRES, the activity of the HAV IRES is strongly inhibited if eIF4G is cleaved by the 2A protease or L-protease (4, 5, 7, 29). This implies that the C-terminal two-thirds fragment of eIF4G, with its associated eIF4A and eIF3, cannot support translation dependent on the HAV IRES. However, it is not clear whether what is required is a larger fragment of eIF4G, or whether the activity of this IRES actually needs eIF4E and eIF4E-eIF4G association. Surprisingly, there appear to have been few attempts, if any, to address these wider questions and to follow up the initial finding of IRES inactivation by cleavage of eIF4G. In this paper we remedy this deficiency by examining the effect on HAV IRES activity of eIF4E-binding protein 1 (4E-BP1), a protein which sequesters eIF4E tightly and as a consequence blocks its association with eIF4G, and of m7GpppG cap analogue, which binds to eIF4E and prevents its interaction with the capped 5′ ends of mRNAs that are translated by the scanning mechanism. We show that internal initiation on the HAV IRES has initiation factor requirements remarkably similar to those for initiation of translation of capped mRNAs by the scanning ribosome mechanism.
MATERIALS AND METHODS
Plasmid constructs.
The dicistronic construct with the HAV IRES used in this work is pXLJ-HAV, described by Borman et al. (5). It has the cDNA sequence of Xenopus laevis cyclin B2 as the upstream cistron, followed by nt 44 to 378 of the HAV sequence as intercistronic spacer, fused directly to the slightly truncated form of the NS1 cDNA described by Borman and Jackson (3), cloned into pGEM-2 (Promega) such that transcription with T7 RNA polymerase generates sense RNA. With one exception, all of the constructs with other viral IRESs are built on similar lines. pXLJHRV10-611 has the complete human rhinovirus type 2 (HRV2) 5′ UTR (except the first 9 nt) fused to the same slightly truncated NS1 cDNA sequence (3, 12). pXLPV1-747 has the complete 5′ UTR and the first 5 nt of coding sequence of poliovirus (PV) type 1 (Mahoney) fused via a 34-nt linker to the full-length NS1 coding sequences (12). The classical swine fever virus (CSFV) construct has the first 423 nt of the CSFV (Alfort) sequence fused, via a 3-nt linker, to the truncated NS1 sequences; the downstream cistron product is therefore the slightly truncated NS protein with an N-terminal extension consisting of the first 17 amino acids of CSFV coding sequence, plus one amino acid encoded by the 3-nt linker. All of the above plasmids were linearized with EcoRI prior to in vitro transcription.
The upstream cistron was deleted from pXLJ-HAV to generate a monocistronic derivative, pHAV-NS. pXLJ-HAV was cut with HindIII and SalI, the ends were filled in, and the plasmid was religated. pHAV-NS was likewise cut with EcoRI prior to in vitro transcription.
The dicistronic construct with the encephalomyocarditis virus (EMCV) IRES was generated from pEMCV L-VP0, which has been described previously (16). The cDNA encoding the slightly truncated form of NS1 and the NS1 3′ UTR was excised from the previously described pJ0 (3) by cutting with SalI and EcoRI, the overhanging ends were filled in, and the fragment was inserted into the blunted EcoRI site of pEMCV L-VP0, upstream of the EMCV IRES sequence. A clone with the correct (sense) orientation of the NS-related cDNA sequence was selected. It was linearized with StuI prior to in vitro transcription.
Two constructs were used to generate monocistronic mRNAs that would be translated by the scanning mechanism. One has the cDNA sequences coding for unr (upstream of N-ras) cloned into pET21d (13) and was linearized with HindIII prior to transcription. The other (pXL4) codes for X. laevis cyclin A (22) and was linearized with BamHI.
In vitro transcription and translation assays.
Transcription of linearized plasmids by bacteriophage T7 RNA polymerase was carried out exactly as described previously (8). In vitro translation assays were carried out as described by Jackson and Hunt (15). Briefly, the reactions consisted of 60 to 70% (by volume) micrococcal nuclease-treated rabbit reticulocyte lysate and the following additional components at the final concentrations stated: 100 or 70 mM KCl (as stated in the figure legends), 0.5 mM MgCl2, 10 mM creatine phosphate, 50 μg of creatine phosphokinase per ml, 15 μM hemin, 0.1 mM each amino acid except methionine, 50 μg of calf liver tRNA (Boehringer) per ml, and 0.5 mCi of [35S]methionine (SJ1515; Amersham Pharmacia Biotech) per ml. Assays supplemented with m7GpppG or GpppG also received additional MgCl2 at 0.8 mol/mol of cap analogue, to counteract the chelating potential of the cap analogue; the ratio was chosen empirically on the basis of the observed shift in Mg2+ optimum caused by these cap analogues. In experiments in which the HRV and PV IRESs were assayed, the reticulocyte lysate was replaced by a mixture (80:20, vol/vol) of reticulocyte lysate and HeLa cell high-salt (HS)S100. The latter is essentially the complete cytoplasmic extract minus salt-washed ribosomes; it is prepared by making HeLa cell S10 postmitochondrial supernatant 0.5 M in KCl and 6 mM in magnesium acetate, centrifuging the ribosomes at 100,000 × g, and dialyzing the supernatant against low-KCl buffer as described previously (12, 13). Translation assays were incubated at 30°C for 60 min, and then the translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography. Hyperfilm βmax (Amersham Pharmacia Biotech) was used for autoradiography, and quantitative densitometry of the films was done using Phoretix software.
To generate FMDV L-protease by in vitro transcription and translation, plasmid pMMl (21) was linearized with XbaI prior to in vitro transcription by T7 polymerase under conditions to generate uncapped transcripts (8). The RNA was translated in vitro at 50 μg/ml for 90 min under the conditions described above except that the added KCl concentration was reduced to 50 mM. The reaction was then made 2 mM in CaCl2, to reactivate the micrococcal nuclease, and left for 15 min at room temperature before addition of 5 mM EGTA. The protease preparation was diluted 100-fold into fresh lysate, which was then incubated for 10 min at 30°C to effect complete cleavage of the endogenous eIF4G.
Recombinant L-protease and 4E-BP1 were expressed in Escherichia coli and purified as described previously (23). Rabbit reticulocyte lysates were depleted of eIF4G by an affinity column depletion method described in detail elsewhere (1); for the experiments described here, the lysates were made 70 mM in KCl prior to addition to the affinity matrix, rather than the 100 mM used previously. The expression in E. coli and subsequent purification of recombinant p100 fragment of eIF4G was as described elsewhere (1).
RESULTS
Assay systems.
The standard approach in this work has been in vitro translation of a dicistronic mRNA (with the HAV IRES as intercistronic spacer) in rabbit reticulocyte lysates. The dicistronic mRNA was that described by Borman et al. (5) and has an upstream cistron coding for X. laevis cyclin B2 and a downstream cistron coding for a slightly truncated version of the influenza virus (A strain) NS1 protein. In view of the previous finding that the optimum monovalent salt concentration for HAV IRES activity is atypically low (5), the translation assays were carried out with 70 mM added KCl rather than the 100 mM normally used for translation of typical capped mRNAs. In several experiments, comparisons were made between this dicistronic mRNA with the HAV IRES and similar mRNAs with other picornavirus IRESs. When the comparison was with the PV or HRV IRES, the reticulocyte lysate was replaced by a mixture of rabbit reticulocyte lysate and HeLa cell HS S100 (80:20, vol/vol) to provide the trans-acting factors which are necessary for the activity of these two IRESs but which are either absent from reticulocyte lysates or present only in very low abundance. We have not observed any major differences between the characteristics of HAV IRES function in this mixed system as opposed to the standard reticulocyte lysate system.
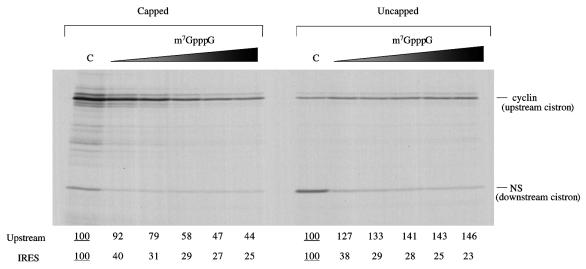
For some experiments, we used monocistronic mRNAs with the HAV IRES linked to the slightly truncated NS1 reporter rather than dicistronic templates. The reason for this is that the HAV IRES is rather weak, and despite the use of a reduced KCl concentration (70 mM), and even though the RNA concentration was rather below the saturating level, there were indications that the apparent activity of the HAV IRES was influenced by the efficiency of the competing translation of the upstream cistron of the dicistronic mRNA. For example, in a comparison of capped and uncapped dicistronic mRNAs, the IRES appears to be more active in the latter case than in the capped mRNA background, presumably because there is much more competition from upstream cistron translation when the mRNA is capped (see, for example, Fig. 4).
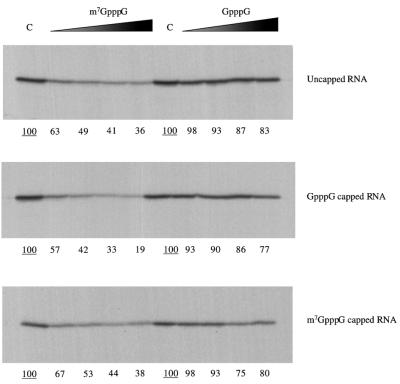
FIG. 4.
The activity of the HAV IRES is inhibited by m7GpppG cap analogue. Capped or uncapped dicistronic mRNAs with the HAV IRES were translated at 25 μg/ml in reticulocyte lysate, in the presence of added KCl at 70 mM. Cap analogue (m7GpppG) was added at 0 (lane C), 0.025, 0.05, 0.1, 0.2, and 0.4 mM, and additional MgCl2 was also added at 0.8 mol/mol of cap analogue. Translation was at 30°C for 60 min, and the translation products were analyzed by SDS-PAGE followed by autoradiography. The positions of the upstream (cyclin) cistron product and downstream, IRES-dependent, NS-related product are shown. The yields of radiolabeled translation products of the upstream and IRES-driven cistrons were determined by scanning densitometry and are expressed relative to the yield in the corresponding control assay, which was set at 100 (the underlined value).
The HAV IRES is inhibited by recombinant FMDV L-protease and L-protease expressed in vitro.
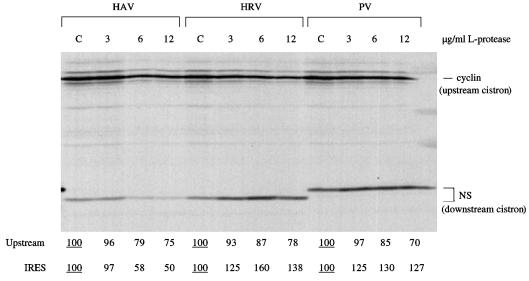
To verify that the HAV IRES behaves in the same way in our system as reported previously by others (4, 5), we first looked at the effect of recombinant L-protease on translation of capped dicistronic mRNAs with the HAV, HRV, or PV IRES. The results (Fig. 1) show that, particularly at the two higher concentrations of protease, there was inhibition of translation of the upstream (capped cistron) and translation driven by the HAV IRES. In complete contrast, translation dependent on the HRV and PV IRESs was significantly stimulated by L-protease, in agreement with previously published results (5). (The larger size of the downstream cistron product translated from the PV IRES is because it has the full-length NS coding sequences, rather than a slightly truncated form, and this sequence is fused to the initiation codon via a short linker sequence rather than being linked directly to the initiation codon.)
FIG. 1.
Recombinant FMDV L-protease inhibits HAV IRES activity. Capped dicistronic mRNAs, each with the designated IRES, were translated at 25 μg/ml in the mixed reticulocyte lysate–HeLa cell HS-S100 system (see Materials and Methods), which had been preincubated for 10 min at 30°C with the indicated concentration of recombinant FMDV L-protease or with buffer (lanes C [control]). The concentration of added KCl in the assays was 70 mM. Translation was for 60 min, and the translation products were analyzed by SDS-PAGE followed by autoradiography. The positions of the upstream (cyclin) cistron product and downstream, IRES-dependent, NS-related product are shown. The NS-related product translated from the PV IRES is larger because this is full-length NS1 protein, rather than the slightly truncated form linked to the HAV and HRV IRESs, and because the NS coding sequences are joined to the authentic PV initiation codon via a short linker. The yields of radiolabeled translation products of the upstream and IRES-driven cistrons were determined by scanning densitometry and are expressed relative to the yield in the corresponding control assay, which was set at 100 (the underlined value).
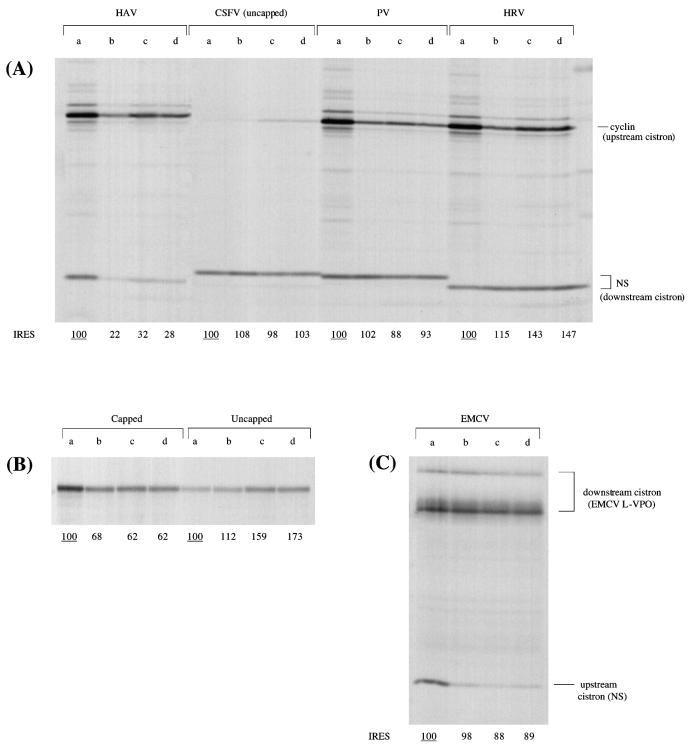
One possible reservation concerning these findings is that very high concentrations of recombinant L-protease are needed to effect the cleavage of eIF4G in the reticulocyte lysate and to obtain the effects seen in Fig. 1. In contrast, L-protease expressed by in vitro translation appears to be at least 100-fold more active (per microgram) in eIF4G cleavage activity (2). Accordingly, we considered it important to test whether these low levels of in vitro-expressed L-protease could also inhibit HAV IRES activity, a question which does not seem to have been addressed previously. Figure 2 shows that, as expected, this preparation of L-protease, which caused complete cleavage of the endogenous eIF4G (data not shown), inhibited the translation of a capped monocistronic RNA (unr mRNA) and the upstream cistron of all capped dicistronic mRNAs. In complete contrast, it stimulated the translation of uncapped monocistronic unr mRNA, entirely consistent with previously published results (22). Not surprisingly, therefore, in the case of the uncapped dicistronic mRNA with the CSFV IRES, there was also a stimulation of translation of the upstream cistron, which was very inefficiently translated not just because the RNA was uncapped but also because of strong competition by the powerful CSFV IRES (Fig. 2). As for IRES-dependent translation, there was no significant effect on the EMCV, CSFV, and PV IRESs, but the HRV IRES was quite strongly stimulated (Fig. 2). In complete contrast, however, the HAV IRES was strongly inhibited by this protease preparation (Fig. 2), just as effectively as when much higher levels of recombinant L-protease were used (Fig. 1). (Here again, the different sizes of the IRES-dependent NS-related translation product in Fig. 2 depend on whether it is the full-length or slightly truncated form of NS coding sequences that is present, and whether these NS-related sequences are fused directly to the initiation codon or fused indirectly, either via linker sequences as in the case of the PV construct or via viral coding sequences as for the CSFV construct).
FIG. 2.
The activity of the HAV IRES is inhibited by 4E-BP1 and by FMDV L-protease expressed in vitro. All RNAs were translated at 25 μg/ml in the mixed reticulocyte lysate–HeLa cell HS-S100 system (see Materials and Methods), which had been preincubated at 30°C as follows: lanes a, 15-min preincubation with buffer (control); lanes b, 10-min preincubation with 4E-BP1 (10 μg/ml); lanes c, 5-min preincubation with FMDV L-protease expressed in vitro; lanes d, 5-min preincubation with FMDV L-protease followed by 10-min preincubation with 4E-BP1 (10 μg/ml). The concentration of added KCl in all of the assays was 70 mM. (A) All dicistronic mRNAs have an upstream cistron coding for X. laevis cyclin B2 and a downstream cistron coding for an influenza virus NS1 derivative, and all except for the RNA with the CSFV IRES were capped. (B) Control assays carried out with monocistronic mRNAs coding for unr in both capped and uncapped forms. (C) The template was a capped dicistronic mRNA with a slightly truncated form of the influenza virus NS1 coding sequence as the upstream cistron, an EMCV IRES, and EMCV sequences coding for viral L-VP0 as the downstream IRES-dependent cistron. In all cases, translation was for 60 min and the translation products were analyzed by SDS-PAGE followed by autoradiography. The positions of the upstream cistron product and downstream, IRES-dependent product are shown. The different sizes of the IRES-dependent NS-related translation product are discussed in Results. The yields of radiolabeled translation products of the IRES-driven cistrons (A and C) and of the single product in the case of the monocistronic RNAs (B) were determined by scanning densitometry and are expressed relative to the yield in the corresponding control assay, which was set at 100 (the underlined value).
The HAV IRES is inhibited by addition of 4E-BP1, and the inhibition is reversed by eIF4E.
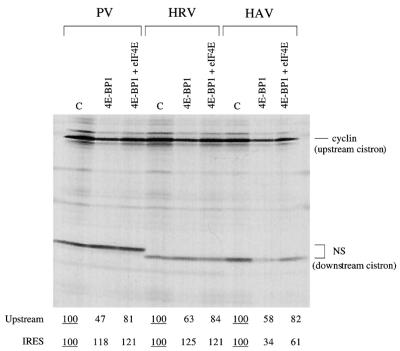
Figure 2 also shows the results of experiments in which the translation assays were supplemented with 4E-BP1, which binds to and sequesters eIF4E, preventing the interaction of eIF4E with eIF4G to constitute the eIF4F holoenzyme complex (25). Preincubation with 4E-BP1 inhibited the translation of a capped monocistronic mRNA (coding for unr) and the upstream cistron of all capped dicistronic mRNAs. At this concentration, it had no marked or significant effect on translation of uncapped monocistronic unr mRNA or on the EMCV, CSFV, HRV, or PV IRES (Fig. 2). The HAV IRES was clearly the exceptional IRES in that it was strongly inhibited (Fig. 2). Inhibition of the upstream cistron of capped dicistronic mRNAs and of the HAV IRES activity could be at least partially reversed by addition of eIF4E (Fig. 3), and the extent of this reversal was dependent on the concentration of added eIF4E (I. K. Ali and R. J. Jackson, unpublished data).
FIG. 3.
The inhibition of HAV IRES activity by 4E-BP1 can be reversed by addition of eIF4E. Mixed reticulocyte lysate–HeLa cell HS-S100 (see Materials and Methods) was preincubated for 10 min at 30°C with 4E-BP1 (10 μg/ml) or with buffer control (C); then KCl was added to 70 mM together with the other components of the translation assay, including capped dicistronic mRNAs, each with the designated IRES, at 25 μg/ml. Where indicated, eIF4E (purified from pig brain) was added at 25 μg/ml. Translation was for 60 min, and the translation products were analyzed by SDS-PAGE followed by autoradiography. The positions of the upstream (cyclin) cistron product and downstream, IRES-dependent, NS-related product are shown. The NS-related product translated from the PV IRES is larger because this is full-length NS1 protein, rather than the slightly truncated form linked to the HAV and HRV IRESs, and because the NS coding sequences are joined to the authentic PV initiation codon via a short linker. The yields of radiolabeled translation products of the upstream and IRES-driven cistrons were determined by scanning densitometry and are expressed relative to the yield in the corresponding control assay, which was set at 100 (the underlined value).
We also examined the effect of pretreating the system first with FMDV L-protease and then with 4E-BP1, this sequence of pretreatments being dictated by the fact that cleavage of eIF4G by the protease is known to be inhibited by 4E-BP1, presumably because 4E-BP1 sequesters eIF4E and thus strips the eIF4E from association with eIF4G, which appears to change the conformation of eIF4G to one that it is uncleavable (11, 24). The results (Fig. 2) indicate that with the upstream citron of capped dicistronic mRNAs, the effect of the protease was epistatic to the influence of 4E-BP1. This is consistent with the fact, mentioned above, that cleavage of eIF4G by the in vitro-expressed protease was virtually complete. If there had been significant amounts of residual uncleaved eIF4G driving the translation of these capped mRNAs, addition of 4E-BP1 to the L-protease-treated lysate would have been expected to cause further inhibition of translation beyond that due to the protease.
The HAV IRES is inhibited by m7GpppG cap analogue but not by GpppG.
Addition of m7GpppG cap analogue to assays of dicistronic mRNAs caused the expected inhibition of translation of the upstream cistron if the mRNA was capped but a stimulation of translation of the 5′-proximal cistron if the RNA was uncapped (Fig. 4). The latter effect, which has been reported previously (9), may be the consequence of relief of competition due to translation of short capped fragments of globin mRNA which will be present in the micrococcal nuclease-treated lysate but will not give rise to detectable translation products. As for translation dependent on the HAV IRES, this was inhibited by cap analogue, irrespective of whether the 5′ end of the dicistronic mRNA was capped or uncapped (Fig. 4).
At first sight it would appear that the HAV IRES is more sensitive than the upstream capped cistron to inhibition by cap analogue (Fig. 4). However, this conclusion needs to be tempered by the facts that capping of in vitro transcripts is never 100% efficient (in fact, it is only about 70% efficient in our hands [8]) and that m7GpppG actually stimulates translation of the upstream cistron of uncapped dicistronic mRNAs (Fig. 4). Thus, the observed effect of cap analogue on the upstream cistron of capped transcripts in fact represents the sum of two opposing effects: an inhibition of translation of the majority capped species, and a stimulation of the minority uncapped transcripts in the preparation. Thus, the data will underestimate the true sensitivity to inhibition by cap analogue of transcripts that are 100% capped.
Another feature of the data of Fig. 4 which deserves comment is the fact that although translation dependent on the HAV IRES seems highly sensitive to inhibition by m7GpppG, there is a residual amount, equivalent to about 25% of the control, which appears to be rather resistant to such inhibition. There are two alternative types of explanation which can be invoked to account for this result. One possibility is that there are two distinct mechanisms of initiation operating: the minority of initiation events occurring via a mechanism that is very resistant to cap analogue, and the majority of initiation occurring by another route that is highly sensitive to such inhibition. The alternative explanation, if all initiation events follow the same single pathway, is that this mechanism must still be able to operate, albeit at somewhat lower efficiency, even when the cap-binding pocket of the eIF4E component of the eIF4F complex is occupied by cap analogue. This would be rather different from the effect of cap analogue on the translation of capped mRNAs: with natural mRNAs that are 100% capped (as opposed to capped RNAs generated by in vitro transcription, which inevitably include some uncapped species), 0.4 mM cap analogue generally causes near-complete inhibition (1).
The data of Fig. 4 give the appearance that in the complete absence of cap analogue, the HAV IRES was more active in the uncapped dicistronic mRNA background than if the mRNA was capped. We believe that the explanation for these differences lies in competition between the 5′-proximal cistron and the IRES-dependent cistron. The more efficient translation of the first cistron in the capped mRNA than the uncapped version results, by competition, in a lower IRES activity in the capped than the uncapped mRNA background.
In view of these complications caused by competition between the two cistrons, we studied the effect of m7GpppG cap analogue, or a GpppG control, on the translation of monocistronic mRNAs with the HAV IRES. These monocistronic mRNAs were generated by in vitro transcription under conditions designed to produce either uncapped, m7GpppG-capped, or GpppG-capped RNAs. The translation assays show that the HAV IRES in these monocistronic mRNAs was quite strongly inhibited by even the lowest concentration of m7GpppG (Fig. 5), irrespective of whether the 5′ end was uncapped, GpppG capped, or m7GpppG capped.
FIG. 5.
The inhibitory effect of m7GpppG cap analogue on HAV IRES activity is independent of the nature of the 5′ end. Monocistronic mRNAs synthesized either as uncapped RNAs or with GpppG or m7GpppG capped 5′ ends, as indicated, were translated at 20 μg/ml in rabbit reticulocyte lysate, in the presence of added KCl at 70 mM. Cap analogues, either m7GpppG or GpppG, as indicated, were added at 0 (lane C), 0.025, 0.05, 0.1 or 0.2 mM, and additional MgCl2 was also added at 0.8 mol/mol of cap analogue. Translation was at 30°C for 60, min and the translation products were analyzed by SDS-PAGE followed by autoradiography. The yields of radiolabeled translation products were determined by scanning densitometry and are expressed relative to the yield in the corresponding control assay, which was set at 100 (the underlined value).
Ability of the C-terminal two-thirds of eIF4G to drive translation dependent on the HAV IRES.
Entero and rhinovirus 2A proteases and FMDV L-protease cleave the eIF4G component of the eIF4F holoenzyme complex into an N-terminal one-third fragment, which has the binding site for the cap-binding factor eIF4E, and a C-terminal two-thirds fragment (hereafter designated p100), which interacts with eIF3 and also has two binding sites for the eIF4A RNA helicase factor (14, 17, 18). Since these viruses shut off translation of capped host cell mRNA, and as the proteases inhibit the translation of capped mRNAs in vitro (4, 22, 23), it has been generally assumed that p100 cannot support the translation of capped mRNAs. However, by the use of a novel eIF4G depletion strategy we have recently shown that capped mRNA translation can be driven by recombinant p100, provided sufficient is added (1); our observations suggest that the shutoff of host cell mRNA translation is due not to an intrinsic inactivity of p100 but to a combination of limiting concentration and affinity for capped mRNAs, coupled with strong competition by the viral RNA for p100.
We have studied the activity of dicistronic mRNAs with the HAV IRES in this eIF4G-depleted reticulocyte lysate system. As we have shown elsewhere, translation in this system is highly dependent on add-back of eIF4G derivatives (e.g., p100) but does not require supplementation with other initiation factors (1). eIF4G is essentially completely (∼95%) depleted, but depletion of other factors is only partial: eIF3, 10 to 20% depleted; eIF4A, 30 to 40% depleted; eIF4B, 10 to 20% depleted; and eIF4E, 20 to 30% depleted (1).
When the dicistronic mRNAs with the HAV IRES were tested in the depleted system, the translation of both cistrons was, not surprisingly, impaired (Fig. 6). Addition of p100 restored the translation of both cistrons, and this rescue was completely resistant to inhibition by m7GpppG cap analogue in the case of both the upstream scanning-dependent cistron and the HAV IRES-dependent cistron (Fig. 6). There was also an increase in the yield of incomplete products of translation of the upstream cistron. However, it should be noted that in general the same set of incomplete products was also seen, albeit in lower yield, in the control translation assays in nondepleted lysate. In these controls, the addition of m7GpppG cap analogue affected the yield of incomplete products and of the major product in the same way: both types of product were inhibited in the case of the capped transcript but stimulated in the case of the uncapped species. In other words, the incomplete products must have been initiated by the same mechanism as the major product. Taken together with other criteria described previously (8), this implies that most of the incomplete products arose from premature termination of translation initiated at the correct site, rather than illegitimate initiation at internal sites.
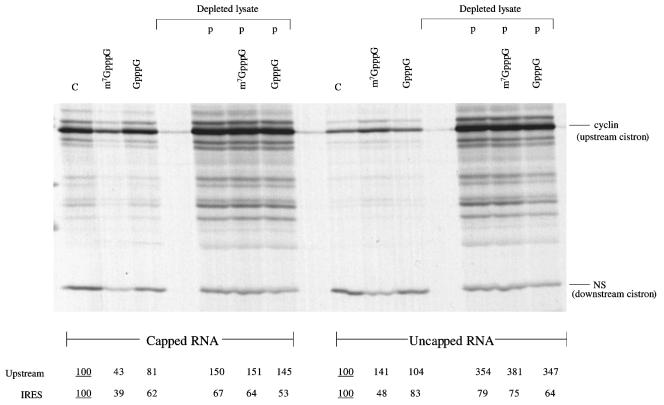
FIG. 6.
Translation driven by the HAV IRES can be supported by the p100 fragment (C-terminal two-thirds) of eIF4G. Capped and uncapped dicistronic mRNAs, as indicated, were translated at 25 μg/ml in either eIF4G-depleted reticulocyte lysate or parent (nondepleted lysate), in the presence of added KCl at 70 mM. Cap analogues, either m7GpppG or GpppG, were added at 0.4 mM, where indicated, together with 0.32 mM additional MgCl2. In lanes labeled “p,”, recombinant p100 was added at 20 μg/ml. Translation was at 30°C for 60 min, and the translation products were analyzed by SDS-PAGE followed by autoradiography. The positions of the upstream (cyclin) cistron product and downstream, IRES-dependent, NS-related product are shown. The yields of radiolabeled translation products of the upstream and IRES-driven cistrons were determined by scanning densitometry and are expressed relative to the yield in the corresponding control assay, which was set at 100 (the underlined value).
It can be seen that the rescue of upstream cistron translation was quite significantly more efficient than restoration of IRES-dependent translation (Fig. 6). This contrasts with what is observed when dicistronic mRNAs with the EMCV or Theiler's murine encephalomyelitis virus (TMEV) IRES are studied in the same system, when it is invariably the case that IRES-dependent cistron translation is rescued very efficiently, but translation of the upstream cistron is rather inefficient (1). These observations suggest that the HAV IRES competes rather poorly against scanning-dependent mRNAs or cistrons for p100, but that p100 interacts preferentially with the EMCV and TMEV IRESs as opposed to scanning-dependent mRNAs. Thus, the hierarchy of the functional interactions of p100 with mRNAs is EMCV or TMEV IRES ≫ scanning-dependent mRNAs > HAV IRES.
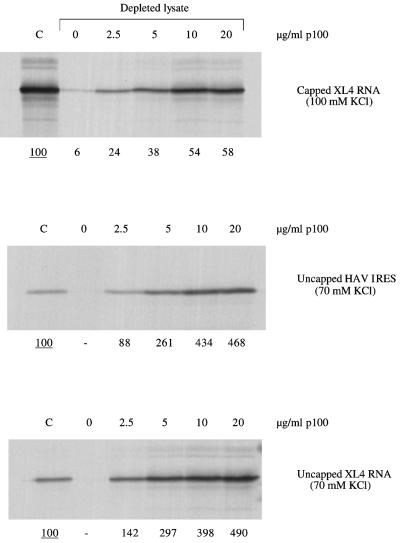
Because of the complication of competition between the two cistrons of the dicistronic mRNA, the experiments with eIF4G-depleted lysate were repeated using a monocistronic mRNA with the HAV IRES. The results show that in the absence of any competition from translation of another cistron, addition of high concentrations of p100 stimulated the HAV IRES activity almost as effectively as the translation of an uncapped mRNA initiated by the conventional scanning mechanism (Fig. 7). Moreover, the dependence of rescue on p100 concentration was not very different for an mRNA with the HAV IRES than for scanning-dependent capped or uncapped mRNAs (Fig. 7). To put these dose-response assays into a physiological perspective, our previous results indicate a concentration of endogenous eIF4F in the lysate equivalent to ∼3.0 μg of p100 per ml (1).
FIG. 7.
Dose response of p100 rescue of HAV IRES activity. Monocistronic XL4 mRNA (coding for X. laevis cyclin A), in either capped or uncapped form, and uncapped monocistronic HAV-NS RNA were translated at 20 μg/ml either in the parent (nondepleted) lysate (lanes C) or in eIF4G-depleted lysate supplemented with the designated concentrations of recombinant p100. Translation was at 30°C for 60 min, and the concentration of added KCl was either 70 mM (both uncapped RNAs) or 100 mM (capped XL4 mRNA), as indicated. The translation products were analyzed by SDS-PAGE followed by autoradiography. The yields of radiolabeled translation products were determined by scanning densitometry and are expressed relative to the yield in the corresponding control assay, which was set at 100 (the underlined value).
In the case of capped mRNAs translated by the scanning mechanism, we have previously shown that addition of p100 to control, (nondepleted) lysate has very little influence apart from a slight increase in the yield of products from those mRNAs which are poorly translated (1). Essentially the same result was seen in the case of translation dependent on the HAV IRES. When the uncapped dicistronic mRNA was translated in the standard, nondepleted lysate, addition of p100 stimulated translation of the upstream cistron, in agreement with previously published results (9), but had no effect on IRES-dependent translation (Ali and Jackson, unpublished). With the capped dicistronic mRNA, addition of p100 stimulated upstream cistron translation only marginally but increased the yield of IRES-dependent product until it became equal to the yield obtained from the uncapped version of the dicistronic template.
We have also previously shown that addition of p100 can reverse the inhibition of translation of capped mRNAs in standard lysate caused by addition of either FMDV L-protease, m7GpppG cap analogue, or 4E-BP1 (1). Here again, p100 also effected a remarkably similar rescue if HAV IRES activity had been inhibited by one of these agents in a nondepleted standard lysate system (Ali and Jackson, unpublished).
DISCUSSION
It is widely recognized that the HAV IRES differs from all other picornavirus IRESs in that it is rendered inactive if the endogenous eIF4G is cleaved by entero- or rhinovirus 2A protease or FMDV L-protease (4, 5, 7, 29). We have shown here that it is also inhibited by m7GpppG cap analogue and by 4E-BP1, which binds to and sequesters eIF4E, preventing its association with eIF4G to generate the complete eIF4F complex (25). This is also in complete contrast to the other picornavirus IRESs which are not inhibited by 4E-BP1 (Fig. 2) or by cap analogue (Ali and Jackson, unpublished). In its sensitivity to inhibition by all three reagents (protease, cap analogue, and 4E-BP1), translation dependent on the HAV IRES resembles the translation of capped mRNAs by the scanning ribosome mechanism. This similarity also extends to the fact that supplementation of the eIF4G-depleted lysate with p100 in sufficient concentrations can support both scanning-dependent translation and initiation on the HAV IRES (Fig. 6 and 7).
A reflection of the close parallel between translation initiation on capped mRNAs and on the HAV IRES is that the translation of both cistrons of a capped dicistronic mRNA with the HAV IRES responds in the same way to protease or 4E-BP1 (Fig. 2) or cap analogue (Fig. 4). This is of some concern because it raises the issue of whether the translation of the downstream cistron really is via an internal initiation mechanism in the normally understood meaning of that term. Is it possible, for example, that the only interaction between eIF4F and the capped dicistronic mRNA is at the 5′ cap (via interaction of the eIF4E subunit with the cap) and that the downstream cistron is translated by the eIF4F “reaching over” to deliver the 40S ribosomal subunit to the intercistronic IRES, which would require a looping out of the whole upstream cistron? A mechanism whereby eIF4F bound solely at the 5′ cap, sometimes delivering the 40S subunits to a cap-proximal site prior to scanning and sometimes delivering the subunits to the IRES, would explain why the translation of both cistrons responds in parallel to agents that perturb eIF4F-cap interactions. It is a matter of semantics whether such a hypothetical mechanism should be classified as internal initiation or pseudo-internal initiation, but it would certainly be quite different from what is believed to be the mechanism of internal initiation of translation of other picornavirus RNAs.
Arguing against such a hypothetical model, there are a number of situations in which the translation of the IRES-dependent cistron does not parallel the translation of the upstream cistron. For example, when uncapped and capped dicistronic mRNAs are compared, the upstream cistron product is synthesized less efficiently from the uncapped species than from the capped mRNA, yet the IRES-dependent cistron translation is more efficient when the mRNA is uncapped rather than capped (Fig. 4). Any interaction of eIF4F with the 5′ end of the uncapped mRNA is likely to be different in nature from its interaction with a 5′ cap, but this difference clearly does not affect the translation of the upstream and downstream cistrons in the same way. Moreover, when m7GpppG is added to translation assays of uncapped dicistronic mRNA, it inhibits IRES-dependent cistron translation but actually stimulates upstream cistron translation (Fig. 4), possibly through relief of the competitive influence of translation of capped fragments of globin mRNA. Finally, it is pertinent that translation of monocistronic mRNAs with the HAV IRES was equally sensitive to inhibition by m7GpppG cap analogue, regardless of the chemical identity of the 5′ end of the RNA, yet the nature of the interaction of eIF4F with the mRNA is strongly influenced by the nature of the 5′ end. All of these observations argue against the notion that the 40S ribosomal subunit is delivered to the initiation codon of the downstream (IRES-dependent) cistron by an eIF4F complex that is bound at the very 5′ end of the mRNA, whether dicistronic or monocistronic.
These considerations suggest that translation driven by the HAV IRES is dependent on some functional relationship between eIF4F and the IRES itself, quite likely a direct physical interaction between eIF4F and the IRES, rather than an interaction of eIF4F with the 5′ end itself. Nevertheless, it is clear that the function of eIF4F in supporting initiation on the HAV IRES is absolutely dependent on the presence of eIF4E in the eIF4F complex, since it is inhibited by 4E-BP1 (Fig. 2 and 3), which in effect strips eIF4E from the eIF4F complex but does not inhibit the ability of the eIF4E to interact with 5′ caps (25); it is further absolutely dependent on the cap-binding pocket (19) of this eIF4E, since it is inhibited by m7GpppG cap analogue. Two alternative models can be advanced to account for these findings. One invokes a direct interaction between the eIF4E component of eIF4F with a specific internal site in the HAV IRES, an interaction which would necessarily involve the cap-binding pocket of eIF4E in order to explain inhibition of the IRES by m7GpppG cap analogue. In this model, the postulated site-specific eIF4E-IRES interaction would, together with perhaps some direct eIF4G-IRES interactions (10), position the eIF4G component of the eIF4F complex in the appropriate proximity and orientation to deliver the 40S ribosomal subunit to the 3′ end of the HAV IRES, at or very close to the initiation codon. The principal strength of this model is that it provides a ready explanation for inhibition of the IRES by m7GpppG cap analogue, but the main problem with it is that the crystal structure of eIF4E-cap analogue complex (19) makes it hard to see how the cap-binding pocket of eIF4E could interact with RNA at an internal site.
The alternative model, which is advanced in the accompanying paper, (7a), is that HAV IRES activity requires the eIF4F complex, complete with associated eIF4E, not because the eIF4E component actually interacts with the IRES but because the eIF4E subunit must be associated with eIF4G in order for the eIF4G to be able to adopt a conformation suitable for direct interaction with the IRES, presumably at a specific site. One advantage of this model is that it does not require an interaction of the cap-binding pocket of the eIF4E component of eIF4F with an internal site in the IRES. Another is that it is indeed quite well established that the withdrawal of eIF4E from the eIF4F complex as a consequence of 4E-BP1–eIF4E interaction does cause a significant change in the conformation of the eIF4G, such that it now cannot be cleaved by picornavirus proteases (11, 24). In addition, structural studies of a peptide representing the site on eIF4G which interacts with eIF4E have shown that this peptide undergoes a considerable conformational change when it binds to eIF4E (20).
On the other hand, this model has difficulty explaining inhibition of the HAV IRES by m7GpppG cap analogue. It needs to postulate that the binding of cap analogue to the cap-binding pocket of eIF4E in the eIF4F complex results in a conformational change in the associated eIF4G. The crystal structure of the eIF4E-m7GDP complex shows that the protein resembles a cupped hand: the cap analogue-binding pocket is on the concave side, and it is the other side, the convex or dorsal face, which interacts with eIF4G (19, 20). Thus, a model which posits that interaction of the eIF4E-4G complex with cap analogue causes a conformational change in the eIF4G moiety would seem to require that the binding of cap analogue to the concave surface of eIF4E induce a conformational change in the diametrically opposite (convex) face, which in turn induces a conformational change in the associated eIF4G. On present evidence this seems improbable. Admittedly we do not know the structure of eIF4E per se, only the crystal structure of the eIF4E-m7GDP complex (19), and it would be fair to say that until we have the difference map between these two states of eIF4E, we cannot definitively rule out the possibility that binding of cap analogue causes a change in the conformation of the opposite (convex) face of eIF4E. However, we can say that if cap analogue binding to the eIF4E moiety does induce a change in the conformation of eIF4G, it must be a conformational change very considerably more subtle than that caused by withdrawal of eIF4E from the complex by interaction with 4E-BP1, since we find that although addition of 4E-BP1 to a lysate prevents cleavage of the endogenous eIF4G by L-protease, as already reported previously by others (11, 24), addition of m7GpppG cap analogue has absolutely no influence on the rate or efficiency of eIF4G cleavage by the protease (Ali and Jackson, unpublished).
In conclusion, we have extended previous work showing that the HAV IRES is inhibited by cleavage of eIF4G by viral proteases (4, 5, 7, 29) to demonstrate that initiation on this IRES is unique among picornavirus IRESs in exhibiting a strong requirement for eIF4E, specifically eIF4E in association with eIF4G. We are left with two alternative models to explain this surprising eIF4E requirement, but current technology cannot at present distinguish between these two explanations.
These results raise interesting questions concerning the evolution of picornaviruses. Current hypothesis envisages that all modern day picornaviruses evolved from a single common ancestral virus. Did the most recent common ancestor have initiation factor requirements resembling those of HAV? Or did it more closely resemble EMCV in having simpler requirements, and the branch which evolved to modern day HAV (re)acquired the need for eIF4E in association with eIF4G? It seems much more likely that the most recent common ancestor had factor requirements similar to those of HAV, and that the requirement for eIF4E and the N-terminal part of eIF4G was lost in all branches of the evolutionary tree except that which has given rise to modern-day HAV.
ACKNOWLEDGMENTS
We thank Andy Borman and Kathie Kean for the gift of pXLJ-HAV; Deirdre Scadden for the gift of pig brain eIF4E; and Simon Fletcher, Ann Kaminski, Sarah Hunt, and Nancy Standart for other constructs. We also thank C. U. T. Hellen (SUNY Health Center, Brooklyn, N.Y.) and J. Lawrence, Jr. (University of Virginia) for providing reagents and Rosemary Farrell for providing technical assistance and infrastructure support to the R.J.J. group.
This work was supported by grants from the Wellcome Trust to R.J.J. (051424) and S.J.M. (040800, 045619, 056778, 057494, and 058915). S.J.M. is a Senior Research Fellow of the Wellcome Trust, and I.K.A. was supported by a Medical Research Council postgraduate research studentship.
REFERENCES
- 1.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 2.Belsham G J, Jackson R J. Translation initiation on picornavirus RNA. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 869–900. [Google Scholar]
- 3.Borman A, Jackson R J. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology. 1992;188:685–696. doi: 10.1016/0042-6822(92)90523-r. [DOI] [PubMed] [Google Scholar]
- 4.Borman A M, Kean K M. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129–136. doi: 10.1006/viro.1997.8761. [DOI] [PubMed] [Google Scholar]
- 5.Borman A M, Bailly J-L, Girard M, Kean K M. Picornavirus internal ribosome entry segments—comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borman A M, Kirchweger R, Ziegler E, Rhoads R E, Skern T, Kean K M. eIF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 7.Borman A M, Le Mercier P, Girard M, Kean K M. Comparison of picornaviral IRES-driven internal initiation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Borman A M, Michel Y M, Kean K M. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J Virol. 2001;75:7864–7871. doi: 10.1128/JVI.75.17.7864-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dasso M C, Jackson R J. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 1989;17:3129–3144. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Gregorio E, Preiss T, Hentze M W. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 11.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt S L, Jackson R J. Polypyrimidine tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt S L, Hsuan J J, Totty N, Jackson R J. Unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson R J, Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 16.Kaminski A, Jackson R J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchweger R, Ziegler E, Lamphear B J, Waters D, Liebig H D, Sommergruber W, Sobrino F, Hohenadl C, Blaas D, Rhoads R E, Skern T. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4 gamma. J Virol. 1994;61:2711–2718. doi: 10.1128/jvi.68.9.5677-5684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 19.Marcotrigiano J, Gingras A C, Sonenberg N, Burley S K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 20.Marcotrigiano J, Gingras A C, Sonenberg N, Burley S K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 21.Medina M, Domingo E, Brangwyn J K, Belsham G J. The 2 species of foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology. 1993;194:355–359. doi: 10.1006/viro.1993.1267. [DOI] [PubMed] [Google Scholar]
- 22.Ohlmann T, Rau M, Morley S J, Pain V M. Proteolytic cleavage of initiation factor eIF-4γ in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 24.Ohlmann T, Pain V M, Wood W, Rau M, Morley S J. The proteolytic cleavage of eukaryotic initiation factor (eIF) 4G is prevented by eIF4E binding protein (PHAS-I; 4E-BP1) in the reticulocyte lysate. EMBO J. 1997;16:844–855. doi: 10.1093/emboj/16.4.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Sonenberg N. Insulin dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′ cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 26.Pestova T V, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor eIF4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts L O, Seamons R A, Belsham G J. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA. 1998;4:520–529. doi: 10.1017/s1355838298971989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whetter L E, Day S P, Elroy-Stein O, Brown E A, Lemon S M. Low efficiency of the 5′ nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J Virol. 1994;68:5253–5263. doi: 10.1128/jvi.68.8.5253-5263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]